Abstract
Several mycoplasma species feature a membrane protrusion at a cell pole, and unknown mechanisms provide gliding motility in the direction of the pole defined by the protrusion. Mycoplasma gallisepticum, an avian pathogen, is known to form a membrane protrusion composed of bleb and infrableb and to glide. Here, we analyzed the gliding motility of M. gallisepticum cells in detail. They glided in the direction of the bleb at an average speed of 0.4 μm/s and remained attached around the bleb to a glass surface, suggesting that the gliding mechanism is similar to that of a related species, Mycoplasma pneumoniae. Next, to elucidate the cytoskeletal structure of M. gallisepticum, we stripped the envelopes by treatment with Triton X-100 under various conditions and observed the remaining structure by negative-staining transmission electron microscopy. A unique cytoskeletal structure, about 300 nm long and 100 nm wide, was found in the bleb and infrableb. The structure, resembling an asymmetrical dumbbell, is composed of five major parts from the distal end: a cap, a small oval, a rod, a large oval, and a bowl. Sonication likely divided the asymmetrical dumbbell into a core and other structures. The cytoskeletal structures of M. gallisepticum were compared with those of M. pneumoniae in detail, and the possible protein components of these structures were considered.
Mycoplasmas are commensal and occasionally pathogenic bacteria that lack a peptidoglycan layer (50). Several species feature a membrane protrusion at a pole; for Mycoplasma mobile, this protrusion is called the head, and for Mycoplasma pneumoniae, it is called the attachment organelle (25, 34-37, 52, 54, 58). These species bind to solid surfaces, such as glass and animal cell surfaces, and exhibit gliding motility in the direction of the protrusion (34-37). This motility is believed to be essential for the mycoplasmas' pathogenicity (4, 22, 27, 36). Recently, the proteins directly involved in the gliding mechanisms of mycoplasmas were identified and were found to have no similarities to those of known motility systems, including bacterial flagellum, pilus, and slime motility systems (25, 34-37).
Mycoplasma gallisepticum is an avian pathogen that causes serious damage to the production of eggs for human consumption (50). The cells are pear-shaped and have a membrane protrusion, consisting of the so-called bleb and infrableb (29), and gliding motility (8, 14, 22). Their putative cytoskeletal structures may maintain this characteristic morphology because M. gallisepticum, like other mycoplasma species, does not have a cell wall (50). In sectioning electron microscopy (EM) studies of M. gallisepticum, an intracellular electron-dense structure in the bleb and infrableb was observed, suggesting the existence of a cytoskeletal structure (7, 24, 29, 37, 58). Recently, the existence of such a structure has been confirmed by scanning EM of the structure remaining after Triton X-100 extraction (13), although the details are still unclear.
A human pathogen, M. pneumoniae, has a rod-shaped cytoskeletal structure in the attachment organelle (9, 15, 16, 31, 37, 57). M. gallisepticum is related to M. pneumoniae (63, 64), as represented by 90.3% identity between the 16S rRNA sequences, and it has some open reading frames (ORFs) homologous to the component proteins of the cytoskeletal structures of M. pneumoniae (6, 17, 48). Therefore, the cytoskeletal structures of M. gallisepticum are expected to be similar to those of M. pneumoniae, as scanning EM images also suggest (13).
The fastest-gliding species, M. mobile, is more distantly related to M. gallisepticum; it has novel cytoskeletal structures that have been analyzed through negative-staining transmission EM after extraction by Triton X-100 with image averaging (45). This method of transmission EM following Triton X-100 extraction clearly showed a cytoskeletal “jellyfish” structure. In this structure, a solid oval “bell,” about 235 nm wide and 155 nm long, is filled with a 12-nm hexagonal lattice. Connected to this bell structure are dozens of flexible “tentacles” that are covered with particles 20 nm in diameter at intervals of about 30 nm. The particles appear to have 180° rotational symmetry and a dimple at the center. The involvement of this cytoskeletal structure in the gliding mechanism was suggested by its cellular localization and by analyses of mutants lacking proteins essential for gliding.
In the present study, we applied this method to M. gallisepticum and analyzed its unique cytoskeletal structure, and we then compared it with that of M. pneumoniae.
MATERIALS AND METHODS
Strains and culture.
The M. gallisepticum S6 and the M. pneumoniae M129 strains were grown in Aluotto medium consisting of 2.1% heart infusion broth, 0.56% yeast extract, 10% horse serum, 0.025% thallium acetate, and 0.005% ampicillin at 37°C (2, 42). The cells were harvested when the optical density at 600 nm reached about 0.1, corresponding to the late exponential phase.
Optical microscopy.
Mycoplasma cells were collected by centrifugation at 12,000 × g for 10 min at room temperature (RT) and suspended in phosphate-buffered saline (PBS)-serum consisting of 10% (non-heat-inactivated) horse serum, 75 mM Na phosphate (pH 7.3), and 68 mM NaCl to a 30-fold density from the original. The cells were poured through a nitrocellulose filter (pore size, 0.45 μm) so they would disperse and were then poured into a tunnel assembled by taping a coverslip cleaned with saturated ethanolic KOH onto a glass slide (28, 59). The tunnel slides containing the cell suspension were kept at 37°C on a microscopic stage equipped with a lens heater (MATS-LH; Tokai Hit, Shizuoka, Japan) and an HT 200 stage heater (Minitube, Verona, WI). After incubation for 10 min, the floating cells were removed by replacing the medium. Static and moving images of cells were observed by phase-contrast microscopy and recorded as previously described (44, 56).
All video data were analyzed by ImageJ 1.37v (http://rsb.info.nih.gov/ij/). To trace the detailed movements of individual cells, the outline of each cell was fitted as an ellipse, and the coordinates of the front end, tail end, and center of the cell were calculated by the analysis functions of ImageJ. The gliding speeds of individual cells were calculated by ParticleTracker (51) (http://weeman.inf.ethz.ch/particletracker/) and MultiTracker (http://rsb.info.nih.gov/ij/plugins/multitracker.html), which are plug-ins for analyzing cell movements running on ImageJ 1.37v.
EM.
Mycoplasma cells were suspended in PBS-serum at a 30-fold density of the original culture, put on a carbon-coated grid, and incubated for 10 min at 37°C. The medium was removed and replaced by Triton X-100 solution containing 0.01% or 0.3% of Triton X-100, 1 mg/ml DNase I, and 5 mM MgCl2 in PBS for 1 min at 37°C or RT. To observe the core structure, mycoplasma cells were suspended in 0.3% Triton X-100. After incubation for 1 min at RT, the suspension was sonicated for 3 s with a Daiseikai sonicator (EK Technology, Chiba, Japan). The sample was put on a grid, kept for 3 min at RT, and stained as described above. Other procedures were done as previously described (38, 45).
RESULTS
Gliding motility of M. gallisepticum.
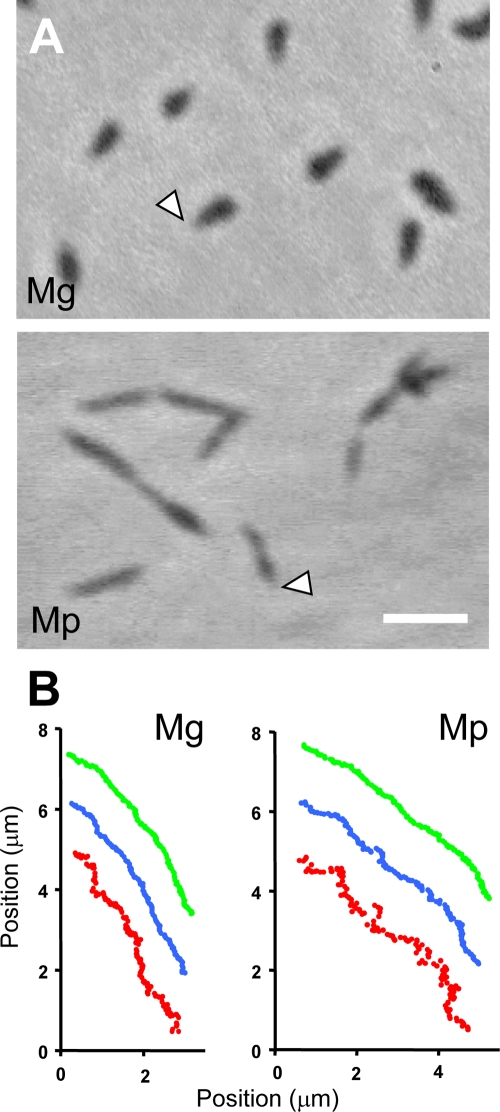
The morphology and motility of M. gallisepticum cells were examined previously (8, 14), but the details have not been studied. We thus examined them in the present study. M. gallisepticum cells suspended in PBS containing 10% serum were put into a tunnel slide constructed by taping a coverslip to a glass slide (28, 59) and were observed by phase-contrast microscopy (Fig. 1A). Pear-shaped cells about 1 μm in length adhered to the glass surface at RT, but no obvious movement was seen. When the temperature was elevated to 37°C, about half of the cells obviously glided in the direction of the tapered end (see video S1 in the supplemental material).
FIG. 1.
Cell images and the motility of M. gallisepticum (Mg) and M. pneumoniae (Mp) observed by phase-contrast microscopy. (A) Images of M. gallisepticum and M. pneumoniae cells are presented in the upper and lower panels, respectively. The cells on glass are gliding in the direction of the tapered end, bleb, and attachment organelle of both species, as marked by open triangles. Bar, 2 μm. (B) Gliding traces of each part of the cell. The left and right panels represent those of M. gallisepticum and M. pneumoniae, respectively. The positions of the cell pole of the tapered end (green), the center (blue), and the other pole end (red) are shown for 13 s at 0.1-s intervals. The relative positions among three traces are rearranged for better presentation.
To judge which part of the cell is responsible for binding to the glass during gliding, we traced the positions of the front end, the center, and the tail end in the microscopic field of a movie (Fig. 1B). In M. pneumoniae cells, the tail end showed more trembling movements than the other two points (Fig. 1B, right panel). The P1 adhesin, responsible for binding and gliding, is clustered at the cell's front end (25, 34-36, 53). A cell's trembling manner can be explained by the fact that binding, which occurs at the front end, reduces trembling by Brownian motion. A similar feature was observed with M. gallisepticum, suggesting that the front of the cell is responsible for glass binding also in this species (Fig. 1B, left panel).
To examine the characteristics that determine gliding direction, we traced the centers of cells at 0.33-s intervals for 90 s. The M. gallisepticum cells basically traced slow curves, occasionally changing their direction (Fig. 2A, upper panels). The tracing patterns were not apparently different from those of M. pneumoniae (Fig. 2B, upper panels).
FIG. 2.
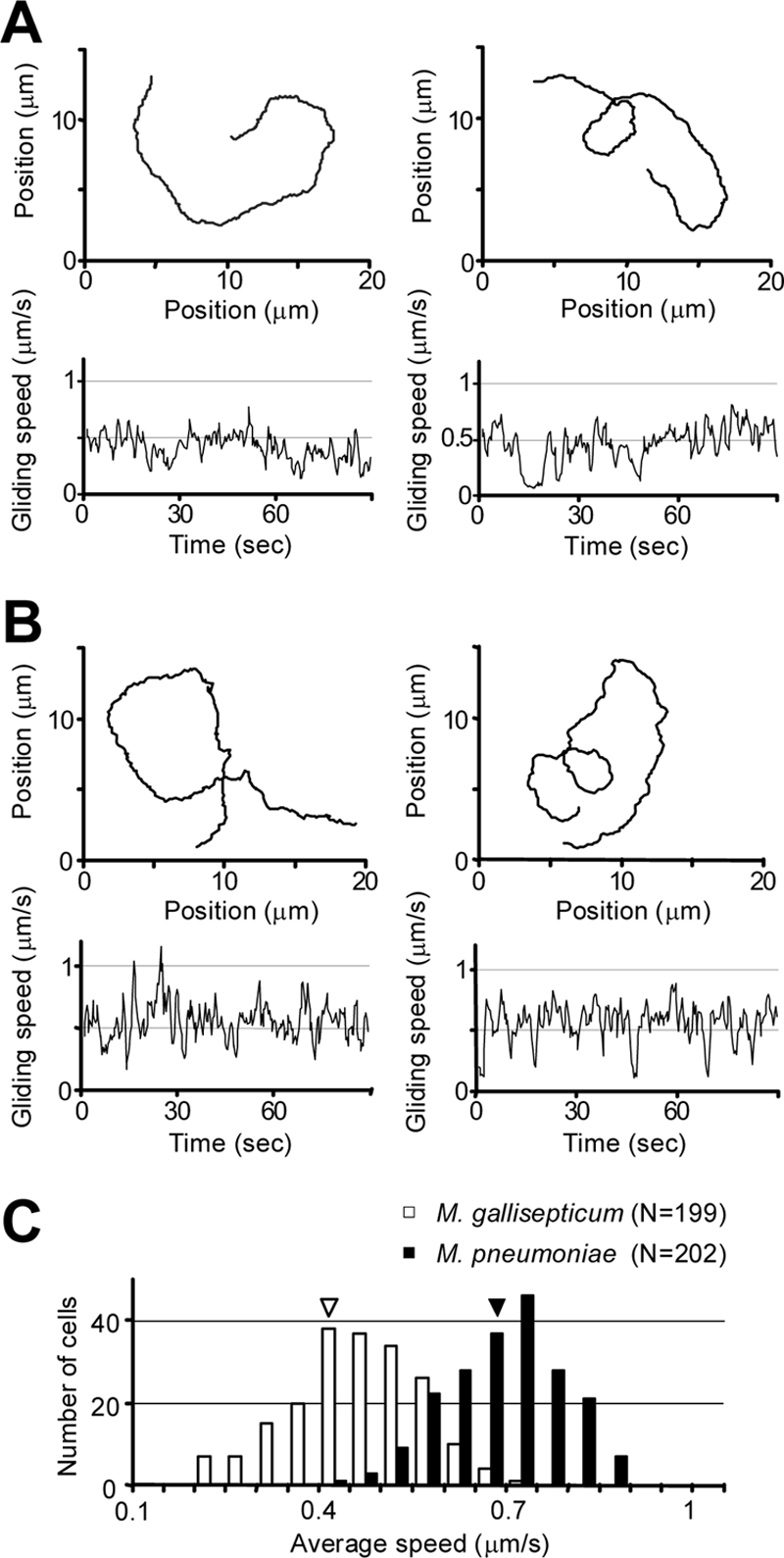
Gliding directions and speeds of M. gallisepticum compared with those of M. pneumoniae. Analyses for M. gallisepticum (A) and M. pneumoniae (B) are presented. The upper panels in each show the representative traces of the center of a gliding cell plotted for 90 s at 0.33-s intervals. Lower panels show the gliding speeds of the corresponding cells calculated for 0.33 s each. (C) Distribution of gliding speeds. The speeds of 200 cells of each species were calculated for 0.33-s periods, averaged for 20 s, and integrated into a histogram with averages represented by triangles.
Next, the gliding speeds of individual cells were analyzed in detail (Fig. 2A and B, lower panels). For a detailed analysis of gliding speed, shorter time periods should be used to calculate speed. However, calculating for short periods generally causes noisy traces. After trying periods of 0.03, 0.10, 0.33, and 1 s, we selected 0.33 s because the small random noises were well reduced in that period and in the 1-s period. The two species showed apparently similar fluctuations in gliding speeds. We analyzed the fluctuation by the Fourier transform but found no obvious oscillations. The average speeds for 20 s were integrated into a histogram for about 200 cells of each species (Fig. 2C). The proportions of gliding cells and the average speeds were 50.9% (total number, 595) and 0.41 μm/s for M. gallisepticum, and 82.4% (total number, 557) and 0.64 μm/s for M. pneumoniae, respectively. These results show that the gliding properties of M. gallisepticum are not far from those of M. pneumoniae, although the speed and frequency of the former are lower. The gliding speed of M. gallisepticum at 41°C, the common body temperature of birds, was 22% faster than its gliding speed at 37°C.
Surface and inside structures of bleb and infrableb.
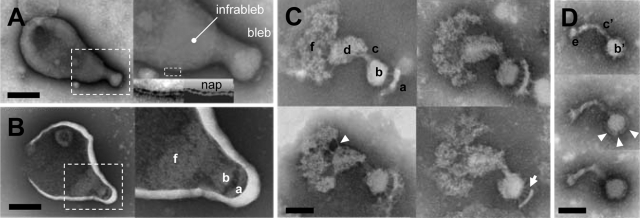
To elucidate the cytoskeletal structure of M. gallisepticum, we treated the cells with Triton X-100 and observed them by negative-staining transmission EM. The intact cell images showed the bleb and infrableb at a cell pole, where nap structures, reminiscent of the raised surfaces of a cloth, were observed on the surface, as was the case in previous studies (Fig. 3A) (23, 43). Next, we observed the cells treated with 0.01% Triton X-100 and DNase on EM grids (Fig. 3B). Under these conditions, the cell membranes were partially damaged and a unique intracellular structure was found inside the bleb and infrableb. When the cell membrane was totally removed by extraction with 0.3% Triton X-100 and DNase, the inside structure, an “asymmetrical dumbbell,” was clearly observable (Fig. 3C). This dumbbell structure is apparently composed of five parts in the following order from the distal end: a cap (structure a), a small oval (structure b), a rod (structure c), a large oval (structure d), and a bowl (structure f). The small oval is linked to the cap by some filaments, and the large oval is linked to the bowl by a few filaments. Such a structure can be found also in the images of cells with partially damaged membranes, as indicated by the corresponding marks in Fig. 3B. To obtain more information, we sonicated the cells suspended in 0.3% Triton X-100, adsorbed the remaining structures to EM grids, and observed (Fig. 3D). We found a small structure composed of a small oval (structure b′) covered by a thin layer, as well as a curved rod (structure c′) featuring a bulge (structure e) at a pole. The dimensions and appearance of this structure suggested that it is probably the core of the cytoskeletal structure, which is resistant to the mechanical shock of sonication. Comparison of the asymmetrical dumbbell and core structures suggested that a rod corresponding to structure c′ can be seen in structures c and d in the asymmetrical dumbbell.
FIG. 3.
Negative-staining EM images of M. gallisepticum cells treated with different concentrations of Triton X-100. (A) Untreated cell. The area indicated by the box with dashed lines is magnified as the neighboring panel. The cell features, namely the bleb, infrableb, and nap, are shown. The nap structures can be observed on the surface of the bleb and infrableb. The inset of the right panel is modified for contrast to show the nap structure and is schematically illustrated in the right neighboring panel. Bar, 300 nm. (B) Cell treated with 0.01% Triton X-100. Cytoskeletal structures were observed ambiguously in the bleb and infrableb, as marked by the characters corresponding to those in panel C. The area indicated by the box with dashed lines is magnified as the neighboring panel. Bar, 300 nm. (C) Cytoskeletal structures remaining on the grid after treatment with 0.3% Triton X-100. Four independent images are presented. The structure is composed of a cap (a), a small oval (b), a connecting rod (c), a large oval (d), and a bowl (f). The large oval is linked to the bowl by a few filaments, as indicated by the white triangle. The small oval is linked to the cap by a larger number of filaments, as indicated by the white arrow. Bar, 100 nm. (D) Core of cytoskeletal structure remained after treatment with 0.3% Triton X-100 and sonication. Three independent images are presented. The structure is composed of a small oval (b′) covered by a thin layer, as marked by white triangles, and a curved rod (c′). The curved rod features a bulge (e) at an end. The bowl, large oval, and cap in the cytoskeletal structure are likely absent from the core structure. Bar, 100 nm.
Outlines and dimensions suggested by image collections of cytoskeletal and core structures.
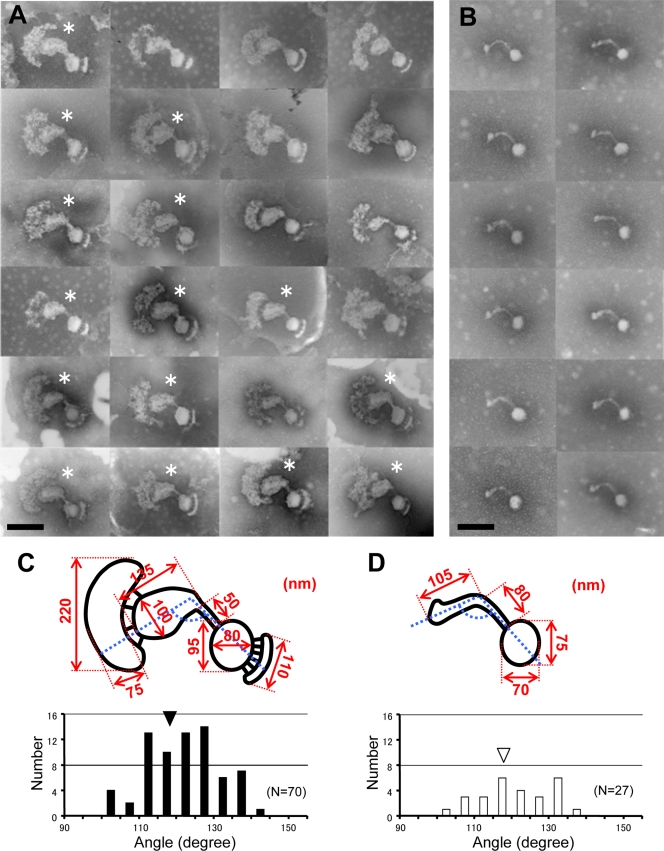
The cytoskeletal structures have some morphological variations that may have been caused by the partial removal of cytoplasm and membrane. However, we did not apply higher concentrations of Triton X-100, because a concentration higher than 1% tended to damage the whole structure. The image collections of the asymmetrical dumbbell and core structures presented in Fig. 4A and B suggested outlines presented in Fig. 4C and D, respectively, with approximate dimensions. In many cases, a structure similar to the core is partially visible in an asymmetrical dumbbell structure, supporting our assumption that the asymmetrical dumbbell can be divided into a core and other parts by sonication. The pointed ends of the rods have a variation in the angle, as defined and summarized in Fig. 4C and D.
FIG. 4.
Variations in images of asymmetrical dumbbell and core structures. The images of asymmetrical dumbbell (A and C) and core (B and D) structures were analyzed. Typical images are presented in panels A and B. Bar, 200 nm. The panels with asterisks show a structure similar to the core partially visible in the asymmetrical dumbbell structure. Schematics are presented in panels C and D with their dimensions obtained as the averages from 25 images in multiples of 5 nm. The distributions of angles defined by dashed lines are shown in histograms of panels C and D, with the averages indicated by triangles.
Branched cytoskeletal and core structures.
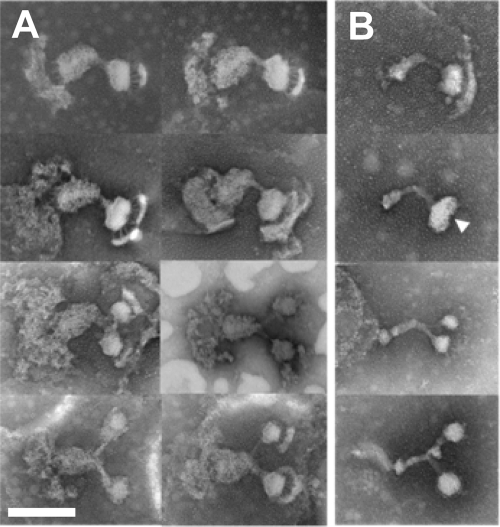
Under EM, branched structures were found for both the asymmetrical dumbbell and core structures at a low frequency, about 1 per 100 structures (Fig. 5), comparable to the frequency of cells with two blebs under optical microscopy. The branched images were classified into two types: one in which the small oval is extended and constricted (Fig. 5, upper panels) and one in which the rod has two branches with equivalent sizes of small ovals for each (Fig. 5, lower panels). These observations may suggest that the duplication of cytoskeletal structures occurs from the distal end and progresses to the proximal side in the cell reproduction cycle.
FIG. 5.
Branched asymmetrical dumbbell and core structures. (A) Asymmetrical dumbbell structures. (B) Core structures. The upper six panels show structures with extended small ovals (a small constriction is observed at the position indicated by the white triangle; panel B). The lower six panels show structures with branched rods. Bar, 200 nm.
DISCUSSION
Gliding motility of M. gallisepticum.
In the present study, M. gallisepticum glided 3 to 10 times faster than reported previously (Fig. 2) (8, 14, 22). The causes of this difference are unclear, but differences in the conditions of the strain, serum, and culture may be involved. We compared the gliding of M. gallisepticum gliding with that of M. pneumoniae and found no critical differences in their properties, such as the direction, the point of the cell where binding occurred, or the frequency and speed of gliding (Fig. 1 and 2). These results may suggest that these two species have similar gliding mechanisms and that the part of M. gallisepticum including the bleb and infrableb can be called the attachment organelle as well.
Comparison of cytoskeletal structures with those previously reported for M. gallisepticum.
Hatchel et al. applied Triton X-100 extraction to M. gallisepticum and observed the cytoskeletal structures by scanning EM (14). Those images are consistent with the transmission EM images presented here, although the surface outlines with lower resolution were presented in the scanning EM images. In 1964, Domermuth et al. observed M. gallisepticum cells by sectioning EM and found an electron-dense structure located in the membrane protrusion (7). The structure is probably identical to the cytoskeletal structure we observed, because the two have very similar outlines and dimensions.
Tubular structures with a diameter of 40 nm have been reported to form loop structures in chemically fixed M. gallisepticum cells, based on serial sectioning EM studies (24, 29, 58). The loops bind to the internal structure in the bleb and line the cell surface of parts other than the protrusion. No such structure was found in the present study, suggesting that this tubular structure is less stable than the asymmetrical dumbbell structure in the bleb. These observations are consistent with previous ones, in that such structures could not be found even in sectioning EM unless the cells were chemically fixed (58).
Effects of Triton X-100 on cytoskeletal structures.
In the present study, we treated the cells with Triton X-100 to remove the structures hiding the cytoskeletal structures. This treatment may have had some effects on biomolecules. However, the cytoskeletal structures found in the present study are probably similar morphologically to those in living mycoplasma cells for the following reasons. (i) We treated M. gallisepticum cells with 0.01% and 0.3% Triton X-100 and continuously observed them under phase-contrast optical microscopy. Although the cell membrane was damaged by the treatment with 0.01% Triton X-100, as detected by the change in the image density, the cells stayed in the same position on the glass. After the treatment with 0.3% Triton X-100, most parts of the cell were gone, while the tip of the bleb remained at the same position on the glass (data not shown). These observations suggest that these treatments do not totally damage the mechanism involved in the binding to a solid surface and also that the cytoskeletal structure faces the original direction for gliding. (ii) The cells of M. mobile retain the activities of binding and gliding after treatment with 0.01% Triton X-100, because the permeabilized cell “ghost” stays on the glass and can be reactivated for normal gliding motility by the addition of nucleotides such as ATP (60). (iii) The asymmetrical dumbbell structure can be seen in the sectioning images of chemically fixed cells (7) as well as in the cells treated with 0.01% and 0.3% Triton X-100. The structures found in these three conditions have similar appearances, suggesting that the structure observed here should reflect the original morphology in a living cell. The images of cytoskeletal structures treated with 0.01% Triton X-100 were slightly different from the images of those treated with 0.3% Triton X-100. Most likely, the cytosol was removed partially in occasional manners by 0.01% Triton X-100, resulting in the differences in appearance. (iv) Generally, the cell architecture, as viewed under cryo-EM, is believed to retain its original structures. The images of the cytoskeleton of M. pneumoniae, which is related to M. gallisepticum, by cryo-EM were found to be consistent with previously reported ones for M. pneumoniae structures treated with Triton X-100 (16, 57), suggesting that the cytoskeleton's original basic morphology can be retained after Triton X-100 treatment.
Comparison of cytoskeletal structures with those of M. pneumoniae and M. mobile.
Here, we compared the cytoskeletal structure of M. gallisepticum with that of M. pneumoniae, the latter of which can be apparently divided into three parts, in the following order from the distal end: a terminal button, a rod, and a bowl (Fig. 6) (25, 34-37). The overall appearance suggests that structures a, b, c, and f of M. gallisepticum correspond to structures g, h, i, and l of M. pneumoniae, but structures d, e, j, and k are difficult to assign (Fig. 6). Perhaps the spoke-like structures in M. gallisepticum may correspond to structures k and h of M. pneumoniae, and structure e of M. gallisepticum may be divided into two parts as observed for structures i and j of M. pneumoniae. The differences in overall dimensions of cytoskeletal structures between the two species may result from the cell thickness, i.e., a M. pneumoniae cell is thinner than a M. gallisepticum cell, probably because of their living environments.
FIG. 6.
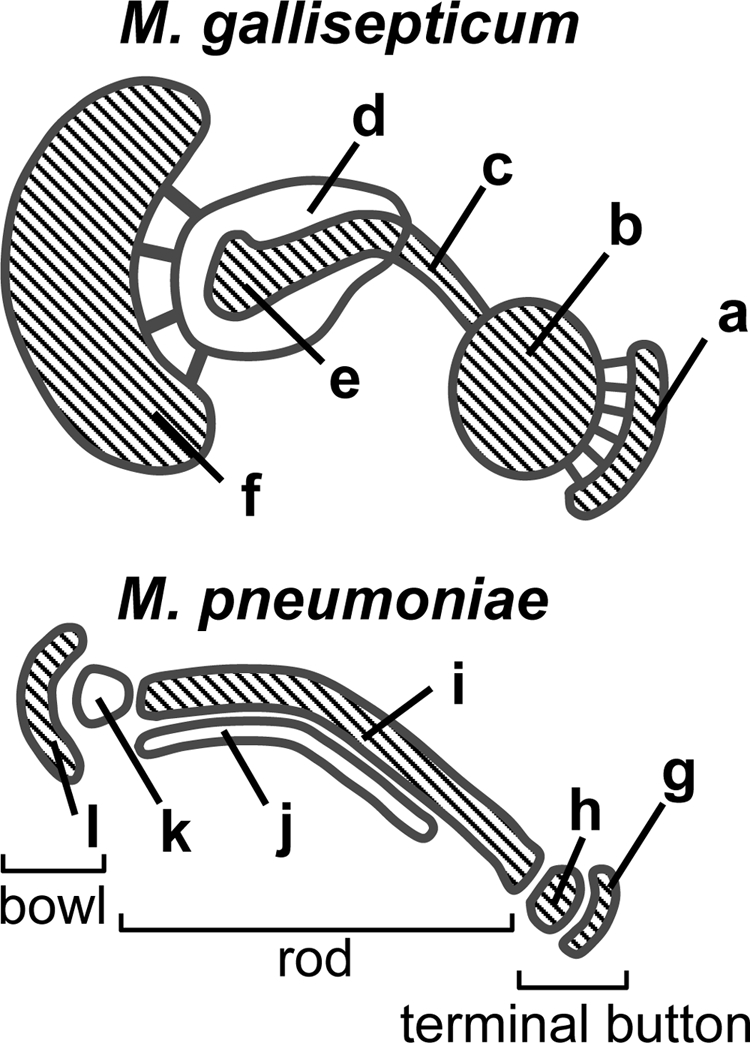
Comparison of cytoskeletal structures of M. gallisepticum and M. pneumoniae. The cytoskeletal structures of M. pneumoniae are summarized from previous reports (15, 16, 25, 34, 36, 57). The parts are marked as follows: a, cap; b, small oval; c, rod; d, large oval; e, bulge; f, bowl; g and h, terminal button; i to k, rod; l, bowl. The distal ends of the cell membrane protrusion are structures a and g. Connecting filaments can be seen at the gaps between structures a and b and between structures d and f. The overall appearance suggests that structures a, b, c, and f of M. gallisepticum correspond to structures g, h, i, and l of M. pneumoniae, as emphasized by the hatching.
In M. pneumoniae, 11 unique proteins have been identified as components of the attachment organelle based on localization by fluorescence microscopy (25, 34-37). P1 adhesin, a 170-kDa protein, clusters across the whole surface of the attachment organelle with the presumably associated proteins P40 and P90. The other proteins are more focused and are aligned from the distal to the proximal ends as (P30, P65)-HMW3-(HMW1, HMW2)-(P24, P41, -P200) (11, 20, 21, 52). These localization patterns suggest that P30 and P65, HMW1 and HMW2, and P24, P41, and P200 are the components of the terminal button, rod, and bowl of the cytoskeletal structure, respectively. HMW3 is around the interface between the terminal button and the rod. Some putative roles have been assigned to the component proteins in the attachment organelles of M. pneumoniae and M. genitalium, a close relative of M. pneumoniae (3, 34, 36). P1 adhesin associated with P90 and P40 is suggested to work as the leg for gliding with repeated binding and releasing with the solid surface (53). HMW1 and HMW2 are responsible for initial stage of formation of the nascent attachment organelle (10, 26, 49, 52). P200 and P65 have roles specific for gliding rather than cytadherence, unlike other protein components (5, 19).
Homologs of these proteins have been found in M. gallisepticum (Table 1; see also Fig. S7 in supplemental material). GapA, CrmA, MGC2, PlpA, and Hlp3 were experimentally characterized as homologs of P1 adhesin, P40/P90, P30, P65, and HMW3 of M. pneumoniae, respectively (18, 30, 47). P40 and P90 of M. pneumoniae are translated as a single peptide and processed into two proteins. CrmA of M. gallisepticum has a clear similarity with this open reading frame product, although CrmA is not processed into two mature proteins. The MGA_0205 and MGA_0306 genes reportedly have sequence similarities to the genes encoding P200 and HMW1 of M. pneumoniae, respectively (48). A BLAST search found that the MGA_1203 gene has similarity to the gene encoding HMW2. The presence of homologs of these genes in the M. gallisepticum genome suggests that these homologs have similar roles also in M. gallisepticum. P41 and P24, responsible for anchoring the attachment organelle to the other part of cell in M. pneumoniae, have not been found in M. gallisepticum (12). The cell architectures of the part other than the attachment organelle may have more variations between these two species than the attachment organelle.
TABLE 1.
Components of cytoskeletal structuresa
|
M. pneumoniae
|
M. gallisepticum
|
Homologous region (no. of aa) | Identity (%) | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|
| MPN code | Protein | No. of aac | Subcellular localizationb | MPN code | Protein | No. of aa | |||
| 141 | P1 | 1,627 | s | 928 | GapA | 1,123 | 165 | 41.2 | 47 |
| 142 | P40/P90 | 1,218 | s | 939 | CrmA | 1,062 | 205 | 44.9 | 47 |
| 309 | P65 | 405 | t | 1199 | PlpA | 856 | 325 | 20 | 30 |
| 310 | HMW2 | 1,818 | r | 1203 | 1,931 | 1,477 | 22.2 | This study | |
| 311 | P41 | 357 | b | ||||||
| 312 | P24 | 218 | b | ||||||
| 447 | HMW1 | 1,018 | r | 306 | 1,976 | 450 | 29.6 | 48 | |
| 452 | HMW3 | 672 | t/r | 928 | Hlp3 | 1,081 | 362 | 31.2 | 30 |
| 453 | P30 | 274 | t | 932 | MGC2 | 297 | 264 | 39.8 | 18 |
| 567 | P200 | 1,036 | b | 205 | 1,942 | 754 | 27.7 | 48 | |
Schematically illustrated in Fig. S7 in the supplemental material.
s, surface of the attachment organelle; t, terminal button; r, rod; b, bowl.
aa, amino acids.
The EM observations obtained in the present study may suggest the components of asymmetrical dumbbell, with MGC2 and PlpA corresponding to the cap (structure a) and small oval (structure b), Hlp3 corresponding to the interface between the small oval and the rod (structure c), the MGA_0306 and MGA_1203 proteins corresponding to the rod and the large oval (structure d), and the MGA_0205 protein corresponding to the bowl (structure f) (Table 1; Fig. 6).
The gliding machinery of M. mobile, the fastest species, is assumed to be composed of four proteins (Gli123, Gli349, Gli521, and P42) around the cell membrane at the cell neck (1, 32, 38, 39, 41, 46, 56, 59, 61, 62). It is also assumed to be supported by the internal cytoskeletal “jellyfish” structure (45). Homologs of the component proteins of these structures were not found in M. gallisepticum (48). Thus, there is not a close relationship between the cytoskeletal structures of these species.
Variations in cytoskeletal structure images.
Recently, the “inchworm” hypothesis was proposed to explain the gliding mechanism of M. pneumoniae, wherein the total length of the cytoskeletal structure oscillates and displaces the putative feet bound to the solid surface (16, 57). Henderson and Jensen detected small variations in the conformation of the segmented rod and the position of the bowl in M. pneumoniae (16). The variation of bend found in the asymmetrical dumbbell and core structures of M. gallisepticum (Fig. 4) may be related to the gliding motility, although the possibility that the variation of apparent angles is caused by the variation of the way that the structures are attached to the grid remains.
Duplication of cytoskeletal structure.
In this study, we obtained images suggesting that the duplication of the asymmetrical dumbbell begins at the distal end and progresses to the proximal side (Fig. 5). In the cultures of M. gallisepticum and M. pneumoniae, all cells have one or two protrusions, suggesting that the formation of the membrane protrusions is well coupled with the cell division cycle (11, 40, 43, 52, 55). Detailed analysis of M. pneumoniae cells by fluorescence microscopy showed that the existing attachment organelle appears to duplicate before cell division (11, 33, 40, 52, 55). This initial event, the duplication of the attachment organelle, can be explained by one of two scenarios: (i) sequentially templated reproduction from the preexisting cytoskeletal structure (15) or (ii) de novo formation near the preexisting one (11). The first model was suggested by an EM observation that the distal side of the cytoskeletal structure was split (15). Our observation here suggests that M. gallisepticum duplicates its bleb through the first scenario.
Supplementary Material
Acknowledgments
This work was supported by a grant-in-aid for scientific research (A) and by a grant-in-aid for scientific research on the priority areas “Applied Genomics,” “Structures of Biological Macromolecular Assemblies,” and “System Cell Engineering by Multi-Scale Manipulation” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M.M.) and by a grant from the Institution for Fermentation, Osaka, Japan (to M.M.).
Footnotes
Published ahead of print on 13 March 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adan-Kubo, J., A. Uenoyama, T. Arata, and M. Miyata. 2006. Morphology of isolated Gli349, a leg protein responsible for glass binding of Mycoplasma mobile gliding revealed by rotary-shadowing electron microscopy. J. Bacteriol. 1882821-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aluotto, B. B., R. G. Wittler, C. O. Williams, and J. E. Faber. 1970. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int. J. Syst. Bacteriol. 2035-58. [Google Scholar]
- 3.Balish, M. F., and D. C. Krause. 2006. Mycoplasmas: a distinct cytoskeleton for wall-less bacteria. J. Mol. Microbiol. Biotechnol. 11244-255. [DOI] [PubMed] [Google Scholar]
- 4.Bredt, W. 1968. Motility and multiplication of Mycoplasma pneumoniae. A phase contrast study. Pathol. Microbiol. 32321-326. [DOI] [PubMed] [Google Scholar]
- 5.Burgos, R., O. Q. Pich, E. Querol, and J. Pinol. 2008. Deletion of the Mycoplasma genitalium MG_217 gene modifies cell gliding behaviour by altering terminal organelle curvature. Mol. Microbiol. 691029-1040. [DOI] [PubMed] [Google Scholar]
- 6.Dandekar, T., M. Huynen, J. T. Regula, B. Ueberle, C. U. Zimmermann, M. A. Andrade, T. Doerks, L. Sanchez-Pulido, B. Snel, M. Suyama, Y. P. Yuan, R. Herrmann, and P. Bork. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 283278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domermuth, C. H., M. H. Nielsen, E. A. Freundt, and A. Birch-Andersen. 1964. Ultrastructure of Mycoplasma species. J. Bacteriol. 88727-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erdmann, T. 1976. Untersuchungen zur Morphologie, Vermehrung und Beweglichkeit von Mycoplasma gallisepticum. M.D. thesis. Johannes Gutenberg Universität, Mainz, Germany.
- 9.Göbel, U., V. Speth, and W. Bredt. 1981. Filamentous structures in adherent Mycoplasma pneumoniae cells treated with nonionic detergents. J. Cell Biol. 91537-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn, T. W., M. J. Willby, and D. C. Krause. 1998. HMW1 is required for cytadhesin P1 trafficking to the attachment organelle in Mycoplasma pneumoniae. J. Bacteriol. 1801270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasselbring, B. M., J. L. Jordan, R. W. Krause, and D. C. Krause. 2006. Terminal organelle development in the cell wall-less bacterium Mycoplasma pneumoniae. Proc. Natl. Acad. Sci. USA 10316478-16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasselbring, B. M., and D. C. Krause. 2007. Cytoskeletal protein P41 is required to anchor the terminal organelle of the wall-less prokaryote Mycoplasma pneumoniae. Mol. Microbiol. 6344-53. [DOI] [PubMed] [Google Scholar]
- 13.Hatchel, J. M., and M. F. Balish. 2008. Attachment organelle, ultrastructure correlates with phylogeny, not gliding motility properties, in Mycoplasma pneumoniae relatives. Microbiology 154286-295. [DOI] [PubMed] [Google Scholar]
- 14.Hatchel, J. M., R. S. Balish, M. L. Duley, and M. F. Balish. 2006. Ultrastructure and gliding motility of Mycoplasma amphoriforme, a possible human respiratory pathogen. Microbiology 1522181-2189. [DOI] [PubMed] [Google Scholar]
- 15.Hegermann, J., R. Herrmann, and F. Mayer. 2002. Cytoskeletal elements in the bacterium Mycoplasma pneumoniae. Naturwissenschaften 89453-458. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, G. P., and G. J. Jensen. 2006. Three-dimensional structure of Mycoplasma pneumoniae's attachment organelle and a model for its role in gliding motility. Mol. Microbiol. 60376-385. [DOI] [PubMed] [Google Scholar]
- 17.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B.-C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 244420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hnatow, L. L., C. L. Keeler, Jr., L. L. Tessmer, K. Czymmek, and J. E. Dohms. 1998. Characterization of MGC2, a Mycoplasma gallisepticum cytadhesin with homology to the Mycoplasma pneumoniae 30-kilodalton protein P30 and Mycoplasma genitalium P32. Infect. Immun. 663436-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan, J. L., H. Y. Chang, M. F. Balish, L. S. Holt, S. R. Bose, B. M. Hasselbring, R. H. Waldo III, T. M. Krunkosky, and D. C. Krause. 2007. Protein P200 is dispensable for Mycoplasma pneumoniae hemadsorption but not gliding motility or colonization of differentiated bronchial epithelium. Infect. Immun. 75518-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenri, T., S. Seto, A. Horino, Y. Arakawa, T. Sasaki, and M. Miyata. 2006. Mapping of localization sites of cytadherence-related and cytoskeletal proteins of Mycoplasma pneumoniae by fluorescent protein tagging. Abstr. 16th Congr. Int. Org. Mycoplasmol., abstr. 245.
- 21.Kenri, T., S. Seto, A. Horino, Y. Sasaki, T. Sasaki, and M. Miyata. 2004. Use of fluorescent-protein tagging to determine the subcellular localization of Mycoplasma pneumoniae proteins encoded by the cytadherence regulatory locus. J. Bacteriol. 1866944-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirchhoff, H. 1992. Motility, p. 289-306. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas—molecular biology and pathogenesis. American Society for Microbiology, Washington, DC.
- 23.Kirchhoff, H., R. Rosengarten, W. Lotz, M. Fischer, and D. Lopatta. 1984. Flask-shaped mycoplasmas: properties and pathogenicity for man and animals. Isr. J. Med. Sci. 20848-853. [PubMed] [Google Scholar]
- 24.Korolev, E. V., A. V. Nikonov, M. S. Brudnaya, E. S. Snigirevskaya, G. V. Sabinin, Y. Komissarchik, P. I. Ivanov, and S. N. Borchsenius. 1994. Tubular structures of Mycoplasma gallisepticum and their possible participation in cell motility. Microbiology 140671-681. [DOI] [PubMed] [Google Scholar]
- 25.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51917-924. [DOI] [PubMed] [Google Scholar]
- 26.Krause, D. C., T. Proft, C. T. Hedreyda, H. Hilbert, H. Plagens, and R. Herrmann. 1997. Transposon mutagenesis reinforces the correlation between Mycoplasma pneumoniae cytoskeletal protein HMW2 and cytadherence. J. Bacteriol. 1792668-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krunkosky, T. M., J. L. Jordan, E. Chambers, and D. C. Krause. 2007. Mycoplasma pneumoniae host-pathogen studies in an air-liquid culture of differentiated human airway epithelial cells. Microb. Pathog. 4298-103. [DOI] [PubMed] [Google Scholar]
- 28.Kusumoto, A., S. Seto, J. D. Jaffe, and M. Miyata. 2004. Cell surface differentiation of Mycoplasma mobile visualized by surface protein localization. Microbiology 1504001-4008. [DOI] [PubMed] [Google Scholar]
- 29.Maniloff, J., H. J. Morowitz, and R. J. Barrnett. 1965. Ultrastructure and ribosomes of Mycoplasma gallisepticum. J. Bacteriol. 90193-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.May, M., L. Papazisi, T. S. Gorton, and S. J. Geary. 2006. Identification of fibronectin-binding proteins in Mycoplasma gallisepticum strain R. Infect. Immun. 741777-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng, K. E., and R. M. Pfister. 1980. Intracellular structures of Mycoplasma pneumoniae revealed after membrane removal. J. Bacteriol. 144390-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metsugi, S., A. Uenoyama, J. Adan-Kubo, M. Miyata, K. Yura, H. Kono, and N. Go. 2005. Sequence analysis of the gliding protein Gli349 in Mycoplasma mobile. Biophysics 133-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyata, M. 2002. Cell division, p. 117-130. In R. Herrmann and S. Razin (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, London, United Kingdom.
- 34.Miyata, M. 2008. Centipede and inchworm models to explain Mycoplasma gliding. Trends Microbiol. 166-12. [DOI] [PubMed] [Google Scholar]
- 35.Miyata, M. 2005. Gliding motility of mycoplasmas—the mechanism cannot be explained by current biology, p. 137-163. In A. Blanchard and G. Browning (ed.), Mycoplasmas: pathogenesis, molecular biology, and emerging strategies for control. Horizon Bioscience, Norfolk, United Kingdom.
- 36.Miyata, M. 2007. Molecular mechanism of mycoplasma gliding—a novel cell motility system, p. 137-175. In P. Lenz (ed.), Cell motility. Springer, New York, NY.
- 37.Miyata, M., and H. Ogaki. 2006. Cytoskeleton of mollicutes. J. Mol. Microbiol. Biotechnol. 11256-264. [DOI] [PubMed] [Google Scholar]
- 38.Miyata, M., and J. Petersen. 2004. Spike structure at interface between gliding Mycoplasma mobile cell and glass surface visualized by rapid-freeze and fracture electron microscopy. J. Bacteriol. 1864382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyata, M., W. S. Ryu, and H. C. Berg. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 1841827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyata, M., and S. Seto. 1999. Cell reproduction cycle of mycoplasma. Biochimie 81873-878. [DOI] [PubMed] [Google Scholar]
- 41.Miyata, M., and A. Uenoyama. 2002. Movement on the cell surface of gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215285-289. [DOI] [PubMed] [Google Scholar]
- 42.Miyata, M., H. Yamamoto, T. Shimizu, A. Uenoyama, C. Citti, and R. Rosengarten. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 1461311-1320. [DOI] [PubMed] [Google Scholar]
- 43.Morowitz, H. J., and J. Maniloff. 1966. Analysis of the life cycle of Mycoplasma gallisepticum. J. Bacteriol. 911638-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagai, R., and M. Miyata. 2006. Gliding motility of Mycoplasma mobile can occur by repeated binding to N-acetylneuraminyllactose (sialyllactose) fixed on solid surfaces. J. Bacteriol. 1886469-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakane, D., and M. Miyata. 2007. Cytoskeletal “jellyfish” structure of Mycoplasma mobile. Proc. Natl. Acad. Sci. USA 10419518-19523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohtani, N., and M. Miyata. 2007. Identification of a novel nucleoside triphosphatase from Mycoplasma mobile: a prime candidate for motor of gliding motility. Biochem. J. 40371-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papazisi, L., S. Frasca, Jr., M. Gladd, X. Liao, D. Yogev, and S. J. Geary. 2002. GapA and CrmA coexpression is essential for Mycoplasma gallisepticum cytadherence and virulence. Infect. Immun. 706839-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papazisi, L., T. S. Gorton, G. Kutish, P. F. Markham, G. F. Browning, D. K. Nguyen, S. Swartzell, A. Madan, G. Mahairas, and S. J. Geary. 2003. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain R(low). Microbiology 1492307-2316. [DOI] [PubMed] [Google Scholar]
- 49.Popham, P. L., T. W. Hahn, K. A. Krebes, and D. C. Krause. 1997. Loss of HMW1 and HMW3 in noncytadhering mutants of Mycoplasma pneumoniae occurs post-translationally. Proc. Natl. Acad. Sci. USA 9413979-13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 621094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sbalzarini, I. F., and P. Koumoutsakos. 2005. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 151182-195. [DOI] [PubMed] [Google Scholar]
- 52.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 1831621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seto, S., T. Kenri, T. Tomiyama, and M. Miyata. 2005. Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J. Bacteriol. 1871875-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seto, S., and M. Miyata. 2003. The attachment organelle formation represented by localization of cytadherence protein and formation of electron-dense core in the wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 1851082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seto, S., and M. Miyata. 1999. Partitioning, movement, and positioning of nucleoids in Mycoplasma capricolum. J. Bacteriol. 1816073-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seto, S., A. Uenoyama, and M. Miyata. 2005. Identification of 521-kilodalton protein (Gli521) involved in force generation or force transmission for Mycoplasma mobile gliding. J. Bacteriol. 1873502-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seybert, A., R. Herrmann, and A. S. Frangakis. 2006. Structural analysis of Mycoplasma pneumoniae by cryo-electron tomography. J. Struct. Biol. 156342-354. [DOI] [PubMed] [Google Scholar]
- 58.Shimizu, T., and M. Miyata. 2002. Electron microscopic studies of three gliding mycoplasmas, Mycoplasma mobile, M. pneumoniae, and M. gallisepticum, by using the freeze-substitution technique. Curr. Microbiol. 44431-434. [DOI] [PubMed] [Google Scholar]
- 59.Uenoyama, A., A. Kusumoto, and M. Miyata. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 1861537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uenoyama, A., and M. Miyata. 2005. Gliding ghosts of Mycoplasma mobile. Proc. Natl. Acad. Sci. USA 10212754-12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uenoyama, A., and M. Miyata. 2005. Identification of a 123-kilodalton protein (Gli123) involved in machinery for gliding motility of Mycoplasma mobile. J. Bacteriol. 1875578-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uenoyama, A., S. Seto, D. Nakane, and M. Miyata. 2009. Regions on Gli349 and Gli521 protein molecules directly involved in movements of Mycoplasma mobile gliding machinery, suggested by use of inhibitory antibodies and mutants. J. Bacteriol. 1911982-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vasconcelos, A. T., H. B. Ferreira, C. V. Bizarro, S. L. Bonatto, M. O. Carvalho, P. M. Pinto, D. F. Almeida, L. G. Almeida, R. Almeida, L. Alves-Filho, E. N. Assuncao, V. A. Azevedo, M. R. Bogo, M. M. Brigido, M. Brocchi, H. A. Burity, A. A. Camargo, S. S. Camargo, M. S. Carepo, D. M. Carraro, J. C. de Mattos Cascardo, L. A. Castro, G. Cavalcanti, G. Chemale, R. G. Collevatti, C. W. Cunha, B. Dallagiovanna, B. P. Dambros, O. A. Dellagostin, C. Falcao, F. Fantinatti-Garboggini, M. S. Felipe, L. Fiorentin, G. R. Franco, N. S. Freitas, D. Frias, T. B. Grangeiro, E. C. Grisard, C. T. Guimaraes, M. Hungria, S. N. Jardim, M. A. Krieger, J. P. Laurino, L. F. Lima, M. I. Lopes, E. L. Loreto, H. M. Madeira, G. P. Manfio, A. Q. Maranhao, C. T. Martinkovics, S. R. Medeiros, M. A. Moreira, M. Neiva, C. E. Ramalho-Neto, M. F. Nicolas, S. C. Oliveira, R. F. Paixao, F. O. Pedrosa, S. D. Pena, M. Pereira, L. Pereira-Ferrari, I. Piffer, L. S. Pinto, D. P. Potrich, A. C. Salim, F. R. Santos, R. Schmitt, M. P. Schneider, A. Schrank, I. S. Schrank, A. F. Schuck, H. N. Seuanez, D. W. Silva, R. Silva, S. C. Silva, C. M. Soares, K. R. Souza, R. C. Souza, C. C. Staats, M. B. Steffens, S. M. Teixeira, T. P. Urmenyi, M. H. Vainstein, L. W. Zuccherato, A. J. Simpson, and A. Zaha. 2005. Swine and poultry pathogens: the complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J. Bacteriol. 1875568-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weisburg, W. G., J. G. Tully, D. L. Rose, J. P. Petzel, H. Oyaizu, D. Yang, L. Mandelco, J. Sechrest, T. G. Lawrence, J. Van Etten, J. Maniloff, and C. R. Woese. 1989. A phylogenetic analysis of the mycoplasmas: basis for their classification. J. Bacteriol. 1716455-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.