Abstract
A multiply antibiotic-resistant Acinetobacter baumannii strain, 3208, contains the aacC1-orfP-orfP-orfQ-aadA1 gene cassette array; sul1, tetA(A), and aphA1b genes; and a mer operon in a large region containing a novel transposon, Tn6020, and segments of Tn1696, Tn21, Tn1721, and Tn5393. This region is part of a genomic resistance island, AbaR5, related to and found in the same chromosomal position as AbaR1. This strain is the first European clone I isolate detected in Australia.
The increase in antimicrobial resistance in Acinetobacter baumannii in hospitals is a major concern, and strains that are resistant to almost all currently available antibacterial agents have been observed (17, 18). Acinetobacter species are naturally transformable and thus can potentially readily acquire resistance genes from the mobile gene pool, and many different antibiotic resistance genes and combinations of resistance genes have been found in clinical isolates (5, 17, 18). However, there is currently very little detailed information on the locations of these genes and their relationships to one another. Until recently, the multiple-antibiotic resistance regions from only two A. baumannii strains had been reported. Strain AYE, which is representative of a clonal line that is epidemic in French hospitals, contains a large cluster of antibiotic and heavy-metal resistance genes in an 86.2-kb resistance island designated AbaR1 (6). The 18 antibiotic resistance genes, some of which are present in multiple copies, confer resistance to most of the known classes of antibiotics. The second strain, ACICU, carries an antibiotic resistance island, AbaR2, consisting of only a small remnant of the AbaR1 region modified by the substitution of an aacA4-orf-oxa1 cassette array for the aacC1-orfP-orfP-orfQ-aadA1 array found in AbaR1 (7). Here, we have examined a cluster of antibiotic resistance genes found in a multiply antibiotic-resistant A. baumannii strain isolated in Australia. After this work was completed, a further sequence containing a related region, designated AbaR3, became available (1).
A. baumannii strain 3208.
Isolate 32080497, herein called 3208, was isolated in 1997 at a Sydney, Australia, hospital from a blood sample. It was identified as A. baumannii with a Vitek 2 compact identification system (bioMérieux, Hazelwood, MO). The sequence of a fragment from the recA gene, amplified as described previously (8, 13), was identical to that of recA of A. baumannii AYE (GenBank accession no. CU459141) (24).
Antibiotic susceptibility for a range of antibiotics was tested using the Vitek 2 system and interpreted using CLSI guidelines (3). 3208 showed intermediate resistance to ceftriaxone, ceftazidime, and cefepime (MIC, 16 μg/ml) as well as ticarcillin-clavulanate (MIC, 32 μg/ml) but was susceptible to piperacillin-tazobactam (MIC, ≤4 μg/ml) and meropenem (MIC, 0.5 μg/ml). It was also resistant to gentamicin (MIC, ≥16 μg/ml) but susceptible to tobramycin (MIC, ≤1 μg/ml). In addition, 3208 was resistant to ciprofloxacin (MIC, ≥4 μg/ml) and trimethoprim-sulfamethoxazole (MIC, ≥320 μg/ml). Additional MICs for antibiotics for which breakpoints for A. baumannii have not been established were ≥32 μg/ml for nalidixic acid, ≥16 μg/ml for norfloxacin, ≥16 μg/ml for trimethoprim, and 1 μg/ml for tigecycline.
Identification of resistance genes.
The intI1 gene from the 5′-conserved segment (5′-CS) and the sul1 sulfonamide resistance gene from the 3′-conserved segment (3′-CS) of a class 1 integron were detected using PCR conditions described previously (19) (see Table S1 in the supplemental material for primers). PCR products were purified using a QIAquick PCR purification kit (Qiagen, Inc., Valencia, CA) or, after separation in an agarose gel, using a QIAquick gel extraction kit. Automated sequencing was performed with an ABI Prism 377 DNA sequencer or a 3130 Exel genetic analyzer (Applied Biosystems, Carlsbad, CA) using the Big Dye system. Amplification of the gene cassettes yielded a 3-kb PCR amplicon that contained the aacC1-orfP-orfP-orfQ-aadA1 gene cassettes (aacC1 confers resistance to gentamicin, and aadA1 confers resistance to streptomycin and spectinomycin). Though found first in Serratia marcescens (2) (GenBank accession no. AF453999), this cassette array, or a variation of it lacking one copy of the orfP gene cassette, has been found most often in A. baumannii isolates (11, 12, 22, 23). It is also present in AbaR1 (GenBank accession no. CT025832 and CU459141) (6).
PCR screening for further resistance genes and several insertion sequences (IS) (see Table S1 in the supplemental material for a complete list) detected the aphA1 (kanamycin and neomycin resistance), tetA(A) (tetracycline resistance), and merA (mercuric ion resistance) genes as well as IS6100 and IS26.
Structure of the 3208 multiple-antibiotic resistance region.
Digestion of the merA amplicon with restriction enzyme RsaI identified it as the Tn1696-associated gene (14), and IS6100 was found to be linked to the mer module and to the sul1 gene, as it is in Tn1696 (GenBank accession no. U12338) (14, 16). However, the gene cassettes could be linked to the intI1 gene and the sul1 gene but not to IS6100, indicating that there are at least two copies of the sul1 gene.
The aphA1 gene was shown to be flanked by directly oriented copies of IS26, as is aphA1a in transposon Tn4352 (26) or Tn4352B (15). However, the sequence revealed an aphA1b gene within a longer central fragment of 1,254 bp, whose sequence diverges from that of Tn4352 immediately downstream of the aphA1 gene, and a complete copy and a partial copy of IS26 were found on one side (Fig. 1A, line 3). This structure was named Tn6020, and the only sequence in GenBank identical to that of Tn6020 was the aphA1 region in AbaR1. In AbaR1, the class 1 integron containing the aacC1-orfP-orfP-orfQ-aadA1 cassette array is next to Tn6020, with a short piece of the Tn21 urf2M gene separating the IRi integron end from Tn6020. PCR was used to link these two regions, followed by sequencing of the PCR amplicon, the same configuration was shown to be present in strain 3208 (Fig. 1A, lines 3 and 4). This finding suggested that there may be a single multiple-antibiotic resistance region in 3208 that is related to or derived from AbaR1. Using the latter part of the AbaR1 sequence as a guide, we mapped and sequenced a continuous region of 36,153 bp from strain 3208 that includes all of the identified antibiotic resistance genes. All of the junctions between adjacent regions (see below for definitions of the regions) were confirmed by PCRs overlapping one or more boundaries, and sequences were assembled using Sequencher, version 4.8 (Gene Codes Corporation, Ann Arbor, MI). The left-hand copy of the sul1 gene, which is separated from a cadmium/zinc resistance transposon, here designated Tn6018, by a region of 29 kb in AbaR1, was linked to Tn6018 by long-range PCR, and a further 5,986 bp of sequence that includes the region marked X in Fig. 1A was obtained. This 3208 sequence (total of 42,139 bp) commences within the left-hand copy of transposon Tn6018 (Fig. 1A) and extends to within the chromosomally located ATPase gene that was interrupted by the insertion of AbaR1 (6) (see below for details).
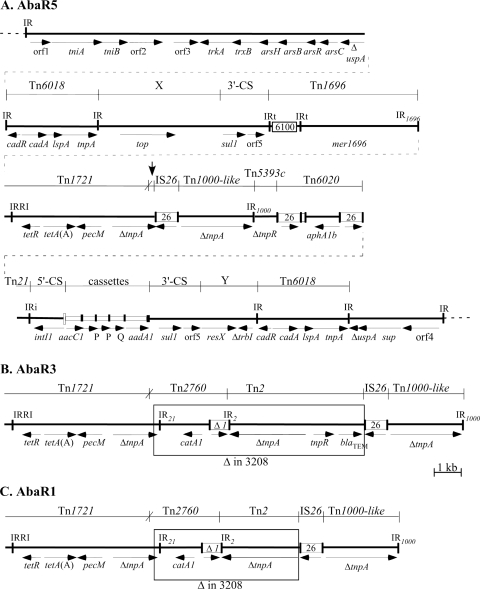
FIG. 1.
Map of AbaR5, showing the origins of different segments. (A) AbaR5. In the central line, the dotted lines represent chromosomal DNA flanking AbaR5, and IR of transposons are indicated with vertical lines. IR within the MARR between the Tn6018 regions have an identifying notation, IRi and IRt for class 1 integron ends and IR1696, IR21, IR2, and IR1000 for IR from Tn1696, Tn21, Tn2, and Tn1000-like, respectively. Numbered open boxes represent the IS, with the number indicating the identity of the IS. The attI1 site of the class 1 integron is shown as a tall open box, and cassettes are represented by an open box with a vertical bar representing the attC site. The lines above indicate the extents of the regions derived from known transposons, with vertical lines indicating the boundaries between segments and X indicating a recombination crossover. The mer region of Tn1696, encoding the merEDACTPR genes, is represented by mer1696. The extents of other genes are shown by horizontal arrows with the gene names underneath. The genes named resX, sup, and orf4 potentially encode a resolvase, a sulfate permease, and a protein of unknown function, respectively. The sup gene was incorrectly identified as sul1 in earlier publications (6, 20). The vertical arrow indicates the position of the deletion. (B and C) Portions of AbaR3 and AbaR1, respectively, that contain the catA1 and blaTEM genes. Only the region flanked by the segments from Tn1721 and Tn1000-like is shown, and the segment missing from AbaR5 is boxed. AbaR3 is otherwise identical to AbaR5, except that only one copy of the orfP cassette is present. AbaR1 contains a large segment replacing the region between Tn6018 and sul1, marked X in the in the top line of panel A. The structure of most of this region can be found in reference 8. Gene Construction Kit (version 2.5; Textco, West Lebanon, NH) was used to create the figure to scale.
The antibiotic resistance genes all lie between the two copies of Tn6018, and we have designated this the multiple-antibiotic resistance region (MARR). To examine how the MARR arose, the various discrete regions of DNA (e.g., gene cassettes, 5′-CS, 3′-CS, and IS6100, etc.) that make up the assembled sequence were delineated by matching the sequence with known complete examples of these regions (listed in Table 1) by using the BLAST paired alignment facility (http://blast.ncbi.nlm.nih.gov). Most of the boundaries between independently derived segments (Fig. 1, vertical lines in top line) coincide with one of the inverted repeats (IR) of one of the transposons, and the overall configuration clearly arose by insertion of one transposon into the next, leaving none of them complete. One boundary (marked with a cross in the top line of Fig. 1A) arose by homologous recombination within a short region of identity in the tnpA genes of Tn1721 and Tn21 to create a hybrid. There are two copies of the sul1 gene, both located within partial copies of the 3′-CS. The regions to the left and right of these 3′-CS segments, marked X and Y in the top and bottom lines of Fig. 1A, are found only in other AbaR regions (see Table 1 and below), and there are no identifiable features at the boundaries.
TABLE 1.
Modules in AbaR5
| Regiona | Length (bp) | Progenitor
|
Region(s)/gene(s) | % Identity | |||
|---|---|---|---|---|---|---|---|
| Name | Length (bp) | GenBank accession no.b | Part present (position, in bp)c | ||||
| 1 | 4,421 | X | 4,421 | CP001182 | top | 100 | |
| 2 | 7,322 | Tn1696 | 16,318 | U12338 | 8675-16318d | Δ3′-CS -IS6100-mer1696 | 100 |
| 3 | 5,157e | Tn1721 | 11,139 | X61367f | 5560-10727 | ΔtnpA-tetR-tetA-pecM-ΔtnpA | 99.7 |
| 4 | 154e | Tn21 | 19,672 | AF071413 | 282-435 | ΔtnpA | 100 |
| 5 | 820 | IS26 | 820 | AY123253 | Complete | IS26 | 100 |
| 6 | 2,883g | Tn1000 | 5,981 | X60200 | 1-2883 | ΔtnpA | 74.9 |
| 7 | 685 | Tn5393c | 5,470 | AF313472 | 2968-3652 | ΔtnpR | 99.9 |
| 8 | 820 | IS26h | 820 | AY123253 | Complete | IS26 | 100 |
| 9 | 175 | IS26h | 820 | AY123253 | 645-820 | ΔIS26 | 100 |
| 10 | 3,069 | Tn6020 | 3,069 | CT025832 | 1-3069 | aphA1b | |
| 11 | 820 | IS26 | 820 | AY123253 | Complete | IS26 | 100 |
| 12 | 446 | Tn21 | 19,672 | AF071413 | 3594-4039 | ΔtnpM | 100 |
| 13 | 1,371 | 5′-CSi | 1,371 | U12338 | Complete | 5′-CS | 99.6 |
| 14 | 2,962 | Cassettes | 2,962 | AF453999 | 1-2962 | aacC1-orfP-orfP-orfQ-aadA1j | 99.6 |
| 15 | 1,987 | 3′-CSi | 2,239 | U12338 | 1-1987 | 3′-CS | 100 |
| 16 | 2,025 | Y | CT025832k | resX-ΔtrbI | |||
| 17 | 3,372 | Tn6018l | 3,372 | AY128707 | Complete | cadR-cadA-lspA-tnpA | 96.7 |
| 18 | 4,003 | CT025832k | ΔuspA-sup-orf4m | ||||
The analysis begins with the first base of the multiple-antibiotic resistance region, i.e., after the left-hand Tn6018 in AbaR5.
Accession numbers used are either the earliest complete sequences or the complete and annotated sequences.
Numbering for IS is with the transposase gene from left to right; for class II Tn, the tnpA gene is on the left, except for Tn1721, which is as in X61367.
The first base of this region corresponds to bp 391 of the 3′-CS, and AbaR5 lacks the partial copy of IS6100 (bp 11527 to 11974) in Tn1696.
Regions 3 and 4 overlap by 23 bp that are identical in Tn21 and Tn1721 and could be derived from either.
This sequence has several single base differences and a short deletion which are not present in other published sequences of this region and might be errors in the Tn1721 complete sequence (X61367).
A portion (1,209 bp) was 99.6% identical to a partially sequenced Tn1000-like element found in GenBank accession no. AY598759.
Part of IS15.
5′-CS and 3′-CS are the 5′ conserved segment and 3′ conserved segment, respectively, of a class 1 integron.
Open reading frames orfP and orfQ have also been called orfX and orfX′, respectively.
Found only in A. baumannii strains AYE and ACICU.
Was described as ISPu12-like but is not an IS and was therefore renamed.
sup (sulfate permease) is incorrectly labeled sul1 (sulfonamide-resistant dihydropteroate synthase) in previous publications (1, 6, 20); orf4 encodes an open reading frame of unknown function.
nChromosomal gene found in A. baumannii strains AYE, ACICU, and ATCC 17978.
The region of 3208 found to the left of the left-hand Tn6018 was mapped using overlapping PCRs with primers (see Table S1 in the supplemental material) designed using the sequence of the corresponding region of AbaR1, which contains an arsenate resistance region (Fig. 1A, line 1). The PCR products were digested with restriction enzyme DdeI or HindIII and yielded fragments of the sizes predicted from the AbaR1 sequence, indicating that this region is the same as that in AbaR1. Sequencing revealed that the boundaries with the left-hand part of the ATPase gene and with the left-hand Tn6018 were identical to those in AbaR1. Because of the similarities to both AbaR1 and AbaR3 (see below), the 3208 region shown between the dashed lines in Fig. 1A was named AbaR5.
Relationship of AbaR5 to AbaR1 and AbaR3.
AbaR1 was the first AbaR region identified, and it was defined as the region that has inserted into an ATPase gene in the A. baumannii genome, creating a 5-bp duplication (6). AbaR1 is actually a large (86.2-kb) composite transposon that is bounded by imperfect IR of 26 bp (19/26 matches). Our analysis of this sequence revealed that the MARR and the two flanking copies of Tn6018 together constitute a compound transposon that creates a duplication of 8 bp when inserted into the uspA gene in the backbone transposon. AbaR5 has the same general structure. However, none of the resistance genes [veb1, oxa10, arr2, aadB, an unnumbered aacA gene, aadA1, strA and strB, cmlA1, cmlA5, cmlA9, tetA(G), dfrA1, and dfrA10] found in the first part of the MARR segment of AbaR1 were detected in 3208. Most of this 29-kb region was reanalyzed recently (9) and was shown to consist of a very complex class 1 integron. AbaR1 also carries a catA1 gene that is missing from AbaR5, and this can be accounted for by an IS26-induced deletion of 5,287 bp relative to AbaR1 (boxed in Fig. 1C).
The configuration of the AbaR5 region from strain 3208 is largely identical to that of AbaR3 (GenBank accession no. CP001182). The only difference is that AbaR5 lacks the catA1 and blaTEM genes found in AbaR3 (Fig. 1B), and this was accounted for by an IS26-induced deletion of 7,324 bp (boxed in Fig. 1B). Thus, it is possible that the AbaR5 configuration arose from AbaR3 or that both have a common ancestor.
Clonality.
In Europe, much of the increase in resistance is due to the spread of a small number of clones, designated European clones I, II, and III (4, 12, 21, 25). However, these clones have not been reported in Australia (17). As the A. baumannii strain AYE, carrying AbaR1, was recently shown to belong to European clone I (21) and as strain AB0057 (AbaR3) is very closely related to it (1) but strain ACICU (AbaR2) belongs to European clone II (7), we examined the possibility that 3208 was a member of one of these clones. By use of a recently described discriminatory PCR assay (21), 3208 was found to belong to European clone I. This is therefore the first report of a European clone I strain in Australia, indicating that the clonal identity of other Australian multiply resistant A. baumannii isolates (10, 23), particularly those that carry the same cassette array as AbaR1, AbaR3, and AbaR5 and also the aphA1b gene, should be investigated.
Nucleotide sequence accession number.
The sequence of the MARR of A. baumannii strain 3208 has been deposited in GenBank under accession no. FJ172370.
Supplementary Material
Acknowledgments
R.M.H. was supported by NHMRC fellowship grant 358713. V.P. was supported by a University of Sydney International Research Scholarship, and the project was supported by NHMRC project grant 352352.
We thank Clarence Fernandez for supplying the strain and John Merlino for the Vitek 2 analysis.
Footnotes
Published ahead of print on 13 April 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Adams, M. D., K. Goglin, N. Molyneaux, K. M. Hujer, H. Lavender, J. J. Jamison, I. J. MacDonald, K. M. Martin, T. Russo, A. A. Campagnari, A. M. Hujer, R. A. Bonomo, and S. R. Gill. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centrón, D., and P. H. Roy. 2002. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 46:1402-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.Dijkshoorn, L., H. Aucken, P. Gerner-Smidt, P. Janssen, M. E. Kaufmann, J. Garaizar, J. Ursing, and T. L. Pitt. 1996. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J. Clin. Microbiol. 34:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dijkshoorn, L., A. Nemec, and H. Seifert. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939-951. [DOI] [PubMed] [Google Scholar]
- 6.Fournier, P.-E., D. Vallenet, V. Barbe, S. Audic, H. Ogata, L. Poirel, H. Richet, C. Robert, S. Mangenot, C. Abergel, P. Nordmann, J. Weissenbach, D. Raoult, and J. M. Claverie. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iacono, M., L. Villa, D. Fortini, R. Bordoni, F. Imperi, R. J. Bonnal, T. Sicheritz-Ponten, G. De Bellis, P. Visca, A. Cassone, and A. Carattoli. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52:2616-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krawczyk, B., K. Lewandowski, and J. Kur. 2002. Comparative studies of the Acinetobacter genus and the species identification method based on the recA sequences. Mol. Cell. Probes. 16:1-11. [DOI] [PubMed] [Google Scholar]
- 9.Levings, R. S., S. P. Djordjevic, and R. M. Hall. 2008. SGI2, a relative of Salmonella genomic island SGI1 with an independent origin. Antimicrob. Agents Chemother. 52:2529-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mak, J. K., M. J. Kim, J. Pham, J. Tapsall, and P. A. White. 2009. Antibiotic resistance determinants in nosocomial strains of multidrug-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 63:47-54. [DOI] [PubMed] [Google Scholar]
- 11.Nemec, A., L. Dolzani, S. Brisse, P. van den Broek, and L. Dijkshoorn. 2004. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J. Med. Microbiol. 53:1233-1240. [DOI] [PubMed] [Google Scholar]
- 12.Nemec, A., L. Krízová, M. Maixnerová, L. Diancourt, T. J. van der Reijden, S. Brisse, P. van den Broek, and L. Dijkshoorn. 2008. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug-resistant strains of European clone II. J. Antimicrob. Chemother. 62:484-489. [DOI] [PubMed] [Google Scholar]
- 13.Nowak, A., and J. Kur. 1996. Differentiation of seventeen genospecies of Acinetobacter by multiplex polymerase chain reaction and restriction fragment length polymorphism analysis. Mol. Cell. Probes 10:405-411. [DOI] [PubMed] [Google Scholar]
- 14.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez, F., A. M. Hujer, K. M. Hujer, B. K. Decker, P. N. Rather, and R. A. Bonomo. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3471-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Post, V., G. D. Recchia, and R. M. Hall. 2007. Detection of gene cassettes in Tn402-like class 1 integrons. Antimicrob. Agents Chemother. 51:3467-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, M. G., T. A. Gianoulis, S. Pukatzki, J. J. Mekalanos, L. N. Ornston, M. Gerstein, and M. Snyder. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21:601-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turton, J. F., S. N. Gabriel, C. Valderrey, M. E. Kaufmann, and T. L. Pitt. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807-815. [DOI] [PubMed] [Google Scholar]
- 22.Turton, J. F., M. E. Kaufmann, J. Glover, J. M. Coelho, M. Warner, R. Pike, and T. L. Pitt. 2005. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J. Clin. Microbiol. 43:3074-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valenzuela, J. K., L. Thomas, S. R. Partridge, T. van der Reijden, L. Dijkshoorn, and J. Iredell. 2007. Horizontal gene transfer in a polyclonal outbreak of carbapenem-resistant Acinetobacter baumannii. J. Clin. Microbiol. 45:453-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vallenet, D., P. Nordmann, V. Barbe, L. Poirel, S. Mangenot, E. Bataille, C. Dossat, S. Gas, A. Kreimeyer, P. Lenoble, S. Oztas, J. Poulain, B. Segurens, C. Robert, C. Abergel, J. M. Claverie, D. Raoult, C. Médigue, J. Weissenbach, and S. Cruveiller. 2008. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS ONE 19:e1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Dessel, H., L. Dijkshoorn, T. van der Reijden, N. Bakker, A. Paauw, P. van den Broek, J. Verhoef, and S. Brisse. 2004. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res. Microbiol. 155:105-112. [DOI] [PubMed] [Google Scholar]
- 26.Wrighton, C. J., and P. Strike. 1987. A pathway for the evolution of the plasmid NTP16 involving the novel kanamycin resistance transposon Tn4352. Plasmid 17:37-45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.