Abstract
The eukaryotic plasma membrane exhibits both asymmetric distribution of lipids between the inner and the outer leaflet and lateral segregation of membrane components within the plane of the bilayer. In budding yeast (Saccharomyces cerevisiae), maintenance of leaflet asymmetry requires P-type ATPases, which are proposed to act as inward-directed lipid translocases (Dnf1, Dnf2, and the associated protein Lem3), and ATP-binding cassette (ABC) transporters, which are proposed to act as outward-directed lipid translocases (Pdr5 and Yor1). The S. cerevisiae genome encodes two other Pdr5-related ABC transporters: Pdr10 (67% identity) and Pdr15 (75% identity). We report the first analysis of Pdr10 localization and function. A Pdr10-GFP chimera was located in discrete puncta in the plasma membrane and was found in the detergent-resistant membrane fraction. Compared to control cells, a pdr10∆ mutant was resistant to sorbate but hypersensitive to the chitin-binding agent Calcofluor White. Calcofluor sensitivity was attributable to a partial defect in endocytosis of the chitin synthase Chs3, while sorbate resistance was attributable to accumulation of a higher than normal level of the sorbate exporter Pdr12. Epistasis analysis indicated that Pdr10 function requires Pdr5, Pdr12, Lem3, and mature sphingolipids. Strikingly, Pdr12 was shifted to the detergent-resistant membrane fraction in pdr10∆ cells. Pdr10 therefore acts as a negative regulator for incorporation of Pdr12 into detergent-resistant membranes, a novel role for members of the ABC transporter superfamily.
Keywords: Pdr10, ABC transporter, Sphingolipid, Lipid raft, Detergent-resistant membrane, Lateral segregation, Pdr12, Lem3, Dnf1, Dnf2
It has been appreciated for quite some time that the two leaflets of a eukaryotic plasma membrane have distinct lipid compositions (Devaux 1991; Gordesky and Marinetti 1973). Now there is much evidence for additional layers of structural complexity in the plasma membrane. For example, fluorescently tagged annexins with different lipid specificities will label distinct regions of the plasma membrane in smooth muscle cells (Draeger et al. 2005). Such distinct subdomains in the plasma membrane are also reminiscent of much more specialized structures, like the rhodopsin-rich photosensory discs in the vertebrate retina. Similarly, the plasma membrane of the yeast Saccharomyces cerevisiae contains at least two microscopically distinct subdomains (Malinska et al. 2003, 2004). One of these compartments contains the plasma membrane ATPase Pma1, whereas the other contains a number of permeases, including Can1 and Fur4 (Bagnat et al. 2000; Dupré and Haguenauer-Tsapis 2003; Hearn et al. 2003; Malinska et al. 2003, 2004). By contrast, the amino acid permease Gap1 is found in both compartments of the yeast plasma membrane (Lauwers and Andre 2006; Lauwers et al. 2007).
It has been proposed that these subdomains of the plasma membrane can be equated with structures variously termed lipid rafts or detergent-resistant membranes (DRMs), which have been proposed in a variety of eukaryotic systems (Bagnat et al. 2000; Simons and Toomre 2000). This proposal remains quite controversial (Kenworthy 2008; Munro 2003). In particular, it is unclear whether there is any relationship between plasma membrane microdomains observed in vivo by fluorescence microscopy and DRM fractions obtained in vitro (Lichtenberg et al. 2005). By definition, a DRM fraction can only be observed after extraction of the membrane with detergent and thus may not correspond to structures in the native membrane (Lichtenberg et al. 2005). Based on studies of model membranes, it has been argued that DRM fractions might be artifactually created by detergent treatment (Heerklotz 2002; Heerklotz et al. 2003; Keller et al. 2005; Lichtenberg et al. 2005), particularly in the presence of sterols or sphingolipids. Attempts to perturb detergent-resistant subdomains in vivo via depletion of sterols are similarly subject to alternative interpretations (Kenworthy 2008).
Despite the questionable correspondence between in vivo observations of lateral segregation or microdomain formation and in vitro examination of DRM components, it is clear that the formation, turnover, and biological significance of membrane microdomains are important topics of current research. Establishment of these unique regions, whether highly specialized or rudimentary, requires lateral segregation of both lipid and protein components within the plane of the plasma membrane (Bogdanov et al. 2008). S. cerevisiae has proven to be a valuable system for studying this phenomenon, due largely to thorough elucidation of lipid biosynthetic pathways in this organism (Carman and Henry 2007), particularly for sphingolipids (Daum et al. 1998; Dickson 2008).
Advances have been made in our understanding of the underlying biophysical basis of the lateral segregation found in plasma membranes. Sphingolipids and sterols seem important both for “raft” assembly in vivo and for formation of the DRM fraction in vitro (Bagnat et al. 2000; Dupré and Haguenauer-Tsapis 2003; Hearn et al. 2003). There is also evidence that certain glycerophospholipids are important in establishment of discrete plasma membrane subdomains, including phosphatidylethanolamine (PE) (Opekarova et al. 2005) and a specialized phosphatidyinositol with a very long fatty acid substituent that is a component of GPI-anchored proteins (Fujita et al. 2006). It has been reported that localization of certain permeases to one of the plasma membrane subdomains in S. cerevisiae depends on maintenance of the transmembrane electrochemical potential, whereas other segregated plasma membrane proteins, including the eisosome component Sur7 (Walther et al. 2006) and the plasma membrane ATPase Pma1, are not affected by membrane depolarization (Grossmann et al. 2007). Pma1 is able to form high molecular weight oligomers in the presence of ceramide (Bagnat et al. 2001; Lee et al. 2002), suggesting that, at least in some cases, protein-protein interactions can contribute to lateral segregation.
In eukaryotic cells, aminophospholipids, like PE, are normally markedly enriched in the inner leaflet of the plasma membrane. Some of the enzymes that contribute to the maintenance of such plasma membrane asymmetry have been elucidated in yeast. For PE and phosphatidylcholine (PC), there are both inward-directed and outward-directed lipid translocases (Kean et al. 1993; Pomorski et al. 2003). Thus, establishment of the observed asymmetric distribution demands balance between these opposing catalytic activities. The inward-directed PE translocases in the plasma membrane of S. cerevisiae are proposed to be Dnf1 and Dnf2, two members of a novel subgroup (class 4) of membrane transporters of the P-type ATPase family (Hua et al. 2002; Pomorski et al. 2003), although there is some ambiguity as to whether these proteins are responsible for translocation of phosphatidylserine (Chen et al. 2006; Stevens et al. 2008). For their proper localization and function, Dnf1 and Dnf2 associate with a noncatalytic subunit, Lem3/Ros3 (Kato et al. 2002; Saito et al. 2004). Dnf1 and Dnf2 also catalyze inward translocation of other compounds, including PC, lysophospholipids, and ether-linked lipid antifungals (Elvington et al. 2005; Hanson et al. 2003; Riekhof et al. 2007). Dnf1, Dnf2, and Lem3 all have paralogues that reside in intracellular membranes and that have been implicated in endocytosis, intracellular vesicular trafficking, and polarized growth (Chen et al. 2006; Furuta et al. 2007; Hua et al. 2002; Pomorski et al. 2003; Saito et al. 2007). Interestingly, in some instances, these biological processes do not seem to require the translocase function of these proteins per se (Natarajan et al. 2004; Noji et al. 2006).
The outward-directed PE translocases in the plasma membrane of S. cerevisiae are thought to be members of a different superfamily of membrane transporters, the ATP-binding-cassette (ABC) transporters (Pomorski et al. 2003). The S. cerevisiae plasma membrane likely contains as many as 10 ABC transporters; of these, Pdr5 and Yor1 have been specifically implicated as outward-directed PE translocases (Pomorski et al. 2003). Both had been identified previously via their ability to mediate resistance to a broad spectrum of antimicrobial drugs (Balzi et al. 1994; Bissinger and Kuchler 1994; Cui et al. 1996; Decottignies et al. 1998; Katzmann et al. 1995; Kralli et al. 1995). The S. cerevisiae genome encodes two other ABC transporters that are closely related to Pdr5, which have been designated Pdr10 (67% identity to Pdr5) and Pdr15 (75% identity to Pdr5). Our work has shown that Pdr15 has considerable functional overlap with Pdr5 and can also mediate drug export (Schüller et al. 2007; Wolfger et al. 2004). Unexpectedly, a pdr5∆ pdr15∆ double mutant strain exhibited increased outer-leaflet PE (Schüller et al. 2007), indicating that loss of the activity of two putative outward-directed translocases was somehow offset, presumably by decreased activity of the inward-directed translocases. Our findings suggest, therefore, that maintenance of plasma membrane PE asymmetry is under some type of feedback control, reminiscent of the interplay observed between glycerophospholipid asymmetry and sphingolipid transport (Kihara and Igarashi 2004).
Pdr10 has been presumed to play a role in pleiotropic drug resistance only by virtue of its resemblance to Pdr5 and Pdr15 (Abolmaali et al. 2008; Decottignies et al. 1998) and to have a function that overlaps with those of Pdr5 and Pdr15 only because expression of the PDR10 gene involves the same transcription factors (Pdr1 and Pdr3) that control PDR5 and PDR15 expression (Wolfger et al. 1997). By contrast, for example, the ABC transporter Pdr12 (39% identity to Pdr5) is under the control of a dedicated transcription factor (War1) and has a well-defined and unique function, namely, export of weak organic acids such as sorbate (Kren et al. 2003; Piper et al. 1998; Schüller et al. 2004). In any event, the properties, subcellular localization, and function of Pdr10 had not heretofore been directly examined experimentally.
Here we describe our analysis of the physiological role of Pdr10. First, we constructed a functional Pdr10-GFP chimera to visualize its distribution in vivo. Second, we also examined the behavior of Pdr10 upon biochemical fractionation of cell extracts. Third, we constructed a null mutation and compared the phenotypic characteristics of pdr10∆ cells to those of otherwise isogenic PDR10+ cells. Fourth, we analyzed various properties of the cell envelope (plasma membrane and cell wall) and performed additional genetic analysis to pinpoint the underlying molecular causes of the defects and phenotypes observed. Our findings demonstrate that Pdr10 contributes in a significant way to maintaining proper lateral segregation of Pdr12 by helping to exclude it from the DRM fraction. To our knowledge, the function of Pdr10 in establishment of the normal plasma membrane microenvironment for another protein is a novel role for an ABC transporter in cell biology.
Materials and Methods
Strains, Plasmids, and Growth Conditions
Yeast strains used in this study are listed in Table 1. The strains generated in the course of this study were constructed either by standard yeast genetic methods (Sherman et al. 1986) or by one-step gene replacement via integration of appropriate products of the polymerase chain reaction (PCR) using short flanking homology (Wach et al. 1994). Standard rich media (YPD) and defined dropout media (SCD) were used for yeast cultivation (Sherman et al. 1986). Escherichia coli strain DH5α (Hanahan 1983) was used for routine construction and propagation of plasmids, and Pfu Turbo polymerase (Stratagene, La Jolla, CA) and Taq Gold polymerase (Roche) were used for PCR.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source/reference |
|---|---|---|
| W303-1A | MATaura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100 | Sherman et al. (1986) |
| W303-1B | MATα ura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100 | Sherman et al. (1986) |
| BYB67 | MATa/MATα ura3-1/ura3-1 leu2-3,112/leu2-3, 112 his3-11,15/his3-11,15 trp1-1/trp1-1 ADE2/ade2-1 LYS2/lys2::hisG can1-100/can1-100 | Shulewitz et al. (1999) |
| WKK7 | W303-1A ste6∆::HIS3 | Kuchler et al. (1989) |
| NRY201 | W303-1A pdr5∆::HIS3MX6 | Schüller et al. (2007) |
| NRY212 | W303-1A pdr15∆::TRP1 | Schüller et al. (2007) |
| NRY230a | W303-1A pdr5∆::URA3MX4 pdr10∆::HIS3MX6 | This study |
| NRY236a | W303-1A pdr10∆::HIS3MX6 pdr15∆::TRP1 | This study |
| NRY243b | W303-1A pdr10∆::HIS3MX6 | This study |
| NRY251b | W303-1A pdr10∆::URA3MX4 | This study |
| NRY301b | W303-1A pdr12∆::HIS3MX6 | This study |
| NRY334b | W303-1A YOL075c∆::URA3MX4 | This study |
| NRY336b | W303-1A pdr18∆::HIS3MX6 | This study |
| NRY366a | NRY251 pdr12∆::HIS3MX6 | This study |
| NRY407b | W303-1B cho1∆::HIS3MX6 | This study |
| NRY427a | W303-1A dnf1∆::URA3MX4 dnf2∆::kanMX4 | This study |
| NRY409b | W303-1A sur1∆::URA3MX4 | This study |
| NRY410b | W303-1A ipt1∆::URA3MX4 | This study |
| NRY526a | NRY243 sur1∆::URA3MX4 | This study |
| NRY534a | NRY243 ipt1∆::URA3MX4 | This study |
| NRY549b | W303-1A lem3∆::URA3MX4 | This study |
| NRY551a | W303-1A pdr10∆::HIS3MX6 lem3∆::URA3MX4 | This study |
| NRY558a | W303-1A pdr10∆::HIS3MX6 dnf1∆::URA3MX4 dnf2∆::kanMX4 | This study |
| NRY601b | NRY243 chs3∆::URA3MX4 | This study |
| NRY602b | W303-1A chs3∆::URA3MX4 | This study |
| NRY608b | W303-1A chs6∆::URA3MX4 | This study |
| NRY610b | NRY243 chs6∆::URA3MX4 | This study |
| NRY802c | W303-1A ADE2 lys2::hisG | This study |
| NRY804c | W303-1B ADE2 lys2::hisG | This study |
| NRY904d | W303-1A LEM3-mh::kanMX6 | This study |
| NRY906d | NRY804 PDR5-GFP::HIS3MX6 | This study |
| NRY908d | W303-1A pdr10∆::HIS3MX6 LEM3-mh::kanMX6 | This study |
| NRY912d | NRY802 PDR10-GFP::HIS3MX6 | This study |
| NRY914d | NRY802 LEM3-GFP::HIS3MX6 | This study |
| NRY915d | NRY804 PDR12-GFP::kanMX6 | This study |
| NRY916d | NRY802 pdr10∆::URA3MX4 PDR5-GFP::HIS3MX6 | This study |
| NRY918d | NRY802 pdr10∆::URA3MX4 LEM3-GFP::HIS3MX6 | This study |
| NRY921d | NRY802 DNF1-GFP::HIS3MX6 | This study |
| NRY923d | NRY802 DNF2-GFP::HIS3MX6 | This study |
| NRY925d | NRY802 pdr10∆::HIS3MX6 PDR12-GFP::kanMX6 | This study |
| NRY929d | NRY802 pdr10∆::URA3MX4 DNF2-GFP::HIS3MX6 | This study |
| NRY930d | W303-1A PDR10-mh::kanMX6 | This study |
| NRY932d | NRY802 pdr10∆::URA3MX4 DNF1-GFP::HIS3MX6 LYS2 | This study |
| YPH499 | MATaade2-101ochis3-∆200 leu2-∆1 lys2-801a trp1-∆63 ura3-52 | Sikorski and Hieter (1989) |
| YHW10 N | YPH499 pdr10∆::URA3 | This study |
aConstructed by sporulation and tetrad dissection of appropriate diploid parents
bConstructed by integration of PCR products amplified with appropriate primers using pFA6aHIS3MX6 or pAG60 as template and verified by PCR on genomic DNA
cConstructed by tetrad dissection of BYB67
dConstructed by integration of PCR products amplified with appropriate primers using pFA6a-GFP(S65T)-HIS3MX6, pFA6a-GFP(S65T)-kanMX6, or pNR99A as template and verified by PCR on genomic DNA
Plasmids pFAkanMX4, pFA6aHIS3MX6, pAG60, pFA6aGFP(S65T)kanMX6, and pFA6aGFP(S65T)HIS3MX6 have been described previously (Goldstein et al. 1999; Wach et al. 1994, 1997). To construct pNR99A, which permits PCR-based C-terminal tagging of yeast proteins with a tandem c-Myc-His6 tag followed by a STOP codon and transcriptional terminator (the “mh” tag), plasmid yGALSte6(∆A-box)-myc-his (gift from E. E. Swartzman, Thorner Lab), which encodes a functional Ste6 with a C-terminal mh tag, was amplified by PCR with appropriate primers to permit cloning of a fragment containing 165 bp from the 3′ end of STE6 followed by the mh tag coding sequence and a STOP codon, which was inserted into pFA6aGFP(S65T)kanMX6 using unique BamHI and AscI sites. The presence of a single, correctly cloned insert was verified by restriction digestion and nucleotide sequencing. This plasmid permits integration of the mh tag with or without linker sequence derived from the C-terminus of Ste6. In the present work, the Lem3-mh fusion lacks any linker, whereas the Pdr10-mh chimera incorporates a pentapeptide (–SNQSS–) sequence derived from the STE6 gene that is situated between the C-terminal residue of Pdr10 and the mh tag. Both Pdr10-mh and Lem3-mh were functional, as judged by a phenotypic test (normal Calcofluor sensitivity compared to untagged strains and pdr10∆ or lem3∆ strains).
Immunoblotting
Rabbit polyclonal antisera used in this study were anti-Ste2 (David et al. 1997), anti-Chs2 and anti-Chs3 (R. Schekman, University of California, Berkeley, USA), anti-Drs2 (T. Graham, Vanderbilt University, Nashville, TN, USA), anti-Pdr12 (Piper et al. 1998), anti-Pdr5 (Egner et al. 1995), anti-Snq2 (Mahé et al. 1996b), anti-Ycf1 and anti-Yor1 (S. Moye-Rowley, University of Iowa, Iowa City, USA), ant-Gas1 (A. Conzelmann, University of Fribourg, Fribourg, Switzerland), and anti-Hxt1 (E. Boles, Goethe-Universität Frankfurt, Frankfurt-am-Main, Germany). Mouse monoclonal antibodies used in this study were anti-Fks1 mAb (Y. Ohya, University of Tokyo, Toyko, Japan), anti-Pma1 mAb (F. Portillo, Universidad Autonoma de Madrid, Madrid, Spain), anti-Vph1 mAb (T. Stevens, University of Oregon, Eugene, OR, USA), anti-ALP/Pho8 mAb (Molecular Probes), and mAb 9E10 directed against the c-Myc epitope used in this work (Evan et al. 1985).
Two techniques were used to quantitate immunoblots. For samples analyzed by chemiluminescence, exposures were taken using Kodak type MS film, which was scanned and quantitated using densitometry and open-source image analysis software (NIH Image, NIH, Bethesda, MD, USA). Alternately, blots were probed with infrared dye-labeled secondary antibodies and scanned using an infrared scanner (Odyssey; Li-Cor, Inc., Lincoln, NE, USA). Results obtained by these two techniques were crosschecked using samples of equivalent protein content from the total membrane fraction of wild-type cells and pdr10∆ mutants, which were blotted with anti-Pdr12 antiserum. When determined by either method, the value obtained for the ratio of Pdr12 content in pdr10∆ mutants to that in wild-type cells was essentially the same. Thus, measurements of relative protein content made using either procedure were comparable.
Growth Assays
All spot-dilution assays were performed using overnight cultures grown in YPD. Aliquots from such cultures were adjusted to an A600 nm = 1 with sterile H2O. Three 10-fold serial dilutions were made in sterile H2O, and aliquots (5 μl) of all four cell suspensions were spotted onto YPD plates lacking or containing the additive to be tested. After the liquid in each spot was absorbed, the plates were inverted, incubated at 30°C for 2 days, and then photographed. The following additives were tested at the indicated concentrations: Calcofluor White (Fluorescent Brightener 28; Sigma Chemical Co., St. Louis, MO, USA), 0 to 8 μg/ml; Congo Red, 100 μg/ml; ketoconazole, 2 μM; rhodamine 6G, 20 μM; and cycloheximide, 100 ng/ml. For assay of sorbate sensitivity, the cell suspensions were spotted onto YPD plates that were buffered using 50 mM sodium succinate (pH 3.5) and contained various concentrations of potassium sorbate (0 to 4 mM), and the resulting plates were photographed after incubation at 30°C for 3 days.
Detergent sensitivity was assessed using a diffusion (“halo”) assay. Cells of the strain to be tested (0.1 A600 nm unit) from overnight cultures grown in YPD were diluted into 3 ml of YPD top agar and spread on the surface of a YPD plate. Sterile filter disks (1-cm diameter; Whatman 3MM) were placed on the seeded cell lawn and samples (10 μl) of solutions (10%, w/v) of the detergents to be tested were spotted onto the disks. Plates were then incubated right-side-up at 30°C for 2 days.
Microscopy
A functional GFP-tagged chitin synthase (Chs3-GFP) was expressed from plasmid pCRP12 with simultaneous overexpression of Chs7 from pRS315-CHS7, as previously described (Valdivia et al. 2002). Exponentially growing cultures of strains W303-1A and NRY251 carrying these plasmids were treated with α-factor mating pheromone to induce cell cycle arrest and shmoo formation, and then pheromone was washed out with fresh, prewarmed medium to permit synchronized re-entry into the cell cycle. The cellular distribution of Chs3-GFP was visualized under a fluorescence microscope (Olympus BH-2) equipped with a 100× objective and appropriate bandpass filters (Chroma Technology). Digital images were captured using an Olympus charge-coupled device (CCD) camera and Magnafire-SP software (Optronics). To visualize the distribution of fluorescently labeled chitin in the cell wall, cells were stained with Calcofluor White following a published procedure (Pringle 1991). Briefly, exponentially growing cells were collected by centrifugation, washed twice in H2O, incubated in aqueous Calcofluor White (0.1 mg/ml) for 5 min at room temperature, washed three times with H2O, recollected by centrifugation, resuspended at an appropriate dilution in H2O, and viewed using appropriate filters under the same fluorescence microscope described above. The intensity of Calcofluor staining in photomicrographs was quantitated using NIH Image; pilot experiments examining chitin levels in wild-type, pdr10∆, and cho1∆ cells by fluorescence-activated cell sorting were in agreement with quantitation of photomicrographs.
To examine uptake of the water-soluble fluorescent dye Lucifer Yellow as a measure of fluid phase endocytosis, cells (0.5 A600 nm unit) from exponentially growing cultures of W303-1A and NRY243 grown in YPD were collected by centrifugation and resuspended in fresh YPD medium (90 μl). An aliquot (10 μl) of an aqueous stock solution (40 mg/ml) of Lucifer Yellow CH (Molecular Probes, Inc., Eugene, OR, USA) was added and the cell suspension was incubated with gentle agitation at 30°C for 1 h. After this incubation, the cells were recollected, washed with 1 ml H2O, and viewed using appropriate filters under the Olympus fluorescence microscope. Similarly, for uptake of the lipophilic fluorescent dye FM 4-64, this compound (16 μM final concentration) was added from a concentrated stock (16 mM) in DMSO directly to cultures of exponentially growing W303-1A and NRY243 cells and the suspension was incubated with gentle agitation at 30°C for 40 min, then washed with H2O and viewed using appropriate filters under the Olympus fluorescence microscope. To examine filipin staining and sterol polarization, log phase cells were resuspended in SCD medium buffered to pH 3.5 with 25 mM sodium succinate. Aliquots were treated with α-factor as described above or with solvent control. After pheromone treatment, cells were washed twice in SCD buffered with 1× lysis buffer (defined below) and stained with 10 μg/ml filipin for 5 min in the dark at room temperature. Cells were then washed with SCD buffered with lysis buffer and examined on the Olympus fluorescence microscope. To visualize actin, 134 μl of 37% formaldehyde (commercial) was added to1-ml log phase cultures of appropriate strains and the mixture incubated for 30 min at room temperature with gentle agitation. Cells were washed twice in phosphate-buffered saline supplemented with 1 mg/ml bovine serum albumin and then resuspended in 50 μl of the same buffer. Ten microliters of rhodamine-phalloidin from a stock of 300 units dissolved in methanol was added, and cells were incubated for 30 min in the dark at 4°C with gentle agitation. Cells were then washed three times with the same buffer, resuspended in 10 μl buffer supplemented with 20 μl Citifluor, and visualized by fluroescence microscopy.
The Pdr10-GFP, Pdr12-GFP, Pdr5-GFP, Lem3-GFP, Dnf1-GFP, and Dnf2-GFP chimeras were constructed by generating in-frame fusions of the coding sequence for GFP to the coding sequence of each protein at its resident locus in its respective chromosome using a PCR-based insertion method and plasmids containing the cognate genes as the template DNA (Wach et al. 1997). The primers used for each construction were designed to retain the native C-terminal sequence of each protein, followed by a nonapeptide linker (–AAGRRIPGL–) and then by GFP. Proper integration was verified by PCR. Retention of biological function of each chimera was assessed by its ability to complement the phenotype of the corresponding mutant: for Pdr5-GFP, restoration of resistance to rhodamine 6G; for Pdr12-GFP, restoration of resistance to sorbate; and for Pdr10-GFP, Lem3-GFP, Dnf1-GFP, and Dnf2-GFP, restoration of resistance to Calcofluor White.
To visualize the subcellular localization of each chimera (except Pdr10-GFP), cells from exponentially growing cultures were washed once in 10 mM Tris–HCl (pH 7.0) and then viewed using a Nikon TE300 fluorescence microscope equipped with a 100× objective and appropriate bandpass filters (Chroma Technology). Digital images were captured with a bottom-ported Orca 100 CCD camera (Hamamatsu, Bridgewater, NJ, USA) and Phase 3 imaging software (Northern Exposure, Glen Mills, PA, USA). To examine Pdr10-GFP, we determined empirically that the most reliable results were obtained for stationary phase cells (i.e., cultures grown in YPD for~2 days), which were then, as for the others, washed once in 10 mM Tris–HCl (pH 7.0) prior to viewing under the fluorescence microscope. For time-lapse photomicroscopy, cells were immobilized by spotting onto polylysine-coated coverslips to minimize their movement and short exposure times (at high gain) were taken to capture as many frames as possible to minimize the loss of signal intensity due to photobleaching. When cells were treated with the actin filament-depolymerizing drug latrunculin A, an aliquot (2 μl) containing this compound was added from a concentrated stock (10 mM) in DMSO to a sample (150 μl) of cells from a culture of strain NRY912 that had grown in YPD for 2 days. After incubation with end-over-end mixing for 10 min at 30°C, the cells were collected by centrifugation, washed in 10 mM Tris–HCl (pH 7.0), and examined using the Nikon fluorescence microscope.
Uptake of fluorescent C6-7-nitrobenz-2-oxa-1,3-diazol-4-yl-PE (NBD-PE) and C6-NBD-ceramide (NBD-Cer) (Avanti Polar Lipids) was monitored in exponentially growing cells from cultures grown in synthetic complete (SC) medium containing glucose (dextrose) as the carbon source (SCD), as follows. Cells were collected by centrifugation and resuspended 10-fold concentrated in 300 μl of prewarmed SCD, and then latrunculin A (4 μl) was added from a concentrated stock (10 mM) stock in DMSO. After incubation by end-over-end mixing for 5–10 min at 30°C, a sample (2 μl) of either NBD-PE or NBD-Cer, each from a concentrated stock (10 mM) in DMSO, was added while the suspension was subjected to vigorous vortex mixing (Grant et al. 2001; Pomorski et al. 2003). After a further incubation with end-over-end mixing for 30 min at 30°C, the cells were recollected and washed three times with ice-cold SC medium containing 2% sorbitol and 20 mM NaN3 (abbreviated SCNaN3) prior to viewing under the Nikon fluorescence microscope adjusted for optimal viewing of fluorescence emission in the same range as for fluorescein.
Preparation of Total Yeast Membranes
For membrane preparations, cells were inoculated into fresh, prewarmed YPD medium and grown at 30°C for one to three doublings. The cells were then chilled on ice at the same time that NaN3 and NaF were added (each at a 20 mM final concentration). After 10 min on ice, cells were harvested by centrifugation, washed with 10 ml ice-cold aqueous 20 mM NaN3–20 mM NaF, and resuspended in aliquots (0.12–0.15 g wet weight) that were flash-frozen in liquid N2 and stored at –80°C until used. For lysis, frozen cell pellets were resuspended in 650 μl ice-cold lysis buffer (50 mM Tris–HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA) containing a protease inhibitor mix (Complete; Roche), 1 mM PMSF, 200 μM pepstatin A, and 5 μM MG-132 and then ruptured by five 1-min pulses of vigorous vortex mixing with glass or silica/zirconium beads (Biospec, Bartlesville, OK, USA) separated by 1 min of cooling on ice between each pulse. The resulting lysate was removed and then combined with the solutions generated by extracting the beads three times with 1-ml portions of ice-cold lysis buffer. The combined crude extract was subjected to centrifugation at 500 g for 5 min at 4°C to remove any broken beads, unlysed cells, cell walls, and other large debris. The clarified extract was subjected to centrifugation at 43,000 rpm for 1 h at 4°C in the TLA 100.3 rotor of a TL100 tabletop ultracentrifuge (Beckman). The resulting total membrane pellets were resuspended in 100 mM Tris–HCl (pH 7) containing 5% SDS, 40 μM pepstatin A, and 1 mM PMSF and warmed briefly to 37°C. The concentration of protein in these membrane preparations was determined using the bicinchoninic acid (BCA) method (Pierce Chemical Co.). Samples (typically 50 μg) were adjusted to 1× SDS-PAGE (sample buffer final concentration) by addition of an appropriate volume of 6×-concentrated SDS-PAGE sample buffer (Laemmli 1970) and then divided into aliquots, which were frozen in liquid N2 and stored at –80°C before use. For analysis, an aliquot was thawed for 5 min at 37°C and then subjected to SDS-PAGE and immunoblotting.
Density Gradient Analysis of Detergent-Resistant Membranes
Analysis of DRMs was carried out using modifications of the Optiprep flotation procedure first developed by Simons and co-workers (Bagnat et al. 2000). Total membrane pellets, prepared as described in the preceding section, were resuspended in 230 μl ice-cold buffer lysis buffer containing 1% Triton X-100 in a 4°C room and kept on ice for 1 h. A sample (170 μl) of this material was then added to LOAD solution (2500 μl Optiprep, 150 μl 10× lysis buffer, 150 μl 10% Triton X-100, containing PMSF, pepstatin A, and MG-132 at the same concentration as in lysis buffer), and the resulting mixture was transferred to ultracentrifuge tubes. The material in the tubes was then overlaid with 45% Optiprep (3 ml), 40% Optiprep (3 ml), 30% Optiprep (2.5 ml), and 0% Optiprep (200 μl); each of these solutions was in 0.5× lysis buffer, 0.5% Triton X-100, plus PMSF, pepstatin A, and MG-132. The resulting step gradient was subjected to centrifugation at 4°C for 18 h at 39,000 rpm in an SW41 rotor (Beckman). After centrifugation, fractions (600 μl) were collected, starting from the top of the gradient, and the material in each was precipitated by the addition of 1.2 ml 15% trichloroacetic acid (TCA) followed by immediate vortex mixing and incubation on ice for 45 min (Dupré and Haguenauer-Tsapis 2003). The resulting precipitates were collected by centrifugation at maximum speed in a microfuge for 15 min, washed twice with ~250 μl ice-cold acetone, and solubilized by resuspension in 100 μl 5% SDS containing 0.1 M Tris base and brief incubation at 37°C. For ease of analysis, adjacent fractions were pooled so as to divide each gradient into nine separate portions. Aliquots of each of the nine portions were flash-frozen in liquid N2, some of which were used for protein determination and the remainder of which were used for analysis by SDS-PAGE and immunoblotting as described in the preceding section.
Lipid Analysis and TNBS Labeling
Cells were grown in 1 ml YPD supplemented with 50–250 μCi 32P-PO43– (10 mCi/ml; Perkin-Elmer) for 15–18 h to label phospholipids to a constant specific activity (Chang et al. 1998). For reaction of intact yeast cells with 2,4,6-trinitrobenzene sulfonate (TNBS), a published procedure (Siegmund et al. 1998) was used with only minor modifications. After radioactive labeling, cells were washed twice (1 ml each) in ice-cold TNBS buffer (120 mM NaHCO3 [pH 8.4], 40 mM NaCl), resuspended in 1 ml of the same buffer, and then aqueous TNBS (31 μl) was added from a concentrated stock (5%; Sigma). After incubation on ice with brief vortex mixing every 20 min over the course of 1 h, the cells were collected by brief centrifugation, washed three times (1 ml each) in ice-cold TNBS buffer, and resuspended in 600 μl CHCl3/CH3OH/0.1 N HCl (1:2:0.8) containing carrier lipid (Chang et al. 1998). To promote lipid extraction after addition of the organic solvent mix, the cells were subjected to vigorous vortexing with glass beads for 3 min, and an additional 200 μl each of CHCl3 and 0.1 N HCl–0.5 M NaCl was added. To achieve good separation of the organic phase from the aqueous phase, the resulting mixture was subjected to centrifugation for 2 min at maximum speed in a microfuge. After withdrawing the organic phase, the lipids present in it were analyzed by two-dimensional thin-layer chromatography (2D-TLC) using CHCl3:MeOH:glacial acetic acid, 65:25:10 (dimension I), and CHCl3:MeOH:88% formic acid, 65:25:10 (dimension II) (Esko and Raetz 1980), with nonradioactive trinitrophenyl-PE (TNP-PE) added as an internal standard. The radioactivity present in each spot on the resulting thin-layer chromatograms was quantitated by using the plates to expose phosphor storage screens, which were then analyzed using PhosphorImager or Typhoon imaging systems and ImageQuant software (Molecular Dynamics). The fraction of PE accessible to TNBS was calculated as TNP-PE/total PE (where total PE = TNP-PE + unreacted PE). TNP derivatives of phosphatidylserine (PS) were not observed. Comparing across independent experiments, this assay procedure yielded somewhat variable backgrounds, presumably due to slight differences in the fraction of inviable cells in cultures. We have established that there is no statistically significant difference in cell viability in strains cultured together, both by measurement of the ratio of colony-forming units to optical density and by methylene blue staining (Siegmund et al. 1998) of TNBS-labeled, nonradioactive control cultures (>500 cells counted per strain). Therefore, other strains should be measured accurately relative to the background TNBS labeling of wild-type cultures. All such measurements were performed with wild-type controls grown side by side. In every such trial, more PE was accessible in pdr10∆ cells than in the control cultures (n = 7). In reporting the results, we have shown the actual labeled percentages in Fig. 6b for comparison to results of other workers, who have reported such data in this fashion (Pomorski et al. 2003), and we have also reported the data as -fold differences in Table 2 (which should be more accurate, in light of the variable background).
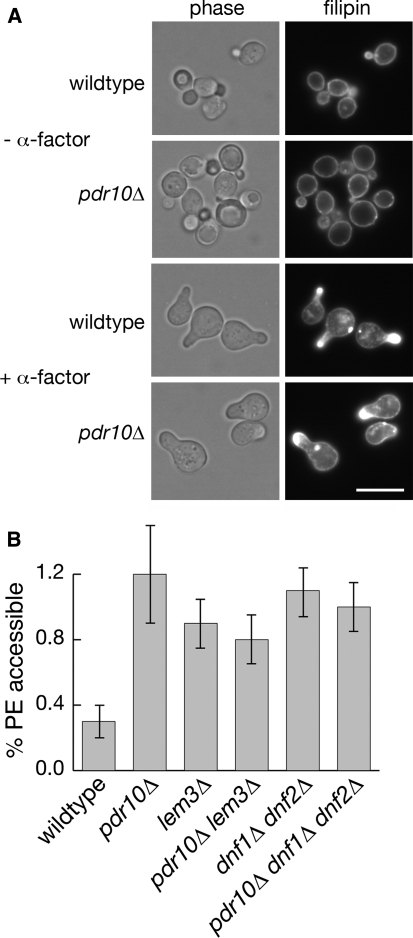
Fig. 6.
Increased phosphatidylethanolamine is accessible to TNBS labeling in pdr10∆ cells. a Strains W303-1A and NRY243 were grown to log phase, incubated with or without α-factor, and then stained with filipin to visualize sterols by fluorescence microscopy. Bar, 5 μm. b Exposure of endogenous aminophospholipids to TNBS was assayed with cells grown overnight in the presence of 32P-phosphate as described under Materials and Methods. Strains used were W303-1A (wild type), NRY243 (pdr10∆), NRY549 (lem3∆), NRY551 (pdr10∆ lem3∆), NRY427 (dnf1∆ dnf2∆), and NRY558 (pdr10∆ dnf1∆ dnf2∆)
Table 2.
Total phospholipid composition of yeast strains
| Straina | % Total phospholipid (as incorporated 32P label) | TNBS labelingb | ||||
|---|---|---|---|---|---|---|
| PS | PE | PC | PI | SPL | ||
| Wild type | 7.6 ± 0.6 | 20.2 ± 1.5 | 35.5 ± 2.1 | 24.1 ± 0.6 | 7.3 ± 0.9 | 1.0 |
| pdr10∆ | 7.7 ± 0.5 | 20.3 ± 1.2 | 34.9 ± 1.1 | 23.9 ± 1.1 | 7.3 ± 1.3 | 2.5 ± 0.6 |
| dnf1∆ dnf2∆ | 7.5 ± 0.5 | 20.5 ± 0.5 | 36.3 ± 0.7 | 22.9 ± 0.5 | 7.8 ± 0.6 | 2.0 ± 0.6 |
| pdr10∆ dnf1∆ dnf2∆ | 7.7 ± 0.5 | 20.0 ± 0.5 | 35.9 ± 0.5 | 23.8 ± 0.5 | 7.4 ± 0.5 | 2.3 ± 1.0 |
| lem3∆ | 7.9 ± 0.1 | 20.7 ± 0.7 | 35.1 ± 1.5 | 23.6 ± 1.2 | 7.3 ± 0.4 | 2.7 ± 0.5 |
| pdr10∆ lem3∆ | 7.8 ± 0.2 | 20.3 ± 1.1 | 35.0 ± 0.5 | 24.1 ± 1.9 | 7.2 ± 0.4 | 2.7 ± 1.0 |
| pdr5∆ | 9.2 ± 0.5 | 22.7 ± 0.5 | 32.5 ± 0.5 | 19.8 ± 0.8 | 7.4 ± 0.5 | 0.9 ± 0.2 |
| pdr5∆ pdr10∆ | 7.9 ± 1.0 | 20.3 ± 2.0 | 38.7 ± 1.7 | 18.7 ± 0.7 | 6.4 ± 1.3 | 0.3 ± 0.2 |
| pdr12∆ | 7.5 ± 0.5 | 19.7 ± 0.5 | 36.0 ± 0.5 | 24.0 ± 0.5 | 7.7 ± 0.8 | 1.6 ± 0.6 |
| pdr10∆ pdr12∆ | 8.1 ± 0.5 | 19.9 ± 0.5 | 35.0 ± 1.1 | 24.0 ± 0.5 | 7.6 ± 0.5 | 1.5 ± 0.6 |
| sur1∆ | 8.2 ± 0.5 | 19.1 ± 0.5 | 34.2 ± 0.6 | 23.3 ± 0.5 | 10.0 ± 0.5 | 1.1 ± 0.2 |
| pdr10∆ sur1∆ | 8.2 ± 0.5 | 19.7 ± 0.5 | 35.6 ± 0.7 | 22.5 ± 1.2 | 9.2 ± 0.6 | 1.1 ± 0.2 |
| cho1∆ | <0.1 | 11.0 ± 3.0 | 52.7 ± 6.2 | 26.9 ± 2.8 | 5.6 ± 1.2 | 2.7 ± 0.8 |
Note: PS phosphatidylserine; PE phosphatidylethanolamine; PC phosphatidylcholine; PI phosphatidylinositol; SPL total phosphosphingolipids. Lipids were labeled and analyzed as described under Materials and Methods. Significant changes relative to wild type are in boldface
aStrains were W303-1A (wild type), NRY243 (pdr10∆), NRY427 (dnf1∆ dnf2∆), NRY558 (pdr10∆ dnf1∆ dnf2∆), NRY549 (lem3∆), NRY551 (pdr10∆ lem3∆), NRY201 (pdr5∆), NRY230 (pdr5∆ pdr10∆), NRY301 (pdr12∆), NRY366 (pdr10∆ pdr12∆), NRY409 (sur1∆), and NRY526 (pdr10∆ sur1∆)
bTNBS labeling is reported relative to the wild type
To analyze the composition of mature sphingolipids, cells were grown and labeled as described above, and sphingolipids were extracted using a previously published procedure with minor modifications (Dickson et al. 1997). To samples (1 ml) of cultures incubated on ice was added 53 μl of 100% TCA. After 15 min, the acid-treated cells were washed twice with water (1 ml each), resuspended in 400 μl solvent E (diethyl ether/95% ethanol/water/pyridine, 5:15:15:1), and incubated at 60°C for 60 min with occasional vortex mixing. To separate the phases, the mixtures were subjected to centrifugation at maximum speed in a microfuge for 2 min and the sphingolipids present in the resulting organic phase were separated by ascending TLC with CHCl3:CH3OH:4.2 N NH4OH (9:7:2), which yields an acceptable resolution of the major sphingolipid species (Hama et al. 2000), which were quantified as described in the preceding paragraph.
Nonradioactive TNP-PE was synthesized according to a published procedure (Gordesky and Marinetti 1973) with minor modifications (Schüller et al. 2007). PE (25 mg) was dissolved in 2.5 ml CHCl3 and added to a solution composed of 2.5 ml CH3OH, 1 ml CHCl3, 250 μl 5% aqueous NaHCO3, and 390 μl 5% aqueous TNBS. This resulting reaction mixture was incubated at room temperature for 2 h under argon gas and then mixed with 2 ml CHCl3 and 20 ml aqueous saturated NaCl (brine). After separating the phases, the organic phase was withdrawn, acidified with aqueous 1 N HCl, and re-extracted with 5 ml brine, and the resulting organic phase was dried over Na2SO4 and stored at –20°C under argon.
Results
Pdr10 is a Plasma Membrane Protein
To confirm that the product of the PDR10 gene is a protein that is expressed at a detectable level and, if so, to establish its subcellular localization, a strain was constructed in which the green fluorescent protein (GFP) was fused in-frame to the C-terminal end of the open reading frame encoded by the chromosomal PDR10 locus. We found that Pdr10-GFP localized to discrete, punctate structures in the plasma membrane (Fig. 1a). This chimera was most easily visualized in cells that had entered stationary phase (after 2 days of growth); however, the same pattern was observed in exponentially growing cells, only with a much weaker signal (data not shown). This finding is in agreement with global microarray analysis showing that transcription of PDR10 is induced when cells undergo the diauxic shift from fermentative to nonfermentative growth as glucose becomes limiting (Roberts and Hudson 2006) or when cells are treated with rapamycin, which mimics conditions of nutrient limitation (Huang et al. 2004). Collectively, these findings suggest that the demand for this ABC transporter is greatest when cells are undergoing the stress of maintaining viability as growth rate slows down. Moreover, because our Pdr10-GFP construct was expressed from the endogenous PDR10 promoter, transcription factors other than (or in addition to) Pdr1 and Pdr3 likely mediate PDR10 expression under the stress of nutrient depletion.
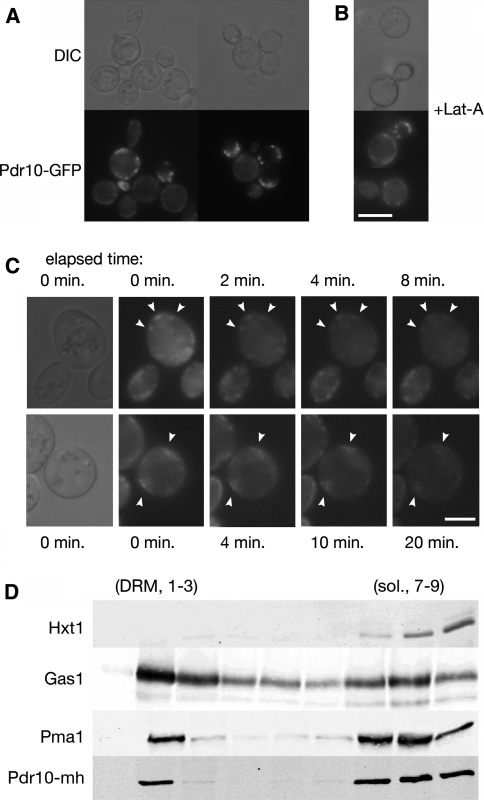
Fig. 1.
Pdr10 localizes to punctate structures in the plasma membrane and to the detergent-resistant fraction. a Strain NRY912 was grown in YPD for 2 days, and Pdr10-GFP was examined by fluorescence microscopy. Pdr10-GFP function was confirmed by Calcofluor sensitivity. b Strain NRY912 was grown for 2 days as in (a) but was treated with latrunculin A prior to microscopy. Bar for (a) and (b), 6 μm. c Cells were repeatedly exposed to look for movement of Pdr10-GFP puncta (arrowheads). Shorter exposure times and higher gain were used to permit the maximum number of exposures prior to loss of signal due to photobleaching. Bar, 3 μm. d Total membranes were isolated from log phase cells of strain NRY930, extracted with cold Triton X-100, and analyzed by flotation though Optiprep density gradients with subsequent immunoblotting as described under Materials and Methods. Strain NRY930 expresses a PDR10 allele with a C-terminal tandem myc-His6 (mh) tag. The function of the resulting Pdr10-mh protein was confirmed by Calcofluor sensitivity. Hxt1, Gas1, and Pma1 were used as markers for detergent-soluble and detergent-resistant membranes
The markedly punctate distribution in the plasma membrane displayed by Pdr10-GFP has been observed for several other integral plasma membrane proteins. In some cases, these puncta are mobile because they represent proteins (for example, the glucan synthase Fks1) associated with sites where actin-mediated endocytosis occurs (“actin patches”) (Utsugi et al. 2002). In other cases, the puncta are immobile because they represent proteins (for example, the arginine permease Can1 and the tetraspanin Sur7) associated with stable, static plasma membrane subdomains such as the MCC compartment (Grossmann et al. 2007; Malinska et al. 2003; Young et al. 2002). Therefore, we examined whether the Pdr10-GFP puncta were stable over time and whether they were affected by inhibition of actin function. We found that treatment of cells with latrunculin A did not disrupt the punctate localization displayed by Pdr10-GFP (Fig. 1b). Some of the Pdr10-GFP puncta in a typical cell were immobile for at least 20 min (Fig. 1c). However, other Pdr10-GFP puncta seemed more mobile, particularly in the bud. There is evidence that the mother and bud compartments have distinct properties. For example, PE exposure is reportedly greater at the bud tip than in the mother cell (Iwamoto et al. 2004), and the collar of septin filaments at the bud neck acts as a diffusion barrier to confine certain plasma membrane proteins uniquely in the bud (Takizawa et al. 2000). However, the presence of mobile patches in the mother cell as well indicates that there is a mobile population of Pdr10 throughout the cell. Pdr10-GFP did not behave like an actin patch-associated protein, but the presence of both mobile and immobile populations argues against its exclusive localization to previously defined subdomains of the yeast plasma membrane.
Can1 and other structurally related permeases not only display a punctate distribution in the plasma membrane, but also are found in the detergent-resistant membrane (DRM) fraction (sometimes equated with lipid rafts) when cells are subjected to biochemical fractionation (Malinska et al. 2003, 2004). To permit facile examination of the behavior of Pdr10 during biochemical fractionation, we tagged the chromosomal allele of PDR10 at its C-terminus with c-Myc and His6 epitopes in tandem (the mh tag) under control of the native PDR10 promoter. We then prepared total membranes from such cells, extracted them with cold detergent (Triton X-100), and fractionated them by density gradient flotation. When subjected to this procedure, the residual lipid associated with the DRM fraction causes it to appear at the top of the gradient, while detergent-soluble or aggregated proteins exhibit higher apparent density and are found at the bottom of the gradient (Bagnat et al. 2000; Malinska et al. 2003, 2004). Previous studies using this assay have identified the plasma membrane H+-ATPase Pma1 and the glycosylphosphatidylinositol (GPI)-anchored protein Gas1 as markers for the DRM fraction (Bagnat et al. 2000; Malinska et al. 2003). Markers for the detergent-soluble fraction of the plasma membrane have proven more controversial: the hexose transporter Hxt1 has been reported to primarily localize to the detergent-soluble fraction by Tanner and colleagues (Malinska et al. 2003, 2004), using anti-Hxt1 antisera for detection, but Hxt1-GFP has instead been found in the detergent-resistant fraction by Lauwers and André (2006) using anti-GFP antisera for detection. These studies also use different strain backgrounds, which may explain the discrepancy. We find that Hxt1 is primarily (~90%) in the detergent-soluble fraction (Fig. 1d), in good agreement with the results reported by Tanner and colleagues (Malinska et al. 2003, 2004).
To detect the inabundant Pdr10 in flotation gradients, we found that it was necessary to load substantially more material than has typically been reported. Unfortunately, the greater amounts loaded resulted in loss of resolution in the small Optiprep (iodixanol) density gradients pioneered by Simons and colleagues (Bagnat et al. 2000). We therefore developed a modified procedure using a larger gradient with a consequently longer centrifugation (described in detail under Materials and Methods). The DRM marker Gas1 exhibits essentially identical behavior in both gradients, as does the chitin synthase Chs3 (data not shown). However, while more than 90% of Pma1 was found in the DRM fraction in the smaller gradients, a substantial amount of Pma1 was instead associated with the soluble fraction in these expanded gradients, with only 25%–50% of Pma1 remaining in the DRM fraction (Fig. 1d). We have not attempted to ascertain whether this reflects slow solubilization or aggregation of Pma1 in the longer ultracentrifugation procedure we employ, because Gas1 and Hxt1 serve as robust markers for the detergent-resistant and detergent-soluble fractions in this gradient.
Using this procedure, we were able to detect Pdr10-mh with antisera directed against the c-myc epitope (Fig. 1d). Tellingly, Pdr10-mh displayed a distribution quite similar to that of Pma1, with substantial Pdr10-mh residing in the DRM fraction. The presence of both detergent-soluble and detergent-resistant Pdr10 is reminiscent of the presence of immobile and mobile populations of Pdr10-GFP as seen by fluorescence microscopy, but the nature of the DRM experiment precludes assignment of the in vitro populations to in vivo populations (Lichtenberg et al. 2005).
Absence of Pdr10 Causes Distinct Phenotypes
Among the ABC transporters encoded in the S. cerevisiae genome, Pdr10 exhibits the closest homology to Pdr5 (67% identity), Pdr15 (68% identity), and Snq2 (40% identity). All three of the latter are capable of mediating the export of various drugs, antimetabolites, membrane perturbing agents, and other noxious compounds (Balzi et al. 1994; Bissinger and Kuchler 1994; Decottignies et al. 1995; Schüller et al. 2007; Servos et al. 1993; Wolfger et al. 2004). Therefore, we examined whether deletion of the PDR10 gene would confer enhanced sensitivity to a spectrum of cytotoxic compounds. We also examined whether loss of Pdr10 would further enhance (or otherwise alter) the drug sensitivity of a pdr5∆ mutant. We found that, in contrast to pdr5∆ cells, pdr10∆ cells were not abnormally sensitive to ketoconazole, rhodamine 6G, or cycloheximide (Fig. 2a), nor did a pdr5∆ pdr10∆ double mutant differ in its sensitivity from a pdr5∆ single mutant (Fig. 2a). The absence of Pdr10 similarly had no effect on the toxicity of itraconazole, oligomycin, amphotericin B, or Crystal Violet, with or without Pdr5 (data not shown). Hence, Pdr10 does not appear to contribute significantly to the capacity of the cell to export such growth-inhibitory compounds.
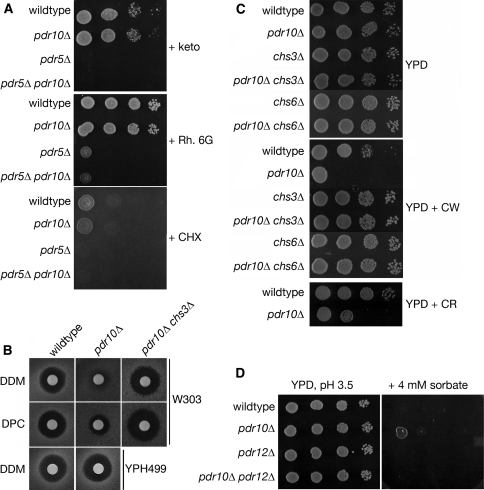
Fig. 2.
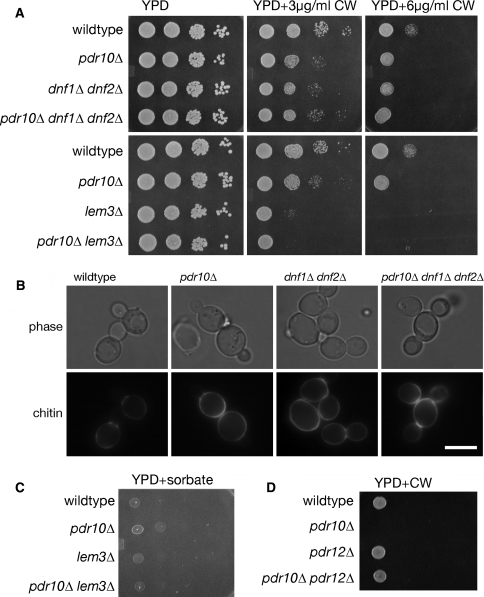
Pdr10 is required for normal response to Calcofluor and sorbate. a Spot assays were performed using serial dilutions prepared from overnight cultures of strains W303-1A (wild type), NRY243 (pdr10∆), NRY201 (pdr5∆), and NRY230 (pdr5∆ pdr10∆). All plates were grown for 2 days at 30°C unless otherwise noted. YPD plates contained either 2 μM ketoconazole (keto), 20 μM rhodamine 6G (Rh. 6G), or 100 ng/ml cycloheximide (CHX). b Halo assays were performed on YPD plates with various detergents. Aliquots from overnight cultures were diluted into YPD top agar as described under Materials and Methods. Top row, strains W303-1A, NRY243, and NRY601 (pdr10∆ chs3∆) were assayed with dodecyl maltoside (DDM). Middle row, the same strains were assayed with dodecyl phosphocholine (DPC). Bottom row, strains YPH499 (wild type) and YHW10 N (pdr10∆) were assayed with DDM. c Spot assays were performed as in A using strains W303-1A, NRY243, and NRY602 (chs3∆), NRY601 and NRY608 (chs6∆), and NRY610 (pdr10∆ chs6∆) on YPD plates (top) or YPD plates containing 6 μg/ml Calcofluor White (CW; middle) or 100 μg/ml Congo Red (CR; bottom). d Similar assays were performed using strains W303-1A (wild type), NRY243 (pdr10∆), NRY301 (pdr12∆), and NRY366 (pdr10∆ pdr12∆) on YPD plates buffered to pH 3.5 with 50 mM sodium succinate with or without 4 mM potassium sorbate
We also challenged pdr10∆ cells with a number of detergents. As judged by a halo assay, a pdr10∆ mutant strain in the W303 genetic background reproducibly exhibited modestly elevated resistance to detergents containing C12 alkyl chains, including dodecyl maltoside and dodecyl phosphocholine (Fig. 2b, upper two rows). However, a pdr10∆ mutant in the YPH499 genetic background did not exhibit this effect (Fig. 2b, lowest row). We therefore used pdr10∆ cells derived from the W303 lineage for further study.
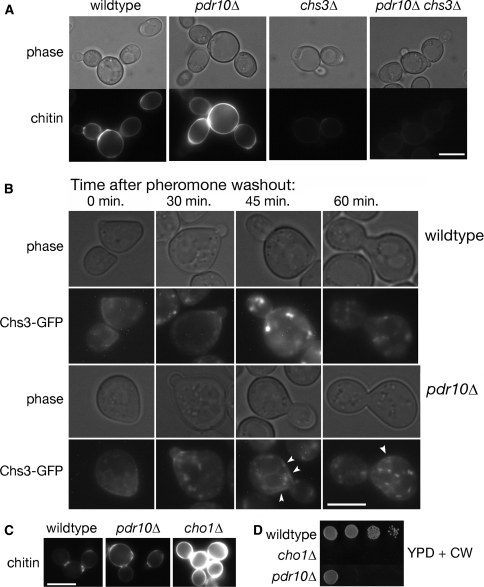
In addition to changes in membrane composition, changes in cell wall structure can affect the detergent sensitivity displayed by S. cerevisiae cells; indeed, cell wall alterations can affect the sensitivity of yeast cells even to a strong anionic detergent like SDS (Bickle et al. 1998; Machi et al. 2004; Santos and Snyder 2000). The yeast cell wall is composed of three types of polymers: mannoproteins, glucan, and chitin (Klis et al. 2006; Lesage and Bussey 2006). We found (Fig. 2c) that pdr10∆ mutants were abnormally sensitive to two agents, Calcofluor White and Congo Red, that inhibit the growth of cells that have abnormally high amounts of chitin (Roncero et al. 1988). In contrast, otherwise isogenic wild-type cells (Fig. 2c), or cells expressing either Pdr10-GFP or Pdr10-mh from the chromosomal PDR10 locus (data not shown), did not display such hypersensitivity (and, thus, both Pdr10-GFP and Pdr10-mh are functional). If the sensitivity of the pdr10∆ cells is due to abnormal chitin, then we expected that sensitivity to Calcofluor White might be ameliorated by compromising chitin production. The chitin synthase responsible for producing the majority of the chitin generated in a vegetatively growing cell is Chs3, but cells can survive in its absence due to the function of the homologous chitin synthase Chs2 (Cabib 2004). Chs3 is normally delivered to the plasma membrane via the action of dedicated escort factors, including Chs6 (Valdivia et al. 2002; Valdivia and Schekman 2003). Consistent with the conclusion that the Calcofluor White sensitivity of pdr10∆ cells arises from elevated cell wall chitin, we found that pdr10∆ chs3∆ and pdr10∆ chs6∆ double mutants were not hypersensitive to Calcofluor White (Fig. 2c). Likewise, in the W303 strain background, a pdr10∆chs3∆ double mutant was not detergent resistant (Fig. 2b, upper rows). These findings indicate that both the Calcofluor White sensitivity and the detergent resistance displayed by pdr10∆ cells are largely due to enhanced chitin production by the integral membrane protein Chs3 in the absence of this ABC transporter.
Although Pdr10 shares its highest degree of homology with Pdr5 and Pdr15, it is also closely related to other ABC transporters in the S. cerevisiae genome, including Pdr12 (36% identity to Pdr10). Pdr12 is located in the plasma membrane and is responsible for the export of weak organic acids (Piper et al. 1998). Consequently, pdr12∆ mutants are hypersensitive to the growth inhibitory action of weak organic acids used as food preservatives (e.g., sorbate or benzoate) (Piper et al. 1998). We found that pdr10∆ cells exhibited weak resistance to sorbate (Fig. 2d); however, this behavior required the function of Pdr12, because the modest sorbate resistance observed was eliminated in a pdr10∆ pdr12∆ double mutant (Fig. 2d). Thus, here, too, the absence of Pdr10 somehow promoted the function of a sister integral membrane protein, Pdr12.
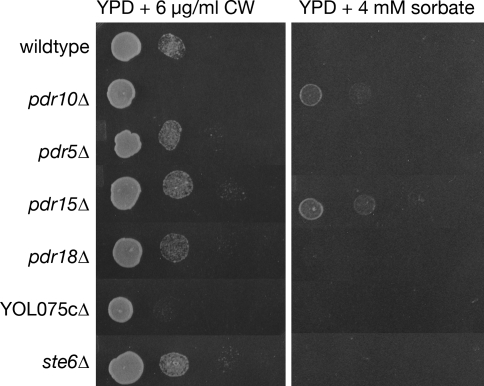
Taken together, these results suggested that the function of Pdr10 contributes to the normal expression, and/or sorting and trafficking, and/or catalytic competence of Chs3 and Pdr12. To determine whether this phenotype was unique to the loss of Pdr10, we examined the Calcofluor White and sorbate sensitivity of yeast strains lacking other members of the S. cerevisiae ABC transporter family known to reside in the plasma membrane. Although comparable sorbate resistance was observed in pdr15∆ cells and comparable Calcofluor White sensitivity was observed in cells lacking the putative, but uncharacterized, ABC transporter encoding by the YOL075c locus, the combination of Calcofluor White sensitivity and sorbate resistance was only observed in pdr10∆ cells (Fig. 3). Thus, Pdr10 uniquely has a role in maintaining the proper level, maturation, localization, and/or function of both Chs3 and Pdr12.
Fig. 3.
Calcofluor sensitivity and sorbate resistance define a specific pdr10∆ phenotype. Serial dilutions of overnight cultures of strains W303-1A (wild type), NRY243 (pdr10∆), NRY201 (pdr5∆), NRY212 (pdr15∆), NRY336 (pdr18∆), NRY334 (YOL075c∆), and WKK7 (ste6∆) were spotted onto YPD plates containing 6 μg/ml Calcofluor White (CW; left column) or 4 mM sorbate (right column)
Pdr10 Is Required for Normal Endocytic Trafficking of Chs3
To distinguish among these possibilities, we first examined the effect of a pdr10∆ mutation on the function and localization of Chs3. To confirm that pdr10∆ cells do produce abnormally high levels of chitin, we examined cells by fluorescence microscopy after staining with Calcofluor White. Indeed, we found that, compared to otherwise isogenic PDR10+ control cells, pdr10∆ cells exhibited brighter fluorescence and therefore excess chitin, while cell wall chitin was almost entirely eliminated when Chs3 was absent (Fig. 4a). Elevated chitin production could arise from increased Chs3, more active Chs3, or aberrant localization of Chs3. As mentioned already, normal anterograde trafficking of Chs3 to the plasma membrane requires additional protein factors, including Chs6 (Valdivia and Schekman 2003), and we found that, like a chs3∆ mutation, a chs6∆ mutation also eliminated the Calcofluor White sensitivity of pdr10∆ cells (Fig. 2c). Therefore, the elevated chitin production of pdr10∆ cells does not occur because the normal pathway for transport of Chs3 to the plasma membrane is bypassed when Pdr10 is absent.
Fig. 4.
Defective endocytosis of Chs3 causes excess chitin deposition in pdr10∆ cells. a Cells from log phase cultures of strains W303-1A (wild type), NRY243 (pdr10∆), NRY602 (chs3∆), and NRY601 (pdr10∆ chs3∆) were stained for chitin and examined by fluorescence microscopy. Bar, 4 μm. b Log phase cells from strains W303-1A and NRY251 (pdr10∆) carrying plasmids pRS315-CHS7 and pCRP12 were treated with 3.6 μM α-factor for 35 min in SCD buffered to pH 3.5 with 25 mM sodium succinate and supplemented with adenine. Pheromone was then washed out at time 0, and the localization of Chs3-GFP was monitored by fluorescence microscopy as described under Materials and Methods. Punctate Chs3-GFP staining of the plasma membrane in pdr10∆ cells at late time points is indicated by arrow heads. Bar, 3 μm. c Strains W303-1A, NRY243, and NRY407 (cho1∆) were grown into log phase, stained for chitin, and examined by fluorescence microscopy. Bar, 5 μm. d Serial dilutions of overnight cultures of strains W303-1A, NRY243, and NRY407 were spotted onto YPD plates containing Calcofluor
In vegetatively growing cells, Chs3 cycles between the trans-Golgi network and endosomal compartments during most of the cell cycle (Valdivia et al. 2002) but displays significant accumulation at the plasma membrane primarily at the small-bud stage (Chuang and Schekman 1996). The intracellular pool of Chs3 can also be redistributed to the plasma membrane after treatment of cells with mating pheromone, an effect we exploited to trap Chs3 at the plasma membrane in wild-type and pdr10∆ cells. Upon washout of pheromone, cells re-entered the cell cycle and Chs3 moved to the bud neck, as expected (Fig. 4b). At later time points, Chs3 was efficiently internalized in wild-type cells, whereas in pdr10∆ cells a small amount of Chs3-GFP persisted in discrete patches at the plasma membrane, even though the majority was intracellular. Thus, in pdr10∆ cells, endocytosis of Chs3 was somewhat less efficient than in wild-type cells (although this block was by no means complete). The prolonged residence time of Chs3 at the plasma membrane in pdr10∆ cells could well explain the increased amount of chitin produced in these cells.
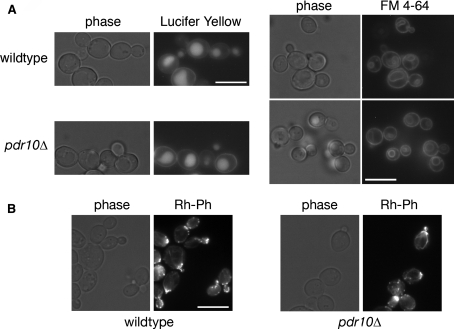
We were interested in determining whether this modest endocytic defect of pdr10∆ cells was specific for Chs3 or was more general. We therefore examined endocytic uptake of two fluorescent dyes, Lucifer Yellow and FM 4-64 (Fig. 5a). These dyes serve as markers for fluid phase endocytosis and bulk membrane internalization, respectively, with both being internalized to the vacuole. Uptake of both of these markers was normal in pdr10∆ cells, demonstrating that the partial block in Chs3 endocytosis is not a general effect on all endocytic processes. We also examined actin morphology in these cells (Fig. 5b), because an intact actin cytoskeleton is a prerequisite for endocytosis in yeast (Ayscough et al. 1997; Kubler and Riezman 1993). Deletion of PDR10 had no apparent effect on actin localization or on its polarization in small-budded cells (Fig. 5b).
Fig. 5.
Pdr10 is not required for bulk endocytosis or actin morphology. a Strains W303-1A (wild type) and NRY243 (pdr10∆) were grown to log phase, incubated with Lucifer Yellow or FM 4-64 as a marker for endocytosis, and examined by fluorescence microscopy. b Strains W303-1A and NRY243 were grown to log phase, permeabilized, and stained with rhodamine-phalloidin to visualize actin by fluorescence microscopy. All bars, 5 μm
Lack of Dnf1 and Dnf2 Results in Defective Endocytosis of Chs3
If the partial defect in endocytosis of Chs3 in a pdr10∆ mutant is responsible for its elevated chitin production and thus its increased sensitivity to Calcofluor White, then other mutations or conditions that influence the efficiency of Chs3 endocytosis should elicit the same phenotype. Interestingly, we found that a cho1∆ strain, which lacks phosphatidylserine synthase (Atkinson et al. 1980a, b; Kiyono et al. 1987; Kohlwein et al. 1988; Letts et al. 1983), displayed even greater chitin deposition (Fig. 4c) and an even higher sensitivity to Calcofluor White than the pdr10∆ strain (Fig. 4d). Quantitation of chitin deposition indicated that pdr10∆ cells had approximately 1.9-fold more chitin in their cell walls than wild-type cells, while cho1∆ cells had approximately 3.8-fold more chitin. We have not yet examined directly the localization of Chs3-GFP in cho1∆ cells; however, these data suggested that membrane composition might be important in Chs3 endocytosis and thus that the absence of Pdr10 might influence plasma membrane lipid distribution. We therefore examined sterol localization in wildtype and pdr10∆ cells by staining with filipin (Fig. 6a), finding little difference between those two strains. Simons and colleagues have reported that treatment with mating pheromone polarizes sterols to the site of polarized growth (the shmoo tip) (Bagnat and Simons 2002), and we find that this process proceeds normally in pdr10∆ cells as well (Fig. 6a).
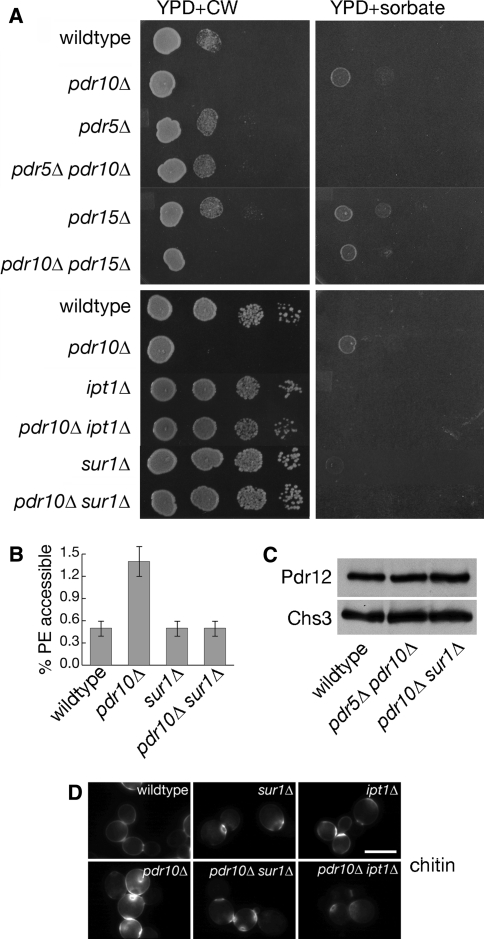
It has been proposed that proper maintenance of membrane asymmetry is important for endocytosis at low temperatures and that Pdr5 and another ABC transporter, Yor1, are candidate outward-directed PE translocases (Pomorski et al. 2003). Moreover, pdr5∆ pdr15∆ cells exhibit increased accessibility of PE to the small-molecule reagent TNBS (Schüller et al. 2007), which is able to pass through the cell wall. TNBS labeling has been used to assess exposure of PE and PS on the outer leaflet of the plasma membrane (Gordesky and Marinetti 1973; Pomorski et al. 2003; Siegmund et al. 1998), although subcellular fractionation data raise the possibility that there is considerable leakage of the reagent into the secretory pathway (Pomorski et al. 2003). We analyzed the total phospholipid composition (Table 2) and accessibility of aminophospholipids to TNBS (Fig. 6b) in wild-type and pdr10∆ cells grown on 32P-phosphate. We found that a pdr10∆ mutation had no discernible effect on overall phospholipid composition (Table 2), but caused a significant increase in the fraction of the PE that was accessible to TNBS labeling (Fig. 6b). Thus, pdr10∆ plasma membranes either are significantly more permeable to TNBS or exhibit a defect in maintenance of plasma membrane asymmetry.
The ABC transporters with established transport activities that are most closely related to Pdr10, including Pdr5 and Snq2, mediate outward-directed lipid transport and/or export of xenobiotics from the cell (Balzi et al. 1994; Bissinger and Kuchler 1994; Decottignies et al. 1995; Egner et al. 1998; Mahé et al. 1996a; Pomorski et al. 2003; Schüller et al. 2007). If Pdr10 had a similar function, then loss of Pdr10 should reduce, not increase, the fraction of PE that is surface-exposed. In contrast, if the absence of Pdr10 somehow indirectly crippled the function of the inward-directed PE translocases, then the biochemical phenotype observed in pdr10∆ cells would be more readily explained. In support of this, we observed that the level of PE accessible to TNBS labeling in pdr10∆ cells was comparable to that in cells in which the genes for both putative inward-directed plasma membrane PE translocases, DNF1 and DNF2, were deleted (Fig. 6b); similar results were also obtained for cells lacking the LEM3 gene, which encodes an integral membrane protein that is required for exit of Dnf1 and Dnf2 from the endoplasmic reticulum (Saito et al. 2004). Furthermore, no additive increase in exposure of PE was observed in pdr10∆ lem3∆ or pdr10∆ dnf1∆ dnf2∆ strains (Fig. 6b). The overall phospholipid compositions of all of these strains were quite comparable (Table 2). The absence of any additive or synergistic effect of the dnf1∆dnf2∆ (or lem3∆) mutations in a pdr10∆ mutant demonstrated that pdr10∆ phenocopied dnf1∆ dnf2∆. That is, regardless of whether TNBS labeling is measuring PE exposure on the outer leaflet or permeability, the loss of Pdr10 function mimicked the loss of Dnf1 and Dnf2 function in these cells.
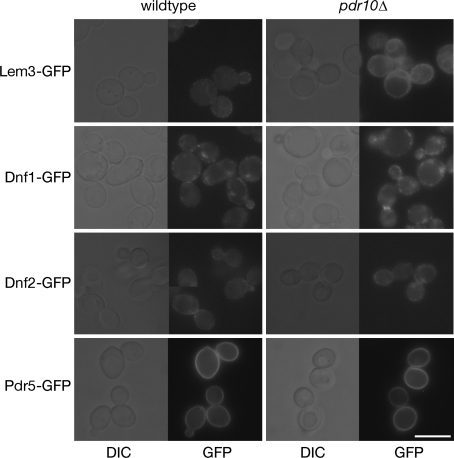
We were interested in whether this might reflect a direct role for Pdr10 in the synthesis, sorting, or activity of the inward-directed translocases. Therefore, we examined the level of expression and the localization of Dnf1, Dnf2, and Lem3 in pdr10∆ cells by constructing functional chromosomal GFP fusions (tests for functionality described under Materials and Methods). Dnf1-GFP, Dnf2-GFP, and Lem3-GFP were all readily detectable by fluorescence microscopy, present at equivalent levels, and exhibited the expected punctate pattern in the plasma membrane in both wild-type and pdr10∆ cells (Fig. 7). There were no discernible differences in the localization of any of these three proteins between wild-type and pdr10∆ cells. Likewise, a functional Pdr5-GFP chimera expressed from the chromosomal PDR5 locus was localized to the plasma membrane indistinguishably in wild-type and pdr10∆ cells (Fig. 7). Putative inward- and outward-directed translocases were thus present at normal levels and were properly localized to the plasma membrane in pdr10∆ cells.
Fig. 7.
Pdr10 is not required for normal localization of putative PE translocases. Strains NRY914 (wild type, Lem3-GFP), NRY918 (pdr10∆, Lem3-GFP), NRY921 (wild type, Dnf1-GFP), NRY932 (pdr10∆, Dnf1-GFP), NRY923 (wild type, Dnf2-GFP), NRY929 (pdr10∆, Dnf2-GFP), NRY906 (wild type, Pdr5-GFP), and NRY916 (pdr10∆, Pdr5-GFP) were grown to log phase, and the indicated proteins were visualized by fluorescence microscopy. Collages of representative images are shown. Bar, 6 μm
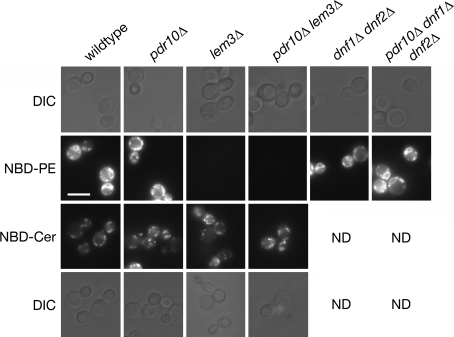
To assess whether inward-directed translocase activity was still present in pdr10∆ cells, we examined the uptake of an exogenously supplied fluorescent PE surrogate, NBD-PE (Kean et al. 1993), in wild-type and pdr10∆ cells (Fig. 8). It has been reported previously that it is possible to observe specific, translocase-dependent internalization of NBD-labeled lipids at low temperature, which prevented background internalization due to endocytosis (Grant et al. 2001). However, in our hands, both wild-type and pdr10∆ cells were still able to internalize modest amounts of NBD-PE to discrete intracellular puncta at low temperature, indicating either that endocytosis was not completely blocked in our hands or that a low level of membrane leakage was occurring (data not shown). We therefore examined NBD-PE uptake in the presence of latrunculin A, which disrupts the actin cytoskeleton and hence blocks endocytosis (Ayscough et al. 1997; Pomorski et al. 2003). We found that pdr10∆ cells exhibited no defect compared to wild-type cells. dnf1∆ dnf2∆ cells also exhibited no defect in NBD-PE internalization under these conditions (Fig. 8), perhaps because the homologous P-type ATPase Drs2 can relocalize to the plasma membrane upon latrunculin treatment (Liu et al. 2007). Strikingly, however, lem3∆ cells were unable to internalize NBD-PE under these conditions, with or without the presence of Pdr10. These data demonstrate that Lem3 must have functions other than its role in the biogenesis of Dnf1 and Dnf2, perhaps in forward transport of Drs2 to the plasma membrane. Neither pdr10∆ nor lem3∆ had any effect on uptake of a fluorescent ceramide surrogate, NBD-Cer (Fig. 8). Taken together, these results demonstrate the presence of normal amounts of Dnf1, Dnf2, and Lem3 in pdr10∆ plasma membranes and demonstrate that pdr10∆ cells are still capable of PE internalization. It is known that mutations in a number of genes implicated in vesicular trafficking produce altered plasma membrane asymmetry as a secondary effect (Chen et al. 2006; Parsons et al. 2006), so it seems likely that increased accessibility of PE to TNBS labeling in pdr10∆ cells is such an indirect consequence.
Fig. 8.
pdr10∆ cells internalize NBD-PE in a Lem3-dependent manner. Strains W303-1A (wild type), NRY243 (pdr10∆), NRY549 (lem3∆), NRY551 (pdr10∆ lem3∆), NRY427 (dnf1∆ dnf2∆), and NRY558 (pdr10∆ dnf1∆ dnf2∆) were grown to log phase, treated with latrunculin A to block endocytosis, and incubated with the fluorescent reporter lipid NBD-PE or NBD-Cer as described under Materials and Methods. Cells were examined by fluorescence microscopy and differential interference contrast microscopy (DIC). Bar, 6 μm
Dnf1 and Dnf2 have been implicated in endocytosis at low temperatures (Furuta et al. 2007; Pomorski et al. 2003), in keeping with a broader role for members of this transporter family in vesicular trafficking (Chen et al. 2006). Given that the PE exposure caused by a pdr10∆ mutation was quite comparable to that caused by dnf1∆ dnf2∆ double deletion, the mild defect in endocytic trafficking we observed for Chs3 in pdr10∆ cells might also be associated with dnf1∆ dnf2∆ or lem3∆ cells. Indeed, we found that cells lacking both Dnf1 and Dnf2 were sensitive to Calcofluor White (Fig. 9a), exhibited increased chitin in their cell wall (Fig. 9b), and were equivalent to pdr10∆ cells in both respects. Moreover, a pdr10∆ dnf1∆ dnf2∆ triple mutant strain did not display any increase in Calcofluor sensitivity or chitin deposition compared to the otherwise isogenic pdr10∆ single mutant or the dnf1∆ dnf2∆ double mutant (Fig. 9a, b), with all mutant strains exhibiting a 1.8- to 1.9-fold increase in chitin deposition. This lack of synergy was not due to any limitation in the capacity of these assays to detect such potentiation, because cho1∆ cells exhibited much higher Calcofluor White sensitivity and chitin deposition than pdr10∆ cells (Fig. 4). Moreover, lem3∆ cells were also more sensitive to Calcofluor White than pdr10∆ or dnf1∆ dnf2∆ cells (Fig. 9a), which is reminiscent of the more profound effect of this mutation on NBD-PE uptake (Fig. 8). Taken together, these results show that pdr10∆ cells exhibit a modest defect in endocytosis of Chs3, that this mimics a phenotype of dnf1∆ dnf2∆ cells, and that Pdr10 is not required for expression or localization of Dnf1, Dnf2, and Lem3. We have not been able to elucidate the molecular basis for this defect in Chs3 trafficking.
Fig. 9.
Loss of Dnf1 and Dnf2 function in pdr10∆ cells is associated with their increased chitin deposition. a Serial dilutions from overnight cultures of strains W303-1A (wild type), NRY243 (pdr10∆), NRY427 (dnf1∆ dnf2∆), NRY558 (pdr10∆ dnf1∆ dnf2∆), NRY549 (lem3∆), and NRY551 (pdr10∆ lem3∆) were spotted onto YPD plates at increasing concentrations of Calcofluor White (CW). b Log phase cultures of strains W303-1A, NRY243, NRY427, and NRY558 were stained for chitin and examined by fluorescence microscopy. Bar, 5 μm. c Serial dilutions of strains W303-1A, NRY243, NRY549, and NRY551 were spotted onto YPD plates buffered to pH 3.5 with sodium succinate and containing potassium sorbate. d Serial dilutions of strains W303-1A, NRY243, NRY301 (pdr12∆), and NRY366 (pdr10∆ pdr12∆) were spotted onto YPD plates containing Calcofluor White
Complex Interplay Among Pdr10, Lem3, and Pdr12
Cells lacking Pdr10 also exhibit a modest increase in their resistance to sorbate that requires the sorbate transporter, Pdr12 (Fig. 2d). We were interested in whether this was also a phenotype of dnf1∆ dnf2∆ cells or lem3∆ cells. Interestingly, we found not only that lem3∆ cells were not sorbate resistant, but that a lem3∆ mutation eliminated the modest sorbate resistance conferred by the pdr10∆ mutation; no such epistasis was found with the dnf1∆ dnf2∆ double mutant (Fig. 9c and data not shown). Thus, the apparent increase in Pdr12 action found in cells lacking Pdr10 does not mimic a defect found in dnf1∆ dnf2∆ or lem3∆ cells but, rather, requires the presence of functional Lem3.
Cells lacking Pdr10 exhibit increased sensitivity to the chitin-directed agent, Calcofluor White (Fig. 2c), and that is not further enhanced by the absence of Dnf1 and Dnf2 (Fig. 9a). Strikingly, as observed for the sorbate resistance of pdr10∆ cells, we found that loss of Pdr12 eliminated the Calcofluor White sensitivity of pdr10∆ (Fig. 9d). We also found that the introduction of a pdr12∆ mutation into pdr10∆ cells substantially reduced the accessibility of PE to TNBS labeling (Table 2). Collectively, these findings indicate complex interactions among Pdr10, Pdr12, and Lem3, which produce secondary effects on Chs3 endocytosis.
Pdr10 Action Excludes Pdr12 from the Detergent-Resistant Membrane Fraction
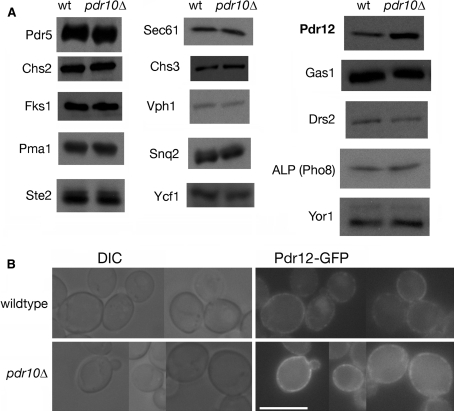
To see whether a clearer molecular function could be assigned to Pdr10, we examined whether loss of Pdr10 affects the level and/or localization of Pdr12. When equivalent amounts of plasma membrane protein were resolved by SDS-PAGE and analyzed by immunoblotting, we found that, compared to otherwise isogenic control cells, pdr10∆ mutant cells exhibited a reproducible, approximately twofold increase in the amount of Pdr12 relative to wild-type cells, whereas there was no significant difference between wild-type and pdr10∆ cells in the content of numerous other membrane proteins in the same samples (Fig. 10a). Visualization of functional Pdr12-GFP expressed from the PDR12 locus in wild-type and pdr10∆ cells confirmed that Pdr12-GFP expression was elevated in the pdr10∆ mutant cells (Fig. 10b), but we could distinguish no obvious difference in localization. In both wild-type and pdr10∆ cells, the Pdr12-GFP was observed throughout the plasma membrane in a readily observable punctate pattern. Approximately 30% of cells in both strains also exhibited some intracellular punctate staining (data not shown), with most such cells also exhibiting plasma membrane Pdr12-GFP. The level of PDR12 mRNA in pdr10∆ cells was not different from that in PDR10+ control cells, as judged by global transcriptome analysis using hybridization to DNA microarrays (S. Shtivelman, N. C. Rockwell, and J. Thorner, unpublished data). Thus, the increase in Pdr12 found in pdr10∆ cells could not be attributed to any change in transcription, and therefore the elevation of plasma membrane Pdr12 in pdr10∆ cells most likely results from a reduced rate of turnover rather than a faster rate of synthesis. The latter might occur if compartmentation of Pdr12 was altered in pdr10∆ cells in a manner that was not revealed at the level of fluorescence microscopy.
Fig. 10.
pdr10∆ cells accumulate excess Pdr12 in the plasma membrane. a Total membranes were prepared from log phase cultures of strains W303-1A (wild type) and NRY243 (pdr10∆). The indicated proteins were detected by immunoblotting, with equal protein loaded for each strain. b Strains NRY915 (wild type) and NRY925 (pdr10∆) were grown to log phase in YPD, and Pdr12-GFP was examined by fluorescence microscopy. A collage of representative images is shown. Pdr12-GFP function was confirmed by sorbate sensitivity. Bar, 6 μm
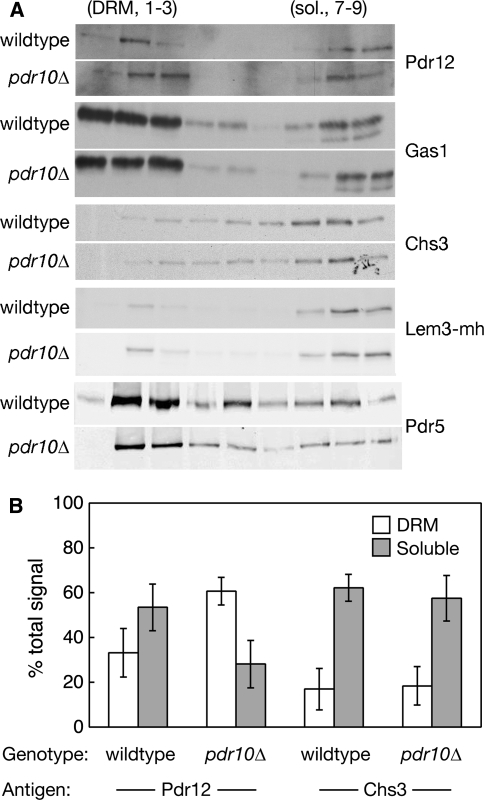
When membranes are subjected to biochemical fractionation, a readily detectable amount of Pdr10 is associated with the DRM fraction (Fig. 1d). Hence, we also examined the distribution of Pdr12 in this assay. Strikingly, we found that approximately 60% of the total pool of Pdr12 was associated with the DRM fraction in membranes from pdr10∆ cells, whereas in the membranes from wild-type cells approximately one-third of the total Pdr12 was present in the DRM fraction and more than 50% was present in the detergent-soluble fraction (Fig. 11 and Table 3). The Pdr10-dependent shift observed for Pdr12 was in contrast to Chs3 and Gas1 (Bagnat et al. 2000; Nuoffer et al. 1991), whose distributions were unaltered by loss of Pdr10 (Fig. 11 and Table 3). Association of Lem3 and Pdr5 with the DRM fraction was slightly higher in pdr10∆ cells than in the wild type (Table 3), but this effect was too small to be statistically significant. Thus, our data indicate a novel role for Pdr10 as a negative regulator of the partitioning of Pdr12 into the DRM fraction; that is, Pdr10 must regulate the local environment of Pdr12 in some manner that alters its solubilization properties in the presence of detergent.
Fig. 11.
pdr10∆ cells accumulate Pdr12 in the detergent-resistant membrane fraction. a Total membranes from log phase cultures of strains W303-1A or NRY904 (wildtype) and NRY243 or NRY908 (pdr10∆) were examined by cold detergent extraction, Optiprep density gradient flotation, and immunoblotting for the indicated proteins. Strains NRY904 and NRY908 contain Lem3 C-terminally tagged with a tandem myc-His6 (mh) tag. In vivo function of the resulting Lem3-mh protein was confirmed by Calcofluor sensitivity. b Immunoblotting data were quantitated, and the percentage of total Pdr12 (left) and Chs3 (right) associated with detergent-resistant and detergent-soluble fractions is presented as the mean value for four independent experiments, with error bars representing the standard deviation. Detergent-resistant and detergent-soluble membranes were defined as fractions 1–3 and 7–9, respectively, in the Optiprep density gradients
Table 3.
Association of membrane proteins with the detergent-resistant membrane fractiona
| Protein | Wild type | pdr10∆ | ||
|---|---|---|---|---|
| % DRM | % Soluble | % DRM | % Soluble | |
| Pdr12 | 33 ± 11 | 54 ± 10 | 61 ± 6 | 28 ± 11 |
| Chs3 | 17 ± 9 | 62 ± 6 | 19 ± 9 | 56 ± 10 |
| Lem3-mh | 19 ± 5 | 75 ± 7 | 24 ± 4 | 72 ± 1 |
| Gas1 | 47 ± 5 | 37 ± 1 | 45 ± 2 | 36 ± 2 |
| Pdr5 | 57 ± 11 | 23 ± 10 | 69 ± 9 | 14 ± 8 |
Note: Membranes were prepared and analyzed by cold Triton X-100 extraction, Optiprep density gradient flotation, and quantitative immunoblotting as described under Materials and Methods. For Pdr12 and Chs3, results are based on four independent experiments. All other proteins were examined at least twice. Strains were W303-1A or NRY904 (wild type with or without chromosomally mh-tagged Lem3) and NRY243 or NRY908 (pdr10∆ with or without chromosomally mh-tagged Lem3)
We were also interested in whether Pdr10 function was redundant with that of its two closest homologues, Pdr5 and Pdr15. As one means to examine possible redundancy of this sort among Pdr10, Pdr5, and Pdr15, we examined their genetic interactions using Calcofluor White sensitivity and sorbate resistance. A pdr5∆ mutation eliminated both the Calcofluor White sensitivity and the mild sorbate resistance of pdr10∆ cells (Fig. 12a). pdr5∆ pdr10∆ cells also exhibited significantly lower TNBS labeling of PE (Table 2) and normal levels of Pdr12 (Fig. 12c). In contrast, a pdr15∆ mutation had no discernible effect on pdr10∆ phenotypes (Fig. 12a).
Fig. 12.
Pdr5 and phosphosphingolipids are required for pdr10∆ phenotypes. a Serial dilutions of overnight cultures of strains W303-1A (wild type), NRY243 (pdr10∆), NRY201 (pdr5∆), NRY230 (pdr5∆ pdr10∆), NRY212 (pdr15∆), NRY236 (pdr10∆ pdr15∆), NRY410 (ipt1∆), NRY534 (pdr10∆ ipt1∆), NRY409 (sur1∆), and NRY526 (pdr10∆ sur1∆) were spotted onto YPD plates containing Calcofluor White (CW; left column) or sorbate buffered with sodium succinate (right column). b Strains W303-1A, NRY243, NRY409, and NRY526 were assayed for PE accessibility to TNBS labeling after overnight labeling with 32P-phosphate as described under Materials and Methods. c Total Pdr12 and Chs3 levels were examined by immunoblotting equal amounts of total membrane proteins isolated from log phase cultures of strains W303-1A, NRY230, and NRY526. d Strains W303-1A, NRY243, NRY409, NRY526, NRY410, and NRY534 were grown to log phase, stained for chitin, and examined by fluorescence microscopy. Bar, 5 μm
Interaction Between Pdr10 Function and Sphingolipids
Sphingolipids are known to be important factors for lateral segregation and DRM formation in yeast membranes (Bagnat et al. 2000). We were therefore interested in the interplay between sphingolipid composition and Pdr10 function, because of its role as a negative regulator of Pdr12 association with the DRM fraction. In S. cerevisiae, the headgroups of mature sphingolipids contain inositol phosphate. Inositol phosphate is added to ceramide by Aur1, producing inositolphosphorylceramide (IPC). AUR1 is an essential gene, demonstrating that IPC or lipids derived from IPC are essential (Nagiec et al. 1997). IPC is converted to mannosyl-inositolphosphoryl-ceramide (MIPC) by the redundant action of Sur1 (also known as Csg1) and Csh1 (Beeler et al. 1997; Uemura et al. 2003). MIPC is then converted to mannosyl-di(inositolphosphoryl)-ceramide (M[IP]2C) by the subsequent action of Ipt1 (Dickson et al. 1997).
Both sur1∆ and ipt1∆ mutations were epistatic to pdr10∆ for both Calcofluor White sensitivity and sorbate resistant (Fig. 12a). Thus, deletion of genes required for synthesis of mature sphingolipid headgroups ablated the phenotypes of pdr10∆ cells, even though pdr10∆ cells did not exhibit significant changes either in the level of total phosphosphingolipid (Table 2) or in the composition of phosphosphingolipids (Table 4). However, sur1∆ and ipt1∆ cells exhibited significant changes in sphingolipid composition whether Pdr10 was present or absent, as expected (Table 4). pdr10∆ sur1∆ cells did not display abnormally high TNBS labeling of PE (Fig. 12b and Table 2) and contained normal levels of Pdr12 (Fig. 12c). Furthermore, both sur1∆ and ipt1∆ eliminated the elevated chitin deposition caused by a pdr10∆ mutation (Fig. 12d). Taken together, these results demonstrate that the aberrant accumulation of Pdr12 and defective endocytosis of Chs3 that occur in pdr10∆ cells require the presence of mannosylphosphosphingolipids.
Table 4.
Composition of mature sphingolipids
| Strain | IPC | MIPC | M(IP)2C |
|---|---|---|---|
| Wild type | 35 ± 4 | 17 ± 2 | 47 ± 4 |
| pdr10∆ | 41 ± 2 | 18 ± 2 | 41 ± 1 |
| sur1∆ | 61 ± 3 | 13 ± 1 | 26 ± 3 |
| ipt1∆ | 46 ± 3 | 48 ± 2 | 7 ± 1 |
Note: Strains were W303-1A (wild type), NRY243 (pdr10∆), NRY409 (sur1∆), and NRY410 (ipt1∆). Sphingolipids were labeled with 32P-phosphate, extracted, and analyzed as described under Materials and Methods
Discussion
We have shown that the ABC transporter Pdr10 is a plasma membrane protein that plays a role in maintaining the proper distribution and function of a subset of other membrane proteins in the yeast S. cerevisiae. In the absence of Pdr10, yeast cells exhibit a modest defect in the endocytosis of the chitin synthase Chs3, resulting in increased cell wall chitin and consequent sensitivity to Calcofluor White. The increased chitin deposition and Calcofluor sensitivity are equivalent to those seen in dnf1∆dnf2∆ cells, which lack the putative inward-directed PE translocases of the yeast plasma membrane (Pomorski et al. 2003). Both pdr10∆ cells and dnf1∆dnf2∆ cells exhibit increased accessibility of PE to TNBS labeling, consistent with a defect in maintenance of plasma membrane asymmetry. However, Dnf1 and Dnf2 are present at normal levels in pdr10∆ cells, and these cells are able to internalize NBD-PE. It thus seems likely that these phenotypes of pdr10∆ cells arise indirectly, perhaps as a consequence of a modest defect in trafficking of plasma membrane proteins.
The absence of Pdr10 function also results in increased resistance to the weak organic acid sorbate. Sorbate is directly exported from yeast cells by the plasma membrane ABC transporter Pdr12, itself a Pdr10 paralogue. We have shown that pdr10∆ cells accumulate higher levels of Pdr12, despite no changes in PDR12 transcription. Additionally, we have shown that the behavior of Pdr12 upon detergent extraction is altered in pdr10∆ membranes: while Pdr12 in wild-type membranes is mainly in the detergent-soluble fraction, it is mainly in the DRM fraction in pdr10∆ membranes. There are complex interactions among PDR10, PDR12, and LEM3, because pdr12∆ is epistatic to pdr10∆ for Calcofluor sensitivity and for TNBS labeling of PE, while lem3∆ is epistatic to pdr10∆ for sorbate resistance. By contrast, the dnf1∆ dnf2∆ double mutant is equivalent to pdr10∆ for Chs3-related phenotypes, with no additive effect in the pdr10∆ dnf1∆ dnf2∆ triple mutant, but does not affect the Pdr12-related phenotype of sorbate resistance. Moreover, pdr5∆ is epistatic to pdr10∆ for both phenotypes. Pdr5, Pdr12, and Lem3 thus seem to be proteins intimately linked to the molecular function of Pdr10. Interestingly, sur1∆ and ipt1∆, which lack genes required for elaboration of sphingolipid headgroups, are epistatic to pdr10∆ for Calcofluor sensitivity, chitin deposition, sorbate resistance, Pdr12 accumulation, and TNBS labeling of PE. The aberrant behavior of pdr10∆ membranes thus requires at least three specific proteins (Pdr5, Pdr12, and Lem3) and also requires mature mannosylphosphosphingolipids.
In the presence of these components, pdr10∆ cells significantly overincorporate Pdr12 into the DRM fraction. Pdr10 thus functions as a negative regulator of Pdr12 incorporation into DRMs. Slight increases in Pdr5 and Lem3 association with the DRM fraction are also seen, while Gas1 and Chs3 distributions are unchanged. Pdr12 has been estimated to be present at approximately 750 molecules per cell (Ghaemmaghami et al. 2003), and approximately one-third of the total is shifted to the DRM fraction in pdr10∆ membranes relative to wild-type membranes (Table 3). Therefore, approximately 250 additional Pdr12 molecules per cell would become incorporated into the DRM fraction in the absence of Pdr10. In the same study, levels of Ste20 were estimated at approximately 260 molecules per cell, and both Dnf1 and Dnf2 exhibited lower but still detectable signals; however, Pdr10 could not be detected at all (Ghaemmaghami et al. 2003). Therefore, Pdr10 levels must be substantially lower than 250 molecules per cell, which means that Pdr10 is substoichiometric with the Pdr12 population that is shifted to the DRM fraction in pdr10∆ membranes. It thus seems unlikely that the overincorporation of Pdr12 in the DRM fraction of pdr10∆ membranes reflects the loss of a stoichiometric association between Pdr10 and Pdr12.
One could also envision the redistribution of Pdr12 into the DRM fraction as arising from an endocytic defect, such that an intracellular pool of Pdr12 would effectively relocalize to the plasma membrane in the absence of Pdr10. The proposed link between endocytosis and exit from the DRM fraction (Lauwers and Andre 2006; Lauwers et al. 2007) would imply that such an intracellular pool would become available for partitioning into the DRM fraction upon such relocalization. In this model, the plasma membrane Pdr12 would be in equilibrium between a microenvironment that gives rise to a DRM fraction and a microenvironment that does not, while intracellular Pdr12 would instead be entirely associated with the detergent-soluble fraction. The absence of Pdr12 endocytosis in pdr10∆ cells would then result in the total Pdr12 population equilibrating between these two microenvironments, rather than just a plasma membrane population doing so. Were this to be the case, the association of approximately 60% of total Pdr12 with the DRM fraction in pdr10∆ cells would reflect the intrinsic affinity of Pdr12 for this fraction, while no intracellular Pdr12 would be found in the DRM fraction. The amount of Pdr12 in the DRM fraction would thus vary between 0% and 60%. The association of approximately 30% of total Pdr12 with the DRM fraction in wild-type cells would thus imply the presence of approximately 50% of total Pdr12 in an intracellular pool. However, we demonstrated that a functional Pdr12-GFP chimera is predominantly localized to the plasma membrane (Fig. 10b). Only ≤ 30% of cells exhibited any detectable intracellular Pdr12-GFP, and the vast majority of such cells also displayed plasma membrane Pdr12-GFP. Moreover, this localization pattern was equivalent in both wild-type and pdr10∆ cells. Therefore, the intracellular population of Pdr12 is likely to be much less than 50% of the total, and there is no evidence that pdr10∆ cells lack this population. These observations are inconsistent with the idea that Pdr12 accumulation in the DRM fraction of pdr10∆ membranes arises from relocalization of Pdr12 to the plasma membrane. We favor, therefore, the idea that the composition of plasma membrane in cells lacking Pdr10 is itself different from that of the wild-type plasma membrane in a way that results in overincorporation of Pdr12 into the DRM fraction.
The gradient flotation assay we used to examine DRM incorporation does not provide information about the actual size of any detergent-resistant structures present in the yeast plasma membrane, but is only sensitive to density during fractionation. It is not rigorously established whether microdomains of native membranes can be stably maintained during the lysis, extraction, and flotation procedure, and it has been argued that detergent treatment can induce DRM formation in homogeneous model membranes (Lichtenberg et al. 2005). However, it is hard to see how such an effect could be specifically enhanced by the absence of such a rare protein as Pdr10. Therefore, the higher occupancy of Pdr12 in the DRM fraction observed in pdr10∆ membranes would represent either its increased partitioning into a specialized membrane subdomain or its increased interaction with boundary lipids that are resistant to removal by detergent. It thus seems that the microenvironment of Pdr12 in the native membrane is altered in the absence of Pdr10.
The most parsimonious model to accommodate all of the observations we have made is that the normal function of Pdr10 is to counteract some of the protein or lipid movements that are needed to establish a specific microdomain in the plasma membrane. Hence, when Pdr10 is absent, there is aberrant or excess formation of this microdomain, and Pdr12 becomes sequestered. Chs3 and Gas1 do not get trapped in this way. Pdr12 sequestered in such a compartment may be more resistant to the processes responsible for its endocytosis, explaining why it accumulates to a higher than normal level in pdr10∆ cells. If Pdr12, Lem3, and Pdr5 are all required for formation of this microdomain, the observed epistasis among these membrane proteins would be explained, because removal of a required microdomain component would disrupt the microdomain and mimic the loss of a negative regulator. Similarly, if the proposed microdomain regulated by Pdr10 requires mature sphingolipids for its formation, then alteration of sphingolipid composition could also ameliorate the effects resulting from the absence of Pdr10, as we observed. Lem3 and Pdr5 are both present at higher levels than Pdr12 (Ghaemmaghami et al. 2003), which provides a possible explanation for why their shift to the DRM fraction (Table 3) is not statistically significant: if Pdr12 is also required for microdomain formation and is less abundant than Lem3 or Pdr5, it will be limiting for microdomain formation, such that the amount of Lem3 and Pdr5 shifted to the DRM fraction is smaller.
The precise nature of the putative sphingolipid-dependent microdomain on which Pdr10 has its effect is unclear. Pdr12, Dnf1, Dnf2, Lem3, and Pdr5 do not accumulate in the ER in pdr10∆ cells, and we have shown that Pdr12 and Pdr5 are active in these cells. The absence of Pdr10 also does not activate the unfolded-protein response pathway (Ron and Walter 2007) (data not shown), providing further evidence that these membrane proteins are not misfolded. Therefore, this microdomain should be viewed as an accretion of functional proteins rather than as an aggregate of unfolded proteins. It has been reported that association of permeases into ~300-nm patches in the membrane requires maintenance of the transmembrane electrochemical potential (Grossmann et al. 2007). Also, it is now generally accepted that segregation of membrane proteins into the DRM fraction in yeast and other eukaryotes depends on the presence of both sterols and sphingolipids (Bagnat et al. 2000; Dupré and Haguenauer-Tsapis 2003; Hearn et al. 2003). Formation of large oligomers of Pma1 is required for the segregation of this protein into the DRM fraction (Bagnat et al. 2001; Lee et al. 2002), and there is ample evidence for certain members of the ABC transporter family that their native state in the plasma membrane is as a large homomultimer (Poruchynsky and Ling 1994; Xu et al. 2007). Thus, both protein-protein and lipid-protein interactions can contribute in a positive sense to microdomain formation, and indeed our epistasis analysis provides indirect evidence for both. Pdr10 acts as a negative regulator of the establishment and/or maintenance of plasma membrane microdomains. This suggests that microdomain assembly and breakdown are quite dynamic processes in vivo.
If microdomains have a critically important role in the physiological functions of the plasma membrane, one might question why a pdr10∆ mutant is viable. However, all of the evidence we have presented indicates that microdomain formation still occurs in cells lacking Pdr10 and that the defect is in the efficiency of microdomain disassembly. Moreover, there are yet other ABC transporters encoded in the S. cerevisiae genome whose function may be semiredundant with that of Pdr10 and, hence, partially mask the true effect of the problems that might be caused if microdomains remained too static. The latter consideration may explain why we observed some strain-background dependence in the phenotypes conferred by a pdr10∆ mutation. In this regard, it is also interesting to note that the noncoding region of the PDR10 gene has been identified as a mutational hotspot in the S. cerevisiae genome (Fay and Benavides 2005). Thus, different lineages of this yeast may have adapted to cope with different demands for Pdr10 function. It is also important to note that the functional significance of the putative Pdr10-regulated microdomain is unclear; it is quite possible that this structure is actually a pathological association of functional Pdr12, Pdr5, and Lem3 in the presence of sphingolipids, and we do not propose to identify it with one of the previously defined compartments of the yeast plasma membrane.
The function of Pdr10 at the molecular level and the means by which its biochemical activity contravenes microdomain formation or maintenance have yet to be elucidated. Possibly, Pdr10 catalyzes outward translocation of an as yet unidentified lipid substrate, by analogy to its paralogous ABC transporter, Pdr5. Alternatively, the fact that Pdr10 itself localizes in discrete puncta in the plasma membrane may be important. For example, the microdomain compartment that appears to accumulate when Pdr10 is absent might be formed from (or beside) the subdomain normally occupied by Pdr10 itself. Hence, Pdr10 could help to occlude or exclude other integral membrane proteins from this same region without invoking any need for it to have transport activity. Another possibility is that, when Pdr10 is embedded in the plasma membrane, it dynamically associates with a specific set of lipids (or even other proteins); in this way, Pdr10 may act as a “detergent” to “solubilize” these components and thus enhance their dispersal within the plane of the membrane.
Acknowledgments
The authors thank Françoise Roelants, Victor Cid, Matthias Versele, Daniel Ballon, and other members of the Thorner and Kuchler laboratories for helpful discussions. We also wish to thank Gustav Ammerer for many stimulating discussions; William Dowhan, Günther Daum, and Dennis Voelker for protocols for lipid analysis and useful advice; and Todd Graham and the reviewers for constructive criticism. We thank Randy Schekman, Scott Moye-Rowley, Todd Graham, John McCusker, Andreas Conzelmann, Paco Portillo and Pilar Eraso, Eckhard Boles, Tom Stevens, and Yoshi Ohya for antibodies and/or plasmids used in the course of these studies. This work was supported by postdoctoral fellowship PF98-141-01 from the American Cancer Society and NCI postdoctoral traineeship CA09041 to N.C.R., by grants from the Austrian Science Foundation (“Fonds zur Förderung der wissenschaftlichen Forschung” project SFB035-04) and the Marie-Curie training network RTN-Flippases (CT-MC-RTN-2004-005330) to K.K., and by NIH Research Grant GM21841 and facilities provided by the Berkeley campus Cancer Research Laboratory to J.T.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Nathan C. Rockwell, Email: ncrockwell@ucdavis.edu
Jeremy Thorner, Email: jthorner@berkeley.edu.
References
- Abolmaali S, Mitterbauer R, Spadiut O, Peruci M, Weindorfer H, Lucyshyn D, Ellersdorfer G, Lemmens M, Moll WD, Adam G (2008) Engineered bakers yeast as a sensitive bioassay indicator organism for the trichothecene toxin deoxynivalenol. J Microbiol Methods 72:306–312 [DOI] [PubMed]
- Atkinson K, Fogel S, Henry SA (1980a) Yeast mutant defective in phosphatidylserine synthesis. J Biol Chem 255:6653–6661 [PubMed]
- Atkinson KD, Jensen B, Kolat AI, Storm EM, Henry SA, Fogel S (1980b) Yeast mutants auxotrophic for choline or ethanolamine. J Bacteriol 141:558–564 [DOI] [PMC free article] [PubMed]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG (1997) High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol 137:399–416 [DOI] [PMC free article] [PubMed]
- Bagnat M, Simons K (2002) Cell surface polarization during yeast mating. Proc Natl Acad Sci USA 99:14183–14188 [DOI] [PMC free article] [PubMed]
- Bagnat M, Keranen S, Shevchenko A, Simons K (2000) Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl Acad Sci USA 97:3254–3259 [DOI] [PMC free article] [PubMed]
- Bagnat M, Chang A, Simons K (2001) Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol Biol Cell 12:4129–4138 [DOI] [PMC free article] [PubMed]
- Balzi E, Wang M, Leterme S, Van Dyck L, Goffeau A (1994) PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J Biol Chem 269:2206–2214 [PubMed]
- Beeler TJ, Fu D, Rivera J, Monaghan E, Gable K, Dunn TM (1997) SUR1 (CSG1/BCL21), a gene necessary for growth of Saccharomyces cerevisiae in the presence of high Ca2 + concentrations at 37 degrees C, is required for mannosylation of inositolphosphorylceramide. Mol Gen Genet 255:570–579 [DOI] [PubMed]
- Bickle M, Delley PA, Schmidt A, Hall MN (1998) Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J 17:2235–2245 [DOI] [PMC free article] [PubMed]
- Bissinger PH, Kuchler K (1994) Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. A yeast ABC transporter conferring mycotoxin resistance. J Biol Chem 269:4180–4186 [PubMed]
- Bogdanov M, Mileykovskaya E, Dowhan W (2008) Lipids in the assembly of membrane proteins and organization of protein supercomplexes: implications for lipid-linked disorders. Subcell Biochem 49:197–239 [DOI] [PMC free article] [PubMed]
- Cabib E (2004) The septation apparatus, a chitin-requiring machine in budding yeast. Arch Biochem Biophys 426:201–207 [DOI] [PubMed]
- Carman GM, Henry SA (2007) Special issue: Regulation of lipid metabolism in yeast. Biochim Biophys Acta 1771:239–240 [DOI] [PMC free article] [PubMed]
- Chang SC, Heacock PN, Clancey CJ, Dowhan W (1998) The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J Biol Chem 273:9829–9836 [DOI] [PubMed]
- Chen S, Wang J, Muthusamy BP, Liu K, Zare S, Andersen RJ, Graham TR (2006) Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic 7:1503–1517 [DOI] [PubMed]
- Chuang JS, Schekman RW (1996) Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J Cell Biol 135:597–610 [DOI] [PMC free article] [PubMed]
- Cui Z, Hirata D, Tsuchiya E, Osada H, Miyakawa T (1996) The multidrug resistance-associated protein (MRP) subfamily (Yrs1/Yor1) of Saccharomyces cerevisiae is important for the tolerance to a broad range of organic anions. J Biol Chem 271:14712–14716 [DOI] [PubMed]
- Daum G, Lees ND, Bard M, Dickson R (1998) Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast 14:1471–1510 [DOI] [PubMed]
- David NE, Gee M, Andersen B, Naider F, Thorner J, Stevens RC (1997) Expression and purification of the Saccharomyces cerevisiae alpha-factor receptor (Ste2p), a 7-transmembrane-segment G protein-coupled receptor. J Biol Chem 272:15553–15561 [DOI] [PubMed]
- Decottignies A, Lambert L, Catty P, Degand H, Epping EA, Moye-Rowley WS, Balzi E, Goffeau A (1995) Identification and characterization of SNQ2, a new multidrug ATP binding cassette transporter of the yeast plasma membrane. J Biol Chem 270:18150–18157 [DOI] [PubMed]
- Decottignies A, Grant AM, Nichols JW, de Wet H, McIntosh DB, Goffeau A (1998) ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J Biol Chem 273:12612–12622 [DOI] [PubMed]
- Devaux PF (1991) Static and dynamic lipid asymmetry in cell membranes. Biochemistry 30:1163–1173 [DOI] [PubMed]
- Dickson RC (2008) Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. J Lipid Res 49:909–921 [DOI] [PMC free article] [PubMed]
- Dickson RC, Nagiec EE, Wells GB, Nagiec MM, Lester RL (1997) Synthesis of mannose-(inositol-P)2-ceramide, the major sphingolipid in Saccharomyces cerevisiae, requires the IPT1 (YDR072c) gene. J Biol Chem 272:29620–29625 [DOI] [PubMed]
- Draeger A, Wray S, Babiychuk EB (2005) Domain architecture of the smooth-muscle plasma membrane: regulation by annexins. Biochem J 387:309–314 [DOI] [PMC free article] [PubMed]
- Dupré S, Haguenauer-Tsapis R (2003) Raft partitioning of the yeast uracil permease during trafficking along the endocytic pathway. Traffic 4:83–96 [DOI] [PubMed]
- Egner R, Mahé Y, Pandjaitan R, Kuchler K (1995) Endocytosis and vacuolar degradation of the plasma membrane-localized Pdr5 ATP-binding cassette multidrug transporter in Saccharomyces cerevisiae. Mol Cell Biol 15:5879–5887 [DOI] [PMC free article] [PubMed]
- Egner R, Rosenthal FE, Kralli A, Sanglard D, Kuchler K (1998) Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Mol Biol Cell 9:523–543 [DOI] [PMC free article] [PubMed]
- Elvington SM, Bu F, Nichols JW (2005) Fluorescent, acyl chain-labeled phosphatidylcholine analogs reveal novel transport pathways across the plasma membrane of yeast. J Biol Chem 280:40957–40964 [DOI] [PubMed]
- Esko JD, Raetz CR (1980) Mutants of Chinese hamster ovary cells with altered membrane phospholipid composition. Replacement of phosphatidylinositol by phosphatidylglycerol in a myo-inositol auxotroph. J Biol Chem 255:4474–4480 [PubMed]
- Evan GI, Lewis GK, Ramsay G, Bishop JM (1985) Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol 5:3610–3616 [DOI] [PMC free article] [PubMed]
- Fay JC, Benavides JA (2005) Hypervariable noncoding sequences in Saccharomyces cerevisiae. Genetics 170:1575–1587 [DOI] [PMC free article] [PubMed]
- Fujita M, Umemura M, Yoko-o T et al (2006) PER1 is required for GPI-phospholipase A2 activity and involved in lipid remodeling of GPI-anchored proteins. Mol Biol Cell 17:5253–5264 [DOI] [PMC free article] [PubMed]
- Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K (2007) Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol Biol Cell 18:295–312 [DOI] [PMC free article] [PubMed]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425:737–741 [DOI] [PubMed]
- Goldstein AL, Pan X, McCusker JH (1999) Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast 15:507–511 [DOI] [PubMed]
- Gordesky SE, Marinetti GV (1973) The asymetric arrangement of phospholipids in the human erythrocyte membrane. Biochem Biophys Res Commun 50:1027–1031 [DOI] [PubMed]
- Grant AM, Hanson PK, Malone L, Nichols JW (2001) NBD-labeled phosphatidylcholine and phosphatidylethanolamine are internalized by transbilayer transport across the yeast plasma membrane. Traffic 2:37–50 [DOI] [PubMed]
- Grossmann G, Opekarova M, Malinsky J, Weig-Meckl I, Tanner W (2007) Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J 26:1–8 [DOI] [PMC free article] [PubMed]
- Hama H, Takemoto JY, DeWald DB (2000) Analysis of phosphoinositides in protein trafficking. Methods 20:465–473 [DOI] [PubMed]
- Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580 [DOI] [PubMed]
- Hanson PK, Malone L, Birchmore JL, Nichols JW (2003) Lem3p is essential for the uptake and potency of alkylphosphocholine drugs, edelfosine and miltefosine. J Biol Chem 278:36041–36050 [DOI] [PubMed]
- Hearn JD, Lester RL, Dickson RC (2003) The uracil transporter Fur4p associates with lipid rafts. J Biol Chem 278:3679–3686 [DOI] [PubMed]
- Heerklotz H (2002) Triton promotes domain formation in lipid raft mixtures. Biophys J 83:2693–2701 [DOI] [PMC free article] [PubMed]
- Heerklotz H, Szadkowska H, Anderson T, Seelig J (2003) The sensitivity of lipid domains to small perturbations demonstrated by the effect of Triton. J Mol Biol 329:793–799 [DOI] [PubMed]
- Hua Z, Fatheddin P, Graham TR (2002) An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol Biol Cell 13:3162–3177 [DOI] [PMC free article] [PubMed]
- Huang J, Zhu H, Haggarty SJ, Spring DR, Hwang H, Jin F, Snyder M, Schreiber SL (2004) Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc Natl Acad Sci USA 101:16594–16599 [DOI] [PMC free article] [PubMed]
- Iwamoto K, Kobayashi S, Fukuda R, Umeda M, Kobayashi T, Ohta A (2004) Local exposure of phosphatidylethanolamine on the yeast plasma membrane is implicated in cell polarity. Genes Cells 9:891–903 [DOI] [PubMed]
- Kato U, Emoto K, Fredriksson C, Nakamura H, Ohta A, Kobayashi T, Murakami-Murofushi K, Umeda M (2002) A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J Biol Chem 277:37855–37862 [DOI] [PubMed]
- Katzmann DJ, Hallstrom TC, Voet M, Wysock W, Golin J, Volckaert G, Moye-Rowley WS (1995) Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol Cell Biol 15:6875–6883 [DOI] [PMC free article] [PubMed]
- Kean LS, Fuller RS, Nichols JW (1993) Retrograde lipid traffic in yeast: identification of two distinct pathways for internalization of fluorescent-labeled phosphatidylcholine from the plasma membrane. J Cell Biol 123:1403–1419 [DOI] [PMC free article] [PubMed]
- Keller S, Tsamaloukas A, Heerklotz H (2005) A quantitative model describing the selective solubilization of membrane domains. J Am Chem Soc 127:11469–11476 [DOI] [PubMed]
- Kenworthy AK (2008) Have we become overly reliant on lipid rafts? Talking Point on the involvement of lipid rafts in T-cell activation. EMBO Rep 9:531–535 [DOI] [PMC free article] [PubMed]
- Kihara A, Igarashi Y (2004) Cross talk between sphingolipids and glycerophospholipids in the establishment of plasma membrane asymmetry. Mol Biol Cell 15:4949–4959 [DOI] [PMC free article] [PubMed]
- Kiyono K, Miura K, Kushima Y, Hikiji T, Fukushima M, Shibuya I, Ohta A (1987) Primary structure and product characterization of the Saccharomyces cerevisiae CHO1 gene that encodes phosphatidylserine synthase. J Biochem (Tokyo) 102:1089–1100 [DOI] [PubMed]
- Klis FM, Boorsma A, De Groot PW (2006) Cell wall construction in Saccharomyces cerevisiae. Yeast 23:185–202 [DOI] [PubMed]
- Kohlwein SD, Kuchler K, Sperka-Gottlieb C, Henry SA, Paltauf F (1988) Identification of mitochondrial and microsomal phosphatidylserine synthase in Saccharomyces cerevisiae as the gene product of the CHO1 structural gene. J Bacteriol 170:3778–3781 [DOI] [PMC free article] [PubMed]
- Kralli A, Bohen SP, Yamamoto KR (1995) LEM1, an ATP-binding-cassette transporter, selectively modulates the biological potency of steroid hormones. Proc Natl Acad Sci USA 92:4701–4705 [DOI] [PMC free article] [PubMed]
- Kren A, Mamnun YM, Bauer BE, Schüller C, Wolfger H, Hatzixanthis K, Mollapour M, Gregori C, Piper P, Kuchler K (2003) War1p, a novel transcription factor controlling weak acid stress response in yeast. Mol Cell Biol 23:1775–1785 [DOI] [PMC free article] [PubMed]
- Kubler E, Riezman H (1993) Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J 12:2855–2862 [DOI] [PMC free article] [PubMed]
- Kuchler K, Sterne RE, Thorner J (1989) Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J 8:3973–3984 [DOI] [PMC free article] [PubMed]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed]
- Lauwers E, Andre B (2006) Association of yeast transporters with detergent-resistant membranes correlates with their cell-surface location. Traffic 7:1045–1059 [DOI] [PubMed]
- Lauwers E, Grossmann G, Andre B (2007) Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: essential role in transport activity and normal control by ubiquitination. Mol Biol Cell 18:3068–3080 [DOI] [PMC free article] [PubMed]
- Lee MC, Hamamoto S, Schekman R (2002) Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. J Biol Chem 277:22395–22401 [DOI] [PubMed]
- Lesage G, Bussey H (2006) Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 70:317–343 [DOI] [PMC free article] [PubMed]
- Letts VA, Klig LS, Bae-Lee M, Carman GM, Henry SA (1983) Isolation of the yeast structural gene for the membrane-associated enzyme phosphatidylserine synthase. Proc Natl Acad Sci USA 80:7279–7283 [DOI] [PMC free article] [PubMed]
- Lichtenberg D, Goni FM, Heerklotz H (2005) Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem Sci 30:430–436 [DOI] [PubMed]
- Liu K, Hua Z, Nepute JA, Graham TR (2007) Yeast P4-ATPases Drs2p and Dnf1p are essential cargos of the NPFXD/Sla1p endocytic pathway. Mol Biol Cell 18:487–500 [DOI] [PMC free article] [PubMed]
- Machi K, Azuma M, Igarashi K, Matsumoto T, Fukuda H, Kondo A, Ooshima H (2004) Rot1p of Saccharomyces cerevisiae is a putative membrane protein required for normal levels of the cell wall 1,6-beta-glucan. Microbiology 150:3163–3173 [DOI] [PubMed]
- Mahé Y, Lemoine Y, Kuchler K (1996a) The ATP binding cassette transporters Pdr5 and Snq2 of Saccharomyces cerevisiae can mediate transport of steroids in vivo. J Biol Chem 271:25167–25172 [DOI] [PubMed]
- Mahé Y, Parle-McDermott A, Nourani A, Delahodde A, Lamprecht A, Kuchler K (1996b) The ATP-binding cassette multidrug transporter Snq2 of Saccharomyces cerevisiae: a novel target for the transcription factors Pdr1 and Pdr3. Mol Microbiol 20:109–117 [DOI] [PubMed]
- Malinska K, Malinsky J, Opekarova M, Tanner W (2003) Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol Biol Cell 14:4427–4436 [DOI] [PMC free article] [PubMed]
- Malinska K, Malinsky J, Opekarova M, Tanner W (2004) Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J Cell Sci 117:6031–6041 [DOI] [PubMed]
- Munro S (2003) Lipid rafts: Elusive or illusive? Cell 115:377–388 [DOI] [PubMed]
- Nagiec MM, Nagiec EE, Baltisberger JA, Wells GB, Lester RL, Dickson RC (1997) Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J Biol Chem 272:9809–9817 [DOI] [PubMed]
- Natarajan P, Wang J, Hua Z, Graham TR (2004) Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc Natl Acad Sci USA 101:10614–10619 [DOI] [PMC free article] [PubMed]
- Noji T, Yamamoto T, Saito K, Fujimura-Kamada K, Kondo S, Tanaka K (2006) Mutational analysis of the Lem3p-Dnf1p putative phospholipid-translocating P-type ATPase reveals novel regulatory roles for Lem3p and a carboxyl-terminal region of Dnf1p independent of the phospholipid-translocating activity of Dnf1p in yeast. Biochem Biophys Res Commun 344:323–331 [DOI] [PubMed]
- Nuoffer C, Jeno P, Conzelmann A, Riezman H (1991) Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol Cell Biol 11:27–37 [DOI] [PMC free article] [PubMed]
- Opekarova M, Malinska K, Novakova L, Tanner W (2005) Differential effect of phosphatidylethanolamine depletion on raft proteins: further evidence for diversity of rafts in Saccharomyces cerevisiae. Biochim Biophys Acta 1711:87–95 [DOI] [PubMed]
- Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, Lorenz TC, Payne GS, Ishihara S, Ohya Y, Andrews B, Hughes TR, Frey BJ, Graham TR, Andersen RJ, Boone C (2006) Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126:611–625 [DOI] [PubMed]
- Piper P, Mahé Y, Thompson S, Pandjaitan R, Holyoak C, Egner R, Muhlbauer M, Coote P, Kuchler K (1998) The Pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J 17:4257–4265 [DOI] [PMC free article] [PubMed]
- Pomorski T, Lombardi R, Riezman H, Devaux PF, van Meer G, Holthuis JC (2003) Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol Biol Cell 14:1240–1254 [DOI] [PMC free article] [PubMed]
- Poruchynsky MS, Ling V (1994) Detection of oligomeric and monomeric forms of P-glycoprotein in multidrug resistant cells. Biochemistry 33:4163–4174 [DOI] [PubMed]
- Pringle JR (1991) Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol 194:732–735 [DOI] [PubMed]
- Riekhof WR, Wu J, Gijon MA, Zarini S, Murphy RC, Voelker DR (2007) Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae: the role of P-type ATPases in transport and a broad specificity acyltransferase in acylation. J Biol Chem 282:36853–36861 [DOI] [PubMed]
- Roberts GG, Hudson AP (2006) Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Mol Genet Genomics 276:170–186 [DOI] [PubMed]
- Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nature Rev Mol Cell Biol 8:519–529 [DOI] [PubMed]
- Roncero C, Valdivieso MH, Ribas JC, Duran A (1988) Isolation and characterization of Saccharomyces cerevisiae mutants resistant to Calcofluor white. J Bacteriol 170:1950–1954 [DOI] [PMC free article] [PubMed]
- Saito K, Fujimura-Kamada K, Furuta N, Kato U, Umeda M, Tanaka K (2004) Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol Biol Cell 15:3418–3432 [DOI] [PMC free article] [PubMed]
- Saito K, Fujimura-Kamada K, Hanamatsu H, Kato U, Umeda M, Kozminski KG, Tanaka K (2007) Transbilayer phospholipid flipping regulates Cdc42p signaling during polarized cell growth via Rga GTPase-activating proteins. Dev Cell 13:743–751 [DOI] [PubMed]
- Santos B, Snyder M (2000) Sbe2p and Sbe22p, two homologous Golgi proteins involved in yeast cell wall formation. Mol Biol Cell 11:435–452 [DOI] [PMC free article] [PubMed]
- Schüller C, Mamnun YM, Mollapour M, Krapf G, Schuster M, Bauer BE, Piper PW, Kuchler K (2004) Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol Biol Cell 15:706–720 [DOI] [PMC free article] [PubMed]
- Schüller C, Mamnun YM, Wolfger H, Rockwell N, Thorner J, Kuchler K (2007) Membrane-active compounds activate the transcription factors Pdr1 and Pdr3 connecting pleiotropic drug resistance and membrane lipid homeostasis in Saccharomyces cerevisiae. Mol Biol Cell 18:4932–4944 [DOI] [PMC free article] [PubMed]
- Servos J, Haase E, Brendel M (1993) Gene SNQ2 of Saccharomyces cerevisiae, which confers resistance to 4-nitroquinoline-N-oxide and other chemicals, encodes a 169 kDa protein homologous to ATP-dependent permeases. Mol Gen Genet 236:214–218 [DOI] [PubMed]
- Sherman F, Fink GR, Hicks JB (1986) Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- Shulewitz MJ, Inouye CJ, Thorner J (1999) Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol 19:7123–7137 [DOI] [PMC free article] [PubMed]
- Siegmund A, Grant A, Angeletti C, Malone L, Nichols JW, Rudolph HK (1998) Loss of Drs2p does not abolish transfer of fluorescence-labeled phospholipids across the plasma membrane of Saccharomyces cerevisiae. J Biol Chem 273:34399–34405 [DOI] [PubMed]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed]
- Simons K, Toomre D (2000) Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 1:31–39 [DOI] [PubMed]
- Stevens HC, Malone L, Nichols JW (2008) The putative aminophospholipid translocases, DNF1 and DNF2, are not required for 7-nitrobenz-2-oxa-1, 3-diazol-4-yl-phosphatidylserine flip across the plasma membrane of Saccharomyces cerevisiae. J Biol Chem 283:35060–35069 [DOI] [PMC free article] [PubMed]
- Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD (2000) Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290:341–344 [DOI] [PubMed]
- Uemura S, Kihara A, Inokuchi J, Igarashi Y (2003) Csg1p and newly identified Csh1p function in mannosylinositol phosphorylceramide synthesis by interacting with Csg2p. J Biol Chem 278:45049–45055 [DOI] [PubMed]
- Utsugi T, Minemura M, Hirata A, Abe M, Watanabe D, Ohya Y (2002) Movement of yeast 1, 3-beta-glucan synthase is essential for uniform cell wall synthesis. Genes Cells 7:1–9 [DOI] [PubMed]
- Valdivia RH, Schekman R (2003) The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc Natl Acad Sci USA 100:10287–10292 [DOI] [PMC free article] [PubMed]
- Valdivia RH, Baggott D, Chuang JS, Schekman RW (2002) The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell 2:283–294 [DOI] [PubMed]
- Wach A, Brachat A, Pohlmann R, Philippsen P (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793–1808 [DOI] [PubMed]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P (1997) Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13:1065–1075 [DOI] [PubMed]
- Walther TC, Brickner JH, Aguilar PS, Bernales S, Pantoja C, Walter P (2006) Eisosomes mark static sites of endocytosis. Nature 439:998–1003 [DOI] [PubMed]
- Wolfger H, Mahé Y, Parle-McDermott A, Delahodde A, Kuchler K (1997) The yeast ATP binding cassette (ABC) protein genes PDR10 and PDR15 are novel targets for the Pdr1 and Pdr3 transcriptional regulators. FEBS Lett 418:269–274 [DOI] [PubMed]
- Wolfger H, Mamnun YM, Kuchler K (2004) The yeast Pdr15p ATP-binding cassette (ABC) protein is a general stress response factor implicated in cellular detoxification. J Biol Chem 279:11593–11599 [DOI] [PubMed]
- Xu J, Peng H, Chen Q, Liu Y, Dong Z, Zhang JT (2007) Oligomerization domain of the multidrug resistance-associated transporter ABCG2 and its dominant inhibitory activity. Cancer Res 67:4373–4381 [DOI] [PubMed]
- Young ME, Karpova TS, Brugger B, Moschenross DM, Wang GK, Schneiter R, Wieland FT, Cooper JA (2002) The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol Cell Biol 22:927–934 [DOI] [PMC free article] [PubMed]