Summary
In a screen for cell-cycle regulators, we identified a Drosophila maternal effect-lethal mutant that we named `no poles' (nopo). Embryos from nopo females undergo mitotic arrest with barrel-shaped, acentrosomal spindles during the rapid S-M cycles of syncytial embryogenesis. We identified CG5140, which encodes a candidate RING domain-containing E3 ubiquitin ligase, as the nopo gene. A conserved residue in the RING domain is altered in our EMS-mutagenized allele of nopo, suggesting that E3 ligase activity is crucial for NOPO function. We show that mutation of a DNA checkpoint kinase, CHK2, suppresses the spindle and developmental defects of nopo-derived embryos, revealing that activation of a DNA checkpoint operational in early embryos contributes significantly to the nopo phenotype. CHK2-mediated mitotic arrest has been previously shown to occur in response to mitotic entry with DNA damage or incompletely replicated DNA. Syncytial embryos lacking NOPO exhibit a shorter interphase during cycle 11, suggesting that they may enter mitosis prior to the completion of DNA replication. We show that Bendless (BEN), an E2 ubiquitin-conjugating enzyme, interacts with NOPO in a yeast two-hybrid assay; furthermore, ben-derived embryos arrest with a nopo-like phenotype during syncytial divisions. These data support our model that an E2-E3 ubiquitination complex consisting of BEN-UEV1A (E2 heterodimer) and NOPO (E3 ligase) is required for the preservation of genomic integrity during early embryogenesis.
Keywords: Drosophila, Embryogenesis, Cell cycle, Mitosis, DNA checkpoint, E3 ubiquitin ligase
INTRODUCTION
To ensure faithful transmission of the genome upon cell division, eukaryotic cells have developed checkpoints, regulatory pathways that delay cell-cycle progression until completion of prior events. The DNA damage/replication checkpoint plays a crucial role in preserving genomic integrity (Branzei and Foiani, 2008). Upon detection of DNA defects, the kinases ATM (ataxia telangiectasia mutated) and ATR (ATM-Rad3-related) are recruited to sites of damage and activated. ATM and ATR substrates include checkpoint kinases CHK1 and CHK2, which phosphorylate proteins that mediate cell-cycle arrest. The ensuing delay, resulting from engagement of this checkpoint, presumably allows cells time to correct defects.
Research over the past decade has highlighted major roles for protein ubiquitination in regulating cellular responses to DNA damage (Harper and Elledge, 2007). This post-translational modification, which involves covalent linkage of one or more ubiquitin molecules to another protein, regulates many fundamental cellular processes (Pickart, 2001). Ubiquitination may alter the fate of a protein in numerous ways, such as targeting it for destruction by the 26S proteasome, changing its subcellular location, or changing its protein-protein interactions.
Ubiquitination is a highly dynamic, multi-step process that requires three components: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2 or Ubc) and ubiquitin ligase (E3). E3s can be divided into two main classes: HECT and RING domain-containing proteins. RING-type E3 ubiquitin ligases (Freemont, 2000; Jackson et al., 2000) contain a specialized motif of 40 to 60 residues that binds two zinc atoms. Many RING-type E3s bind to partnering E2 conjugating enzymes via their RING domains (Passmore and Barford, 2004). Database searches of the Drosophila genome predict that it contains one E1, 36 E2s and ∼130 E3s, which represents ∼40% of the ubiquitination machinery in humans (Ditzel and Meier, 2005).
Significant insights into the roles of many cell-cycle regulators have come from studying their functions in Drosophila. Drosophila is well suited for studying cell-cycle regulation during the formation of a multicellular organism, in large part because of its developmental use of cell cycles that differ in structure from canonical G1-S-G2-M cycles and the availability of genetic tools (Garcia et al., 2007; Lee and Orr-Weaver, 2003). The first thirteen cell cycles of Drosophila embryogenesis involve nearly synchronous nuclear divisions driven by stockpiles of maternally expressed mRNA and protein (Foe et al., 1993). These rapid cycles (∼10 minutes in length) consist of oscillating S-M (DNA replication-mitosis) phases without intervening gap phases or cytokinesis. Minimal gene transcription occurs during this developmental stage, so cell cycles are regulated by post-transcriptional mechanisms. At cycle 14, the embryo cellularizes and initiates zygotic transcription at the midblastula transition (MBT).
We report here the identification and characterization of a Drosophila maternal-effect lethal mutant that we have named `no poles' (nopo). Embryos from nopo females undergo mitotic arrest with acentrosomal, barrel-shaped spindles during syncytial divisions. Our results indicate that this arrest is secondary to the activation of a CHK2-mediated DNA checkpoint in early embryos. We show that NOPO, a predicted E3 ubiquitin ligase, interacts with an E2 component, BEN. ben females are sterile, producing embryos with nopo-like defects. We propose that BEN-UEV1A and NOPO function together as an E2-E3 complex required for genomic integrity during Drosophila embryogenesis.
MATERIALS AND METHODS
Drosophila stocks
Flies were maintained at 25°C using standard techniques. y w was used as wild type unless otherwise indicated. cn Z2-1447 bw/CyO was a gift from Charles Zuker (UC San Diego); ben1 and mnk6006 stocks were from Mark Tanouye (UC Berkeley) and Bill Theurkauf (UMass Worcester), respectively; and the EYG5845 stock was from GenExel (Seoul, Korea). Other fly stocks were from the Bloomington or Szeged stock centers.
Quantification of egg hatch rates
Five newly eclosed females of the indicated genotype and five wild-type males were incubated in yeast-pasted vials for two days and transferred to egg-collection chambers at 25°C. Eggs were collected daily over five days and scored for hatching ∼40 hours post-collection (>500 eggs per genotype). Hatch rate is the ratio of hatched to total eggs expressed as a percentage.
Genetic and molecular mapping of nopo
We screened a second chromosome deficiency collection for non-complementation of female sterility of nopoZ1447. Females carrying nopoZ1447 in trans to any of several overlapping deficiencies (Df(2R)Pcl-11B, Df(2R)Pcl-XM82, Df(2R)Pcl-7B or Df(2R)PC4) were sterile, placing nopo in the 55A1-C1 interval.
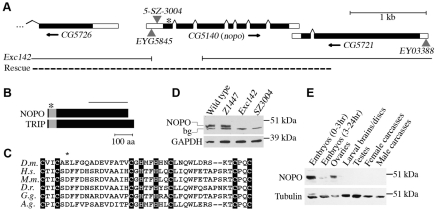
We further mapped nopoZ1447 by P-element-induced male recombination (Chen et al., 1998) relative to several insertions: lolalEP2169, Dgp-1BG00396, CG5721EY03388, fjKG03419 and EP(2)1081. Multiple independent recombinant chromosomes were recovered for each P-element tested. We narrowed nopo to five candidates in the 55B11-12 region (Dgp-1, CG10916, CG5726, CG5140 and CG5721) distal to Dgp-1BG00396 and proximal to CG5721EY03388, as annotated on FlyBase (Grumbling and Strelets, 2006).
For each candidate gene, coding regions were sequenced as described (Rickmyre et al., 2007). nopoZ1447 is a missense mutation in CG5140 causing a glutamic acid to lysine change at residue 11 of the predicted protein. Df(2R)Exel7153, which deletes 15 genes in this region, was subsequently found to uncover nopo. Putative nopo homologs were identified using HomoloGene (release 56), and the RING domain of NOPO was identified using ScanProsite.
Generation of the nopo-null allele
A nopo-null allele was generated by imprecise excision of P-element EYG5845. The 771-bp deletion nopoExc142 lacks part of the 5′-UTR and exons encoding residues 1-181.
cDNA clones
cDNA encoding NOPO, BEN and UEV1A (GH03577, LD24448 and LD28904, respectively) were from the Drosophila Gene Collection. Human TRIP cDNA (ID 2821007) was from Open Biosystems.
Transgenesis
A 3.8-kb genomic fragment containing CG5140 and flanking regions (Fig. 2A) was PCR-amplified from BAC clone BACR15G20 (Drosophila Genomics Resource Center) and subcloned into pCaSpeR4. A transgenic line carrying pCaSpeR4-CG5140 was generated by P-element-mediated transformation via embryo injection (Rubin and Spradling, 1982).
Fig. 2.
CG5140 is the nopo gene. (A) nopo structure. Coding regions are represented by black boxes, 5′- and 3′-UTRs by white boxes, splicing events by lines. Arrows indicate transcription direction. Asterisk marks position of E11K mutation in nopoZ1447. Triangles represent P-elements. EYG5845 imprecise excision generated nopoExc142 (gap represents deleted region). Dashed line represents genomic region used to create rescue construct. (B) Comparison of Drosophila NOPO and human TRIP. Gray boxes represent RING domains. Asterisk marks mutation in nopoZ1447. Line indicates NOPO region used for antibody production. (C) Alignment of RING domains of putative NOPO/TRIP homologs in Drosophila melanogaster, Homo sapiens, Mus musculus, Danio rerio, Gallus gallus and Anopheles gambiae. Residues 6-46 of Drosophila NOPO are shown. Crucial RING-domain cysteines and histidines are highlighted. Asterisk marks residue mutated in nopoZ1447. (D,E) NOPO immunoblots. (D) NOPO levels in embryos (1-2 hours) of wild-type or nopo females. Anti-NOPO antibodies recognize a specific band the predicted size of NOPO (48 kDa) and a non-specific band (bg). (E) NOPO developmental western. Anti-GAPDH or anti-α-tubulin was used as a loading control.
Embryo immunostaining and microscopy
Methods for fixation, staining and fluorescence microscopy of embryos (1.5-2.5 hours unless otherwise indicated) and live-image analysis were previously described (Rickmyre et al., 2007). P-values for live-image data were obtained using a two-tailed, unpaired Student's t-test.
NOPO polyclonal antibodies
A fusion consisting of an N-terminal MBP tag and C-terminal NOPO was used to generate anti-NOPO antibodies. DNA encoding C-terminal NOPO (residues 224 to 435) was PCR amplified and subcloned into pMAL (New England Biolabs). MBP-C-NOPO was produced in bacteria, purified using amylose resin, and injected into guinea pigs (Covance).
Protein extracts and immunoblots
Protein extracts were made by homogenizing embryos (1-2 hours) or dissected tissues in urea sample buffer (Tang et al., 1998). Proteins were transferred to nitrocellulose for immunoblotting using standard techniques. Antibodies used were as follows: guinea pig anti-NOPO (1:1000), mouse anti-GAPDH (1:1000, Abcam), mouse anti-α-tubulin (DM1a, 1:5000, Sigma), mouse anti-Cyclin B (F2F4, 1:200, Developmental Studies Hybridoma Bank), and rabbit anti-pY15-CDK1 (1:1000, Upstate).
Mammalian cell transfection, staining and microscopy
HeLa cells were maintained in Dulbecco's modified Eagle Medium (DMEM) containing 10% fetal bovine serum. Plasmids encoding N-terminally tagged (eGFP or mCherry) versions of NOPO, TRIP and BEN generated by subcloning into pCS2 were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's directions.
Cells were plated on fibronectin-coated coverslips 21 hours post-transfection and fixed three hours later. For direct fluorescence and centromere staining, cells were fixed for 20 minutes with 4% formaldehyde in CBS [10 mM MES (pH 6.1), 138 mM KCl, 3 mM MgCl2, 2 mM EGTA, 0.32 M sucrose]. For PCNA staining, cells were fixed for 5 minutes in 70% methanol/30% acetone. For Cyclin A staining, cells were fixed for 20 minutes in 3% paraformaldehyde/20% sucrose in phosphate-buffered saline. Cells were permeabilized for 10 minutes with 0.5% Triton X-100 in Tris-buffered saline. Primary antibodies used were as follows: human autoimmune (CREST) serum (1:1000, ImmunoVision), Cyclin A (H-432, 1:100, Santa Cruz Biotechnology), and PCNA (PC10, 1:200 Santa Cruz Biotechnology). To visualize actin, cells were stained for one hour with fluorescently conjugated phalloidin (1:1000, Invitrogen). Fluorescently conjugated secondary antibodies were used at a dilution of 1:5000. Slides were mounted in Vectashield with DAPI (Vector Laboratories). Images were acquired using a Nikon Eclipse 80i microscope equipped with a CoolSNAP ES camera (Photometrics) and Plan-Apo 60× objective. For experiments involving quantification, at least 400 cells per condition were scored.
Yeast two-hybrid assays
Yeast two-hybrid assays were performed as described (James et al., 1996). Plasmids expressing wild-type and mutant versions of NOPO, BEN and UEV1A fused to the Gal4 DNA-binding domain (`bait' vector pGBD-C) or Gal4-activation domain (`prey' vector pGAD-C) were transformed into Saccharomyces cerevisiae strain PJ69-4A. Cells containing both bait and prey plasmids were selected by growth on synthetic complete (SC) plates lacking tryptophan and leucine, and spotted onto SC plates lacking tryptophan, leucine and histidine; growth on the latter (scored after two days at 30°C) indicates physical interaction between the fusion proteins tested.
DNA damage-response assays
The sensitivity of nopo larvae to hydroxyurea or irradiation was tested as described (Rickmyre et al., 2007).
Behavioral assays and TDT morphology
To assess the visually mediated jump response, white-eyed control (w1118) and mutant flies (two days old) were dark adapted, transferred without anesthesia to a Petri dish covered in vellum, and exposed to a `lights off' stimulus using an LED light apparatus as described (Fayyazuddin et al., 2006). Ten males per genotype were each tested in 10 trials separated by 30 seconds. The climbing ability of adult males was assessed as described (Silva et al., 2004), with three replicates per genotype. P-values were obtained using two-tailed, unpaired Student's t-tests. To visualize TDT muscle attachment sites, adult males (30 per genotype) were ventrally trans-illuminated with a dissecting microscope lamp as described (Edgecomb et al., 1993).
Innate immunity assay
Adult males (5- to 7-days old) were injected using a Drummond Nanoject with ∼50 nl of an overnight culture of Escherichia coli resuspended in phosphate-buffered saline. Six hours later, RNA was isolated by homogenizing flies in STAT-60 buffer according to the manufacturer's directions (Isotex Diagnostics). Following DNase treatment, cDNA was prepared by reverse transcription using Superscript II (Invitrogen). A diptericin-specific LUX primer (Invitrogen) was used to perform quantitative real-time PCR with the 7300 Real-Time PCR System (Applied Biosystems). diptericin levels were normalized to Rp49 levels as an endogenous control. Results from three independent experiments were averaged and further normalized against buffer-injected Canton S flies. P-values were obtained using a two-tailed, unpaired Student's t-test.
RESULTS
The nopo phenotype in the early embryo
We previously screened the maternal-effect lethal subset of the Zuker collection to identify genes that regulate S-M cycles of Drosophila early embryogenesis (Koundakjian et al., 2004; Lee et al., 2003; Rickmyre et al., 2007). We identified an allele (Z1447) of a gene that we have named `no poles' (nopo), based on the phenotype of acentrosomal mitotic spindles in mutant-derived embryos (see below). nopoZ1447 females are completely sterile (Table 1). DNA staining of the egg chambers of nopoZ1447 females revealed no obvious oogenesis defects, and the presence of polar bodies in their unfertilized eggs indicated that meiosis was completed (data not shown).
Table 1.
nopo allelic series and transgenic rescue
| Temperature (°C) | Genotype | Cortical (%)* | Gastrulation (%)† | Hatch rate (%) |
|---|---|---|---|---|
| 25 | Wild type | 92 | 91 | 89 |
| nopoSZ3004 | 62 | 65 | 12 | |
| nopoSZ3004/Df(2R)Exel7153 | 67 | 12 | 0 | |
| nopoZ1447 | 72 | 0 | 0 | |
| nopoZ1447/Df(2R)Exel7153 | 68 | 0 | 0 | |
| pCaSpeR4-CG5140/+; nopoZ1447 | 95 | 92 | 87 | |
| nopoExc142 | 15 | 0 | 0 | |
| nopoExc142/Df(2R)Exel7153 | 7 | 0 | 0 | |
| pCaSpeR4-CG5140/+; nopoExc142 | 93 | 90 | 83 | |
| 18 | Wild type | 97 | 97 | 89 |
| nopoSZ3004 | 78 | 86 | 76 | |
| nopoZ1447 | 67 | 1 | 0 |
Embryos collected from females of the indicated genotypes were fixed for DNA and tubulin staining (see Materials and methods for details).
Percentage of embryos that develop to cycle 10 or beyond. For each genotype, at least 200 embryos (2-2.5 hours) were scored.
Percentage of embryos that develop to the initiation of gastrulation or beyond. For each genotype, at least 200 embryos (3-4 hours) were scored.
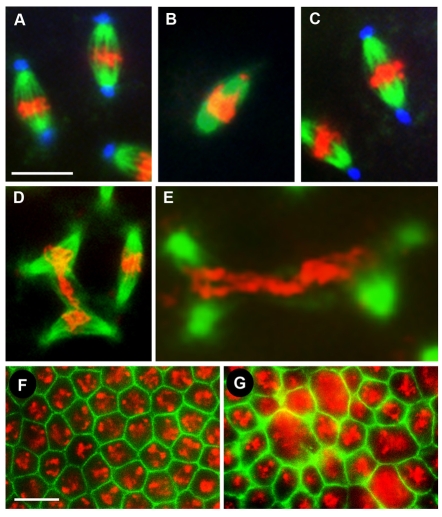
We found that nopoZ1447-derived embryos undergo mitotic arrest during syncytial divisions with none developing to cellularization or gastrulation (Tables 1, 2; data not shown). Nuclei were unevenly spaced (compare Fig. 1B with 1A), and centrosome duplication before telophase was occasionally evident (Fig. 1F), both of which are consistent with failed mitotic divisions. nopo spindles are barrel-shaped, lack tubulin foci, and have misaligned chromosomes (Fig. 1D-F; Table 2); the lack of tubulin foci correlates with the loss of centrosomes at the poles, as revealed by staining for Centrosomin, a core component (Fig. 1I,J) (Li and Kaufman, 1996). Approximately 10% of nopo spindles are tripolar (Fig. 1K; Table 2). Bipolar nopo spindles often appear to be wider and to contain more than the wild-type complement of chromosomes (compare Fig. 1E with 1C). Similar results were obtained for nopoZ1447 in trans to Df(2R)Exel7153, which deletes nopo (Table 1).
Table 2.
The nopo phenotype is suppressed by mnk
|
Mitotic spindle defects (%
spindles)†
|
|||||||
|---|---|---|---|---|---|---|---|
|
Bipolar spindles
|
|||||||
|
Abnormal centrosome number
|
Tripolar spindles | Gastrulation (% embryos)¶ | Hatch rate (%) | ||||
| Genotype | MI* | Decreased‡ | Increased§ | Barrel-shaped | |||
| Wild type | 0.63 | <1 | 0 | <1 | <1 | 100 | 89 |
| nopoZ1447 | 0.95 | 62 | 13 | 64 | 10 | 0 | 0 |
| mnk6006 | 0.64 | 1 | <1 | 1 | <1 | 99 | 80 |
| mnk6006 nopoZ1447 | 0.68 | <1 | <1 | <1 | <1 | 77 | 0 |
Embryos collected from females of the indicated genotypes were used to determine hatch rates or were fixed for DNA and tubulin staining (see Materials and methods for details).
Mitotic index (MI)=% embryos in mitosis/total number of embryos. More than 300 embryos were scored per genotype. Chromosome condensation and the presence of a mitotic spindle were used as the criteria for mitosis.
All mitotic spindles (>500 total) in a single focal plane were scored using at least 25 embryos per genotype.
Spindles with centrosomal detachment at one or both poles.
Spindles with >1 centrosome per pole (one or both poles). Telophase spindles were not scored because centrosome duplication occurs during this phase in the early embryo.
Stained embryos (3-4 hours) were scored for development to the initiation of gastrulation (or beyond). More than 200 embryos were scored per genotype.
Fig. 1.
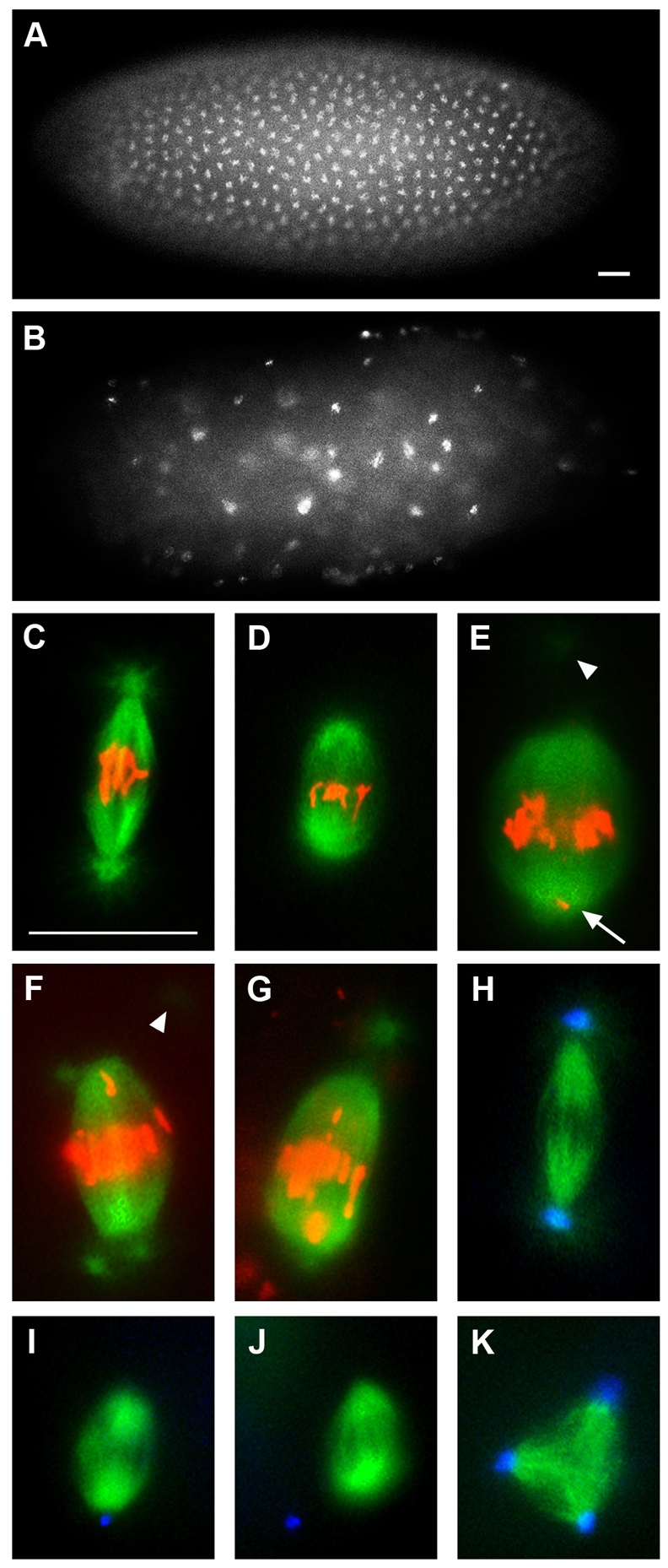
The nopo phenotype. Representative syncytial embryos and mitotic spindles in embryos from wild-type or nopoZ1447 females. (A,B) Staining of nopo-derived embryos reveals developmental arrest with condensed, unevenly spaced DNA (B) compared with wild type (A). (C-G) Microtubules are in green and DNA in red. (C) Wild-type spindle. (D-F) Shortened, barrel-shaped nopo spindles with detached centrosomes and misaligned chromosomes. Arrowheads indicate detached centrosomes out of focal plane; arrow, DNA at pole. Metaphase-like spindle with two centrosomes per pole (F) reveals an asynchrony of nuclear and centrosome cycles. (G) Similar defects are observed in an nopoExc142/Df(2R)Exel7153-derived embryo. (H-K) Microtubules are in green; centrosomes in blue. (H) Wild-type spindle. (I,J) nopo spindles with detached and/or missing centrosomes. (K) Tripolar nopo spindle. Scale bars: 20 μm.
nopo encodes a RING domain-containing protein
We identified CG5140 as the nopo gene, using a combination of genetic mapping and molecular biology approaches (Fig. 2A; see Materials and methods for details). A wild-type copy of CG5140 carried as a transgene fully restored fertility to nopo females (Table 1), confirming that CG5140 is, indeed, the nopo gene. nopo encodes a predicted protein of 435 amino acids containing an N-terminal RING domain (Fig. 2B) (Saurin et al., 1996). The putative mammalian homolog of NOPO was named `TRAF-interacting protein' (TRIP) based on its ability to bind tumor necrosis factor (TNF) receptor-associated factors (TRAFs) (Lee et al., 1997). Mammalian TRIP was recently demonstrated to have RING-dependent E3 ubiquitin ligase activity in an auto-ubiquitination assay (Besse et al., 2007). Drosophila NOPO and human TRIP are 20% identical and 34% similar overall, with 47% identity and 65% similarity in their RING domains. Importantly, nopoZ1447 causes a glutamic acid to lysine change in the RING domain at position 11 of the predicted protein, a residue that is invariantly negatively charged across species (Fig. 2C).
nopo is maternally provided and essential solely in the early embryo
To gain further insights into the functions of nopo, we obtained additional alleles. 5-SZ-3004 (abbreviated as SZ3004) and EYG5845 are P-element insertions in the 5′-UTR of nopo (Fig. 2A). nopoSZ3004 females have decreased embryonic hatch rates that are completely restored by precise P-element excision (Table 1; data not shown). nopoSZ3004 is weaker than nopoZ1447, based on the percentage of mutant-derived embryos that develop to gastrulation and embryonic hatching, and its phenotype is strongly temperature dependent. We generated a null allele of nopo (Exc142) via imprecise excision of EYG5845 (Fig. 2A; see Materials and methods). nopoExc142 adults are viable and appear normal, except that females are sterile, producing embryos with the nopo phenotype (Fig. 1G); a CG5140 transgene fully restored fertility (Table 1). nopoExc142 is stronger than nopoZ1447 with 15% versus 72%, respectively, of their embryos reaching cortical divisions, suggesting that nopoZ1447 has residual function.
We assessed NOPO levels in mutant embryonic extracts by immunoblotting using anti-NOPO antibodies that we generated (Fig. 2D). Consistent with the predicted size of NOPO (435 residues), these antibodies recognize an ∼48 kDa band in wild-type embryos that is absent in nopoExc142-derived (null) embryos. NOPO was not detected in nopoSZ3004-derived embryos, although we occasionally observed trace amounts (data not shown). Wild-type levels of NOPO were found in nopoZ1447-derived embryos, suggesting that the E11K mutation alters the function of NOPO, but not its stability.
We assessed NOPO levels throughout Drosophila development (Fig. 2E). NOPO is abundant in ovaries and early (0- to 3-hour) embryos; trace amounts are present in older (3- to 24-hour) embryos. We did not detect NOPO in larval brains, imaginal discs, testes or adult carcasses lacking germline tissues. Subsequent experiments, however, revealed roles for NOPO outside of early embryonic development, suggesting that our antibodies might not be sufficiently sensitive to detect its expression during other stages (see below). Using the UAS/Gal4 system, we found NOPO overexpression in the female germline causes severely reduced egg-laying and hatch rates, whereas broad overexpression of NOPO in somatic cells causes lethality, which suggests that NOPO levels must be tightly regulated (data not shown).
The nopo phenotype is suppressed by mutation of the checkpoint kinase MNK (CHK2)
In Drosophila syncytial embryos, mitotic entry with incompletely replicated or damaged DNA triggers a CHK2-mediated protective mechanism known as centrosomal inactivation (Sibon et al., 2000; Takada et al., 2003). This damage-control system senses DNA defects and elicits localized changes in spindle structure that block mitotic progression, presumably to prevent the propagation of defective DNA. We previously reported that embryos from microcephalin (mcph1) females arrest in mitosis with acentrosomal, barrel-shaped spindles similar to those that we now observe in nopo-derived embryos (Rickmyre et al., 2007). We demonstrated that these mcph1 defects were suppressed by mutation of maternal nuclear kinase (mnk), also known as loki, which encodes Drosophila CHK2 (Abdu et al., 2002; Brodsky et al., 2004; Masrouha et al., 2003; Xu et al., 2001). mnk nulls exhibit increased sensitivity to ionizing radiation, but are viable and fertile. Suppression of mcph1 by mnk revealed that centrosomal inactivation significantly contributes to the mcph1 phenotype.
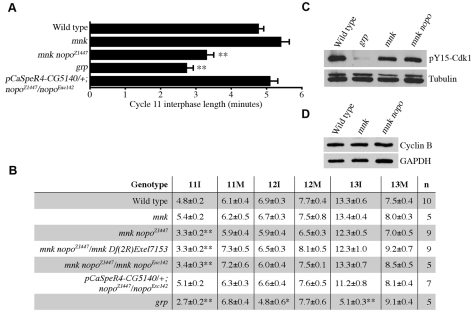
To determine whether the mitotic defects of nopo-derived embryos are, like those of mcph1, due to CHK2-mediated centrosomal inactivation, we created lines doubly mutant for nopo and mnk. The nopo phenotype of acentrosomal, barrel-shaped mitotic spindles was strongly suppressed by mnk, as evidenced by the restoration of normal spindles with attached centrosomes (Fig. 3A-C; Table 2). DNA defects, however, were common in embryos from mnk nopo females, particularly during cortical divisions. We frequently observed abnormal DNA aggregates, some of very large size, shared by more than one spindle (Fig. 3D,E).
Fig. 3.
Suppression of nopo by mnk. (A-E) Representative mitotic spindles in syncytial embryos from wild-type (A), nopoZ1447 (B) and mnk nopoZ1447 females (C-E). (A-C) Microtubules are in green, DNA in red, and centrosomes in blue. nopo (B) is suppressed by mnk, as evidenced by the restoration of elongated spindles with attached centrosomes (C). (D,E) Microtubules are in green and DNA in red. Aberrant mitotic figures with DNA shared by two spindles are observed in mnk nopo-derived embryos. (F,G) Cellularized embryos (2-3 hours). Actin is in green and DNA in red. Developmental arrest of nopo is suppressed by mnk. Cellularized mnk nopoZ1447-derived embryos show large DNA masses (G) compared to wild type (F). Scale bars: 10 μm in A-E; 20 μm in F,G.
Like mcph1, we found that the developmental arrest of nopo mutants is suppressed by mnk (Fig. 3F,G; Table 2). In contrast to nopo-derived embryos, which arrest in syncytial divisions, most embryos from mnk nopo females complete syncytial divisions, cellularize, and arrest with aberrant morphology upon the initiation of gastrulation. Cellularized embryos from mnk nopo females contain unusually large DNA masses within irregularly sized cells compared with wild type. Thus, mnk suppresses the spindle/centrosomal defects and the developmental arrest of nopo mutants, but DNA defects appear to accumulate.
Based on these findings, we propose that nopo is required for the preservation of genomic integrity during syncytial embryogenesis. Lack of nopo activity leads to the occurrence of DNA defects, which then trigger CHK2-mediated centrosomal inactivation, thereby causing widespread mitotic arrest and the blockade of embryonic development. Mutation of mnk (Chk2) allows further nuclear divisions and developmental progression in nopo-derived embryos, despite the accumulation of extensive DNA defects that eventually lead to their arrest at the onset of gastrulation.
nopo-derived embryos exhibit decreased interphase length
The DNA-replication checkpoint mediated by MEI-41 and Grapes, orthologs of ATR and CHK1, respectively, is developmentally activated in late syncytial embryos of Drosophila (Sibon et al., 1999; Sibon et al., 1997). Checkpoint activation, which may be triggered by titration of a maternal replication factor, leads to inhibitory phosphorylation of CDK1 and a gradual slowing of mitotic entry, presumably to allow sufficient time to complete DNA replication. At MBT (cycle 14), the first G2 gap phase is introduced. Embryos from mei-41 or grapes (grp) mutant females fail to lengthen the interphases of late syncytial cycles and are thought to enter mitosis without completing DNA replication; a secondary damage-control system, CHK2-mediated centrosomal inactivation, then becomes operational (Sibon et al., 2000; Takada et al., 2003).
Mitotic entry with incompletely replicated DNA can cause CHK2-mediated centrosomal inactivation in syncytial embryos (Sibon et al., 2000; Takada et al., 2003). Control mechanisms to ensure completion of DNA replication prior to mitosis may be particularly critical during rapid S-M cycles. Oscillating CDK1-Cyclin B activity plays a key role in coordinating these cycles (Edgar et al., 1994; Su et al., 1998). S-M transitions appear to be controlled by Cyclin B levels prior to cycle 10, and by both Cyclin B levels and a DNA-replication checkpoint in cycles 10-13 (Ji et al., 2004; Sibon et al., 1997).
Using a previously described approach, we monitored the timing of nuclear envelope breakdown and reformation in cycles 11-13 by differential interference contrast (DIC) microscopy to test whether nopo mutants, like mei-41 and grp mutants, fail to lengthen interphase (see Fig. S1 in the supplementary material) (Rickmyre et al., 2007; Takada et al., 2007). Live imaging of nopo-derived embryos during cortical divisions was not feasible because the majority arrest either before or during these cycles (Table 1; data not shown); yolk proteins obscure the interior nuclei of precortical embryos, making imaging of these earlier divisions technically difficult. We analyzed the timing of cortical cell cycles in embryos from mnk nopo females (lacking a CHK2-mediated checkpoint) because they develop further than nopo-derived embryos; we reasoned that primary defects in cell-cycle kinetics due to nopo mutation would still be apparent. In support of this line of reasoning, embryos from mnk grp females have been shown to retain the cell-cycle timing defects of grp-derived embryos (Takada et al., 2007). Embryos from mnk nopoZ1447 females exhibited significantly shorter interphases during cycle 11 (mean of 3.3 minutes) compared to wild type with mnk (4.8 or 5.4 minutes, respectively; Fig. 4A,B). Essentially identical results were obtained for mnk nopoZ1447/Df(2R)Exel7153 and mnk nopoZ1447/nopoExc142 females. Importantly, interphase 11 length was restored by transgenic rescue. A shortened interphase 11 (2.7 minutes) was observed in grp-derived embryos, as expected. Unlike grp, however, mnk nopo-derived embryos exhibited normal cycle 12 and 13 interphase lengths.
Fig. 4.
Shortened cycle 11 interphase of mnk nopo-derived embryos. (A,B) Cell-cycle timing during cortical embryonic divisions. Bar graph (A) shows mean cycle 11 interphase lengths for various genotypes. Table (B) summarizes mean cycle 11-13 interphase (I) and mitosis (M) lengths. n, number of embryos. Error bars (A) and ±values (B) represent s.e.m. Single and double asterisks mark interphases significantly shorter than wild type (P<0.01 and <0.001, respectively). (C,D) Immunoblotting reveals normal pY15-CDK1 (C) and Cyclin B (D) levels in mnk nopoZ1447-derived embryos (1-2 hours). Control grp-derived embryos have reduced pY15-CDK1 levels. Anti-α-tubulin or anti-GAPDH was used as a loading control.
Based on our observations of shorter cycle 11 interphases in mnk nopo-derived embryos, we infer that interphases of earlier (precortical) syncytial cycles may be relatively short in nopo-derived embryos. We hypothesize that DNA replication is not completed during these truncated interphases, resulting in mitotic entry with unreplicated DNA, the triggering of CHK2-mediated centrosomal inactivation, and mitotic arrest with failure of further embryonic development.
Most embryos from nopo-null females arrest prior to the onset at cycle 11 of a detectable DNA-replication checkpoint effect (Table 1) (Crest et al., 2007). Thus, we reasoned that nopo is unlikely to regulate interphase length via mei-41/grp. Nonetheless, we tested the intactness of the MEI-41/GRP-mediated DNA-replication checkpoint by assessing levels of CDK1 inhibitory phosphorylation in mnk nopo-derived embryos and found them to be comparable to wild type (Fig. 4C). We also observed an intact DNA damage response by nopo larvae treated with hydroxyurea, a DNA replication inhibitor, or irradiation (see Table S1 in the supplementary material). We observed no genetic interactions between nopo and mei-41 (data not shown). We detected wild-type levels of Cyclin B and Cyclin A in mnk nopo-derived embryos and observed no genetic interactions between nopo and cyclin B (Fig. 4D; data not shown). These results suggest that nopo regulates the S-M transition independent of the MEI-41/GRP-dependent checkpoint and mitotic cyclin levels.
Drosophila NOPO and human TRIP co-localize to nuclear puncta in cultured mammalian cells
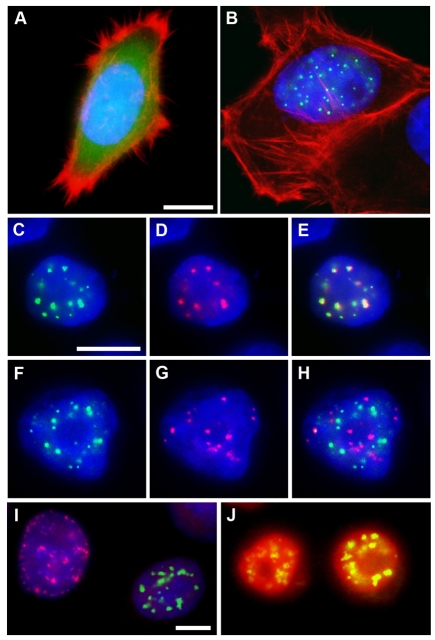
To determine the subcellular localization of NOPO, we used transfected mammalian cells because our anti-NOPO antibodies did not work for immunofluorescence, and epitope-tagged forms of NOPO expressed via transgenesis were not stable in Drosophila embryos (data not shown). We transfected HeLa cells with fluorescently tagged versions of Drosophila NOPO and human TRIP (candidate homolog of NOPO) and assessed their localization by immunofluorescence microscopy. Whereas eGFP (control) was homogeneously distributed, eGFP-NOPO localized to nuclear puncta in the majority of interphase cells (compare Fig. 5A with 5B); a similar pattern was observed for Myc-tagged NOPO (data not shown). mCherry-TRIP also exhibited a punctate distribution in nuclei (Fig. 5D). Co-expression of eGFP-NOPO and mCherry-TRIP in HeLa cells confirmed their essentially identical localization patterns, underscoring the likelihood that NOPO and TRIP are functional homologs (Fig. 5C-E).
Fig. 5.
Nuclear localization of NOPO. Immunofluorescence microscopy of transfected HeLa cells. DNA is in blue. (A,B) eGFP is in green and actin in red. eGFP-Drosophila NOPO (B) localizes to nuclear puncta; eGFP (A) is homogeneously distributed. (C-E) eGFP is in green and mCherry in red. eGFP-Drosophila NOPO (C) and mCherry-human TRIP (D) co-localize in nuclear puncta (E, merge). (F-H) eGFP is in green and CREST in red. eGFP-NOPO (F) is not at the centromeres (G; H, merge). (I) Cells with eGFP-NOPO puncta (green) are negative for PCNA puncta (red). (J) Cells with eGFP-NOPO puncta (green) are positive for nuclear Cyclin A (red). Scale bars: 10 μm.
CREST staining of HeLa cells expressing eGFP-NOPO revealed that NOPO/TRIP localizes to nuclear regions distinct from the centromeres. To assess whether eGFP-NOPO localizes to nuclear puncta in a cell cycle-dependent manner, we immunostained transfected HeLa cells for PCNA or Cyclin A. We found that >99% of cells positive for eGFP-NOPO puncta were negative for insoluble PCNA foci in the nucleus, a marker of S-phase (Somanathan et al., 2001). By contrast, ∼97% of cells positive for eGFP-NOPO puncta were positive for nuclear Cyclin A, a marker of both S and G2 phases (Girard et al., 1991). Taken together, these data indicate that eGFP-NOPO specifically localizes to nuclear puncta in transfected HeLa cells during G2 phase.
NOPO associates with BEN, an E2 heterodimer component
The presence of a RING domain in NOPO suggested that it might function as an E3 ubiquitin ligase (Lorick et al., 1999). In a high-throughput yeast two-hybrid screen (Giot et al., 2003), NOPO interacted with an E2 ubiquitin-conjugating enzyme, Bendless (BEN), the Drosophila homolog of Ubc13 (Muralidhar and Thomas, 1993; Oh et al., 1994; Zhou et al., 2005).
To confirm and extend these observations, we tested for interactions between combinations of wild-type and mutant NOPO and BEN proteins in a yeast two-hybrid assay (Fig. 6A). We used mutant NOPO and BEN forms encoded by nopoZ1447 (E11K in the RING domain; Fig. 2A-C) and ben1 (proline to serine change at position 97) (Muralidhar and Thomas, 1993). We found that both wild-type and mutant NOPO self-interact in this assay. When used as bait, wild-type BEN strongly interacted with wild-type NOPO; we occasionally observed weak interaction in the reverse direction (data not shown). By contrast, wild-type BEN and mutant NOPO do not interact, and mutant BEN interacts only marginally with wild-type or mutant NOPO.
Fig. 6.
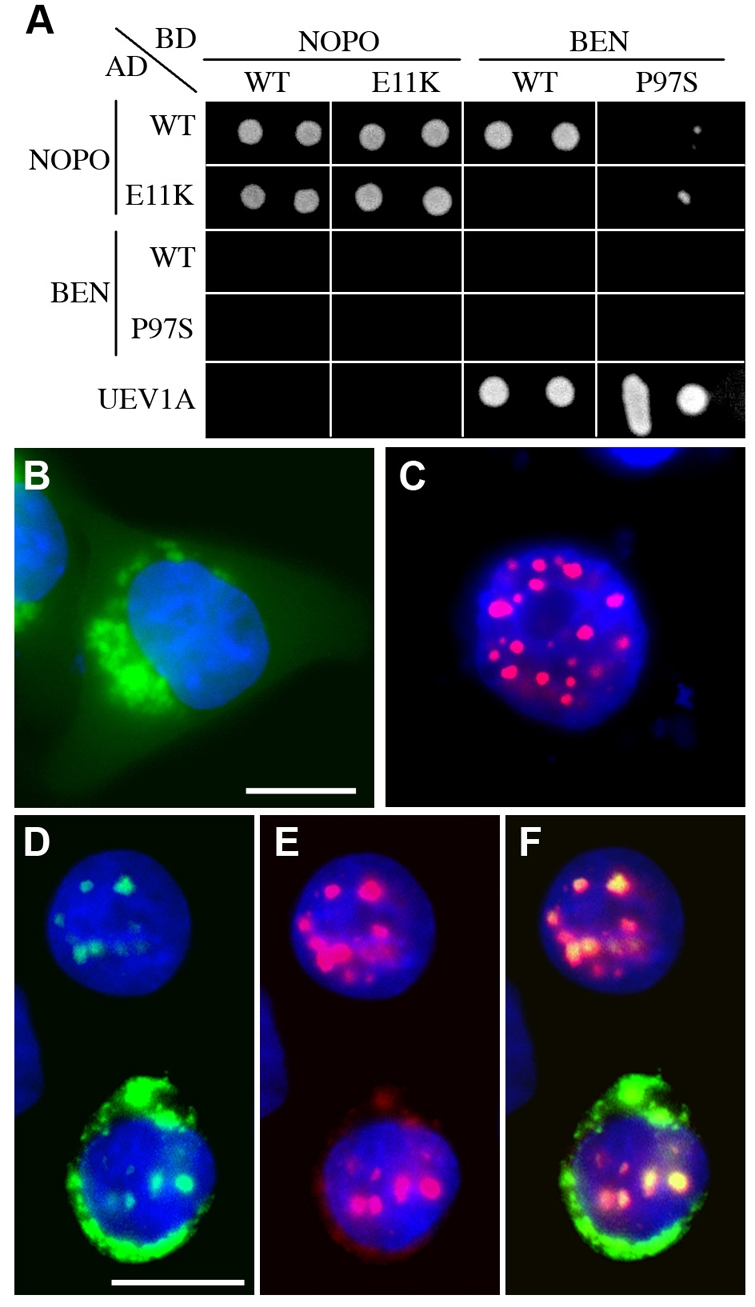
NOPO/TRIP, BEN and UEV1A interactions and co-localization. (A) Yeast two-hybrid assay. Yeast cells expressing combinations of NOPO, BEN and UEV1A fused to the Gal4 DNA-binding domain (BD, `bait') or activation domain (AD, `prey') were spotted onto selective media. Growth on SC-Trp-Leu-His media (shown) indicates physical interaction between the fusion proteins. Wild-type and mutant versions of NOPO and BEN (E11K and P97S, respectively) were tested. A representative plate spotted in duplicate is shown; identical results were obtained for three independent Trp+Leu+ colonies per plasmid combination tested. (B-F) Immunofluorescence microscopy of transfected HeLa cells. eGFP-BEN is in green, mCherry-TRIP in red, and DNA in blue. eGFP-BEN (B) and mCherry-TRIP (C) localize distinctly when transfected alone. (D-F) Co-transfection of eGFP-BEN (D) with mCherry-TRIP (E) promotes its localization into nuclear puncta (F, merge). Scale bars: 20 μm.
We detected comparable levels of mutant and wild-type fusion proteins (both NOPO and BEN) in transformed yeast. Furthermore, nopoZ1447-derived embryos have wild-type NOPO levels, and ben1 ovaries have wild-type BEN levels (Fig. 2D; data not shown). Thus, the lack of two-hybrid interactions observed for mutant NOPO and BEN is likely to reflect changes in protein-protein interactions rather than decreased stability.
To obtain further evidence that NOPO and BEN interact, we compared the localization patterns of fluorescently tagged versions of these proteins in transfected HeLa cells. Both eGFP-NOPO and mCherry-TRIP accumulate in nuclear puncta (Fig. 5E, Fig. 6C). When transfected alone, eGFP-BEN distributes throughout cells with a perinuclear concentration but no obvious nuclear puncta (Fig. 6B). When eGFP-BEN and mCherry-TRIP were co-transfected, however, eGFP-BEN localized to nuclear puncta in the majority (56%) of cells positive for mCherry-TRIP nuclear puncta; furthermore, all puncta positive for eGFP-BEN were positive for mCherry-TRIP (Fig. 6D-F). These data suggest that NOPO/TRIP can recruit BEN/Ubc13 to chromatin, and provide further evidence for in vivo interactions between these proteins.
The E2 activity of Ubc13 has been shown in other systems to require heterodimerization with a UEV (ubiquitin-conjugating E2 enzyme variant) family member (Pickart, 2001). UEV proteins (Mms2p in budding yeast; Uev1A and Mms2 in mammals) resemble E2s but lack an active site cysteine (Broomfield et al., 1998; Sancho et al., 1998). Our BLAST searches revealed a single UEV homolog in Drosophila encoded by Uev1A. In our two-hybrid assay, UEV1A interacted strongly with wild-type and mutant BEN, but not with NOPO (Fig. 6A). Our attempts to detect BEN-NOPO complexes in Drosophila embryos were unsuccessful, however, possibly due to transience of this interaction (data not shown). Our yeast two-hybrid data suggest that the RING domain of NOPO interacts directly with BEN to promote the formation of a UEV1A-BEN-NOPO (E2-E3) complex.
ben-derived embryos have nopo-like defects
ben was identified in a screen for Drosophila mutants with neuronal connectivity defects (Thomas and Wyman, 1982). Its yeast two-hybrid interaction with NOPO suggested that BEN might regulate embryonic development. We found that embryos from ben1 homozygotes or hemizygotes fail to develop, revealing a new function for ben (Table 3).
Table 3.
Quantification of defects in ben-derived embryos
|
Number of nuclei/embryo (% embryos)
|
nopo-like spindles (% embryos)‡ | |||||
|---|---|---|---|---|---|---|
| Genotype | MI* | Hatch rate (%) | 1 | 2-8 | >8 | |
| Wild type | 0.63 | 89 | − | − | − | <1 |
| ben1 | 0.92 | 2† | 72 | 12 | 16 | 80 |
| ben1/Df(1)KA10 | 0.80 | 0 | 40 | 43 | 17 | 50 |
Embryos collected from females of the indicated genotypes were used to determine hatch rates or were fixed for DNA and tubulin staining (see Materials and methods for details). At least 50 stained embryos were analyzed per genotype. Eggs/embryos in which DNA and tubulin were not visualized by staining were excluded from analysis.
Mitotic index (MI) = % embryos in mitosis/total number of embryos. Chromosome condensation and the presence of a mitotic spindle were used as the criteria for mitosis.
For ben1 homozygotes, only 100 embryos were scored for hatching owing to low egg-laying rates.
Embryos in which the majority of mitotic spindles were barrel-shaped and/or lacking centrosomes were scored as being `nopo-like'.
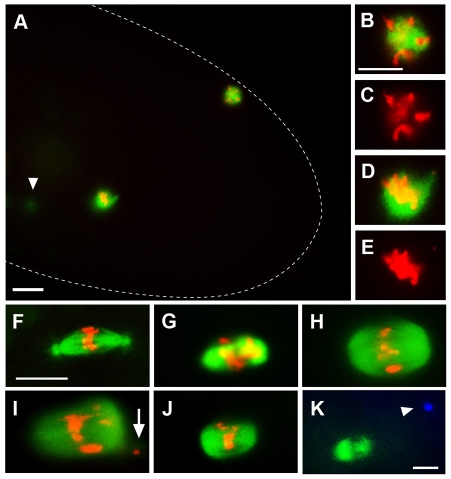
Immunostaining of ben1-derived embryos revealed that they arrest early in syncytial development with a small number (1-8) of mitotic nuclei (Fig. 7; Table 3). Most (72%) ben1-derived embryos contain one acentrosomal spindle. In Drosophila females, meiotic spindles lack centrosomes, which are provided by sperm (Foe et al., 1993). Several lines of evidence suggest that ben1 acentrosomal spindles are mitotic rather than meiotic. Their presence requires fertilization, and they are positioned deep within the egg interior where the first mitotic spindle resides (meiotic spindles are positioned near the cortex); furthermore, the presence of polar bodies indicates completion of meiotic divisions, and centrosomes are occasionally seen near the spindle (Fig. 7A-E; data not shown).
Fig. 7.
ben phenocopies nopo. Representative mitotic spindles in syncytial embryos from wild-type, nopo and ben females. (A-J) Microtubules are in green and DNA in red. (A-E) Single mitotic spindle and polar body in a ben1-derived embryo. (A) Dashed line outlines the embryo; arrowhead indicates detached centrosome out of focal plane. (B-E) Magnified images of the polar body (B,C) and mitotic spindle (D,E) from A. (F-K) Mitotic spindles in embryos from wild-type (F), nopoZ1447 (G), ben1 (H,I) and ben1/Df(1)HA92 (J,K) females. ben-derived embryos exhibit nopo phenotypes, including barrel-shaped, acentrosomal spindles and displaced DNA (I, arrow). (K) Microtubules are in green and centrosomes in blue. A ben1/Df(1)HA92 spindle with a detached centrosome (arrowhead). Scale bars: 20 μm in A; 10 μm in B-K.
The spindle defects of ben1-derived embryos strikingly resemble those of nopo mutants (compare Fig. 7H,I with 7G). ben1 spindles are often acentrosomal, barrel-shaped, variable in width, and have misaligned chromosomes; similar phenotypes were observed in ben1 hemizygotes (Fig. 7J,K; Table 3). We were unable to test whether CHK2-mediated centrosomal inactivation causes mitotic arrest in ben-derived embryos because doubly homozygous adults were not viable. Taken together, the yeast two-hybrid interactions, co-localization, and similar mutant phenotypes that we have observed suggest that BEN-UEV1A and NOPO function together as an E2-E3 complex required to preserve genomic integrity during early embryonic development in Drosophila.
Assessment of ben-mediated functions in nopo mutants
Because a given E2 can act in concert with multiple E3 ubiquitin ligases (Pickart, 2001), we sought to determine which activities of BEN are mediated by NOPO. We assayed our nopo mutants for four additional biological functions previously ascribed to BEN. The Drosophila giant fiber system (GFS) is a simple neural circuit that mediates an escape response to visual stimuli (Allen et al., 2006). Because ben is required for proper synaptic connectivity in the GFS, ben mutant adults fail to elicit a normal jump response to a light-off stimulus (Thomas and Wyman, 1982; Thomas and Wyman, 1984). ben mutant adults have also been reported to exhibit abnormalities in thoracic musculature and impaired mobility, and a role in innate immunity has been ascribed to ben (Edgecomb et al., 1993; Zhou et al., 2005).
We assessed the intactness of the GFS of nopo flies by testing their jump response to visual stimuli and found them to be defective, like ben flies (see Fig. S2A in the supplementary material) (Oh et al., 1994; Thomas and Wyman, 1982). Unlike ben, however, nopo flies exhibited normal mobility in a climbing assay and had normal sites of attachment of tergal depressor of the trochanter (TDT) thoracic muscles to the scutellum (see Fig. S2B,C in the supplementary material). We tested the innate immune responses of nopo and ben males and found that both exhibited slightly reduced levels of diptericin induction after infection (see Fig. S2D in the supplementary material). These results suggest that BEN and NOPO act as an E2-E3 complex that regulates GFS synapse formation and innate immunity, whereas BEN regulates mobility and muscle attachment sites via a different E3 ligase; thus, NOPO mediates a subset of BEN's functions.
DISCUSSION
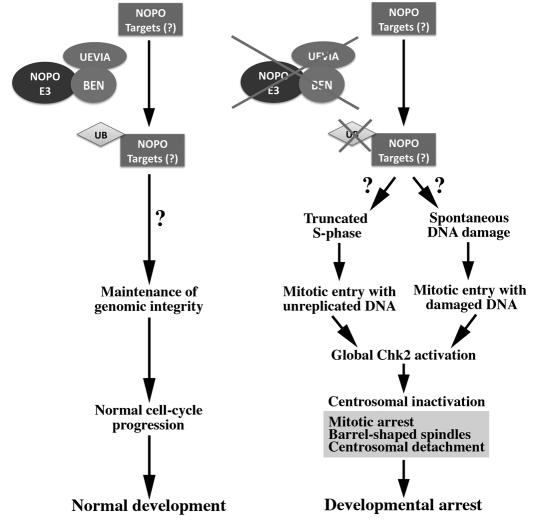
We propose a model in which NOPO, a RING domain-containing protein, interacts with the BEN-UEV1A heterodimer to form a functional E2-E3 ubiquitin ligase complex required during syncytial embryogenesis for genomic integrity, cell-cycle progression, and the continuation of development (Fig. 8). In the absence of NOPO, a lack of ubiquitination of, as yet unidentified, NOPO targets results in the truncation of S-phase and/or spontaneous DNA damage. Mitotic entry with unreplicated and/or damaged DNA triggers the activation of a CHK2-mediated checkpoint that leads to changes in spindle morphology, mitotic arrest and failure of nopo-derived embryos to develop to cellularization.
Fig. 8.
Model of the function of NOPO in the early embryo. We propose a model in which BEN-UEV1A-NOPO (E2-E3) complexes ubiquitinate unidentified target proteins to ensure the maintenance of genomic integrity in the early Drosophila embryo. In the absence of NOPO-mediated ubiquitination, truncation of S-phase and/or spontaneous DNA damage result in mitotic entry with unreplicated and/or damaged DNA, respectively. A CHK2-mediated DNA checkpoint is then triggered that causes mitotic arrest and a block in embryonic development.
We favor a model in which NOPO regulates the timing of S-M transitions in syncytial embryos to ensure that S-phase is of sufficient length to allow the completion of DNA replication prior to mitotic entry. The inhibition of DNA replication in syncytial embryos (e.g. via aphidicolin injection) leads to chromatin bridging in subsequent mitoses and CHK2 activation, both of which occur in nopo-derived embryos, presumably because of mitotic entry with unreplicated chromosomes (Raff and Glover, 1988; Takada et al., 2003). The mechanism by which NOPO coordinates S-M transitions is unknown. Our data suggest that nopo may alter the timing of these transitions independently of CDK1-Cyclin B, although localized changes in the levels and/or activities of these regulators not detectable by immunoblotting of whole-embryo lysates could play a crucial role. It is unclear why the MEI-41/GRP-dependent checkpoint, which appears to be functional in nopo-derived embryos, is not sufficient to slow mitotic entry.
The punctate nuclear localization observed for NOPO and its human homolog, TRIP, expressed in HeLa cells may indicate a direct role for these proteins in the regulation of chromatin structure. Furthermore, the G2 phase-specific localization that we observe for NOPO/TRIP in transfected HeLa cells may be consistent with a role for NOPO in slowing S-M transitions in syncytial embryos; in the absence of nopo, embryos that enter mitosis prematurely would probably do so without finishing DNA replication because of a lack of gap phases.
An alternative explanation for CHK2 activation in nopo-derived embryos is that they might incur elevated levels of spontaneous DNA damage. Syncytial embryos are considered to be unusual in that they activate CHK2 but not CHK1 in response to DNA-damaging agents (Fogarty et al., 1997; Sibon et al., 2000; Takada et al., 2007). Thus, spontaneous DNA damage would not be predicted to elicit the MEI-41/GRP-mediated replication checkpoint but would cause CHK2-dependent centrosomal inactivation during mitosis. Such a model would be consistent with the apparent lack of activation of the MEI-41/GRP-dependent checkpoint in nopo-derived embryos, although it would not explain why interphase 11 is shortened.
We previously reported that syncytial embryos from microcephalin (mcph1) mutant females undergo mitotic arrest with a phenotype similar to that described herein for nopo (Rickmyre et al., 2007). Like nopo, CHK2-mediated centrosomal inactivation causes mitotic arrest in embryos lacking mcph1. nopo and mcph1 are unique among maternal-effect lethal mutants in which CHK2-mediated centrosomal inactivation has been reported (e.g. grp, mei-41, wee1) in that their phenotypes appear to be more severe: centrosomes typically detach from spindles, and mitotic arrest occurs earlier, during precortical syncytial divisions (Rickmyre et al., 2007; Sibon et al., 2000; Stumpff et al., 2004; Takada et al., 2003). The underlying defects in nopo and mcph1 mutants may be distinct, however, because mnk mcph1-derived embryos exhibit normal cycle 11 interphase length, which is truncated in mnk nopo-derived embryos (Rickmyre et al., 2007). Furthermore, we did not detect a genetic interaction between nopo and mcph1 (J.L.R., J.A.M. and L.A.L., unpublished).
Mammalian TRIP was identified in a yeast two-hybrid screen for tumor necrosis factor (TNF) receptor-associated factor (TRAF) interactors (Lee et al., 1997). TRAFs transduce signals from members of the tumor necrosis factor (TNF)/tumor necrosis factor receptor (TNFR) superfamily, which elicit diverse cellular responses in the immune and inflammatory systems (Hehlgans and Pfeffer, 2005). TRIP has been reported to inhibit TRAF2-mediated NFκB activation; the RING domain of TRIP, however, was not required for inhibition (Lee et al., 1997). By contrast, our analysis of nopoZ1447 indicates that this motif is essential for NOPO function in Drosophila embryogenesis, probably by mediating its interactions with E2 components, as has been shown for other E3 ligases (Passmore and Barford, 2004). Drosophila Eiger (TNF ligand) and Wengen (TNF receptor) play roles in dorsal closure, neuroblast divisions, and the response to fungal pathogens (Kauppila et al., 2003; Schneider et al., 2007; Wang et al., 2006). A role for TNF signaling in early Drosophila embryogenesis has not been reported to our knowledge.
TRIP was recently reported to be an essential factor in mice (Park et al., 2007). TRIP-deficient mice die soon after implantation as a result of defects in early embryonic development. Compared with wild-type littermates, TRIP-/- embryos are smaller in size with a reduced cell number. TRAF2 does not appear to be required until later in development, suggesting that TRIP has TRAF2-independent roles in early embryos (Nguyen et al., 1999). It will be interesting to see whether mammalian TRIP, by analogy to Drosophila NOPO, is required for genomic integrity during embryonic development.
Our data support a model in which NOPO ubiquitin ligase acts in concert with BEN-UEV1A heterodimers to regulate Drosophila syncytial embryogenesis. The yeast two-hybrid interaction and co-localization of NOPO and BEN led us to identify an unanticipated role for BEN in early embryogenesis and additional roles for NOPO in synapse formation and innate immunity. Although the spindle defects of ben-derived embryos are strikingly similar to those of nopo mutants, they typically arrest earlier in syncytial development, suggesting that another E3 ligase that requires BEN may function in parallel with NOPO. Although nopo egg chambers appear normal, we have not ruled out a possible requirement for BEN-UEV1A-NOPO complexes during oogenesis; some defects in nopo- and ben-derived embryos could be a secondary consequence of previous defects during oogenesis.
K63-linked ubiquitin chains are thought to act as non-proteolytic signals (e.g. affecting protein localization and/or interactions), whereas K48-linked ubiquitin chains have established roles in targeting proteins for proteasome-mediated degradation (Pickart and Fushman, 2004). BEN-UEV1A E2 homologs in budding yeast (Ubc13-Mms2p) mediate K63-linked polyubiquitination of PCNA during postreplicative repair (Andersen et al., 2005). In mammalian cells, the E2 heterodimer Ubc13-Mms2 mediates DNA damage repair, while Ubc13-Uev1A promotes NFκB activation; both E2 complexes regulate these processes by mediating K63 ubiquitin chain assembly on target proteins. We propose that BEN-UEV1A-NOPO (E2-E3) complexes mediate the assembly of K63-linked ubiquitin chains on proteins that preserve genomic integrity in early Drosophila embryogenesis.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/3/449/DC1
Supplementary Material
We thank Charles Zuker, Bill Theurkauf and Mark Tanouye for fly stocks and antibodies; Amir Fayyazuddin and Prashant Mishra for jump assay advice; Amir Fayyazuddin and Hugo Bellen for LED light apparatus; Abel Alcazar-Roman for yeast two-hybrid advice; and Audrey Frist for technical assistance. Andrea Page-McCaw and Byeong Cha provided critical comments on the manuscript. We thank Terry Orr-Weaver in whose lab this study was initiated (NIH grant GM39341 and NSF grant MCB0132237 to T. Orr-Weaver). This study was supported by NIH grant GM074044 and American Cancer Society grant IRG-58-009-46 to L.A.L. Deposited in PMC for release after 12 months.
References
- Abdu, U., Brodsky, M. and Schupbach, T. (2002). Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr. Biol. 12, 1645-1651. [DOI] [PubMed] [Google Scholar]
- Allen, M. J., Godenschwege, T. A., Tanouye, M. A. and Phelan, P. (2006). Making an escape: development and function of the Drosophila giant fibre system. Semin. Cell Dev. Biol. 17, 31-41. [DOI] [PubMed] [Google Scholar]
- Andersen, P. L., Zhou, H., Pastushok, L., Moraes, T., McKenna, S., Ziola, B., Ellison, M. J., Dixit, V. M. and Xiao, W. (2005). Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J. Cell Biol. 170, 745-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse, A., Campos, A. D., Webster, W. K. and Darnay, B. G. (2007). TRAF-interacting protein (TRIP) is a RING-dependent ubiquitin ligase. Biochem. Biophys. Res. Commun. 359, 660-664. [DOI] [PubMed] [Google Scholar]
- Branzei, D. and Foiani, M. (2008). Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 9, 297-308. [DOI] [PubMed] [Google Scholar]
- Brodsky, M. H., Weinert, B. T., Tsang, G., Rong, Y. S., McGinnis, N. M., Golic, K. G., Rio, D. C. and Rubin, G. M. (2004). Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell. Biol. 24, 1219-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broomfield, S., Chow, B. L. and Xiao, W. (1998). MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl. Acad. Sci. USA 95, 5678-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B., Chu, T., Harms, E., Gergen, J. P. and Strickland, S. (1998). Mapping of Drosophila mutations using site-specific male recombination. Genetics 149, 157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crest, J., Oxnard, N., Ji, J. Y. and Schubiger, G. (2007). Onset of the DNA replication checkpoint in the early Drosophila embryo. Genetics 175, 567-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel, M. and Meier, P. (2005). Ubiquitylation in apoptosis: DIAP1's (N-)en(d)igma. Cell Death Differ. 12, 1208-1212. [DOI] [PubMed] [Google Scholar]
- Edgar, B. A., Sprenger, F., Duronio, R. J., Leopold, P. and O'Farrell, P. H. (1994). Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev. 8, 440-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgecomb, R. S., Ghetti, C. and Schneiderman, A. M. (1993). Bendless alters thoracic musculature in Drosophila. J. Neurogenet. 8, 201-219. [DOI] [PubMed] [Google Scholar]
- Fayyazuddin, A., Zaheer, M. A., Hiesinger, P. R. and Bellen, H. J. (2006). The nicotinic acetylcholine receptor Dalpha7 is required for an escape behavior in Drosophila. PLoS Biol. 4, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe, V. E., Odell, G. M. and Edgar, B. A. (1993). Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint. In The Development of Drosophila Melanogaster (ed. M. Bate and A. Martinez Arias), pp. 149-300. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Fogarty, P., Campbell, S. D., Abu-Shumays, R., Phalle, B. S., Yu, K. R., Uy, G. L., Goldberg, M. L. and Sullivan, W. (1997). The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr. Biol. 7, 418-426. [DOI] [PubMed] [Google Scholar]
- Freemont, P. S. (2000). RING for destruction? Curr. Biol. 10, R84-R87. [DOI] [PubMed] [Google Scholar]
- Garcia, K., Duncan, T. and Su, T. T. (2007). Analysis of the cell division cycle in Drosophila. Methods 41, 198-205. [DOI] [PubMed] [Google Scholar]
- Giot, L., Bader, J. S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y. L., Ooi, C. E., Godwin, B., Vitols, E. et al. (2003). A protein interaction map of Drosophila melanogaster. Science 302, 1727-1736. [DOI] [PubMed] [Google Scholar]
- Girard, F., Strausfeld, U., Fernandez, A. and Lamb, N. J. C. (1991). Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell 67, 1169-1179. [DOI] [PubMed] [Google Scholar]
- Grumbling, G. and Strelets, V. (2006). FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34, D484-D488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J. W. and Elledge, S. J. (2007). The DNA damage response: ten years after. Mol. Cell 28, 739-745. [DOI] [PubMed] [Google Scholar]
- Hehlgans, T. and Pfeffer, K. (2005). The intriguing biology of the tumour necrosis factor/tumour necrosis factor receptor superfamily: players, rules and the games. Immunology 115, 1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, P. K., Eldridge, A. G., Freed, E., Furstenthal, L., Hsu, J. Y., Kaiser, B. K. and Reimann, J. D. (2000). The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10, 429-439. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J. and Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, J. Y., Squirrell, J. M. and Schubiger, G. (2004). Both cyclin B levels and DNA-replication checkpoint control the early embryonic mitoses in Drosophila. Development 131, 401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppila, S., Maaty, W. S., Chen, P., Tomar, R. S., Eby, M. T., Chapo, J., Chew, S., Rathore, N., Zachariah, S., Sinha, S. K. et al. (2003). Eiger and its receptor, Wengen, comprise a TNF-like system in Drosophila. Oncogene 22, 4860-4867. [DOI] [PubMed] [Google Scholar]
- Koundakjian, E. J., Cowan, D. M., Hardy, R. W. and Becker, A. H. (2004). The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics 167, 203-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, L. A. and Orr-Weaver, T. L. (2003). Regulation of cell cycles in Drosophila development: intrinsic and extrinsic cues. Annu. Rev. Genet. 37, 545-578. [DOI] [PubMed] [Google Scholar]
- Lee, L. A., Van Hoewyk, D. and Orr-Weaver, T. L. (2003). The Drosophila cell cycle kinase PAN GU forms an active complex with PLUTONIUM and GNU to regulate embryonic divisions. Genes Dev. 17, 2979-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. Y., Lee, S. Y. and Choi, Y. (1997). TRAF-interacting protein (TRIP): a novel component of the tumor necrosis factor receptor (TNFR)- and CD30-TRAF signaling complexes that inhibits TRAF2-mediated NF-kappaB activation. J. Exp. Med. 185, 1275-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K. and Kaufman, T. C. (1996). The homeotic target gene centrosomin encodes an essential centrosomal component. Cell 85, 585-596. [DOI] [PubMed] [Google Scholar]
- Lorick, K. L., Jensen, J. P., Fang, S., Ong, A. M., Hatakeyama, S. and Weissman, A. M. (1999). RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96, 11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masrouha, N., Yang, L., Hijal, S., Larochelle, S. and Suter, B. (2003). The Drosophila chk2 gene loki is essential for embryonic DNA double-strand-break checkpoints induced in S phase or G2. Genetics 163, 973-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidhar, M. G. and Thomas, J. B. (1993). The Drosophila bendless gene encodes a neural protein related to ubiquitin-conjugating enzymes. Neuron 11, 253-266. [DOI] [PubMed] [Google Scholar]
- Nguyen, L. T., Duncan, G. S., Mirtsos, C., Ng, M., Speiser, D. E., Shahinian, A., Marino, M. W., Mak, T. W., Ohashi, P. S. and Yeh, W. C. (1999). TRAF2 deficiency results in hyperactivity of certain TNFR1 signals and impairment of CD40-mediated responses. Immunity 11, 379-389. [DOI] [PubMed] [Google Scholar]
- Oh, C. E., McMahon, R., Benzer, S. and Tanouye, M. A. (1994). bendless, a Drosophila gene affecting neuronal connectivity, encodes a ubiquitin-conjugating enzyme homolog. J. Neurosci. 14, 3166-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, E. S., Choi, S., Kim, J. M., Jeong, Y., Choe, J., Park, C. S., Choi, Y. and Rho, J. (2007). Early embryonic lethality caused by targeted disruption of the TRAF-interacting protein (TRIP) gene. Biochem. Biophys. Res. Commun. 363, 971-977. [DOI] [PubMed] [Google Scholar]
- Passmore, L. A. and Barford, D. (2004). Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem. J. 379, 513-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart, C. M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503-533. [DOI] [PubMed] [Google Scholar]
- Pickart, C. M. and Fushman, D. (2004). Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610-616. [DOI] [PubMed] [Google Scholar]
- Raff, J. W. and Glover, D. M. (1988). Nuclear and cytoplasmic mitotic cycles continue in Drosophila embryos in which DNA synthesis is inhibited with aphidicolin. J. Cell Biol. 107, 2009-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickmyre, J. L., Dasgupta, S., Ooi, D. L., Keel, J., Lee, E., Kirschner, M. W., Waddell, S. and Lee, L. A. (2007). The Drosophila homolog of MCPH1, a human microcephaly gene, is required for genomic stability in the early embryo. J. Cell Sci. 120, 3565-3577. [DOI] [PubMed] [Google Scholar]
- Rubin, G. M. and Spradling, A. C. (1982). Genetic transformation of Drosophila with transposable element vectors. Science 218, 348-353. [DOI] [PubMed] [Google Scholar]
- Sancho, E., Vila, M. R., Sanchez-Pulido, L., Lozano, J. J., Paciucci, R., Nadal, M., Fox, M., Harvey, C., Bercovich, B., Loukili, N. et al. (1998). Role of UEV-1, an inactive variant of the E2 ubiquitin-conjugating enzymes, in in vitro differentiation and cell cycle behavior of HT-29-M6 intestinal mucosecretory cells. Mol. Cell. Biol. 18, 576-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin, A. J., Borden, K. L., Boddy, M. N. and Freemont, P. S. (1996). Does this have a familiar RING? Trends Biochem. Sci. 21, 208-214. [PubMed] [Google Scholar]
- Schneider, D. S., Ayres, J. S., Brandt, S. M., Costa, A., Dionne, M. S., Gordon, M. D., Mabery, E. M., Moule, M. G., Pham, L. N. and Shirasu-Hiza, M. M. (2007). Drosophila eiger mutants are sensitive to extracellular pathogens. PLoS Pathog. 3, e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibon, O. C., Stevenson, V. A. and Theurkauf, W. E. (1997). DNA-replication checkpoint control at the Drosophila midblastula transition. Nature 388, 93-97. [DOI] [PubMed] [Google Scholar]
- Sibon, O. C., Laurencon, A., Hawley, R. and Theurkauf, W. E. (1999). The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr. Biol. 9, 302-312. [DOI] [PubMed] [Google Scholar]
- Sibon, O. C., Kelkar, A., Lemstra, W. and Theurkauf, W. E. (2000). DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat. Cell Biol. 2, 90-95. [DOI] [PubMed] [Google Scholar]
- Silva, E., Tiong, S., Pedersen, M., Homola, E., Royou, A., Fasulo, B., Siriaco, G. and Campbell, S. D. (2004). ATM is required for telomere maintenance and chromosome stability during Drosophila development. Curr. Biol. 14, 1341-1347. [DOI] [PubMed] [Google Scholar]
- Somanathan, S., Suchyna, T. M., Siegel, A. J. and Berezney, R. (2001). Targeting of PCNA to sites of DNA replication in the mammalian cell nucleus. J. Cell. Biochem. 81, 56-67. [DOI] [PubMed] [Google Scholar]
- Stumpff, J., Duncan, T., Homola, E., Campbell, S. D. and Su, T. T. (2004). Drosophila Wee1 kinase regulates Cdk1 and mitotic entry during embryogenesis. Curr. Biol. 14, 2143-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, T. T., Sprenger, F., DiGregorio, P. J., Campbell, S. D. and O'Farrell, P. H. (1998). Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation. Genes Dev. 12, 1495-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S., Kelkar, A. and Theurkauf, W. E. (2003). Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell 113, 87-99. [DOI] [PubMed] [Google Scholar]
- Takada, S., Kwak, S., Koppetsch, B. S. and Theurkauf, W. E. (2007). grp (chk1) replication-checkpoint mutations and DNA damage trigger a Chk2-dependent block at the Drosophila midblastula transition. Development 134, 1737-1744. [DOI] [PubMed] [Google Scholar]
- Tang, T. T., Bickel, S. E., Young, L. M. and Orr-Weaver, T. L. (1998). Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes Dev. 12, 3843-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J. B. and Wyman, R. J. (1982). A mutation in Drosophila alters normal connectivity between two identified neurones. Nature 298, 650-651. [DOI] [PubMed] [Google Scholar]
- Thomas, J. B. and Wyman, R. J. (1984). Mutations altering synaptic connectivity between identified neurons in Drosophila. J. Neurosci. 4, 530-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Cai, Y., Chia, W. and Yang, X. (2006). Drosophila homologs of mammalian TNF/TNFR-related molecules regulate segregation of Miranda/Prospero in neuroblasts. EMBO J. 25, 5783-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Xin, S. and Du, W. (2001). Drosophila Chk2 is required for DNA damage-mediated cell cycle arrest and apoptosis. FEBS Lett. 508, 394-398. [DOI] [PubMed] [Google Scholar]
- Zhou, R., Silverman, N., Hong, M., Liao, D. S., Chung, Y., Chen, Z. J. and Maniatis, T. (2005). The role of ubiquitination in Drosophila innate immunity. J. Biol. Chem. 280, 34048-34055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.