Abstract
In normal human cells treated with interferons (IFNs), the concentration of tyrosine-phosphorylated STAT1 (YP-STAT1), which drives the expression of a large number of genes, increases quickly but then decreases over a period of several hours. Because the STAT1 gene is activated by YP-STAT1, IFNs stimulate a large increase in the concentration of unphosphorylated STAT1 (U-STAT1) that persists for several days. To test the significance of high U-STAT1 expression, we increased its concentration exogenously in the absence of IFN treatment. In response, the expression of many immune regulatory genes (e.g., IFI27, IFI44, OAS, and BST2) was increased. In human fibroblasts or mammary epithelial cells treated with low concentrations of IFN-β or IFN-γ, the expression of the same genes increased after 6 h and continued to increase after 48 or 72 h, long after the concentration of YP-STAT1 had returned to basal levels. Consistent with its activity as a transcription factor, most U-STAT1 was present in the nuclei of these cells before IFN treatment, and the fraction in nuclei increased 48 h after treatment with IFN. We conclude that the nuclear U-STAT1 that accumulates in response to IFNs maintains or increases the expression of a subset of IFN-induced genes independently of YP-STAT1, and that many of the induced proteins are involved in immune regulation.
Keywords: innate immunity, interferon response, STAT1 gene
Interferons (IFNs), which play key roles in antiviral and antigrowth responses and in modulating immune responses, are classified as type I (e.g., IFN-α and IFN-β) or type II (IFN-γ) (1). Their signaling pathways are mediated by the sequential phosphorylation of Janus family kinases (JAKs) and STATs. Type I IFNs phosphorylate STATs 1 and 2, which form IFN-stimulated gene factor-3 (ISGF-3), a ternary complex that also includes IFN response factor-9 (IRF9). IFN-γ induces the phosphorylation of STAT1, which forms STAT1 homodimers. These activated transcription factors translocate into the nucleus, where they bind to distinct conserved sequences in the promoters of target genes. However, recent studies have shown that IFN signaling is much more complex (2). IFNs activate several different kinases in addition to the JAKs, other STATs in addition to STAT1 and STAT2, and even other transcription factors (3–9).
Our previous work has shown that STAT1 drives the constitutive expression of some genes without phosphorylation (10, 11). For example, the complex of unphosphorylated STAT1 (U-STAT1) and IRF1 mediates the constitutive expression of the low-molecular mass polypeptide 2 (LMP2) gene (11). Similarly, Cui et al. (12) have shown that unphosphorylated STAT6 cooperates with p300 to increase transcription of the cyclooxygenase-2 gene. Initially, unphosphorylated STATs were considered to be latent transcription factors in the cytoplasm, entering the nucleus to induce gene expression only in response to cytokine stimulation. However, consistent with our previous results, STAT1 and STAT3 have been found to be present in nuclei independently of tyrosine phosphorylation, in a cell type-specific manner, and the nuclear import mechanism of U-STAT1 is completely distinct from that of phosphorylated STAT1 (13, 14). The nuclear import of tyrosine-phosphorylated STAT1 dimers depends on importins and metabolic energy. On the other hand, U-STAT1 is transported via a carrier-free mechanism that involves direct interaction with nucleoporins (15).
In response to IFNs, the phosphorylation of STAT1 can last for several hours, but U-STAT1, newly synthesized in response to tyrosine-phosphorylated STAT1 (YP-STAT1) persists for several days (16), raising the possibility that the increased concentration of U-STAT1 might play an important role in IFN-dependent signaling. In a similar way, STAT3 expression is elevated in response to IL-6, and the increased level of U-STAT3 induces the expression of many genes that are quite distinct from those induced by phosphorylated STAT3 (17). A subset of these genes, including RANTES and several oncogenes, is induced by a complex of U-STAT3 and unphosphorylated NF-κB (18). Therefore, U-STAT3 and phosphorylated STAT3 are independently important in responses to IL-6, including IL-6-mediated oncogenesis.
Here, we have investigated the role of the U-STAT1 that is increased in response to IFNs in 2 different nontumorigenic human cell lines, BJ fibroblasts and hTERT-HME1 mammary epithelial cells. In contrast to U-STAT3, which drives the expression of a set of genes that is quite different from those induced in response to phosphorylated STAT3, the results presented here show that U-STAT1 prolongs the expression of a subset of IFN-induced genes, many of which are involved in immune regulation.
Results
IFN-β and IFN-γ Increase STAT1 Expression in BJ and hTERT-HME1 Cells.
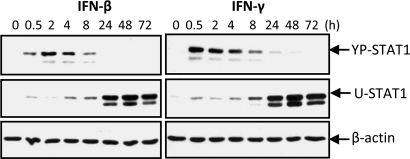
IFN-β and IFN-γ induced the phosphorylation of STAT1 on Y701 within 30 min in hTERT-HME1 cells, and the concentration of YP-STAT1 then decreased over the next several hours (Fig. 1). As YP-STAT1 decreased, there was a reciprocal increase in the concentration of newly synthesized U-STAT1, beginning at about 8 h, which persisted for at least 3 days (Fig. 1). A wide range of IFN concentrations, as low as 5 units/mL IFN-β or 0.1 ng/mL IFN-γ, led to the induction of similar levels of U-STAT1 after 48 h, in contrast to the induced levels of YP-STAT1, which were roughly proportional to the IFN concentrations used (Fig. S1). BJ human fibroblasts showed similar time- and dose-dependent responses to IFN-β and IFN-γ.
Fig. 1.
IFN-β or IFN-γ increases the expression of U-STAT1. hTERT-HME1 cells were treated with IFN-β (50 units/mL) or IFN-γ (1 ng/mL). The amounts of YP-STAT1 and U-STAT1 were measured by the Western blotting method.
Increased STAT1 Is Not Completely Phosphorylated in Response to IFN-γ or IFN-β.
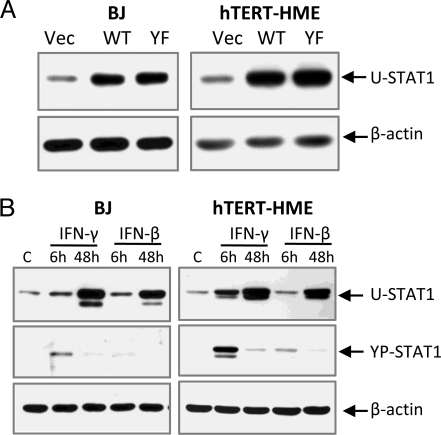
To investigate the role of IFN-induced U-STAT1, we examined gene expression in stable pools of lentivirus-infected cells that express high levels of wild-type STAT1 or Y701F-STAT1, which cannot be phosphorylated (Fig. 2A). Without IFN stimulation, the increased levels of wild-type STAT1 were not phosphorylated (Fig. S2A). When these cells were treated with IFN-γ (100 ng/mL for 30 min), more YP-STAT1 was detected in cells expressing high levels of wild-type STAT1, but the increase was only about 2-fold. Because the concentration of STAT1 increased by 4- to 8-fold, only a relatively small portion of the increased amount of wild-type STAT1 was phosphorylated (Fig. S2A). The increased level of STAT1 did not augment IFN-β-induced STAT1 phosphorylation. When hTERT-HME1 cells expressing different amounts of wild-type STAT1 were treated with various concentrations of IFN-β, cells expressing more STAT1 did not respond more strongly than vector-transfected cells. No phosphorylation of STAT1 was detected in response to 0.5 or 5 units/mL IFN-β, and YP-STAT1 levels were similar when the 2 types of cells were compared at 50 units/mL, even though the U-STAT1 levels were substantially different (Fig. S2B). These results indicate that the increased concentrations of STAT1 formed in response to IFNs might play roles in signaling other than by increasing signal transduction through increased phosphorylation.
Fig. 2.
The expression of U-STAT1 and YP-STAT1 in cells used in microarray analyses. (A) hTERT-HME1 or BJ cells were infected with lentiviruses expressing wild-type (WT) or Y701F-STAT1 (YF), or with empty vector (Vec). The U-STAT1 protein expression levels were measured by the Western blotting method. (B) Cells were treated with IFN-β or IFN-γ for 6 or 48 h to establish a condition in which the maximum levels of U-STAT1 were induced at 48 h, with the minimum levels of YP-STAT1. The selected concentrations of IFNs (0.3 ng/mL IFN-γ or 3 units/mL IFN-β for BJ cells, and 0.1 ng/mL IFN-γ or 5 units/mL IFN-β for hTERT-HME1 cells) were applied, and the amounts of U-STAT1 and PY-STAT1 were analyzed by the Western blotting method.
Increased U-STAT1 Regulates Gene Expression.
A microarray analysis was carried out to analyze gene expression in BJ cells with high levels of wild-type STAT1 (WT) or Y701F-STAT1 (YF) (Fig. 2A). Of the 22,184 probes on the arrays, 46 were changed in response to wild-type STAT1 (40 up, 6 down), 122 were changed in response to Y701F-STAT1 (109 up, 13 down), and 35 were changed in response to both (33 up, 2 down). We include in these numbers only signals that changed by >2-fold, with differential P ≤ 0.05 in WT and/or YF compared with vector controls, and with average signals greater than 25 in WT, YF, or controls.
The 35 probes regulated by both wild-type STAT1 and Y701F-STAT1 represent 30 distinct genes. More than half of these (17 of 30) function in immune responses (Table 1). The functions of the other genes, including HERC6, FLJ20035, and EPSTI1, are mostly unknown. The signal for STAT1 itself was also increased in response to high STAT1 expression. One of the STAT1 probes in Table 1 (ILMN_1777325) is located in the 3′ UTR of endogenous STAT1 mRNA and is not included in the STAT1 mRNA expressed from the lentiviral vector, so it detects only endogenous STAT1 mRNA. Therefore, the increased expression of exogenous U-STAT1 leads to increased expression of endogenous STAT1, showing that this gene responds both to YP-STAT1 and U-STAT1, as do the other genes noted above. The results for the 2 genes whose expression was most highly increased in response to high levels of STAT1, IFI27 and BST2, were confirmed by using real-time PCR (Fig. S3A): IFI27 was increased by 6-fold in WT and 31-fold in YF cells, and BST2 was increased by 7-fold in WT and 25-fold in YF cells.
Table 1.
Genes induced by U-STAT1
| Gene symbol | PROBE_ID | BJ |
BJ |
hTERT-HME |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | YF | IFN-β |
IFN-γ |
IFN-β |
IFN-γ |
||||||
| 6 h | 48 h | 6 h | 48 h | 6 h | 48 h | 6 h | 48 h | ||||
| IFI27* | ILMN_1661581 | 4.3 | 8.1 | 6.1 | 46 | 1.9 | 15 | (2.1) | (51) | 0.0 | (1.0)** |
| BST2 | ILMN_1723480 | 3.7 | 8.1 | 12 | 22 | 5.5 | 25 | 18 | 150 | 24 | 33 |
| OAS1* | ILMN_1658247 | 3.1 | 6.3 | 10 | 7.9 | 2.4 | 2.3 | (26) | (31) | (1.5) | (1.0)** |
| ILMN_1675640 | 5.1 | 6.9 | 35 | 22 | 4.6 | 4.6 | (22) | (25) | 0.0 | 0.0 | |
| OAS2* | ILMN_1674063 | 4.1 | 6.2 | 9.8 | 11 | 4.5 | 4.0 | (11) | (23) | (2.3) | (1.0)** |
| OAS3 | ILMN_1745397 | 2.5 | 2.6 | 6.3 | 7.7 | 2.5 | 2.7 | 8.4 | 13 | 3.7 | 2.8 |
| STAT1 | ILMN_1690105 | 2.2 | 2.8 | 3.2 | 2.8 | 4.0 | 4.3 | 6.8 | 7.9 | 16 | 7.2 |
| ILMN_1777325 | 1.7 | 2.0 | 2.0 | 2.0 | 2.4 | 2.8 | 4.9 | 5.1 | 9.5 | 3.9 | |
| IFI44 | ILMN_1760062 | 2.6 | 3.3 | 6.2 | 4.1 | 2.6 | 2.1 | 130 | 180 | 9.5 | 8.8 |
| IFI44L | ILMN_1723912 | 5.2 | 7.7 | 13 | 13 | 5.5 | 6.3 | 43 | 73 | 3.6 | 2.4 |
| IFIH1 | ILMN_1781373 | 2.7 | 4.0 | 12.8 | 3.9 | 5.3 | 2.5 | 28 | 27 | 22 | 3.9 |
| IFITM1 | ILMN_1801246 | 2.0 | 2.5 | 2.1 | 3.4 | 1.7 | 2.9 | 22 | 76 | 6.4 | 4.7 |
| IFI35 | ILMN_1745374 | 2.1 | 2.2 | 3.9 | 3.0 | 3.4 | 3.3 | 3.7 | 5.2 | 9.4 | 3.6 |
| IFIT3* | ILMN_1701789 | 2.9 | 2.4 | 5.8 | 3.0 | 4.8 | 3.0 | (12) | (3.7) | (15) | (1.0)** |
| MX1* | ILMN_1662358 | 3.6 | 5.3 | 6.7 | 5.9 | 2.5 | 1.7 | (53) | (85) | (1.0)** | 0.0 |
| IRF7 | ILMN_1798181 | 3.0 | 4.3 | 6.1 | 3.3 | 1.6 | 1.2 | 12 | 11 | 2.4 | 1.6 |
| G1P2 | ILMN_1813289 | 2.7 | 3.0 | 3.6 | 4.2 | 1.5 | 1.6 | 27 | 41 | 5.0 | 2.7 |
| IFIT1* | ILMN_1707695 | 2.3 | 2.9 | 5.1 | 2.9 | 1.4 | 0.8 | (45) | (21) | (1.0)** | 0.0 |
| PLSCR1 | ILMN_1752889 | 2.4 | 3.2 | 6.3 | 3.2 | 3.1 | 1.8 | 4.8 | 4.7 | 2.4 | 1.0 |
| HERC6 | ILMN_1654639 | 3.4 | 3.7 | 5.4 | 6.3 | 1.7 | 2.4 | 35 | 100 | 9.9 | 7.8 |
| FLJ20035 | ILMN_1795181 | 2.1 | 2.9 | 3.5 | 4.0 | 3.0 | 3.4 | 32 | 80 | 22 | 17 |
| EPSTI1 | ILMN_1688566 | 2.5 | 3.4 | 4.2 | 3.4 | 3.7 | 4.0 | 7.3 | 11 | 12 | 4.9 |
The expressions of 22,184 probes (a gene is represented by 1–3 probes) in an Illumina Sentrix HumanRef-8 Expression Bead Chip were analyzed by using mRNA from BJ cells transfected with empty vector (Vec), wild-type STAT1 (WT), or Y701F-STAT1 (YF). BJ cells were treated with IFN-β (3 units/mL) or IFN-γ (0.3 ng/mL) for 6 or 48 h; hTERT-HME1 cells were treated with IFN-β (5 units/mL) or IFN-γ (0.1 ng/mL) for 6 or 48 h. The numbers in the table are fold inductions compared with the signals from the mRNAs of control cells (empty vector-transfected BJ cells for WT and YF, untreated BJ or hTERT-HME1 cells for IFN-treated cells). The average signal for each probe was used to determine expression levels. The genes with average signals below 25 in the control and the treated cells, or with detection P values more than 0.05 in the treated cells compared with the control, were excluded from the analyses.
*IFI27, OAS1, OAS2, IFIT3, Mx1, and IFIT1 were not detected in untreated control hTERT-HME1 cells but were induced in response to IFN. In these cases, the numbers in parentheses are fold inductions compared with the levels of expression in cells treated with IFN-γ for 6 or 48 h (denoted by double asterisks in the last column) instead of untreated cells. All of the genes in this table except HERC6, FLI20035, and EPSTI1 are known to play roles related to immune responses.
Some of the Genes Induced at Late Times by IFNs Respond to U-STAT1.
U-STAT1 is substantially increased in response to IFNs after 24 h, suggesting that it might be responsible for delayed gene expression in IFN signaling. We explored this possibility by treating BJ or hTERT-HME1 cells with IFNs, followed by microarray analysis. Because persistent IFN-induced YP-STAT1 might be responsible for gene expression at late times, we first determined concentrations of IFNs that induce substantial levels of U-STAT1 but minimal levels of YP-STAT1 at late times. BJ cells treated with various concentrations of IFN-β or IFN-γ for 6 or 48 h (Fig. S4) provided the optimal concentrations of IFNs for this experiment: 3 units/mL IFN-β or 0.3 ng/mL IFN-γ (Fig. 2B and arrowheads in Fig. S4). Similarly, we determined the optimal concentrations for hTERT-HME1 cells: 5 units/mL IFN-β or 0.1 ng/mL IFN-γ (Fig. 2B).
In BJ cells, the signals for 139 (112 up, 27 down) or 166 (140 up, 26 down) probes were changed in response to treatment for 48 h with IFN-β or IFN-γ, respectively (criteria as above). Among these, 56 were changed in BJ cells in response to either IFN-β or IFN-γ. Many of these genes were also regulated by U-STAT1 (Table 1). In hTERT-HME1 cells, the expression of 86 (75 up, 11 down) or 131 (89 up, 42 down) probes were changed in response to treatment for 48 h with IFN-β or IFN-γ, respectively, and 42 (39 up, 3 down) were changed by either of the IFNs (criteria as above). The immune regulatory genes induced in response to U-STAT1—for example, IFI27, BST2, OAS2, and IFI44—were also highly increased in response to IFNs, especially by IFN-β, both in hTERT-HME1 and BJ cells (Table 1). IFI27, OAS1, OAS2, IFIT3, MX1, and IFIT1 were not expressed detectably in untreated hTERT-HME1 cells, but their expression was also increased by IFNs, especially by IFN-β (marked with an asterisk in Table 1). MX1, IRF7, GIP2, IFIT1, and PLSCR1 were strongly induced in response to IFN-β but induced very little in response to IFN-γ, in both BJ and hTERT-HME1 cells. The expression of the representative genes (IFI27, BST2, and OAS2) was confirmed by using real-time PCR (Fig. S3B), which showed induction levels similar to those determined in the microarray experiment.
U-STAT1-Induced Immune Regulatory Genes Are Also Induced by STAT1 Phosphorylation.
Many of the genes in Table 1 are known to be induced by IFNs, especially by type I IFNs, at early times. We analyzed gene expression in cells treated with IFNs for 6 or 48 h. The increase in U-STAT1 protein can be detected only after about 8 h (Fig. 1). Therefore, the genes induced by IFNs at 6 h can be induced only by YP-STAT1 and not by newly synthesized U-STAT1. As reported by many others, more than 100 genes were induced by IFNs after 6 h: the expression of 187 or 228 probes was changed by IFN-β or IFN-γ in BJ cells, and the expression of 69 or 195 probes was changed by IFN-β or IFN-γ in hTERT-HME1 cells, respectively (criteria as above). In Table 1, most genes expressed at a high level after 48 h were also induced after 6 h of IFN treatment. The levels after 48 h were the same as or more than the levels after 6 h, and the expressions of some were much higher after 48 h. The mRNA synthesized in response to YP-STAT1 might be detected after 48 h if it were to have a long half-life. However, even if this mRNA were not degraded at all over a period of several days, its concentration would decrease because of dilution by cell growth and division. Cells treated with these low concentrations of IFNs grew similarly to untreated cells. Therefore, our results show that some mRNAs synthesized in response to YP-STAT1 at early times are induced further in response to newly synthesized U-STAT1 after about 8 h.
U-STAT1 Is Required to Prolong the Expression of IFN-Induced Immune Regulatory Genes.
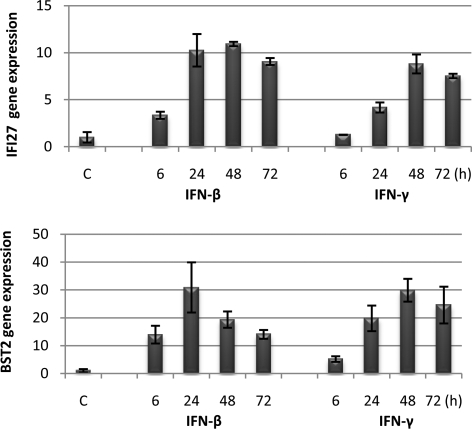
We examined the expression of IFI27 and BST2 in BJ cells for up to 72 h after IFN treatment (Fig. 3). The U-STAT1 induced by IFNs remains high even after 72 h (Fig. 1). The expression of IFI27 and BST2 mRNAs was induced after 6 h of treatment with IFN-β or IFN-γ, and it was further increased, by 3- to 4-fold, after 24 h, suggesting that the mRNA levels are probably the sums of those induced by YP-STAT1 and U-STAT1. The mRNA levels were further increased after 48 h and were sustained similarly after 72 h. As a result, IFI27 mRNA was 9.0- or 7.8-fold higher, and BST2 mRNA was 14- or 25-fold higher after 72 h of IFN-β or IFN-γ treatment, respectively, compared with the levels in untreated cells. The expressions of IFI44 and OAS2 were not increased above the levels induced by YP-STAT1 6 h after IFN treatment, but these levels were sustained after 72 h. The expression of all of these genes was regulated similarly in hTERT-HME1 cells, especially by IFN-β. These results confirm that a group of IFN-induced immune regulatory genes, including IFI27, BST2, IFI44, and OAS2, was highly expressed after several days in response to U-STAT1, the secondary transcriptional regulator that is newly synthesized in response to IFNs.
Fig. 3.
U-STAT1 prolongs the expression of some IFN-induced immune regulatory genes. BJ cells were treated with IFN-β (3 units/mL) or IFN-γ (0.3 units/mL), and the expression of IFI27 and BST2 was measured by real-time PCR. The figure shows the mean values, with standard deviations, of triplicate experiments.
Most U-STAT1 Is in the Nuclei of BJ and hTERT-HME1 Cells.
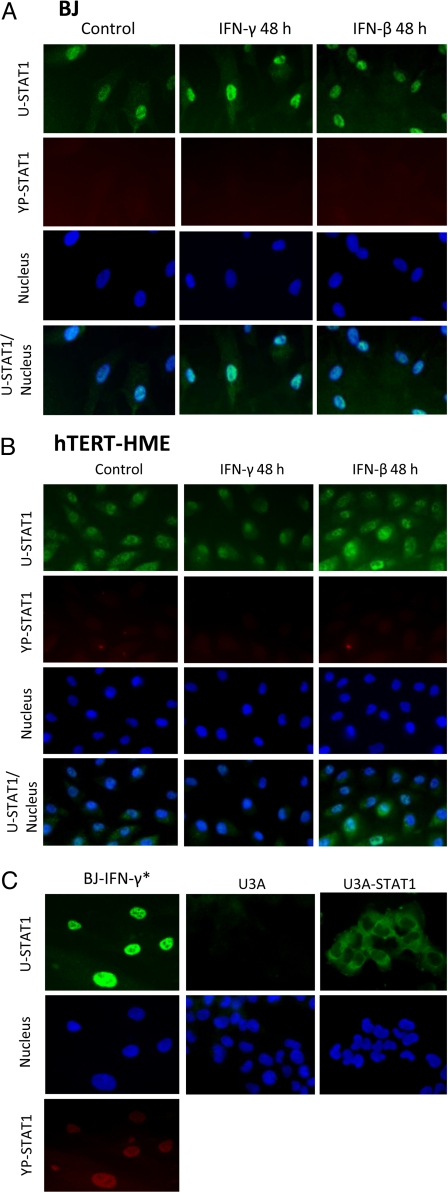
U-STAT1 must be in the nucleus to induce gene expression and, interestingly, most STAT1 is seen in the nuclei of BJ cells even without IFN treatment (Fig. 4A, Control). When BJ or hTERT-HME1 cells were treated for 6 h with the same concentrations of IFN-β or IFN-γ used in Fig. 2B, small amounts of YP-STAT1 could be detected in the nuclei. After 48 h, no YP-STAT1 signal was detected (Fig. 4 A and B), as expected from the data of Fig. 2B. As a positive control, the cells were treated with a higher concentration of IFN-γ (3 ng/mL) for 1 h, and the signal was clearly seen in the nuclei (Fig. 4C). After 48 h, most of the U-STAT1 that had been increased in response to IFN-β or IFN-γ was localized in nuclei. (The monoclonal anti-STAT1 reagent used detects only U-STAT1 and not YP-STAT1.) In hTERT-HME1 cells, U-STAT1 was distributed between cytoplasm and nuclei, but with more in the nuclei (Fig. 4B). After 48 h of treatment with low concentrations of IFNs, U-STAT1 was also mainly in the nuclei. These results show that the U-STAT1 induced by IFNs moves into nuclei, where it can function to increase the expression of immune regulatory genes. As a negative control, we stained STAT1-null U3A cells with the same antibodies and did not detect any signal (Fig. 4C). As further confirmation that the nuclear staining with U-STAT1 antibody is not an artifact, in U3A cells transfected with wild-type STAT1, the protein was clearly localized in the cytoplasm (Fig. 4C). Interestingly, the relative amount of U-STAT1 in nuclei compared with cytoplasm varied, even from cell to cell in the same culture dish.
Fig. 4.
Most U-STAT1 is located in nuclei of untreated and IFN-treated BJ or hTERT-HME1 cells. (A) BJ cells were treated with 0.3 ng/mL IFN-γ or 3 units/mL IFN-β for 48 h. Immunocytochemistry was performed with antibodies against U-STAT1 (green) or YP-STAT1 (red). Nuclei were stained with DAPI. YP-STAT1 was not seen in untreated cells or in cells treated with IFNs for 48 h. (B) hTERT-HME1 cells were treated with 0.1 ng/mL IFN-γ or 5 units/mL IFN-β for 48 h. YP-STAT1 was not seen in untreated cells or in cells treated with IFNs for 48 h. (C) As a positive control for PY-STAT1 staining, cells were treated with 3 ng/mL IFN-γ (IFN-γ*) for 1 h, which shows PY-STAT1 staining inside nuclei. U3A cells (STAT1-null 2fTGH cells) were used as a negative control, with same antibodies used in A and B. Untreated U3A cells transfected with wild-type STAT1 show clear cytoplasmic localization of U-STAT. The magnification of the images is ×400.
Discussion
YP-STAT1 induces transcription rapidly in response to type I or type II IFNs. The tyrosine phosphorylation of STAT1 is transient, and the induction of the genes that respond to it, such as SOCS1, also decreases as the concentration of YP-STAT1 decreases. However, as shown here, the induced levels of more than 100 genes remain high even 48 h after stimulation by IFN. Some of these genes are induced strongly at early times, but the levels of their mRNAs gradually decrease. However, the levels of some induced mRNAs are low at early times and gradually increase, or are high at early times and remain high later, suggesting that there are secondary mechanisms to transcribe those genes. In this study, we studied the role of U-STAT1 as one of the secondary mechanisms that induce late gene expression because the expression of STAT1 is substantially increased by IFN treatment (16). Our previous work suggested that U-STAT1 induces constitutive gene expression (10, 11), but its role in IFN signaling has not been studied. Here, we used the “normal” human cell lines BJ and hTERT-HME1 instead of STAT1-null U3A cells, which are derived from the human fibrosarcoma line HT1080.
High levels of U-STAT1, obtained by transfection with a vector encoding STAT1, increased the expression of many genes in BJ fibroblasts, including IFI27, BST2, OAS1, OAS2, OAS3, IFI44, and STAT1 itself. All of these proteins are involved in immune responses. For unknown reasons, we did not see an increase in the expression of these genes in hTERT-HME1 cells that express high levels of U-STAT1. However, the same genes were highly expressed 48 h after stimulation of either BJ or hTERT-HME1 cells with IFN-γ or IFN-β.
All of the genes regulated by U-STAT1 are well known to be induced also by IFNs, and in the current study IFN-γ and IFN-β did increase their expression after 6 h, as well as after 48 h. However, most of these U-STAT1-induced genes were expressed more after 48 h than after 6 h, or were expressed similarly. If an mRNA were not degraded at all after its synthesis had ceased, its amount would decrease over a period of 48 h because of dilution by continued cell growth. Therefore, we conclude that the mRNAs induced at late times in IFN-treated cells must have been synthesized after YP-STAT1 had disappeared, in response to newly synthesized U-STAT1.
In addition to the rapid activation of STAT1 gene expression in response to YP-STAT1, the STAT1 gene is also induced by U-STAT1. The STAT1 protein remains stable for several days (Fig. 1). The expression of OAS1, OAS2, OAS3, and Mx1, essential for a complete antiviral response to IFNs, was also increased by U-STAT1. OAS enzymes catalyze the synthesis of 2′, 5′-linked oligoadenylates, which bind to RNase L, an endoribonuclease, leading to the cleavage of single-stranded mRNA and rRNA, thereby inhibiting protein synthesis (19). Mx proteins block replication of the infecting virus soon after cell entry (20). The fact that IFN-induced U-STAT1 prolongs the expression of these effector proteins may help cells to clear viruses completely, even after the initial response to IFN and the production of IFN have been down-regulated. IFI27 (also called ISG12) and BST2 are the genes most highly induced by U-STAT1, and their expression is also dramatically increased after 48 h of IFN treatment, compared with their expression after 6 h. IFI27 belongs to a family of small IFN-α-inducible genes, the function of which is unknown. Primary human breast carcinoma cells express high levels of IFI27. It was originally cloned as an estrogen-inducible gene in the human epithelial cell line MCF-7 (21). The expression level did not correlate with the presence of estrogen receptor (21), and the cause of its overexpression in cancer cells has not been understood. Perou et al. (22) showed that breast tumor tissues express high levels of IFN-regulated genes, including OAS1 and STAT1, compared with the human mammary epithelial cell line HME1. From results presented here, it is reasonable to propose that up-regulated STAT1 may enhance IFI27 and OAS1 expression in breast tumors. The function of BST2 also remains unknown but may be important in sorting membrane and secreted proteins in the Golgi apparatus, localized on both cell surface and intracellular compartments (23). The expression of BST2 was predominantly specific for type I IFN-producing cells in the naïve mouse and was up-regulated in most cell types after stimulation with type I IFNs or IFN-γ (24).
In our array data, TAP1 and IRF1, which are also well-known immune regulatory genes, were highly induced by IFN-γ after 6 h but decreased after 48 h. High concentration of U-STAT1 did not change the expression of those genes. Many genes in MHC classes I and II are induced by IFN after 6 h, and their expression is further increased after 48 h. However, they are not induced by exogenous STAT1 expression, suggesting these genes are induced late through mechanisms other than the ones mediated by U-STAT1. Only the group of immune regulatory genes is induced by U-STAT1 at late times after IFN stimulation.
Because unphosphorylated STATs were initially assumed to be latent transcription factors, becoming active only after cytokine stimulation, it has been thought that increased levels of STAT1 might intensify the effects of subsequent IFN challenge by increasing the amount of STAT1 phosphorylation. However, in our experiments, the amount of U-STAT1 seems not to have affected the magnitude of response to IFN stimulation: The increased STAT1 was not completely phosphorylated in response to IFN stimulation and did not make the cells more sensitive to IFN (Fig. S2). Other studies have confirmed this conclusion. The increase in STAT1 levels due to prior exposure to a low dose of IFN (priming) changed the effects of subsequent IFN stimulation not only positively but also negatively, in a cell type-specific manner (9). Priming with IFN-γ increased the levels of IFN-α-induced STAT1 homodimers or ISGF3 but decreased IFN-γ-induced STAT1 homodimers (16). Tassiulas et al. (25) showed that a high level of STAT1 was not sufficient to enhance IFN-α-induced STAT1 activation and gene expression after IFN-γ-priming, but the tyrosine kinase Syk and immunoreceptor tyrosine activation motifs were required. In clinical data, the expression level of STAT1 does not influence the response to IFN-α adjuvant therapy for cancer patients, and STAT1 levels were even greater in recurrent tumors compared with original tumors (26). Only a high level of activation of STAT1 (phosphorylation and DNA-binding activity) in primary tumors serves as a significant indicator of good prognosis and longer survival (27). These results also suggest that U-STAT1 has functions in addition to the traditional ones that are mediated through its phosphorylation, not only in IFN-dependent signaling but also in tumorigenesis.
Our previous studies have already shown that unphosphorylated STAT3 is an active transcription factor (17, 18). The work presented here reveals interesting similarities and differences between U-STAT1 and U-STAT3. First, both U-STAT1 and U-STAT3 are functional transcription factors independent of their phosphorylation. U-STAT3 induces the expression of well-known oncogenes, such as MRAS and MET, which are late-phase genes induced by IL-6. Similarly, U-STAT1 induces many immune regulatory genes, which are also genes strongly induced late by IFN-γ and IFN-β. However, U-STAT1 and U-STAT3 induce completely different sets of genes, and the genes induced by each are closely related to the functions of their inducer cytokines. Second, both U-STAT1 and U-STAT3 induce genes through more than one mechanism. Among the genes induced by U-STAT3, RANTES is induced by binding of a U-STAT3/U-NF-κB complex to a κB site on its promoter, but NF-κB is not involved in the expression of MRAS in response to U-STAT3 (18). We have not yet investigated the detailed mechanism of U-STAT1-induced gene expression, but according to our previous data, a complex of U-STAT1 and IRF1 binds to overlapping IFN consensus sequence 2 (ICS2) and GAS elements on LMP2 promoter to induce the expression of this gene (11). We found an element (TTCNNGGAAANTGAAAC), in which a GAS (TTCNNGGAA) and IRF1 (GGAAANTGAAACN) consensus sequence overlap, on the promoters of the IFI27, the gene most strongly induced by U-STAT1, suggesting the possibility that this gene may be induced by a U-STAT1/IRF1 complex. However, we do not see a similar GAS/IRF1 element in the promoters of other U-STAT1-induced genes. Third, both U-STAT1 and U-STAT3 must be localized in nuclei to function as transcription factors. U-STAT3 is distributed between both nuclei and cytoplasm, to an extent that depends on the cell type, and its import is independent of tyrosine phosphorylation (28). Here, we found that U-STAT1 is also localized extensively in nuclei, more than in the cytoplasm in resting normal human fibroblasts and epithelial cells, and IFN-induced U-STAT1 is also located in nuclei. Our previous study also showed that U-STAT1 predominates in the nuclei of 2fTGH or U3A cells that have been transfected with Y701F-STAT1 (11), and recent repeats of this experiment gave the same results. Meyer et al. (13) extensively studied the localization of U-STAT1 in many transformed cell lines and primary cell cultures, finding that U-STAT1 is present in nuclei independently of tyrosine phosphorylation, in a cell type-specific manner. The same group also showed that YP-STAT1 and U-STAT1 shuttle via independent pathways to distinct sets of target genes (14). We observed that the relative amounts of U-STAT1 in nuclei are different even within a given cell type, depending on the cell density, suggesting that the translocation of U-STAT1 is regulated by unknown factors. YP-STAT1 and U-STAT1 seem to be almost completely different transcription factors, binding to different elements in the promoters of different sets of genes. Nuclear U-STAT1 did not bind to an oligonucleotide to which YP-STAT1 bound, but cytoplasmic YP-STAT1 did bind to the same probe (13). In our previous study (11), we concluded that U-STAT1 binds to the composite ICS-2/GAS element of the LMP2 gene, but that only YP-STAT1 binds to the simple GAS element of the IRF1 gene.
In summary, we have elucidated a novel mechanism that regulates IFN-induced late gene expression: the U-STAT1 that is highly induced in response to IFNs participates in a positive feedback loop that enhances immune responses and the clearance of viruses.
Materials and Methods
Cells and Reagents.
BJ cells were grown in DMEM supplemented with 5% FBS penicillin (100 units/mL) and streptomycin (100 μg/mL). hTERT-HME1 cells were grown in mammary epithelium growth media containing bovine pituitary extract, hydrocortisone, insulin, epithelial growth factor, and gentamicin/amphotericin-B (Clonetics). Human IFN-γ was from Genentech, and human IFN-β was from Biogen Idec. Mouse monoclonal antibody against STAT1 (C-terminal; BD Transduction Laboratories) and rabbit polyclonal antibody against tyrosine-phosphorylated STAT1 (Cell Signaling Technology) were used for Western blot analyses and immunocytochemistry.
Constructs and Gene Transfection.
Lentiviral vectors expressing human wild-type STAT1 or Y701F-STAT1 were kindly provided by Ganes Sen's laboratory (Cleveland Clinic). The coding region of STAT1 was cloned in the lentiviral vector pLV-tetO-CMV-SV40-Puro-LoxP, developed in Andrei Gudkov's laboratory at the Cleveland Clinic (details available upon request). To produce infectious virus, each construct was transfected into 293T packaging cells by using Lipofectamine Plus (Invitrogen). The supernatant medium, collected twice every 24 h, was used to infect cells. To select stably transfected cells, they were treated with 1 μg/mL puromycin for more than 2 weeks. Proteins or RNAs were purified from these cell pools for Western blot or microarray analyses.
Gene Expression Analysis.
Total RNA from BJ or hTERT-HME1 cells was purified with TRIzol (Invitrogen) and RNeasy Mini Kit (Qiagen), and 1 μg of this RNA was used for microarray analysis on an Illumina Sentrix Human Ref-8 Expression Bead Chip. The analysis was carried out in duplicate with each kind of RNA, and the mean of the duplicates was used for further analysis only if the expression patterns between the duplicate were close. The data were normalized by the quantile method, and differential expression analysis was run with references of vector-transfected cells or untreated cells. Genes were selected according to the criteria of differential P values ≤0.05 and average signals >25.
Real-Time PCR.
cDNA was synthesized from total RNA by using a modified manufacturer's protocol with random hexamer and SuperScript II (Invitrogen). Real-time PCR was performed with SYBR Green qPCR master mix (USB) in an iCycler iQ real-time PCR detection system (Bio-Rad). The PCR protocol: initial activation at 95 °C for 5 min, 40 cycles at 95 °C for 15 sec, and 60 °C for 1 min. Aliquots of standard cDNA were included in each PCR run, and standard curves for each gene were generated by linear regression. Ct values were converted to gene expression levels by using standard curves. Each gene expression value was normalized to the expression level of GAPDH. Each PCR run also included nontemplate controls containing all reagents except cDNA, which generated no amplification. The specificity was confirmed by analysis of the melting curves of the PCR products.
Western Blot Analysis.
Cells were resuspended in lysis buffer (250 mM Tris, pH 8.0; 150 mM NaCl; 1% Triton; and 0.1% SDS) containing protease inhibitors (1 mM PMSF, 100 μg/mL aprotinin, and 1 μg/mL leupeptin) and phosphatase inhibitors (10 mM sodium fluoride, 5 mM sodium pyrophosphate, and 1 mM sodium orthovanadate). After incubation on ice for 10–20 min, cell debris was removed by centrifugation. Protein (10–20 μg) was loaded onto 8% SDS/PAGE gels. The separated proteins were transferred onto PVDF membranes (Millipore). The membranes were incubated with primary antibody for 1–2 h, followed by incubation with secondary antibody for 1 h at room temperature.
Immunocytochemistry.
Cells grown on coverslips in 6-well plates were fixed with 4% paraformadehyde for 10 min and with methanol for 5 min. The cells were incubated in blocking solution (10% FBS and 0.3% Triton X-100 in PBS), and the primary antibody, diluted in blocking solution, was applied. After incubation at 4 °C overnight, the cells were incubated with Alexa488-conjugated goat anti-mouse and Alexa594-conjugated goat anti-rabbit antibodies (Invitrogen) diluted in blocking solution for 1 h. Mounting was done with Vectashield mounting medium with DAPI (Vector Laboratories), and the cells were examined with a fluorescence microscope (Leica DMR).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903487106/DCSupplemental.
References
- 1.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 2.Stark GR. How cells respond to interferons revisited: From early history to current complexity. Cytokine Growth Factor Rev. 2007;18:419–423. doi: 10.1016/j.cytogfr.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uddin S, et al. Activation of the p38 mitogen-activated protein kinase by type I interferons. J Biol Chem. 1999;274:30127–30131. doi: 10.1074/jbc.274.42.30127. [DOI] [PubMed] [Google Scholar]
- 4.Sizemore N, et al. Inhibitor of κB kinase is required to activate a subset of interferon γ-stimulated genes. Proc Natl Acad Sci USA. 2004;101:7994–7998. doi: 10.1073/pnas.0401593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qing Y, Stark GR. Alternative activation of STAT1 and STAT3 in response to interferon-γ. J Biol Chem. 2004;279:41679–41685. doi: 10.1074/jbc.M406413200. [DOI] [PubMed] [Google Scholar]
- 6.Gil MP, et al. Biologic consequences of Stat1-independent IFN signaling. Proc Natl Acad Sci USA. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klampfer L, et al. STAT1-independent inhibition of cyclooxygenase-2 expression by IFN γ; a common pathway of IFN γ-mediated gene repression but not gene activation. Oncogene. 2007;26:2071–2081. doi: 10.1038/sj.onc.1210015. [DOI] [PubMed] [Google Scholar]
- 8.Wang K, et al. Inhibition of neutrophil apoptosis by type 1 IFN depends on cross-talk between phosphoinositol 3-kinase, protein kkinase C-δ, and NF-κB signaling pathway. J Immunol. 2003;171:1035–1041. doi: 10.4049/jimmunol.171.2.1035. [DOI] [PubMed] [Google Scholar]
- 9.Van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Kumar A, Commane M, Flickinger TW, Horvath CM, Stark GR. Defective TNF-α-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science. 1997;278:1630–1632. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- 11.Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. How Stat1 mediates constitutive gene expression: A complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui X, et al. Unphosphorylated STAT6 contributes to constitutive cyclooxygenase-2 expression in human non-small cell lung cancer. Oncogene. 2007;26:4253–4260. doi: 10.1038/sj.onc.1210222. [DOI] [PubMed] [Google Scholar]
- 13.Meyer T, Gavenis K, Vinkemeier U. Cell type-specific and tyrosine phosphorylation-independent nuclear presence of STAT1 and STAT3. Exp Cell Res. 2002;272:45–55. doi: 10.1006/excr.2001.5405. [DOI] [PubMed] [Google Scholar]
- 14.Meyer T, Begitt A, Lodige I, van Rossum M, Vinkemeier U. Constitutive and IFN-γ-induced nuclear import of STAT1 proceed through independent pathway. EMBO J. 2002;21:344–354. doi: 10.1093/emboj/21.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer T, Vinkemeier U. Nucleocytoplasmic shuttling of STAT transcription factors. Eur J Biochem. 2004;271:4606–4612. doi: 10.1111/j.1432-1033.2004.04423.x. [DOI] [PubMed] [Google Scholar]
- 16.Lehtonen A, Matikainen S, Julkunen I. Interferons up-regulate STAT1, STAT2, and IRF family transcription factor gene expression in human peripheral blood mononuclear cells and macrophages. J Immunol. 1997;159:794–803. [PubMed] [Google Scholar]
- 17.Yang J, et al. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–947. [PubMed] [Google Scholar]
- 18.Yang J, et al. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFκB. Genes Dev. 2007;21:1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haller O, Kochs G, Weber F. Interferon, Mx, and viral countermeasures. Cytokine Growth Factor Rev. 2007;18:425–433. doi: 10.1016/j.cytogfr.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen UB, et al. Identification of a new interferon-α-inducible gene (p27) on human chromosome 14q32 and its expression in breast carcinoma. Cancer Res. 1993;53:4096–4101. [PubMed] [Google Scholar]
- 22.Perou CM, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kupzig S, et al. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic. 2003;4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 24.Blasius AL, et al. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J Immunol. 2006;177:3260–3265. doi: 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- 25.Tassiulas I, et al. Amplification of IFN-α-induced STAT1 activation and inflammatory function by Syk and ITAM-containing adaptors. Nat Immunol. 2004;5:1181–1189. doi: 10.1038/ni1126. [DOI] [PubMed] [Google Scholar]
- 26.Lesinski GB, et al. Expression of STAT1 and STAT2 in malignant melanoma does not correlate with response to interferon-alpha adjuvant therapy. Cancer Immunol Immunother. 2005;54:815–825. doi: 10.1007/s00262-004-0649-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Widschwendter A, et al. Prognostic significance of signal transducer and activator of transcription 1 activation in breast cancer. Clin Cancer Res. 2002;8:3065–3074. [PubMed] [Google Scholar]
- 28.Liu L, McBride KM, Reich NC. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-α3. Proc Natl Acad Sci USA. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.