Abstract
The biogenesis and function of chloroplast are controlled both by anterograde mechanisms involving nuclear-encoded proteins targeted to chloroplast and by retrograde signals from plastid to nucleus contributing to regulation of nuclear gene expression. A number of experimental evidences support the implication of chlorophyll biosynthesis intermediates on the retrograde signaling, albeit an earlier-postulated direct link between accumulation of chlorophyll intermediates and changes in nuclear gene expression has recently been challenged. By characterization of Arabidopsis mutants lacking the chloroplast localized NADPH-thioredoxin reductase (NTRC) we have recently proposed that imbalanced activity of chlorophyll biosynthesis in developing cells modifies the chloroplast signals leading to alterations in nuclear gene expression. These signals appear to initiate from temporal perturbations in the flux through the pathway from protoporphyrin to protochlorophyllide rather than from the accumulation of a single intermediate of the tetrapyr-role pathway.
Key words: chloroplast biogenesis, NADPH-thioredoxin reductase, porphyrins, ROS, signaling, tetrapyrrole, thioredoxin
Orchestrated regulation of gene expression in the nucleus and plastids is crucial for the proper biogenesis of the organelle during the development and for the acclimation of plants to environmental cues. Multiple potential candidates for initiating plastidial signals have been recognized, including intermediates of the tetrapyrrole biosynthetic pathway, redox state of chloroplast electron transfer components and reactive oxygen species (ROS). These multiple signaling pathways are likely to interact with each others, resulting in a complex signaling network between plastid and nucleus (reviewed in ref. 1).
Control of Nuclear Gene Expression by the Intermediates of Plastidial Tetrapyrrole Pathway
The tetrapyrrole biosynthetic pathway resides in plastids and produces a number of important cofactors for cellular apoproteins, including chlorophylls, heme and the chromophore for the red light photoreceptors.2,3 The pathway is regulated on multiple levels in cells to ensure coordinated productions of ligands and apoproteins and to avoid accumulation of harmful phototoxic intermediates of the pathway. Both the mutations in the genes of tetrapyrrole biosynthesis and the treatment with herbicide Norflurazon have evidenced that the intermediates of tetrapyrrole pathway are involved in retrograde signaling from chloroplast to nucleus.4–8 Photobleaching of chloroplasts by Norflurazon has shown to repress concomitantly the expression of photosynthetic genes in the nucleus.6,7 Norflurazon treatments were used to screen Arabidopsis mutants with defects in retrograde signaling pathway, which resulted in the isolation of genome uncoupled (GUN) mutants. In four out of five gun mutants, the mutated genes encode the tetrapyrrole biosynthetic enzymes or their regulatory components (Fig. 1, reviewed in ref. 7). Characterization of the gun mutants led to the construction of a model of plastid-to-nucleus retrograde signaling, in which the accumulation of Mg-protoporphyrin IX (MgProto) in plastids initiates the signal repressing the expression of nuclear photosynthetic genes.1,6,7 MgProto has also been reported to exit plastid and translocate to cytoplasm9 and the AP2-type transcription factor ABI4 has been shown to act downstream of MgProto in nucleus.10
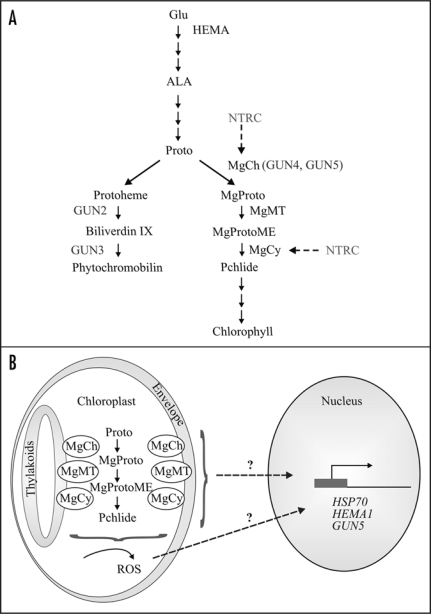
Figure 1.
(A) Schematic pathway of tetrapyrrole biosynthesis in higher plants. The enzymes and gun mutants discussed in the text are marked in the pathway. Glu, glutamate; ALA, 5-aminolevulinic acid; Proto, protoporhyrin IX; MgProto, Mg-protoporhyrin; MgProtoME, Mg-protoporhyrin monomethyl ester; Pchlide, divinyl protochlorophyllide; HEMA1, glytamyl-tRNA reductase; MgCh, Mg-chelatase; MgMT, MgProto methyltransferase; Mg-Cy, Mg-protoporphyrin IX-monomethyl ester cyclase. (B) The tentative model for chloroplast retrograde signaling involving intermediates of chlorophyll biosynthesis. The porphyrin intermediates between protoporphyrin IX and protochlorophyllide may initiate the retrograde signal controlling the gene expression in nucleus. Alternatively, the intermediates may induce production of ROS that in turn activates the signaling cascade. For other details, see the text.
Recently this model has been challenged by extended analyses of tetrapyrrole intermediates in wild type and gun mutants treated with and without Norflurazon under various growth conditions.11,12 In these studies, no correlation between the expression of photosynthetic nuclear genes and the accumulation of tetrapyrrole intermediates was detected. Neither did the loss-of-function mutants of the enzymes catalyzing the biosynthetic reactions from Proto to divinyl protochlorophyllide (Pchlide) show any changes in photosynthetic gene expression, as would have been predictable from the model.3,8 Nevertheless, results obtained upon incubation of algal and plant cells with porphyrins speak for the involvement of tetrapyrrole pathway on the chloroplast retrogarde signaling. In Chlamydomonas, the treatments of cells with MgProtoandMgProtomethylester(MgProtoME) substituted for light in the induction of nuclear HSP70s and HEMA genes encoding chaperones and glytamyl-tRNA reductase of tetrapyrrole pathway, respectively.13,14 von Gromoff et al.15 identified a cis-acting sequence in the promoter region of HSP70A that is employed for induction by both MgProto and light in Chlamydomonas. Furthermore, a recent elegant paper by Kobayashi et al.16 provides substantial experimental evidences for regulation of nuclear DNA replication by a tetrapyrrole signal in plant cells. It was shown that in synchronized cultures of unicellular red alga and tobacco BY-2 cells, the organelle DNA replication preceded and was a prerequisite for nuclear DNA replication in the cell cycle. Additions of Proto and MgProto, however, substituted for the requirement of organelle DNA replication before the nuclear DNA replication. They also showed that organelle DNA replication, as well as porphyrins in the absence of organelle DNA replication, activated an A-type cyclin-dependent kinase that controls the nuclear DNA replication in the G1-S phase transition of the cell cycle. These reports indicate that plastidial retrograde signals may not coordinate only nuclear genes for the photosynthetic machinery but also the cell cycle and stress responses in plant cells.
Distorted Chlorophyll Biosynthesis Causes Modifications in Nuclear HSP Gene Expression and Cell Division
Our studies on knockout lines of nuclear gene encoding chloroplast NADPH-thioredoxin reductase (NTRC) give support to the role of porphyrins as retrograde signals in Arabidopsis.17 Thioredoxin reductases are members of cellular thioredoxin systems that control the dithiol-disulphide exchange in proteins.18 Besides the severe defects in growth, the ntrc knockout lines had strongly reduced chlorophyll content in leaves.17 It is thus conceivable that NTRC regulates the tetrapyrrole pathway in chloroplasts, and that the knockout of this enzyme modifies the flux through the biosynthetic pathway from Proto to Pchlide (Fig. 1). Further, we propose that the imbalanced production of tetrapyrrole intermediates induces a plastidial signal that modifies nuclear gene expression in ntrc plants either directly or indirectly via production of ROS17 (Fig. 1B). This assumption is based on the regulatory mechanisms of the first enzymes in chlorophyll biosynthetic branch and on the structural and biochemical phenotypes of knockout ntrc plants. These aspects can be summarized by the following points. (i) Feeding of 5-aminolevulinic acid to Arabidopsis in darkness resulted in increased accumulation of Proto, MgProto and MgProto methyl ester and reduced amount of Pchlide in ntrc compared to wild type,19 suggesting an imbalanced flux through Proto to Pchlide in ntrc leaf cells. (ii) This may result from reduced activity of Mg-chelatase in the absence of NTRC because this enzyme is regulated via thioredoxin-mediated disulphide/dithiol exchange.20 Furthermore, in the presence of chloroplast 2-Cys peroxiredoxin NTRC has been shown to stimulate the activity of Mg-protoporphyrin IX-monomethyl ester cyclase (MgCy), the enzyme catalyzing the reaction of chlorophyll biosynthesis downstream from Mg-chelatase.19 Interestingly, these enzymes together with MgProto methyltransferase are dually localized both in thylakoid membranes and in the inner chloroplast envelope membrane (reviewed in ref. 2), which provides a natural and rapid route for transporting signals from plastid to the cytoplasm. (iii) Four nuclear genes encoding enzymes in the tetrapyrrole pathway (including GUN5 and HEMA1) and six genes encoding cytoplasmic heat shock proteins were found to be upregulated in ntrc leaves.17 GUN5 transcripts, encoding the H subunit of Mg chelatase, showed the highest differential accumulation in ntrc line compared to wild type. HEMA gene and genes encoding HSP70 proteins were also induced in Chlamydomonas after treatment with MgProto and MgProtoMe in darkness.13,14 The Arabidopsis genome contains several HSP70 genes encoding cytoplasmic HSP70 proteins, but it is intriguing to note that the upregulated genes in ntrc (At3g12580 and At3g09440) encode cytoplasmic proteins that are the closest homologues to the single cytoplasmic HSP70A gene in Chlamydomonas.21 (iv) The pale green ntrc leaves have higher number of cells per area than wild type Arabidopsis and significantly reduced cell size with low number of chloroplasts,17 suggesting that the plastidial and nuclear divisions are uncoupled in ntrc plants. This might be explained by the substitution of plastidial DNA replication with tetrapyrrole signal in the control of nuclear division, like suggested recently by Kobayashi et al.16
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/8711
References
- 1.Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet. 2008;9:383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckhardt U, Grimm B, Hörtensteiner S. Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol Biol. 2004;56:1–14. doi: 10.1007/s11103-004-2331-3. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka R, Tanaka A. Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol. 2007;58:321–346. doi: 10.1146/annurev.arplant.57.032905.105448. [DOI] [PubMed] [Google Scholar]
- 4.Papenbrock J, Mock H-P, Tanaka R, Kruse E, Grimm P. Role of Magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol. 2000;122:1161–1169. doi: 10.1104/pp.122.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brusslan JA, Peterson MP. Tetrapyrrole regulation of nuclear gene expression. Photosynth Res. 2002;71:185–194. doi: 10.1023/A:1015539109209. [DOI] [PubMed] [Google Scholar]
- 6.Strand A, Asami T, Alonso J, Ecker JR, Chory J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature. 2003;421:79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- 7.Nott A, Jung H, Koussevitzky S, Chory J. Plastid to nucleus retrograde signalling. Annu Rev Plant Biol. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- 8.Pontier D, Albrieux C, Joyard J, Lagrange T, Block MA. Knock-out of the magnesium protoporphyrin IX methyltransferase gene in Arabidopsis. Effects on chloroplast development and on chloroplast-to-nucleus signalling. J Biol Chem. 2007;282:2297–2304. doi: 10.1074/jbc.M610286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ankele E, Kindgren P, Pesquet E, Strand A. In vivo visualization of Mg-protoporphyrin IX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell. 2007;19:1964–1979. doi: 10.1105/tpc.106.048744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- 11.Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. The steady-state level of mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signalling in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:15184–15189. doi: 10.1073/pnas.0803245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moulin M, McCormac AC, Terry MJ, Smith AG. Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signalling is not due to Mg-protoporphyrin IX accumulation. Proc Natl Acad Sci USA. 2008;105:15178–15183. doi: 10.1073/pnas.0803054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kropat J, Oster U, Rüdiger W, Beck CF. Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc Natl Acad Sci USA. 1997;94:14168–14172. doi: 10.1073/pnas.94.25.14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasileuskaya Z, Oster U, Beck CF. Involvement of tetrapyrroles in inter-organellar signaling in plants and algae. Photosynth Res. 2004;82:289–299. doi: 10.1007/s11120-004-2160-x. [DOI] [PubMed] [Google Scholar]
- 15.von Gromoff ED, Schroda M, Oster U, Beck CF. Identification of a plastid response element that acts as an enhancer within the Chlamydomonas HSP70A promoter. Nucl Acids Res. 2006;34:4767–4779. doi: 10.1093/nar/gkl602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashia Y, Kanesakia Y, Tanakab A, Kuroiwac H, Kuroiwac T, Tanaka K. Tetrapyrrole signal as a cell cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc Natl Acad Sci USA. 2009;106:803–807. doi: 10.1073/pnas.0804270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lepistö A, Kangasjärvi S, Luomala EM, Brader G, Sipari N, Keränen M, et al. Chloroplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis. Plant Physiol. 2009;149:1261–1276. doi: 10.1104/pp.108.133777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchanan BB, Balmer Y. Redox regulation: A broadening horizon. Annu Rev Plant Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- 19.Stenbaek A, Hansson A, Wulff RP, Hansson M, Dietz KJ, Jensen PE. NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg-protoporphyrin monomethyl ester cyclase. FEBS Lett. 2008;582:2773–2778. doi: 10.1016/j.febslet.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Ikegami A, Yoshimura N, Motohashi K, Takahashi S, Romano PG, Hisabori T, et al. The CHLI1 subunit of Arabidopsis thaliana magnesium chelatase is a target protein of the chloroplast thioredoxin. J Biol Chem. 2007;282:19282–19291. doi: 10.1074/jbc.M703324200. [DOI] [PubMed] [Google Scholar]
- 21.Renner T, Waters ER. Comparative genomic analysis of the Hsp70s from five diverse photosynthetic eukaryotes. Cell Stress Chap. 2007;12:172–185. doi: 10.1379/CSC-230R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]