Abstract
Various means employed to solve problems associated with the ends (telomeres) of linear DNA chromosomes exhibit one common feature: generation of both intra- and intercellular heterogeneity of telomeres at the level of their structural and functional states. We argue that this heterogeneity is not a simple by-product of molecular pathways mediating telomere maintenance. Instead, we propose that these mechanisms were selected because they generate heterogeneity. Similarly as noise in gene expression, stochastic events at telomeres may have an adaptive value allowing cells to sustain viable and flexible populations, with implications for fields ranging from evolutionary biology to molecular medicine.
Keywords: Telomere, stochasticity, heterogeneity
1. Innumerable telomeres in the ‘endless’ Universe
There are approximately 6 × 109 people currently living on Earth. Each individual consists of about 1013 cells, each cell containing 46 chromosomes carrying two specialized terminal nucleoprotein structures (telomeres; [1]). Based on these assumptions, there are currently 5.5 × 1024 human telomeres. If the average length of a human telomeric fragment is approximately 10,000 bp and if the distance between two nucleotides in DNA is 0.3 × 10−12 km, the total length of all currently existing human telomeres would reach 1.6 × 1016 km. If there are about 107 eukaryotic species presently living on Earth, each species consisting of thousands to billions individuals each individual containing thousands to trillions of cells and each cell holding a nucleus with several linear chromosomes carrying two telomeres, whose size can vary from few hundred to several thousands of base pairs. Although the exact number and total length of eukaryotic nuclear telomeres cannot be reasonably estimated it can be expressed in terms of hundreds thousands of light years. Even if we have exact numbers for the above parameters, the total number of telomeres would still be underestimated by several orders of magnitude, because it would ignore linear DNA genomes of numerous viruses, prokaryotes, plasmids and organellar genomes. A wide occurrence of linear DNA genophores indicates that it may bring its host a selective advantage, although its nature is far from being understood [2,3]. Clearly, having chromosomes with defined physical ends can provide qualitatively new opportunities not available for circular DNA genomes, including meiotic recombination [4], novel control mechanisms for genophore segregation [2], compartmentalization [5] or gene expression [6].
2. Different types of telomeres provide alternative mechanisms of their maintenance
Linearity of DNA molecules generates problems concerning their termini including the end-replication problem, protection against nucleolytic attacks and inappropriate DNA repair [1]. There are many types of telomeric structures representing solutions to these obstacles. Telomeres consisting of double-stranded DNA tract of short repeats terminating with single-stranded 3′ overhang that is elongated through the activity of the reverse transcriptase telomerase, is the most promiscuous solution to the end-replication problem in eukaryotic nuclei [1]. However, there are several other independently evolved types of telomeres found in eukaryotic (both nuclear and organellar), prokaryotic and viral genomes including telomeric arrays (t-arrays) of tandem repeats, telomeric hairpins/palindromes (t-hairpins/t-palindromes), telomeric proteins (t-proteins) covalently attached to the 5′ ends, or telomere-associated retrotransposons (t-posons) [2].
3. Heterogeneity in state and/or length is a common feature of various types of telomeres
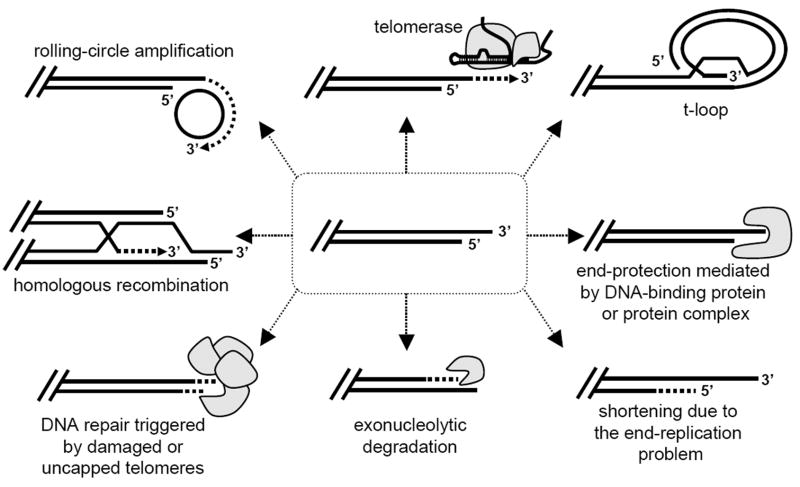
The architecture of various types of telomeres represents a basis for their ability to acquire different states (Figure 1). In general terms, telomeres may adopt various types of open or closed (capped) conformations differing in their susceptibility to the machineries involved in their maintenance [7]. A formation of a particular state is governed by a number of proteins, whose access to the chromosomal end is regulated by various means including gene expression, protein-protein interactions, or posttranslational modifications [8].
Figure 1. Telomeres exist in multiple structurally and functionally different states.
A typical verterbrate telomere is composed of tandem array of short (e.g. 6 bp) repeat motifs terminating with a 3′ single-stranded overhang (central part of the scheme) is subject of diverse transactions inherently including stochastic phenomena. The stochastic character of telomere transactions results in intra- and intercellular heterogeneity in their structural and functional states with potential adaptive value for clonal cell populations (see text and Table 1 for details).
One of the unifying characteristics of several types of telomeres is their ability to loop back and following the invasion of a 3′ single-stranded overhang into the double-stranded telomeric region form a telomere loop (t-loop) [9,10]. The formation of t-loops is mediated by telomere-binding proteins like mammalian TRF2 [11], or fission yeast Taz1 [12]. T-loops have been visualized not only in nuclei of a variety of eukaryotic organisms [10], but also in mitochondria of Candida parapsilosis harboring t-array type of linear mitochondrial telomeres [13]. A tendency to form fold-back structures is apparent even in cases when the formation of a true t-loop might not be possible like in the case of Saccharomyces cerevisiae telomeres [14].
Different states of a telomere differ in their susceptibility to recombination. Especially in the absence of telomerase, telomeres may be maintained by recombinational mechanisms representing the main means of alternative lengthening of telomeres (ALT) found in telomerase-negative tumors [15]. In numerous situations this appears to involve the generation of duplex DNA circles formed from telomeric repeat sequences (telomeric circles; t-circles), which can undergo rolling-circle dependent replication and then recombine back into telomeres to add new telomeric DNA to eroded telomeres [16]. The first evidence for this phenomenon came from work with the yeast species harboring linear mitochondrial genomes [17], where the mitochondrial DNA was found to be maintained by t-circle amplification [18]. Since then similar mechanism has been observed in a wide variety of systems including mammalian cells [19].
Regardless of the mechanism involved in telomere maintenance, the important point is that the states of individual telomeres within the same cell, or within a clonal population of isogenic cells may greatly differ, which may be crucial for generating flexibility on cells as a population to adjust to the sudden changes in the internal or external environment [20]. The ability to acquire different states is partly due to the differences in lengths between individual telomeres. Intra- and intercellular heterogeneity of lengths of telomeric fragments is a common property of all types of nuclear telomeres [21,22] and a very frequent feature of telomeres associated with extrachromosomal linear DNA genophores [2,23]. Lengths of nuclear telomeric tracts may vary not only between individual chromosomal ends, but the ends of the same chromosome may have substantially different number of tandem repeats in the members of clonal cell population [24]. The lengths of individual telomeres within a population of cells substantially vary (+/− 10–80%) regardless of the average length of telomeric tract (e.g., 300–500 bp in S. cerevisiae [25] versus 10–80 kbp in mouse cells [26]). Stochastic events are inherently associated with telomeric length regardless of their evolutionary origin as demonstrated by a multimodal distribution of mitochondrial telomeric repeats that fluctuates during growth of Tetrahymena cells [27] as well as variability in the number of tandem repeats at the mitochondrial telomeres of C. parapsilosis [28]. Even in Drosophila the number of t-posons varies between the chromosomes [22]. Clearly, although the mean lengths of telomeric fragments can be very different between species (or even between strains or individuals of the same species) indicating a genetic determination, the intra-and intercellular heterogeneity in telomere length within the single individual seems to be a common theme for many types of chromosomal ends.
4. Does the stochastic character of telomere state and/or length represent an adaptive trait?
There are two principal sources of telomere heterogeneity: deterministic and stochastic. Deterministic mechanisms, responsible for species-, individual-, strain-, or tissue-specific differences in telomere state and/or length, include regulated recruitment of telomerase to chromosomal ends, or regulated access of single- and double-stranded DNA binding proteins to the telomere [8]. On the other hand, intra- and intercellular heterogeneity of telomere state and/or length is in large part due to stochastic processes (Table 1). For example, when mammalian TRF2 or fission yeast Taz1 proteins are incubated with a model telomere in vitro, and majority of the DNA molecules are protein-bound, only a fraction (~15%) adopts a t-loop conformation [11,12]. In both cases, the reaction mixtures contained supposedly homogeneous population of DNA and protein molecules and yet the reaction produced at least two qualitatively very different categories of molecules. Most likely Brownian motion of the molecules results in random invasion of the 3′ telomeric overhang into the double-stranded region of the telomere and this transient state is stabilized by TRF2 or Taz1, similarly as ratcheting proposed to play an essential role for functioning of molecular motors [29].
Table 1. Examples of stochastic sources of telomere heterogeneity in eukaryotic nuclei.
(see text for details).
| Molecular component or process | Mechanism |
|---|---|
| DNA polymerase | Slippage along the telomeric repeats |
| DNA repair | Random triggering of DNA damage response (at telomeric loci) |
| Fluctuations in DNA repair enzymes activities on uncapped (eroded) telomeres | |
| Exonucleases | Random association/dissociation from the substrate DNA leading to single-stranded overhangs of variable lengths |
| Recombination | Nonreciprocal recombination within the same or between two telomeres leading to telomere rapid deletion (TRD) as well as sudden dramatic lengthening of a telomere |
| Telomerase | Fluctuations of telomerase activity between members of clonal cell populations caused by both deterministic (e.g. regulation of gene expression) and stochastic (e.g. unequal distribution of TERT and/or TER during cell division due to their low abundance) events |
| Misincorporation of deoxyribonucleotides | |
| Random association/dissociation from the substrate DNA leading to 3′ single-stranded overhangs of variable lengths | |
| Random reactivation of telomerase gene expression (e.g. during carcinogenesis) | |
| Sequence heterogeneity of telomeric repeats generated by two alleles of TER with different template domains | |
| Telomere associated retrotransposons (t-posons) | Fluctuations in a number of t-posons integrated into the telomere |
| Telomere-binding proteins | Fluctuations in DNA binding due to Brownian movements and relatively low abundance |
| Variable accessibility of telomeric binding sites | |
| Telomeric circles (t-circles) | Amplification of telomeric tract of various lengths through rolling-circle-formation |
| Excision from differently sized t-loops or by intrachromosomal recombination between the repeats leading to telomere rapid deletion (TRD) | |
| Fluctuations in sizes | |
| Telomeric DNA array (t-array) | Brownian movements |
| Telomere shortening due to the end-replication problem generating microheterogeneity | |
| Variability of the anchorage sites within a nuclear membrane | |
| Telomeric loops (t-loops) | Displacement DNA synthesis of telomeric tract of various lengths |
| Fluctuations in sizes and stability | |
| Stochastic events mediating their formation result in various frequency of occurrence |
Analogous stochastic events may be the main causes for telomere length heterogeneity. Telomerase may add one to several tandem repeats to the 3′ telomeric overhang per one reaction cycle and then fall off the substrate due to stochastic reasons [30]. In several yeast and ciliate species telomerase does not generate only length, but also telomeric sequence heterogeneity [25,31], further extending the repertoire of differences between chromosomal ends. These variant telomeric repeats are often caused by stochastic events like template infidelity of S. cerevisiae telomerase resulting from its accidental premature dissociation from the template [32] or random misincorporation of deoxyribonucleotides [31].
In addition to telomerase, there are other sources of telomere length heterogeneity including processing of telomeres by exonucleases and DNA repair machinery involved in end-resection and protection. Furthermore, during a rolling-circle replication, t-circles can turn around different number of times and then be separated from the linear DNA product of the reaction, so a t-circle with a defined length can produce heterogeneous population of linear DNA molecules. Similarly, t-loop-dependent replication at the telomeres can result in telomeric fragments of various lengths due to random dissociation of DNA polymerase from the displacement loop. Length heterogeneity can be a result of DNA polymerase slippage along the telomeric repeats similarly as in case of triplet expansion [33]. Randomness is also involved in localization of telomeric clusters within different areas of nuclear periphery affecting gene expression of telomere-associated genes and access of DNA-modifying proteins [5] as well as recombination events mediating non-reciprocal telomere deletion called telomere rapid deletion (TRD) in S. cerevisiae [34]. Moreover, random variations resulting from deterioration of mitochondrial functions seem to contribute to stochastic heterogeneity of telomeres and telomere-dependent cellular senescence in clonal population of human fibroblasts [35]. Clearly, no matter how accurately the events occurring at the telomeres are controlled, stochastic events may fundamentally affect the perspectives of a state and/or length of a particular telomere.
If we look at the telomere heterogeneity traditionally, the above remarks may seem trivial. The mechanisms involved in telomere maintenance are not precise, but the heterogeneity in telomere state and/or length resulting from this imperfection may simply be just a by-product of their action. However, we would like to provide an alternative view. We suggest that it is equally possible that these mechanisms have been selected to maintain telomeres (also) because they generate heterogeneity. In other words, it is likely that heterogeneous nature of telomeres within a single cell, or within a population of clonal isogenic cells has an adaptive value. In this respect it is tempting to speculate that the promiscuity of t-arrays (and means responsible for their maintenance including telomerase, t-loops, t-circles and other recombination-dependent mechanisms) among eukaryotes can be explained by the fact that they provide higher degree of heterogeneity than other types of telomeres (e.g., t-proteins or t-hairpins/t-palindromes). What are the indications that there was a positive selection in favor of heterogeneity-generating mechanisms? Nuclei of contemporary cells possess mechanisms generating telomeres of homogeneous size, yet they act only as back-ups for the main heterogeneity-generating telomere-maintaining system. For example, when both telomerase-dependent and recombination-dependent mechanisms of telomere maintenance are knocked-out in a strain lacking telomere-associate exonuclease Exo1, S. cerevisiae can employ t-palindromes to solve telomere-associated problems [36]. The fact that this mechanism is used only after both heterogeneity-generating mechanisms are defective argues that although the wild-type cell has an option to produce homogeneous telomeres (t-palindromes), heterogeneity-generating mechanisms (telomerase, recombination) are preferred. Additional indications come from the studies of yeast linear mitochondrial genomes [23], where t-palindromes enable interconversion between linear and circular genome form. Recent results from our laboratory indicate that t-palindromes (homogeneous terminal structures) may represent evolutionary intermediates leading to t-array (heterogeneous) type of mitochondrial telomeres (Valach, M. and Nosek, J. unpublished).
Isogenic populations of cells, in spite of their genetic homogeneity, greatly vary in many phenotypic characteristics. The fact that genetically uniform populations of bacteria exhibit single cell individuality is essential for understanding phenomena like enzyme induction, radiation, antibiotic as well as phage resistance [37–39]. Randomness, noise, and fluctuations are phenomena tightly associated with systems consisting with a limited number of its components and are inversely proportional to the square root of the number of particles [40]. DNA, many mRNA, and enzymes are present at exceedingly low intracellular concentrations (<1–100 molecules per cell) causing widely distributed times of biochemical reactions [41]. Stochastic effects arising due to the inherent nature of biochemical interactions were termed as intrinsic noise, as opposed to extrinsic noise originating from the fluctuations in the intra- and extracellular environment [39]. Intrinsic noise caused by a low abundance of molecules involved in the maintenance of chromosomal ends is also a source for intra- and intercellular telomere heterogeneity. The number of telomeres per cell is relatively low. The number of telomerase RNA (TER) molecules per both haploid (29 molecules) and diploid (37 molecules) S. cerevisiae cell is lower than the number of telomeres [42]. Interestingly, overexpression of S. cerevisiae TER leads to stable, shorter and apparently less heterogeneous telomeres [43], indicating that keeping the expression of TER low benefits the host. The number of TER molecules per mammalian cell is relatively high (>10,000), but the number of mRNA molecules for catalytic subunit of telomerase (TERT) is between 1–30 per cell and correlates with low abundance of the protein [44]. Finally, the estimated number of molecules of major mammalian telomere-binding proteins TRF1 and TRF2 does not exceed several thousands per cell [45], which is much lower than the number of potential DNA binding sites. Although similar measurements are not available for all molecular components involved in telomere maintenance, the above examples illustrate that low abundance of molecular players is one of the sources of telomere heterogeneity caused by stochastic events.
It becomes evident that randomness has been employed by natural selection as an adaptive trait. There are numerous examples illustrating that phenotypic variability of genetically equivalent cells (microbial population, multicellular eukaryotic organisms) resulting from random processes can be of a high value for the long-term survival of the cell population and/or during development [41,46].
Can heterogeneity of telomeres resulting from stochastic events at the ends of chromosomes also be an adaptive trait? Did evolution take advantage of the imperfection of mechanisms stabilizing and replicating ends of linear chromosomes? Although these questions have not been addressed systematically, there are experimental results falling within this theoretical framework. For example, different states of a telomere may affect expression of a gene located nearby a chromosomal end, a process called telomere position effect [6]. In Trypanosoma brucei there are hundreds of silent variant surface glycoprotein (VSG) genes but only one of these is transcriptionally expressed in a given cell at a time. Which VSG gene will be expressed in a particular cell is partly due to random events at the telomeres thus leading to a production of a clonal population of isogenic cells exhibiting antigenic variability. Similarly, many adhesin genes affecting antigenic and cell adherence properties of yeast pathogens are subject to TPE [47] leading to phenotypic variability of a potentially high adaptive value for the cell population and in case of pathogenic species. Finally, most of SUC genes in S. cerevisiae encoding invertase are located near telomeres. Transition between repressible and derepressible state of the corresponding telomeres may generate a heterogeneous population of cells “pre-adapted” for the situation when sucrose would become available as a carbon source [48].
The importance of stochastic events at the telomeres resulting in their heterogeneity may not be limited to regulation of gene expression. Telomeres attract several components of the DNA repair machinery, whose association with the chromosomal ends is dependent on a state of a particular telomere [49]. Telomeres may thus act as loci regulating the availability of some enzymes involved in DNA repair (or chromatin modification (e.g., Sir proteins)) for the rest of the genome. Interestingly, it was recently observed that telomeres in human cells exhibit heterogeneous rapid motions, whose degree is dependent upon telomere length and state [50]. Furthermore, it was shown that intercellular telomere heterogeneity contributes to a different replicative potential of members of a clonal population of cells within a tissue and subsequently their different chances of acquiring oncogenic mutations [51]. Importantly, stochastic telomere lengthening by a telomerase-independent mechanism takes part during normal early embryonic development both in mammalian [52] and plant [53] cells. These examples demonstrate that in addition to affecting expression of telomere-adjacent genes, heterogeneity of telomeres is of a great importance in a wide variety of microevolutionary processes and understanding its sources can lead to models useful for a broad range of fields from evolutionary biology to molecular medicine.
Acknowledgments
We wish to thank Ladislav Kovac (Comenius University) for inspirations, continuous support, Jack Griffith (University of North Carolina) for a long-term collaboration, and Gottfried Schatz (University of Basel) for helpful comments and suggestions and members of our laboratory for discussions. We apologize to those colleagues whose work was not cited due to the space limitations. Our work related to telomere biology is supported by grants from the Fogarty International Research Collaboration Award (2-R03-TW005654-04A1 (L.T.)), Howard Hughes Medical Institute (55005622 (J.N.)), the Slovak grant agencies APVT (20-001604 (L.T.) and 0024-07 (J.N.)) and VEGA (1/0132/09 (L.T.) and 1/0219/08 (J.N.)).
List of abbreviations
- ALT
alternative telomere lengthening
- TER
telomerase RNA
- TERT
telomerase reverse transcriptase
- t-array
telomeric array
- t-circle
telomeric circle
- t-hairpin
telomeric hairpin
- t-loop
telomeric loop
- t-poson
telomere associated retrotransposon
- t-protein
terminal/telomeric protein covalently attached to the 5′ ends
- t-palindrome
telomeric palindrome
- VSG
variant surface glycoprotein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu Rev Biochem. 2000;255:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 2.Nosek J, Kosa P, Tomaska L. On the origin of telomeres: A glimpse at the pre-telomerase world. BioEssays. 2006;28:182–190. doi: 10.1002/bies.20355. [DOI] [PubMed] [Google Scholar]
- 3.Nosek J, Tomaska L. Mitochondrial telomeres: An evolutionary paradigm for the emergence of telomeric structures and their replication strategies. In: Nosek J, Tomaska L, editors. Origin and Evolution of Telomeres. Landes Bioscience; Austin, Texas, USA.: 2008. pp. 162–171. [Google Scholar]
- 4.Ishikawa F, Naito T. Why do we have linear chromosomes? A matter of Adam and Eve. Mutat Res. 1999;434:99–107. doi: 10.1016/s0921-8777(99)00017-8. [DOI] [PubMed] [Google Scholar]
- 5.Taddei A, Gasser SM. Multiple pathways for telomere tethering: functional implications of subnuclear position for heterochromatin formation. Biochim Biophys Acta. 2004;1677:120–128. doi: 10.1016/j.bbaexp.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 6.Magdinier F, Ottaviani A, Gilson E. Telomere position effect and the evolution of the genome. In: Nosek J, Tomaska L, editors. Origin and Evolution of Telomeres. Landes Bioscience; Austin, Texas, USA.: 2008. pp. 128–142. [Google Scholar]
- 7.Blackburn EH. Telomere states and cell fates. Nature. 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- 8.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 9.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 10.de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 11.Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomaska L, Willcox S, Slezáková J, Nosek J, Griffith JD. Taz1 binding to a fission yeast model telomere: formation of telomeric loops and higher order structures. J Biol Chem. 2004;279:50764–50772. doi: 10.1074/jbc.M409790200. [DOI] [PubMed] [Google Scholar]
- 13.Tomaska L, Makhov AM, Griffith JD, Nosek J. t-loops in yeast mitochondria. Mitochondrion. 2002;1:455–459. doi: 10.1016/s1567-7249(02)00009-0. [DOI] [PubMed] [Google Scholar]
- 14.Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. EMBO J. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cesare AJ, Reddel RR. Alternative lengthening of telomeres in mammalian cells. In: Nosek J, Tomaska L, editors. Origin and Evolution of Telomeres. Landes Bioscience; Austin, Texas, USA.: 2008. pp. 45–57. [Google Scholar]
- 16.Tomaska L, McEachern MJ, Nosek J. Alternatives to telomerase: Keeping linear chromosomes via telomeric circles. FEBS Lett. 2004;567:142–146. doi: 10.1016/j.febslet.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 17.Tomaska L, Nosek J, Makhov A, Pastorakova A, Griffith JD. Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: Novel players in the mitochondrial telomere maintenance? Nucleic Acids Res. 2000;28:4479–4487. doi: 10.1093/nar/28.22.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nosek J, Ry ovska A, Makhov AM, Griffith JD, Tomaska L. Amplification of telomeric arrays via rolling-circle mechanism. J Biol Chem. 2005;280:10840–10845. doi: 10.1074/jbc.M409295200. [DOI] [PubMed] [Google Scholar]
- 19.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makovets S, Williams TL, Blackburn EH. The telotype defines the telomere state in Saccharomyces cerevisiae and is inherited as a dominant non-Mendelian characteristic in cells lacking telomerase. Genetics. 2008;178:245–257. doi: 10.1534/genetics.107.083030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pardue M-L, De Baryshe. Drosophila telomeres: a variation on the telomerase theme. In: Nosek J, Tomaska L, editors. Origin and Evolution of Telomeres. Landes Bioscience; Austin, Texas, USA.: 2008. pp. 27–44. [DOI] [PubMed] [Google Scholar]
- 23.Nosek J, Tomaska L, Fukuhara H, Suyama Y, Kovac L. Linear mitochondrial genomes: 30 years down the line. Trends Genet. 1998;14:183–188. doi: 10.1016/s0168-9525(98)01443-7. [DOI] [PubMed] [Google Scholar]
- 24.Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little MT, Dirks RW, Raap AK, Tanke HJ. Heterogeneity in telomere length of human chromosomes. Hum Mol Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 25.Cohn M, McEachern MJ, Blackburn EH. Telomeric sequence diversity within the genus Saccharomyces. Curr Genet. 1998;33:83–91. doi: 10.1007/s002940050312. [DOI] [PubMed] [Google Scholar]
- 26.Zijlmans JMJM, Martens UM, Poon SSS, Raap AK, Tanke HJ, Ward RK, Lansdorp PM. Telomeres in the mouse have large interchromosomal variations in the number of T2AG3 repeats. Proc Natl Acad Sci USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morin GB, Cech TR. Telomeric repeats of Tetrahymena malaccensis mitochondrial DNA: a multimodal distribution that fluctuates erratically during growth. Mol Cell Biol. 1988;8:4450–4458. doi: 10.1128/mcb.8.10.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nosek J, Dinouël N, Kovac L, Fukuhara H. Linear mitochondrial DNAs from yeasts: telomeres with large tandem repetitions. Mol Gen Genet. 1995;247:61–72. doi: 10.1007/BF00425822. [DOI] [PubMed] [Google Scholar]
- 29.Kovac L. Fundamental principles of cognitive biology. Evol Cognit. 2000;6:51–69. [Google Scholar]
- 30.Brault M-E, D’Souza Y, Autexier C. Telomerase: evolution, structure and function. In: Nosek J, Tomaska L, editors. Origin and Evolution of Telomeres. Landes Bioscience; Austin, Texas, USA.: 2008. pp. 1–26. [Google Scholar]
- 31.McCormick-Graham M, Haynes WJ, Romero DP. Variable telomeric repeat synthesis in Paramecium tetraurelia is consistent with misincorporation by telomerase. EMBO J. 1997;16:3233–3242. doi: 10.1093/emboj/16.11.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohn M, Blackburn EH. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 33.Compton SA, Cesare AJ, Fouche N, Ozgur S, Griffith JD. T-Loops, T-Circles and Slippery Forks. In: Nosek J, Tomaska L, editors. Origin and Evolution of Telomeres. Landes Bioscience; Austin, Texas, USA.: 2008. pp. 58–70. [Google Scholar]
- 34.Lustig AJ. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat Rev Genet. 2003;4:916–923. doi: 10.1038/nrg1207. [DOI] [PubMed] [Google Scholar]
- 35.Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, Schaeuble K, Birch-Machin MA, Kirkwood TB, von Zglinicki T. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 2007;5:e110. doi: 10.1371/journal.pbio.0050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maringele L, Lydall D. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 2004;18:2663–2675. doi: 10.1101/gad.316504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novick A, Weiner M. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci USA. 1957;43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spudich JL, Koshland DE., Jr Non-genetic individuality: chance in the single cell. Nature. 1976;262:467–471. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- 39.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 40.Schrödinger E. What is Life? With Mind and Matter and Autobiographical Sketches. Reprint edition. Cambridge University Press; Cambridge, UK.: 1992. [Google Scholar]
- 41.Raj A, van Oudenaarden A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mozdy AD, Cech TR. Low abundance of telomerase in yeast: Implications for telomerase haploinsufficiency. RNA. 2006;12:1721–1737. doi: 10.1261/rna.134706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singer MS, Gottschling DE. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science. 266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 44.Yi X, Shay JW, Wright WE. Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res. 2001;29:4818–4825. doi: 10.1093/nar/29.23.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okabe J, Eguchi A, Masago A, Hayakawa T, Nakanishi M. TRF1 is a critical trans-acting factor required for de novo telomere formation in human cells. Hum Mol Genet. 2001;9:2639–2650. doi: 10.1093/hmg/9.18.2639. [DOI] [PubMed] [Google Scholar]
- 46.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halme A, Bumgarner S, Styles C, Fink GR. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–415. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- 48.Greig D, Travisano M. The Prisoner’s dilemma and polymorphism in yeast SUC genes. Proc R Soc London B. 2002;271:S25–S26. doi: 10.1098/rsbl.2003.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanaar R, Wyman C, Rothstein R. Quality control of DNA break metabolism: in the ‘end’, it’s a good thing. EMBO J. 2008;27:581–588. doi: 10.1038/emboj.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Kam Z, Carlton PM, Xu L, Sedat J, Blackburn EH. Rapid telomere motions in live human cells analyzed by highly time-resolved microscopy. Epigenet Chromatin. 2008;1:4. doi: 10.1186/1756-8935-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin-Ruiz C, Saretzki G, Petrie J, Ladhoff J, Jeyapalan J, Wei W, Sedivy J, von Zglinicki T. Stochastic variation in telomere shortening rate causes heterogeneity of human fibroblast replicative life span. J Biol Chem. 2004;279:17826–17833. doi: 10.1074/jbc.M311980200. [DOI] [PubMed] [Google Scholar]
- 52.Liu L, Bailey SM, Okuka M, Munoz P, Li C, Zhou L, Wu C, Czerwiec E, Sandler L, Seyfang A, Blasco MA, Keefe DL. Telomere lengthening early in development. Nat Cell Biol. 2007;9:1436–1441. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- 53.Ruckova E, Friml J, Prochazkova Schrumpfova P, Fajkus J. Role of alternative telomere lengthening unmasked in telomerase knock-out mutant plants. Plant Mol Biol. 2008;66:637–646. doi: 10.1007/s11103-008-9295-7. [DOI] [PubMed] [Google Scholar]