Abstract
Micro (mi)RNAs are emerging as important regulators of cellular differentiation, their importance underscored by the fact that they are often dysregulated during carcinogenesis. Two evolutionary conserved families, let-7 and miR-200, regulate key differentiation processes during development. Loss of let-7 in cancer results in reverse embryogenesis and dedifferentiation, and miR-200 has been identified as a powerful regulator of epithelialto-mesenchymal transition (EMT). Recent findings have connected let-7 with stem cell maintenance and point at a connection between EMT and stem cell formation. A part of tumor progression can be viewed as a continuum of progressive dedifferentiation (EMT) with a cell at the endpoint that has stem cell-like properties. I propose that steps of this process are driven by specific changes in the expression of let-7 and miR-200 family members.
Keywords: microRNAs, stem cells, EMT, tumor progression, pluripotency, tumor initiating cells
miRNAs Are Emerging as Powerful Master Regulators of Differentiation and Cancer
Micro (mi)RNAs are small, 19−22 nucleotide (nt) long, non-coding RNAs that inhibit gene expression at the posttranscriptional level. They are first transcribed as parts of longer molecules, up to several kilobases in length (pri-miRNA), that are processed in the nucleus into hairpin RNAs of 70−100 nt by the double-stranded RNA-specific ribonuclease, Drosha.1 The hairpin pre-miRNAs are then transported to the cytoplasm by exportin 5 where they undergo final processing by a second, double-strand specific ribonuclease, known as Dicer. In animals, single-stranded miRNAs are incorporated into RNA induced silencing complexes (RISC) that bind primarily to specific messenger RNA (mRNA) at specific sequence motifs within the 3' untranslated region (3'UTR) of the transcript, which are significantly, although not completely, complementary to the miRNA. The mRNA/miRNA duplex then inhibits translation either through a (mRNA 5') cap-dependent mechanism affecting initiation2,3 or through increased degradation of the mRNA.4 Given the frequency with which miRNA target motifs are conserved within 3'UTRs, it is estimated that 20 to 30% of all human genes are targets of miRNAs, and that for each miRNA hundreds of genes exist that carry conserved sequence motifs within the 3'UTR.5-7
A strong link between miRNA dysregulation and human cancer has been established. A comparison of miRNA expression in normal and tumor tissues demonstrated global changes in miRNA expression in various human malignancies.8 In addition, mapping of 186 human miRNA genes has revealed that they are frequently located at fragile sites and other cancer-associated chromosomal regions.9 Consequently miRNAs have been demonstrated to act either as oncogenes (e.g., miR-155, miR-17−5p, miR-21)10,11 or tumor suppressors (e.g., miR-34, miR-15a, miR-16−1, let-7).12-22
Let-7 and miR-200, Global Regulators of Differentiation
Specific miRNAs have been shown to regulate mammalian cellular differentiation as well as developmental patterning and morphogenesis in a tissue specific fashion.23-29 In addition to these specific regulators, there exist classes of miRNAs that have universal functions which are dependent not on the tissue, per se, but rather on the differentiation state of the tissue. Two of the largest miRNA families, let-7 and miR-200 seem to have such activities.
The ubiquitously expressed let-7/miR-98 family was one of the first mammalian miRNAs to be identified.13-16,30,31 The let-7 family is comprised of 12 family members (let-7-a1, a2, a3, b, c, d, e, f1, f2, g, i and miR-98) located on 8 different chromosomes.32 These 12 family members represent 9 distinct let-7 sequences with identical seed sequences (the 5' sequence of the miRNA responsible for initating binding) and, very likely, overlapping sets of targets. Let-7 is expressed late in mammalian embryonic development and plays an evolutionarily conserved role from Caenorhabditis elegans to Drosophila to mammals.31,33-35 The let-7 targets that have been identified include cell cycle regulators such as CDC25A and CDK6,36 promoters of growth including RAS and c-myc16,37,38 and a number of early embryonic genes including HMGA2, Mlin-41 and IMP-1.39-44
The miR-200 family is comprised of 5 members (miR-200a, b, c, miR-141 and miR-429). They are located within two clusters on separate chromosomes. Interestingly the 5 members can be subdivided into two subgroups according to their seed sequences. MiR-200a and miR-141 comprise group I and miR-200b, c and miR-429 comprise group II. Although target prediction algorithms predicted little overlap in the targets of these groups, experimental approaches suggested that their sets of targets are highly overlapping.45 The most prominent targets of the miR-200 family are two E box binding transcription factors, ZEB1 (also known as TCF8 and δEF1) and ZEB2 (also known as ZFXH1B and SMAD interacting protein 1 (SIP1)).45-48 Both are key regulators within a complex network of transcriptional repressors that regulate the expression of E-cadherin and a number of master regulators of epithelial polarity.49-52 Consistent with their function, the miR-200 family was recently identified as both a marker and a powerful regulator of the epithelial-to-mesenchymal transition (EMT).45,48,53,54 For simplicity I will refer to these families of miRNAs as either let-7 or miR-200 unless specific activities of individual miRNA species are being discussed.
Core Transcription Factors that Regulate Embryonic Stem Cells
Embryonic stem (ES) cells have been heralded as the great hope for cures for many degenerative diseases in the future. ES cells have an enormous potential to differentiate into many different tissues (reviewed in ref. 55). This pluripotent property of ES cells is subject to regulation by the homeobox transcription factors, Oct4 and Nanog, which are essential regulators of early development and ES identity.56-59 Disruption of either of these factors causes loss of pluripotency.56,59,60 It has been suggested that Oct4 initiates the pluripotency state whereas Nanog maintains it.61 The HMG-box transcription factor, Sox2, heterodimerizes with Oct4 in ES cells and regulates expression of Oct4.62 Oct4, Sox2 and Nanog are part of an autoregulatory loop. Each binds to its own promoter and to the promoter of the other two genes.63 Autoregulatory loops such as this typically maintain stability of gene expression.64 In ES cells Oct4, Sox2 and Nanog act in concert to maintain the pluripotent state.62 They are often called master regulators of cell state.65 These three transcription factors co-occupy promoters of hundreds of genes, activating some genes and silencing others, in a coordinated fashion.63,66 Among genes that are activated are transcription factors, signaling molecules and chromatin-modifying enzymes, that together promote ES cell self-renewal.63,66 Among the silenced genes are genes that determine lineage commitment. A number of groups have recently reported derivation of induced pluripotent stem cells (iPS) from human somatic cells. Cells were reprogrammed from either differentiated progeny of stem cells,67 fetal and neonatal fibroblasts68 or adult fibroblasts.69 Reprogramming was accomplished using a combination of Oct4, Sox2, c-myc and Klf-4, or a combination of Oct4, Sox2, Nanog and Lin28, omitting the potentially oncogenic gene, c-myc, from the cocktail.70 Oct4 and Sox2 were determined to be essential for the reprogramming process.57,62,71
miRNAs and Stem Cells
miRNAs have been shown to regulate embryonic development72 and to play a role in regulating stem cells. Evidence for this activity comes from experiments demonstrating that ES cells that were deficient in miRNA processing enzymes exhibited defects in their capacity for differentiation and self renewal.73-76 In addition, Dicer deficiency is embryonic lethal, and Dicer deficient embryos exhibit greatly reduced expression of Oct4 suggesting a stem cell defect.77 Furthermore, cells from mice deficient for expression of the Drosha cofactor, DGCR8, proliferate and form colonies, but cannot differentiate.76 These data suggest that upregulation of miRNAs is required for various differentiation processes. Little is known with respect to mechanisms by which miRNA function in controlling the developmental potential of ES cells. A number of ES cell/early development specific miRNAs have been described,78-81 as have miRNAs that are expressed in mouse and human ES cells.80,82 However, it is still largely unknown how ES cell-specific transcription factors and miRNAs work together. The three stem factors (Oct4, Sox2 and Nanog) were recently found to occupy the promoters of many transcription factors and of 14 miRNAs.63 A more direct link between miRNAs and these genes was recently suggested by the report that miR-134, miR-296 and miR-470 target Oct4, Nanog and Sox2 within their open reading frames.83 Because miRNAs are presumed to preferentially target the 3'UTR of genes rather than the coding region, this points to an unusual level of regulation. Clearly, the function of these miRNAs in maintaining embryonic cells in vivo needs to be established. Recently miR-302 was demonstrated to convert human skin cancer cells into a pluripotent ES-like state.84 miR-302 is comprised of a cluster of 8 related miRNAs all of which are regulated by the binding of Oct4 and Sox2 to the miR-302 promoter.85 It appears, therefore, that a substantial portion of the activity of stem cell fate-specific transcription factors can be mimicked by miRNAs, which points at the power of miRNAs in maintaining cells and differentiated tissues.
Cancer Stem Cells/Tumor-Initiating Cells
A small number of cells within a tumor have properties that resemble those of stem cells. These cells have the ability to self-renew, to reproducibly form the same tumor phenotype, and to undergo multipotent differentiation into nontumorigenic cells. They are characterized by expression of distinctive cell surface markers permitting consistent enrichment. Chronic myeloid leukemia (CML) was the first cancer shown to be derived from a cancer stem cell.86 In the chronic phase of the disease the fusion protein BCR-ABL is present at high concentrations in hematopoietic stem cells (HSCs). After transition to blast crisis BCR-ABL was found to be amplified in granulocyte-macrophage progenitors (GMPs), which gain self renewal activity through aberrant activation of the Wnt/β-catenin pathway.87 The first tumor initiating cells (TICs) in a solid tumor were isolated from breast cancer.88 These cells efficiently formed anchorage-independent mammospheres. Mammospheres are known to be derived from mammary epithelial stem cells. It has been shown that a single mammosphere can give rise to an entire mammary ductal tree when implanted in mice.89 When isolated from breast cancer these CD24low CD44high cells have tumor initiating activity as indicated by the fact that they can differentiate and are highly tumorigenic when injected into immunodeficient mice. In addition to breast cancer, TICs have now been identified in melanoma, brain, liver, lung, head and neck, ovarian, pancreas, prostate and colon cancers.90-100
Epithelial-to-Mesenchymal Transition
EMT-like processes occur as part of embryonic development, wound healing101 and during carcinogenesis102 when tumor cells undergo a change from a differentiated to a dedifferentiated, more invasive tumor.101,103 During the process of EMT cells lose epithelial features and acquire mesenchymal characteristics, including vimentin filaments and a flattened morphology. They may start migrating and expressing proteases, gaining invasive activity that allows them to pass through the underlying basement membrane. Adherens junctions and desmosomes become at least partially dissociated. At the same time, a massive cytoskeletal reorganization takes place and a remodeling of the actin microfilament mesh occurs. An analysis estimated that changes in expression levels of about 4,000 genes (representing about 10% of the human genome) could be associated with EMT.104 The final effectors controlling the cell phenotype, including the cytoskeleton and the cell-cell and cell-substratum adhesion systems, appear to be similar or identical in all forms of EMT-related differentiation processes. A family of E-box-binding, zinc finger-containing transcription factors have been associated with EMT. Members of this family, Snail, Slug, ZEB1 and ZEB2/ SIP-1, have been extensively studied with respect to their function both in tumor progression and in embryogenesis.105-110 EMT plays a profound role during embryonic development111,112 in the formation of the primary mesoderm from upper epiblast epithelium, in neural crest cell formation from part of the ectoderm, and in palatal formation. In adults EMT occurs during adult placenta formation, and in the formation of fibroblasts during inflammation113 and wound healing.114 The reverse of EMT, mesenchymal to epithelial transition (MET) also occurs. One instance of MET is the formation of the nephron epithelium in the developing kidney from nephric mesenchymal cells.115 Another instance of EMT/MET is carcino-genesis. Many of the EMT inducing transcription factors such as Snail1, Snail2, ZEB1, ZEB2, TWIST1, FOXC2 and Goosecoid have been associated with tumor invasion and metastasis.112 While EMT during cancer progression is well characterized, MET is more difficult to observe in vivo. In one study, GFP labeled mammary cancer cells were shown to exit the primary tumor and were demonstrated to lose their mesenchymal nature when forming micrometastases in the lungs.116 An example of re-epithelialization of cancer cells during metastasis formation was also shown for the bladder cancer cell line, TSU-PrI,117 which showed increased expression of epithelial markers during metastatic deposits in scid mice. The expression of the epithelial marker E-cadherin is often detected in distant metastases in cells that may have passed through EMT.118 A number of reports recently identified the miR-200 family of miRNAs as a fundamental marker and regulator of EMT.45,48,119 miR-200 family members were found to be downregulated during TGFβ induced EMT induction of MDCK cells, whereas their expression was highly enriched in the epithelial cell lines of the NCI60 cell lines. Most importantly, altering their expression in cancer cells forced a number of cell lines to undergo either EMT or MET.45,48
EMT and Stem Cells
Evidence is mounting that the dedifferentiation process, EMT, may result in the generation of stem cells. The reversion of mesenchymal cells such as fibroblasts to iPS colonies expressing cadherins, mimics the MET characteristic of malignant transformation suggesting that reprogramming and tumor progression may act on similar dedifferentiation programs. Recently it was reported that poorly differentiated breast, glioma and bladder cancer cells express an ES cell signature.120 In addition, targets that are transcriptionally activated by Oct4, Sox2, Nanog and c-myc are frequently upregulated in these cancers. Breast cancers expressing an ES cell signature often belong to the basal type, which is characterized by poor clinical outcome. The fact that Oct4, Sox2 and Nanog can be used to reprogram somatic cells into pluripotent stem cells makes it likely that the combined expression of these genes in cancer cells could also contribute to the dedifferentiation observed during cancer progression. In fact, ectopic expression of the master regulator, Oct4, is sufficient to induce tumor growth in mice.121 The key regulators of stem cell pluripotency have been detected in various human cancers.122-125 Recently more direct evidence of a connection between stem cells and EMT was provided by the demonstration that induction of EMT in human mammary epithelial cells resulted in upregulation of a number of stem cell markers.126,127 In addition, cells isolated according to their stem cell markers had undergone EMT. These cells were found to overexpress classical EMT markers such as FOXC2, ZEB2, TWIST and SNAIL. Functionally, it was shown that induction of EMT resulted in the generation of cancer stem cells with the formation of markedly increased numbers of mammospheres by these cells.
There are additional parallels between the induction of dedifferentiation during the process of EMT and the generation of iPS cells. In both processes the achievement of substantial dedifferentiation requires treatment for weeks rather than days or hours. We and others observed that the induction of EMT required that cells be treated for 10 to 20 days.45,48,126 The induction of EMT in HCT116 cells required that cells be repeatedly transfected with miR-200 for up to three weeks.45 During this time cells gradually acquired a mesenchymal phenotype. Although the reason for this delay is unclear, it is consistent with the observation that TGFβ induced EMT requires a similarly long time in certain cells.48,126 The delay in the acquisition by cells of miRNA-induced changes is remarkably similar to the timing observed in reprogramming somatic fibroblasts to become iPS cells. It was shown that for complete reprogramming to occur the four factors Oct4, Sox2, Klf4 and c-Myc had to be expressed in the cells for at least 12 days.128 Current thinking is that a series of stochastic events leads to the formation of iPS cells.129 The notion is that ectopic expression of the four factors may trigger a series of epigenetic events, such as chromatin modification or remodeling, that results in only about 0.5% of the infected mouse embryonic fibroblasts giving rise to iPS cells within 3−4 weeks. Similar mechanisms may be involved in EMT.
Negative Feedback Loops Ensure Switch-Like Regulation by let-7 and miR-200
Both let-7 and miR-200 family members are differentially expressed in tumor cell lines representing the two differentiation stages we called Type I and Type II cells.42,45,130 We suggested that Type I cells have a less differentiated, mesenchymal phenotype and represent more advanced cancer, whereas Type II cells have a more differentiated, epithelial phenotype and represent less advanced cancer. Both let-7 and miR-200 are almost absent in Type I cells consistent with the assumption that these cells represent more advanced forms of cancer.45 Interestingly the regulation of expression of both miRNA families involves negative feedback loops (Fig. 1). Hammond and colleagues demonstrated that early during embryonic development at a time when mature let-7 levels are undetectable, the primary transcripts of let-7 are highly expressed. Based on this observation, it was postulated that the amount of let-7 would be regulated at a posttranscriptional level.131 Three studies shed some light on this mechanism by showing that the let-7 targets, Lin28 and Lin28B, are both inhibitors of let-7 processing and their expression is restricted to early embryonic development. Both proteins were shown to bind to the loop region of let-7 precursors, which results in blocking of processing of let-7 at either the Drosha132,133 or the Dicer level134 (Fig. 1A). Recently, it was shown that Lin28 and Lin28B act as posttranscriptional repressors through an additional mechanism by mediating terminal uridylation of let-7 precursors, which results in their degradation.135 Hence, the two proteins seem to suppress let-7 expression by acting at multiple levels.
Figure 1.
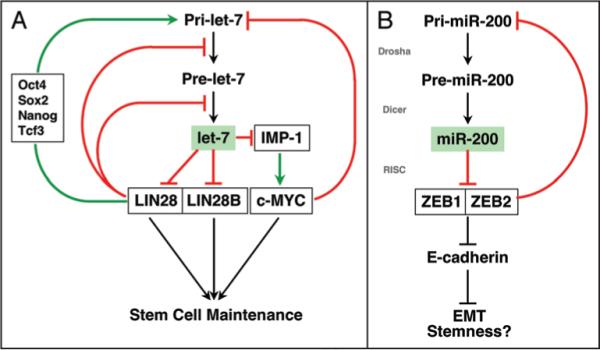
Expression of let-7 and miR-200 Is Regulated by Negative Feedback Loops. (A) Regulation of let-7 processing by the let-7 targets Lin28/Lin28B. (B) Regulation of miR-200 expression by the miR-200 targets ZEB1/ZEB2. For details see text.
In contrast to let-7, miR-200 family members are regulated at the transcriptional level. miR-200 negatively regulates the expression of the E-box binding transcription factors ZEB1 and ZEB2,42,45 (Fig. 1B). Interestingly however, the promoters of both miR-200 clusters, one on chr 1 and the other on chr 12, contain E-boxes and are negatively regulated by ZEB1 or ZEB2.53,136 Both the let-7 and the miR-200 miRNA families, are comprised of multiple members and each member is predicted to regulate a largely overlapping set of targets. In order to alter the expression of these targets without having to regulate expression of each miRNA individually, regulatory mechanisms are in place such that reducing the amount of only one abundant member of each family is thought to result in the reduction of the total amount of miRNA below a threshold at which suppression of expression of the inhibitors is abrogated. This results in the expression of inhibitors and the consequent shutting down of the expression of all miRNA family members. These kinds of negative feedback regulatory mechanisms ensure that the differentiation processes regulated by entire families of miRNAs behave in a switch-like fashion. This phenomenon is especially obvious for the epithelial marker E-cadherin and the mesenchymal marker vimentin. Cells either express large amounts of either of these markers or express none. Very little intermediate expression is observed.
Let-7 and miR-200 are Upregulated during Differentiation of Stem Cells
In a recent study, breast TICs were isolated from patients prior to and after receiving chemotherapy.137 Cells after chemotherapy were found not only to be more resistant to drugs, in vitro, but were also to express surface markers characteristic of TICs. These cells were devoid of let-7 expression but, when plated under conditions favoring differentiation, strongly upregulated let-7 over a course of 5 days. Interestingly, all miR-200 family members were also upregulated, albeit slightly delayed compared to let-7, indicating that stem-like cancer cells lack both let-7 and miR-200 expression. This finding is not restricted to cancer cells. A recent study using a deep sequencing approach identified expression of 14 miRNAs (9 known and 5 unknown) in undifferentiated normal human embryonic stem cells (HEC).81 Chromatin IP data demonstrated binding of Nanog, Sox2 and Oct4 to the promoters of 9 of the expressed miRNAs. It was also shown that both let-7 and miR-200 were upregulated when HECs were induced to differentiate. The miRNA showing the highest upregulation was let-7e (927 fold). In addition, in an analysis to identify ES-associated transcriptional regulators, not only were Oct4, Sox2, Nanog and Lin28 identified, but also identified was the miR-200 target, ZEB2.120 These data suggest that both let-7 and miR-200 are involved in regulating normal and cancer stem cells.
The Role of let-7 and miR-200 in Cancer
If one views cancer formation as a general dedifferentiation phenomenon, one would not be surprised if cancer cells displayed global downregulation of miRNAs. Consistently a large number of miRNAs that are downregulated during various forms of cancer have emerged.138 However, some miRNAs are upregulated during tumor progression. Among them are many that exhibit tissue specific expression. Prominent examples are miR-10b and miR-335 which promote breast cancer metastases.139,140 However, expression of certain miRNAs is altered during tumor progression in a more universal fashion. miR-21 has been shown to be upregulated during EMT141 and in many human cancers when compared to matching normal tissue.142 In addition, two large miRNA families have emerged as universal markers of advanced cancer. The first of these is let-7, the loss of which occurs early during neoplastic transformation, setting in motion a process that is similar to reversed embryogenesis.32 The second, miR-200, the fundamental regulator of EMT is often deregulated as tumors acquire invasive behavior. In studies of various human cancers including lung, colon, ovarian, gastric cancer, leiomyoma and melanoma, let-7 family members have been described as being downregulated during cancer progression.13-16,42,143-148 The situation with respect to miR-200 is more complex. The early stage of the process of metastasis is similar to EMT, but at later stages following extravasation, metastasizing cells start to settle down in their target tissue and undergo a differentiation process similar to MET.111 This, in theory, should again result in upregulation of miR-200. It may therefore not surprise that there is no general consensus as to whether or not miR-200 is upregulated or downregulated in advanced or metastasizing cancers. MiR-200 has been reported to be upregulated,146,147 downregulated48,149-151 or to remain unchanged152,153 in human cancers. In tumors from primary ovarian cancer (OvCa) we found a positive correlation between the expression of E-cadherin and miR-200. Goodall and colleagues reported an intriguing loss of expression of miR-200 in advanced breast cancer.48 In clear cell carcinoma both miR-141 and miR-200c were the most downregulated miRNAs.149 In OvCa the situation seems to be more complex. Three recent studies reported that miR-200 is a marker for more advanced OvCa. When OvCa tumors were compared to normal ovaries or ovarian surface epithelial cells miR-200 was found to be upregulated.146-148 In some of these reports the miR-200 family was found to be upregulated among all four histologic types of OvCa as compared to normal tissue. However, another recent study reported downregulation of miR-200 in OvCa as compared to normal tissue.154 Finally, to further confound the issue, in another recent analysis of the expressed miRNAs in OvCa none of the miR-200 miRNAs were found to be changed.152
An apparent upregulation of miR-200 during OvCa could have two reasons: (1) MiR-200 becomes upregulated during advanced OvCa. It was shown previously that early during OvCa progression cancer cells upregulate E-cadherin.155 (2) However, in some of the studies whole ovaries were taken rather than the single cell layer of epithelium on the surface of the ovaries from which the cancer is derived. The majority of cells inside of ovaries are of stromal nature and we demonstrated that stromal cells are completely devoid of miR-200 expression.45 It is therefore reasonable to assume that this mesenchymal cell mass contains less miR-200 than the epithelial cells or the cancer cells which were derived from the epithelial layer. Alternatively, ovarian surface epithelial cells were isolated and cultured before the analysis146 and the culturing or immortalization of these cells might have changed their miRNA composition. While such technical issues may have contributed to the contradicting results, it could be in the nature of miR-200 to be both up or downregulated during cancer depending on the stage of progression. Early when cancer cells acquire invasive behavior miR-200 may be downregulated, but it may also be upregulated again during the re-epithelialization of distal metastases when cells undergo MET. Such stage specific regulation of miR-200 has been demonstrated for liver cancer.156 miR-200 was found to be down-regulated in comparison to normal liver tissue with benign lesions, but was upregulated in advanced carcinomas. To clarify the role of miR-200 in cancer progression it will be necessary to improve the methods of isolating both cancer cells and normal cells and/or to develop better methods to quantify miRNA expression levels in single cells.
The Role of let-7 and its Targets in Stem Cells
In humans let-7 has been shown to act as a tumor suppressor, most likely through targeting a number of genes with oncogenic activity such as RAS13-16 or high mobility group (HMG) A2.38-40,42 Based on the evolutionary conserved role let-7 plays during development, its main function may lie in suppressing early embryonic genes some of which may have activities in stem cells. Using a genome-wide bioinformatics approach we recently identified 12 let-7 regulated oncofetal genes (LOGs) that are suppressed by let-7 in most adult tissues, but which are re-expressed in various forms of cancer when let-7 expression is lost. In the next section I discuss the function of the top three LOGs in cancer and in stem cells.
LOG1/HMGA2
Stem cells gradually lose their proliferative activity and capacity for self renewal with age.157 Recently, in a screen to identify genes that are selectively expressed in young versus old HSCs, Morrison and colleagues identified 26 genes that were differentially expressed between young and old stem cells.158 More recently using higher stringency they focused on HMGA2 as the only factor that is highly expressed in HSC and fetal neuronal stem cells but not expressed in any adult CD45+ cell.159 HMGA2 expression declined in aging stem cells and was undetectable in stem cells from 2-year-old mice. It was found that HMGA2 does not generally promote the growth or proliferation of cells but rather selectively promotes self renewal of stem cells.
Interestingly, the phenotype of the HMGA2 knock out mice is somewhat similar to that of mice deficient for the somatic stem cell factor, Bmi1.160 It was shown that the deficiency of Bmi1 could be compensated by crossing the mice with mice lacking expression of p16Ink4a and p19Arf,161 suggesting that Bmi1 suppresses expression of these cell cycle regulators. Similarly, deletion of either Ink4a or Arf partially rescued the defects of the HMGA2 deficient stem cells.159 The data establish HMGA2 as a stem cell factor that acts in young stem cells by repressing p16Ink4a and p19Arf, whereas Bmi1 acts by repressing expression of these cell cycle regulators postnatally in stem cells. The behavior of HMGA2 is similar to that of Oct4, which has also been shown to be important in embryonic stem cells, but which is entirely dispensable for the self-renewal and maintenance of adult stem cells.162 Recently it was demonstrated that in cancer stem cells let-7 regulates HMGA2,137 and it was suggested that in stem cells that lack let-7 expression, HMGA2 was involved in the maintenance of the undifferentiated state.163 Consistently, in HMGA2 containing mammospheres containing large numbers of TICs, Oct4 was found to be highly expressed.137
Evidence supporting the conclusion that HMGA2 is a stem cell factor controlled by let-7 also came from three large studies demonstrating that HMGA2 was associated with height,164-167 and that a particular single nucleotide polymorphism (SNP) associated with height mapped close to a predicted let-7 seed match in the 3'UTR of HMGA2.164 Assuming that HMGA2 in stem cells is under control of let-7, these data suggested that during aging let-7 expression in stem cells would increase. Consistent with this prediction, Nishino et al. reported that in aging stem cells the decline in HMGA2 levels was partly caused by let-7b upregulation, and confirmed that HMGA2 expression in aged stem cells was under control of let-7.159 Interestingly, it had previously been shown that HMGA2 together with INK4a contributed to induction of cellular senescence168 by suppressing expression of genes that drive proliferation. These data suggest that the low level of proliferation seen in stem cells could in part be driven by HMGA2/INK4a and be regulated by expression of let-7.
LOG2/IMP-1
IMP-1/CRD-BP is a member of a family of RNA binding proteins including IMP-2 (=LOG6) and IMP-3, that bind to the 5'UTR of the Insulin-like Growth Factor II leader 3 mRNA species.169 IMP-1 recognizes c-myc, IGF-II, tau, FMR1, semaphorin and βTrCP1 mRNAs, as well as H19 RNA. It acts to stabilize target RNAs by shielding them from degradation.169 IMP-1 has a classic oncofetal expression pattern, with expression only early during fetal life170,171 and re-expression in many human cancers.172 Some of the mRNAs that are stabilized by IMP-1 such as IGF-II and c-myc can guide stem cell function.173,174 In addition IMP-1 has been shown to be selectively expressed in cord blood derived CD34+ stem cells but not in adult CD34+ cells.175 Consistent with a proposed function of IMP-1 in stem cells is the fact that the phenotype of mice lacking IMP-1 expression is very similar to that of mice deficient for HMGA2,171,176 which are also small and have a defect in gut development. Finally, it was previously shown that IMP-1 is one of 26 genes that were found to be selectively expressed in young, but not old, HSC.158
LOG3/Lin28B
Interestingly Lin28B is a structural and functional homolog of Lin28.177 Recently it was found by two groups that Lin28/Lin28B act as selective inhibitors of let-7 processing in early embryonic cells132,133,178 demonstrating their function as both regulators of let-7 expression as well as being let-7 targets. Interestingly a combination of Lin28, SOX2, NANOG and OCT4 was demonstrated to reprogram somatic cells to become pluripotent stem cells.68 Lin28B as a stem cell factor responsible for pluripotency is, therefore, a good candidate gene to become upregulated in cancer cells when let-7 expression is lost. In this regard it is not surprising that Lin28B upregulation has been reported for human hepatocarcinoma cells.177 The two founding members of the miRNA family identified in C. elegans, lin-4 and let-7, have recently been linked to stem cells. It was shown that in neuronal stem cells both let-7 and miR-125 (which is the mouse ortholog of lin-4), negatively regulate expression of Lin28.134
In summary, the three let-7 targets, HMGA2, IMP-1 and Lin28 can be connected to stem cell function, thus making let-7 and its oncofetal targets fundamental regulators of stem cells, and therefore, highly relevant to cancer development and therapy.
Crosstalk between the Pathways Regulated by let-7 and miR-200
While loss of let-7 may precede loss of miR-200 during carcinogenesis, I propose that the two processes can be overlapping in many cases. Consistent with this hypothesis, a number of examples of crosstalk between these two pathways regulated by these miRNAs have emerged (Fig. 2). Although both HMGA2 and IMP-1 are regulated by let-7 directly, the expression of IMP-2 was also shown to be dependent on HMGA2 expression.179,180 Mutant mice lacking HMGA2 expression show a marked reduction in IMP-2 expression at day E12.5.179 HMGA2 is required for TGFβ induced EMT of mouse mammary epithelial NMuMG cells. HMGA2 was shown to regulate transcription of Snail, Slug, Twist and Id2.181 In addition RAS, which is another target of let-7 and a driving force for EMT, has been reported to induce expression of HMGA2.182,183 More recently it was demonstrated that ZEB1 and ZEB2 are regulated by HMGA2, and that the regulation of the Snail promoter by HMGA2 involves binding of HMGA2 to Smad3 and Smad4. HMGA2 was found to enhance the binding of Smad3 to the Snail promoter.184 Interestingly ZEB2 was identified through its interaction with Smad1, 2 and 5.185 These pathways could, therefore, interact and the dedifferentiation processes regulated by these pathways could lead certain cells to acquire stem cell-like properties. This hypothesis is consistent with a recent report demonstrating that signaling through the TGFβ/Smad pathway can cause the stem cell factor Nanog to be upregulated.186
Figure 2.
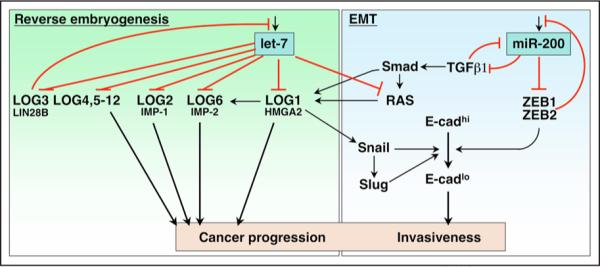
Model to Illustrate how let-7 and miR-200 Could Each Contribute to Tumor Progression, One by Controlling let-7 Regulated Oncofetal Genes (LOGs) and the Other by Regulating EMT and Metastasis. Crosstalk exists between the pathways that regulate reverse embryogenesis and EMT. For details see text.
Conclusion
Mounting evidence links two miRNA families, let-7 and miR-200, which are significantly correlated with dedifferentiation of cancer cells, to stem cells (Fig. 3). In addition to experimental evidence linking let-7 to the stem cell factors Lin28 and Lin28B, Sox2 and Klf4 are predicted to be targets of miR-200b/c/429 (TargetScan. org). Expression of both let-7 and miR-200 was correlated with more differentiated cancer using genome wide miRNA arrays or real time PCR.45,48,187 Interestingly in all of these studies additional miRNAs that were (less significantly) associated with the dedifferentiated phenotype were identified. These “runners up” can also be connected to stem cell regulation. In the study by Goodall and colleagues, in addition to miR-200, miR-205 was identified as a marker for mesenchymal cells, and miR-205 was shown to target the EMT regulators ZEB1 and ZEB2.48 In a recent genome wide analysis of the connections between the core transcriptional machinery and miRNAs, both Sox2 and Nanog were found to be bound to the promoter of miR-205, suggesting a direct link between the EMT regulator, miR-205 and ES cell regulators.63 In our own study in which we identified miR-200 as a regulator of EMT, the miRNA that followed the miR-200 family member in significance was miR-203.45 miR-203 was shown to negatively regulate stemness during skin development by suppressing expression of p63.188,189 Finally, in the screen to identify miRNAs that are preferentially expressed in Type II cells we identified miR-128a as more highly expressed in Type II cells.42 Interestingly, it was shown that the stem cell factors (and let-7 targets), Lin28 and Lin28B, bind not only to the loop region of let-7, but also to the loop region of miR-128,132 suggesting that processing of let-7 and miR-128 is coregulated. Of note, both let-7 and miR-128 are predicted to target Lin28, and miR-128 is also predicted to target Nanog (TargetScan.org). In summary, although other miRNAs have similar functions that need to be experimentally explored, I propose that let-7 and miR-200 are major guardians against inappropriate inclinations toward stemness. One of their normal roles may be to keep differentiated cells in their differentiated state.
Figure 3.
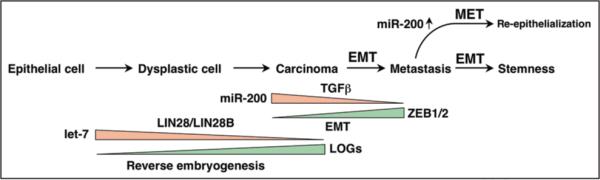
Deregulation of let-7 and miR-200 during carcinogenesis.
Acknowledgements
This work was supported by NIH grant R01 GM61712. I would like to apologize to all whose work could not been cited due to space limitations.
References
- 1.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–5. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–6. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 3.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA. 2005;102:16961–6. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–9. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 7.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–41. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–81. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 12.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and downregulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–6. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 14.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 15.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 16.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–43. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 18.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–93. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 20.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 22.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–7. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 24.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–4. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 27.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–64. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–83. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 29.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 30.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–21. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 31.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 32.Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, et al. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585–90. doi: 10.4161/cc.6.21.4845. [DOI] [PubMed] [Google Scholar]
- 33.Schulman BR, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn. 2005;234:1046–54. doi: 10.1002/dvdy.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–6. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol. 2008;18:943–50. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 37.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, et al. MicroRNA let-7a downregulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–70. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 38.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 39.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007;104:11400–5. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JA, et al. Identification of Let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–91. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 44.Maller Schulman BR, Liang X, Stahlhut C, Delconte C, Stefani G, Slack FJ. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle. 2008;7:3935–42. doi: 10.4161/cc.7.24.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors, ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsamiR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–6. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 47.Christoffersen NR, Silahtaroglu A, Orom UA, Kauppinen S, Lund AH. miR-200b mediates post-transcriptional repression of ZFHX1B. RNA. 2007;13:1172–8. doi: 10.1261/rna.586807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshild G, et al. The microRNA-200 family and miR-205 regulate epithelial-mesenchymal transition by targeting the E-cadherin repressors, ZEB1 and SIP1. Nat Cell Biol. 2008 doi: 10.1038/ncb1722. in press. [DOI] [PubMed] [Google Scholar]
- 49.Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–78. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 50.Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–85. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 51.Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Mol Biol Cell. 2007;18:3533–44. doi: 10.1091/mbc.E07-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007 doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–9. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008 doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donovan PJ, Gearhart J. The end of the beginning for pluripotent stem cells. Nature. 2001;414:92–7. doi: 10.1038/35102154. [DOI] [PubMed] [Google Scholar]
- 56.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 57.Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–60. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- 58.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeo-protein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–42. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 59.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 60.Ying QL, Nichols J, Evans EP, Smith AG. Changing potency by spontaneous fusion. Nature. 2002;416:545–8. doi: 10.1038/nature729. [DOI] [PubMed] [Google Scholar]
- 61.Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–4. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 62.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–35. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 63.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–61. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 65.Odom DT, Dowell RD, Jacobsen ES, Nekludova L, Rolfe PA, Danford TW, et al. Core transcriptional regulatory circuitry in human hepatocytes. Mol Syst Biol. 2006;2:17. doi: 10.1038/msb4100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 67.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–6. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 68.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 69.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluri-potent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 70.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–6. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 71.Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–8. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 72.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–30. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 73.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 74.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–40. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–5. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–50. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 78.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific microRNAs. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 79.Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–71. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 81.Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496–505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:18097–102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008 doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 84.Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. Rna. 2008;14:2115–24. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–38. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wong S, Witte ON. The BCR-ABL story: bench to bedside and back. Annu Rev Immunol. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- 87.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–67. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 88.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moraes RC, Zhang X, Harrington N, Fung JY, Wu MF, Hilsenbeck SG, et al. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development. 2007;134:1231–42. doi: 10.1242/dev.02797. [DOI] [PubMed] [Google Scholar]
- 90.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–51. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 91.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–63. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65:9328–37. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 93.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–35. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 94.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 95.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973–8. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sell S, Leffert HL. Liver cancer stem cells. J Clin Oncol. 2008;26:2800–5. doi: 10.1200/JCO.2007.15.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311–20. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–9. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Keshet GI, Goldstein I, Itzhaki O, Cesarkas K, Shenhav L, Yakirevitch A, et al. MDR1 expression identifies human melanoma stem cells. Biochem Biophys Res Commun. 2008;368:930–6. doi: 10.1016/j.bbrc.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 100.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–66. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 101.Savagner P. Leaving the neighborhood: molecular mechanisms involved during epithelial-mesenchymal transition. Bioessays. 2001;23:912–23. doi: 10.1002/bies.1132. [DOI] [PubMed] [Google Scholar]
- 102.Dvorak HF. Tumors: Wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650–9. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 103.Fuchs IB, Lichtenegger W, Buehler H, Henrich W, Stein H, Kleine-Tebbe A, et al. The prognostic significance of epithelial-mesenchymal transition in breast cancer. Anticancer Res. 2002;22:3415–9. [PubMed] [Google Scholar]
- 104.Zavadil J, Bitzer M, Liang D, Yang YC, Massimi A, Kneitz S, et al. Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc Natl Acad Sci USA. 2001;98:6686–91. doi: 10.1073/pnas.111614398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hemavathy K, Ashraf SI, Ip YT. Snail/slug family of repressors: slowly going into the fast lane of development and cancer. Gene. 2000;257:1–12. doi: 10.1016/s0378-1119(00)00371-1. [DOI] [PubMed] [Google Scholar]
- 106.Duband JL, Monier F, Delannet M, Newgreen D. Epithelium-mesenchyme transition during neural crest development. Acta Anat (Basel) 1995;154:63–78. doi: 10.1159/000147752. [DOI] [PubMed] [Google Scholar]
- 107.Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–66. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 109.Yokoyama K, Kamata N, Fujimoto R, Tsutsumi S, Tomonari M, Taki M, et al. Increased invasion and matrix metalloproteinase-2 expression by Snail-induced mesenchymal transition in squamous cell carcinomas. Int J Oncol. 2003;22:891–8. [PubMed] [Google Scholar]
- 110.Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 111.Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, et al. Epithelial—mesenchymal and mesenchymal—epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374–83. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 112.Yang J, Weinberg RA. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–29. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 113.Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest. 2002;110:341–50. doi: 10.1172/JCI15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Desmouliere A. Factors influencing myofibroblast differentiation during wound healing and fibrosis. Cell Biol Int. 1995;19:471–6. doi: 10.1006/cbir.1995.1090. [DOI] [PubMed] [Google Scholar]
- 115.Davies JA. Mesenchyme to epithelium transition during development of the mammalian kidney tubule. Acta Anat (Basel) 1996;156:187–201. doi: 10.1159/000147846. [DOI] [PubMed] [Google Scholar]
- 116.Xue C, Plieth D, Venkov C, Xu C, Neilson EG. The gatekeeper effect of epithelial-mesenchymal transition regulates the frequency of breast cancer metastasis. Cancer Res. 2003;63:3386–94. [PubMed] [Google Scholar]
- 117.Chaffer CL, Brennan JP, Slavin JL, Blick T, Thompson EW, Williams ED. Mesenchymal-to-epithelial transition facilitates bladder cancer metastasis: role of fibroblast growth factor receptor-2. Cancer Res. 2006;66:11271–8. doi: 10.1158/0008-5472.CAN-06-2044. [DOI] [PubMed] [Google Scholar]
- 118.Bukholm IK, Nesland JM, Borresen-Dale AL. Re-expression of E-cadherin, alpha-catenin and beta-catenin, but not of gamma-catenin, in metastatic tissue from breast cancer patients. J Pathol. 2000;190:15–9. doi: 10.1002/(SICI)1096-9896(200001)190:1<15::AID-PATH489>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 119.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–8. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 120.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–77. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 122.Gidekel S, Pizov G, Bergman Y, Pikarsky E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell. 2003;4:361–70. doi: 10.1016/s1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 123.Santagata S, Ligon KL, Hornick JL. Embryonic stem cell transcription factor signatures in the diagnosis of primary and metastatic germ cell tumors. Am J Surg Pathol. 2007;31:836–45. doi: 10.1097/PAS.0b013e31802e708a. [DOI] [PubMed] [Google Scholar]
- 124.Li XL, Eishi Y, Bai YQ, Sakai H, Akiyama Y, Tani M, et al. Expression of the SRY-related HMG box protein SOX2 in human gastric carcinoma. Int J Oncol. 2004;24:257–63. [PubMed] [Google Scholar]
- 125.Rodriguez-Pinilla SM, Sarrio D, Moreno-Bueno G, Rodriguez-Gil Y, Martinez MA, Hernandez L, et al. Sox2: A possible driver of the basal-like phenotype in sporadic breast cancer. Mod Pathol. 2007;20:474–81. doi: 10.1038/modpathol.3800760. [DOI] [PubMed] [Google Scholar]
- 126.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE. 2008;3:2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–9. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–81. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 130.Algeciras-Schimnich A, Pietras EM, Barnhart BC, Legembre P, Vijayan S, Holbeck SL, et al. Two CD95 tumor classes with different sensitivities to antitumor drugs. Proc Natl Acad Sci USA. 2003;100:11445–50. doi: 10.1073/pnas.2034995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–7. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Newman MA, Thomson JM, Hammond S. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008 doi: 10.1261/rna.1155108. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin-28. Science. 2008 doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–93. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 135.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 136.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–54. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 137.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 138.Lu J, Getz G, Miska EA, varez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 139.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 141.Zavadil J, Narasimhan M, Blumenberg M, Schneider RJ. Transforming growth factor-beta and microRNA:mRNA regulatory networks in epithelial plasticity. Cells Tissues Organs. 2007;185:157–61. doi: 10.1159/000101316. [DOI] [PubMed] [Google Scholar]
- 142.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Motoyama K, Inoue H, Nakamura Y, Uetake H, Sugihara K, Mori M. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res. 2008;14:2334–40. doi: 10.1158/1078-0432.CCR-07-4667. [DOI] [PubMed] [Google Scholar]
- 144.Peng Y, Laser J, Shi G, Mittal K, Melamed J, Lee P, et al. Antiproliferative effects by let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res. 2008;6:663–73. doi: 10.1158/1541-7786.MCR-07-0370. [DOI] [PubMed] [Google Scholar]
- 145.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–57. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 146.Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–33. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 147.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–5. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 148.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 149.Nakada C, Matsuura K, Tsukamoto Y, Tanigawa M, Yoshimoto T, Narimatsu T, et al. Genome-wide microRNA expression profiling in renal cell carcinoma: significant down-regulation of miR-141 and miR-200c. J Pathol. 2008 doi: 10.1002/path.2437. [DOI] [PubMed] [Google Scholar]
- 150.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and onco-gene/tumor suppressor gene mutations. Hepatology. 2008 doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 151.Nakajima G, Hayashi K, Xi Y, Kudo K, Uchida K, Takasaki K, et al. Non-coding MicroRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer Genomics Proteomics. 2006;3:317–24. [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 154.Dahiya N, Sherman-Baust CA, Wang TL, Davidson B, Shih Ie M, Zhang Y, et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS ONE. 2008;3:2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sundfeldt K, Piontkewitz Y, Ivarsson K, Nilsson O, Hellberg P, Brannstrom M, et al. E-cadherin expression in human epithelial ovarian cancer and normal ovary. Int J Cancer. 1997;74:275–80. doi: 10.1002/(sici)1097-0215(19970620)74:3<275::aid-ijc7>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 156.Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Downregulation of the microRNAs miR-34a, miR-127 and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol Carcinog. 2008 doi: 10.1002/mc.20484. [DOI] [PubMed] [Google Scholar]
- 157.Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells and senescence regulation. J Clin Invest. 2004;113:175–9. doi: 10.1172/JCI20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kiel MJ, Iwashita T, Yilmaz OH, Morrison SJ. Spatial differences in hematopoiesis but not in stem cells indicate a lack of regional patterning in definitive hematopoietic stem cells. Dev Biol. 2005;283:29–39. doi: 10.1016/j.ydbio.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 159.Nishino J, Kim I, Chada K, Morrison SJ. HMGA2 promotes neural stem cells self-renewal in young, but not old, mice by reducing p16 Ink4a and p19 Arf expression. Cell. 2008;135:227–39. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, et al. Posterior transformation, neurological abnormalities and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8:757–69. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 161.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–7. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, et al. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–15. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Droge P, Davey CA. Do cells let-7 determine stemness? Cell Stem Cell. 2008;2:8–9. doi: 10.1016/j.stem.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 164.Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, Perry JR, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–50. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–83. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–91. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Narita M, Krizhanovsky V, Nunez S, Chicas A, Hearn SA, Myers MP, et al. Cell. 2006:503–14. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 169.Noubissi FK, Elcheva I, Bhatia N, Shakoori A, Ougolkov A, Liu J, et al. CRD-BP mediates stabilization of betaTrCP1 and c-myc mRNA in response to beta-catenin signalling. Nature. 2006;441:898–901. doi: 10.1038/nature04839. [DOI] [PubMed] [Google Scholar]
- 170.Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–70. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hansen TV, Hammer NA, NielseN J, Madsen M, Dalbaeck C, Wewer UM, et al. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24:4448–64. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yisraeli JK. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol Cell. 2005;97:87–96. doi: 10.1042/BC20040151. [DOI] [PubMed] [Google Scholar]
- 173.Sun Y, Li H, Liu Y, Mattson MP, Rao MS, Zhan M. Evolutionarily conserved transcriptional co-expression guiding embryonic stem cell differentiation. PLoS ONE. 2008;3:3406. doi: 10.1371/journal.pone.0003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Knoepfler PS. Why myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell. 2008;2:18–21. doi: 10.1016/j.stem.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 175.Ioannidis P, Mahaira LG, Perez SA, Gritzapis AD, Sotiropoulou PA, Kavalakis GJ, et al. CRD-BP/IMP1 expression characterizes cord blood CD34+ stem cells and affects c-myc and IGF-II expression in MCF-7 cancer cells. J Biol Chem. 2005;280:20086–93. doi: 10.1074/jbc.M410036200. [DOI] [PubMed] [Google Scholar]
- 176.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–4. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 177.Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 178.Piskounova E, Viswanathan SR, Janas M, Lapierre RJ, Daley GQ, Sliz P, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008 doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 179.Brants JR, Ayoubi TA, Chada K, Marchal K, Van de Ven WJ, Petit MM. Differential regulation of the insulin-like growth factor II mRNA-binding protein genes by architectural transcription factor HMGA2. FEBS Lett. 2004;569:277–83. doi: 10.1016/j.febslet.2004.05.075. [DOI] [PubMed] [Google Scholar]
- 180.Cleynen I, Brants JR, Peeters K, Deckers R, Debiec-Rychter M, Sciot R, et al. HMGA2 regulates transcription of the Imp2 gene via an intronic regulatory element in cooperation with nuclear factor-kappaB. Mol Cancer Res. 2007;5:363–72. doi: 10.1158/1541-7786.MCR-06-0331. [DOI] [PubMed] [Google Scholar]
- 181.Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol. 2006;174:175–83. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Teramoto H, Malek RL, Behbahani B, Castellone MD, Lee NH, Gutkind JS. Identification of H-Ras, RhoA, Rac1 and Cdc42 responsive genes. Oncogene. 2003;22:2689–97. doi: 10.1038/sj.onc.1206364. [DOI] [PubMed] [Google Scholar]
- 183.Tchernitsa OI, Sers C, Zuber J, Hinzmann B, Grips M, Schramme A, et al. Transcriptional basis of KRAS oncogene-mediated cellular transformation in ovarian epithelial cells. Oncogene. 2004;23:4536–55. doi: 10.1038/sj.onc.1207585. [DOI] [PubMed] [Google Scholar]
- 184.Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. HMGA2 and Smads coregulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem. 2008 doi: 10.1074/jbc.M802016200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, et al. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5’-CACCT sequences in candidate target genes. J Biol Chem. 1999;274:20489–98. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- 186.Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, et al. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shell S, Park SM, Radjabi AR, Schickel R, Kistner E, Jewell DA, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007;104:11400–5. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lena AM, Shalom-Feuerstein R, di Val Cervo PR, Aberdam D, Knight RA, Melino G, et al. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–95. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 189.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–9. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]


