Abstract
Phospholipase D (PLD) hydrolyzes phosphatidylcholine to generate phosphatidic acid and choline. Studies in cultured cells and Drosophila melanogaster have implicated PLD in the regulation of many cellular functions, including intracellular vesicle trafficking, cell proliferation and differentiation. However, the function of PLD in vertebrate development has not been explored. Here we report cloning and characterization of a zebrafish PLD1 (pld1) homolog. Like mammalian PLDs, zebrafish Pld1 contains two conservative HKD motifs. Maternally contributed pld1 transcripts are uniformly distributed in early embryo. Localized expression of pld1 is observed in the notochord during early segmentation, in the somites during later segmentation and in the liver at the larval stages. Studies in intact and cell-free preparations demonstrate evolutionary conservation of regulation. Inhibition of Pld1 expression using antisense morpholino oligonucleotides (MO) interfering with the translation or splicing of pld1 impaired intersegmental vessel (ISV) development. Incubating embryos with 1-butanol, which diverts production of phosphatidic acid to a phosphatidylalcohol, caused similar ISV defects. To determine where pld1 is required for ISV development we performed transplantation experiments. Analyses of the mosaic pld1 deficient embryos showed partial suppression of ISV defects in the segments containing transplanted wild-type somitic and notochord cells, or notochord cells alone. These results provide the first evidence that function of Pld1 in the developing notochord is essential for vascular development in vertebrates.
Keywords: Phospholipids, phospholipases, notochord, somites, angiogenesis
Introduction
Phospholipase D (PLD) catalyzes the hydrolysis of phosphatidylcholine (PC) to generate phosphatidic acid (PA) and choline. PA and its metabolites, diacylglycerol (DAG) and lysophosphatidic acid (LPA), participate in multiple cellular activities, including intracellular vesicle trafficking, endocytosis, secretion, cell proliferation, differentiation, migration, and survival (Jenkins and Frohman, 2005; McDermott et al., 2004). In the presence of primary alcohols, PLD catalyzes a transphosphatidylation reaction to generate phosphatidylalcohols with high efficiency (Morris et al., 1997; Yang et al., 1967). This attribute has been utilized to track the activity of PLD in cells. Two mammalian genes encoding PLD, PLD1 and PLD2, have been identified and characterized. Both PLD enzymes require phosphatidylinositol 4,5-bisphosphate (PIP2) as a cofactor. However, PLD1 and PLD2 exhibit quite different regulatory properties and subcellular localization. PLD1 has a low basal activity and is activated by small G proteins (ARF, Rho and Rac) and PKC, whereas PLD2 is constitutively active, and insensitive to PLD1 activators in vitro (Colley et al., 1997; Hammond et al., 1995). Subcellular fractionation and immunocytochemistry studies suggest that PLD1 is localized in intracellular membranes and vesicular compartments, including Golgi, endosomes, lysosomes, and secretory granules. By contrast, PLD2 is associated with plasma membrane. PLD activation has been implicated in the actions of a number of growth factors, cytokines, hormones and neurotransmitters, including those that activate both heterotrimeric G protein-coupled receptors and receptor tyrosine kinases (Brown et al., 2007; Buchanan et al., 2005; Exton, 2002; Zhao et al., 2007). More recently, PLD was implicated in the regulation of neurite outgrowth (Cai et al., 2006; Watanabe et al., 2004). By analyzing Drosophila melanogaster mutants deficient in a pld homolog, Lalonde and collaborators recently suggested that PLD participates in phototransduction by maintaining an adequate level of PIP2 (LaLonde et al., 2005). Due to the difficulty of applying genetic strategies in dissecting PLD function in vertebrate model systems, most of the studies have been conducted at the cellular level using biochemical approaches. Therefore, the function of PLD in vertebrate embryogenesis remains undefined.
Partial cloning of a zebrafish gene encoding Pld1 homolog (amino acids 380-916), and its expression pattern during gastrulation stages was previously reported (Ghosh et al., 2003). However the developmental role of Pld1 was not directly investigated. In this study, we cloned a full-length cDNA encoding the zebrafish Pld1 homolog and demonstrated that it is expressed maternally and in ubiquitous fashion at blastula stages, but at a very low level during gastrulation. Later pld1 transcripts are confined to the notochord during early segmentation stages, to the somites during later segmentation, and are detected in the liver at larval stages. Blocking Pld1 function with antisense morpholino oligonucleotides (MO), designed to interfere with either pld1 RNA translation or splicing, impaired the formation of intersegmental vessels (ISV). Embryos incubated with 1-butanol (0.3%), which diverts the production of phosphatidic acid, exhibited similar ISV defects. Transplantation experiments support the notion that Pld1 promotes ISV development in non-autonomous fashion. The ISV defects were partially restored in pld1 deficient chimeric embryos containing transplanted wild-type cells in their notochord but not somites. This first study of loss of Pld1 function in a vertebrate organism reveals an essential role for PLD in the vascular development.
Methods and materials
Cloning of zebrafish pld1 and RT-PCR
Four zebrafish-expressed sequence tag (EST) clones (GenBank accession numbers: CK237875, CK029229, CD590334 and CF997495) encoding protein fragments with sequence similarity to human PLD1 were identified using NCBI tblastn. The 5′- and 3′-UTR regions of zebrafish pld1 were determined by 5′- and 3′-RACE PCR (Clontech), respectively. The total RNA from 2 dpf embryos was isolated using Trizol® reagent (Invitrogen). SuperScript™ First-Strand Synthesis System for RT-PCR (Invitrogen) was used to synthesize cDNAs. The full length ORF of zebrafish pld1 was amplified by PCR using a forward primer (5′-CACCATGAGTGATTCGGTGGAGAACCTGGACACC-3′) and a reverse primer (5′ -TCAGGTCCAGATCTCGGTGGGCACCA - 3′). The resulting PCR product was directly cloned into pENTR™/SD/D-TOPO® entry vector by Gateway® BP recombination reaction (Invitrogen). Subsequently, pld1 ORF was transferred into pCS2 destination vector (a gift from Lawson lab) for in vitro transcription. Sequencing of the resulting plasmids confirmed the zebrafish pld1 ORF and the deduced amino acid sequences of the pld1 ORF match those in the GenBank database with the exception of M116L and I503M.
To measure the zebrafish Pld1 activity in mammalian cells, the pld1 ORF was amplified by PCR using a forward primer (5′-AACTGCAGTCACCATGAGTGATTCGGTG-3′) and a reverse primer (5′-GGGGTACCTCAGGTCCAGATCTCGGTG-3′) and the PCR product was subcloned into the Pst1 and Kpn1 sites of pEGFP-C1 vector (Clontech). The catalytic inactive mutant K846R was generated by a point mutation in the 2nd HKD motif using Quick Change II Site-directed mutagenesis kit (Stratagene). The resulting mutation was confirmed by sequencing.
The total RNA from control or antisense morpholino oligonucleotides (MOs) injected embryos was isolated using Trizol® reagent (Invitrogen). The first strand cDNAs were synthesized using Thermoscript RT-PCR System (Invitrogen) according to the manufacturer's instructions. pld1 cDNA was then amplified using the first strand cDNA. The following primers were used in the PCR reactions: forward primer 1: 5′-gcagacatgagtgattcggt-3′; forward primer 2: 5′-ctgagccctgagatctttctga-3′; reverse primer 1: 5′-tcttgacaaaggctggatagt-3′; reverse primer 2: ctcccactcctgtctgaagact-3′.
Measurement of Pld activity in vivo and in vitro
TREx HEK293 cells were plated at 5×105 cells/well on poly-L-lysine coated 6-well plates the day before transfection. Cells were transfected with vectors encoding hPLD1 wt, hPLD1 K898R, zPld1 wt, zPld1 K846R, or pEGFP-C1 vector alone (Clontech) using lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. All of the PLD1 expression plasmids were cloned in pEGFP-C1 vector. To measure PLD activity in vivo, 30h after transfection, cells were radiolabeled with 3H-18:1 fatty acid in the serum free medium for 18h. The majority of the label is incorporated as phosphatidylcholine, which is the major substrate of PLD. After removing the labeling medium, cells were treated with either vehicle (basal) or 100 nM PMA (phorbol ester) for 30 min in the presence of 0.3% 1-butanol at 37°C. The assays were conducted as described (Brown et al., 2007; Henage et al., 2006). The in vitro assay is a well-established biochemical assay (Brown et al., 1993) used to measure PLD activity in reconstituted lipid vesicles. The details for making the lipid vesicles, purification of recombinant proteins, and measurements of kinetic parameters are described in detail in (Brown et al., 2007).
The analysis of lipids by mass spectrometry was performed essentially as previously described. Lipids were extracted from zebrafish embryos, prepared for mass spectrometry analysis, and statistically analyzed as detailed in (Ivanova et al., 2007). The references and examples of lipid fragmentation used to identify molecular species can be found (along with downloads of relevant methods chapters) at http://www.lipidmaps.org/.
Zebrafish strains and maintenance and chemical treatment
The AB* and TL WT zebrafish strains were maintained as described (Solnica-Krezel et al., 1994). Embryos were obtained from natural spawnings and staged according to morphology as described (Kimmel et al., 1995). A transgenic line expressing green fluorescent protein (GFP) under the control of the flk-1 promoter TG(flk1:GFP) was described by (Cross et al., 2003). A transgenic line expressing green fluorescent protein (GFP) under the control of the fli-1 promoter TG(fli1:GFP) was described by (Lawson and Weinstein, 2002). To inhibit PLD signaling, embryos were incubated with embryo media containing 0.3% 1-butanol or t-butanol at the indicated stages.
in vitro transcription and translation
In vitro transcription and translation of zebrafish Pld1 was performed by using the TNT Quick Coupled Reticulocyte Lysate system (Promega), according to the manufacturer's protocol. 0.5 μg flag-Pld1-pCS2 and various amount of ATGMO (0, 10 nM, 0.5 μM, and 5 μM) were added for each 25 μl reaction. The reactions were incubated at 30°C for 90 minutes. 6 μl of the reaction samples were denatured, loaded and separated on 8% SDS-PAGE gel and transferred to Immuno-P membrane. The synthesized zebrafish Pld1 protein was detected by anti-flag antibody.
Whole mount in situ hybridization, sectioning, immunostaining and microscopy
Whole mount in situ hybridization was performed as described previously (Thisse et al., 1993). pld1 antisense digoxigen-labeled in situ probe was synthesized using T7 RNA polymerase (Ambion) and precipitated with LiCl and EtOH and re-suspended in RNAase free water. Probe quantity was measured by spectro-photometry and quality was assayed using gel electrophoresis. Photographs of embryos were captured with a Zeiss Discovery V.12 microscope equipped with an Axiocam digital camera. For histological sections, the embryos processed by in situ hybridization were embedded in JB-4 embedding solution and sectioned using Leica RM2265 Rotary Microtome. Whole mount immunohistochemistry was performed as described previously (Topczewska et al., 2001). Monoclonal Anti-Titin (Mouse) and Anti-Laminin (Rabbit) primary antibodies were purchased from Sigma®. Znp1 was obtained from the Developmental Studies Hybridoma Bank. TG(flk1:GFP)/TG(fli1:GFP) embryos were mounted laterally in 2% methylcellulose in glass bottom culture dishes (MatTek). The images were acquired using a Zeiss LSM 510 laser scanning inverted confocal microscope.
Morpholino oligonucleotides and microinjections
Three antisense morpholino oligonucleotides (MOs) (Gene Tools), MO1-pld1 (5′-CCAGGTTCTCCACCGAATCACTCAT-3′), MO2-pld1 (5′-TGTCTCATCACCTCTTAAGAAAGAG-3′), and MO3-pld1 (5′-GGTCCATCATACAAACCTGCTCTAT-3′), were designed to specifically target the ATG start codon and inhibit translation, the intron-exon junction, and the exon-intron junction of the exon coding the first HKD motif respectively. A 5-mismatch MO (5mismatchMO2-pld1: 5′-TGTCTGATGACCTCTTAACAAACAG-3′) corresponding to HKD1a was also designed as a control. Indicated dosages of MOs were injected into the yolk of one to two-cell stage embryos using a pneumatic picopump (WPI) or Eppendorf FemtoJet. Microinjection was performed as described previously (Zeng et al., 2007).
Transplantation
Genetic mosaic analyses were performed essentially as described (Yamashita et al., 2002). WT donor embryos were injected with 1% rhodamine-dextran (Molecular Probes) at the one-cell stage. pld1 morphant host embryos were generated by injecting 6 ng MO2-pld1 into TG(fli1:GFP) transgenic embryos at the one-cell stage. For notochord transplantations, Between 5.7 and 6 hpf (hours post fertilization), when the embryonic shield became morphologically distinct, 30–50 cells from the shield region were aspirated from one WT donor embryo using a transplantation needle. The group of the donor cells was immediately transplanted into the shield of a host embryo. To transplant somitic cells, at the similar stage, 30–50 deep cells were aspirated from the blastoderm margin next to the shield of one donor embryo and immediately transplanted into the equivalent region of a host embryo to ensure that the initial positions of the donor and host embryos were identical. The host embryos were analyzed at 2 dpf.
Statistical analysis
Calculations were made in Microsoft Excel. We report mean and standard error of mean (SEM), and the probability associated with Student's T-Test (with 2-tailed distribution) and two samples of unequal variance.
Results
Cloning of zebrafish pld1
To clone a zebrafish Pld homolog, the human PLD1 amino acid sequence was used to search the zebrafish EST and genomic sequence databases. Four ESTs sharing similarity with human PLD1 gene were found. The full-length pld1 cDNA was amplified by using RT-PCR and 5′-RACE (Rapid Amplification of cDNA Ends, Clontech), and maintained in Gateway® Cloning system (Invitrogen). Multiple sequence alignments revealed a 64-68% shared identity at the protein level between zebrafish Pld1 and mammalian PLD1s (hPLD1, rPLD1, mPLD1) and a 50% shared identity between zebrafish Pld1 and mammalian PLD2s (hPLD2, rPLD2, mPLD2) (Supplementary Figs. 1A, C). The phylogenetic tree of Pld1 and PLDs from other species is shown in Supplemental data (Supplementary Fig. 1B). Similar to mammalian PLDs, Pld1 contains two conserved HxKxxxxD motifs (Supplementary Fig. 1A, blue dotted rectangles) and also putative interaction sites for protein kinase C (PKC) and RhoA (Cai and Exton, 2001; Kook and Exton, 2005) (Supplementary Fig. 1A, the PKC binding site of zebrafish Pld1 is on 1-314 amino acids region; RhoA binding site is on 859-1010 amino acids region). The C-terminal portion of Pld1 is highly similar to mammalian PLD1, with more variation occurring in the N-terminal portion and the loop region of the protein. A 20-amino acid insertion at the extreme N-terminal region of mammalian PLD1 is absent in Pld1 (Supplementary Fig. 1A).
We subsequently performed biochemical assays to compare activity of zebrafish Pld1 with the human enzyme. Similar to hPLD1, zebrafish Pld1 exhibited a low basal activity when expressed in the HEK293 TREx cells and it was robustly stimulated by phorbol ester (PMA), which is widely used as an activator of PKC and PLD catalytic activities (Fig. 1A). A mutation K898R in human PLD1 HKD domain has been shown to produce a catalytically inactive protein (Sung et al., 1997). Likewise, an HKD-mutant construct harboring a corresponding mutation K846R in the zebrafish Pld1 lacked activity in this assay (Fig. 1A). To further characterize the biochemical regulation of pld1 gene product, we next performed in vitro PLD activity assays testing whether its activation is regulated by PKC and small GTPases, as reported for the mammalian enzymes (Brown et al., 1993; Colley et al., 1997; Singer et al., 1996). These assays demonstrated that zebrafish Pld1, similar to mammalian PLD1 enzymes, can be activated by both PKCα and Arf1 (Fig. 1B). The combination of PKCα and Arf1 resulted in an additive activation of wild-type Pld1. In contrast, both human K898R and zebrafish K846R mutants did not reveal additional activity beyond that exhibited by the endogenous PLD in the control HEK293 cells transformed with the empty vector (Fig. 1B). The western blot in Fig. 1C indicates the relative amounts of wild-type GFP-Pld1/PLD1 and mutated GFP-Pld1/PLD1 that were used in the activity assays. Taken together, these cell culture and in vitro data demonstrate that the zebrafish pld1 gene encodes a bona fide Pld1.
Fig. 1. Activation of zebrafish Pld1 in HEK293 cells.
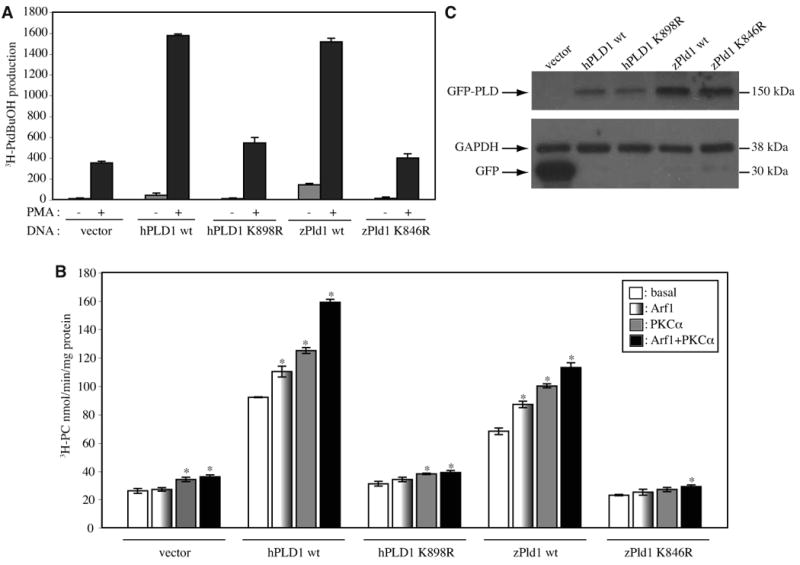
Recombinant human PLD1 (hPLD1) or zebrafish Pld1 (zPld1), was transiently expressed in HEK293 cells, and activity was measured, in vivo (A) and in vitro (B). Cells were transfected with an empty pEGFP-C1 vector, or a vector encoding wild-type hPLD1 (hPLD1 wt), or zPld1 wt. In addition, catalytically inactive mutants of hPLD1 K989R and zPld1 K846R were transfected. (A) in vivo activity was measured as the transphosphatidylation of radiolabeled endogenous substrate to 1- butanol, in response to 100 nM PMA compared to basal. (B) in vitro PLD activity was measured using 20 μg total cell lysates for either basal or with the following recombinant protein additions: Arf1 (275 nM), PKCα (1 μM), or Arf1 (275 nM) + PKCα (1 μM) for each transfection condition. Data are expressed as mean ± SEM (*, p<0.03 when compared to the basal activity of each transfection) of triplicate samples from three independent experiments. (C) Representative Western blot analysis of the transiently expressed N′-GFP tagged PLD1 proteins (GFP-PLD) or GFP vector control (GFP) used in the PLD activity assay. Equal loading of the protein was standardized by GAPDH.
The dynamic expression pattern of pld1 during early zebrafish development
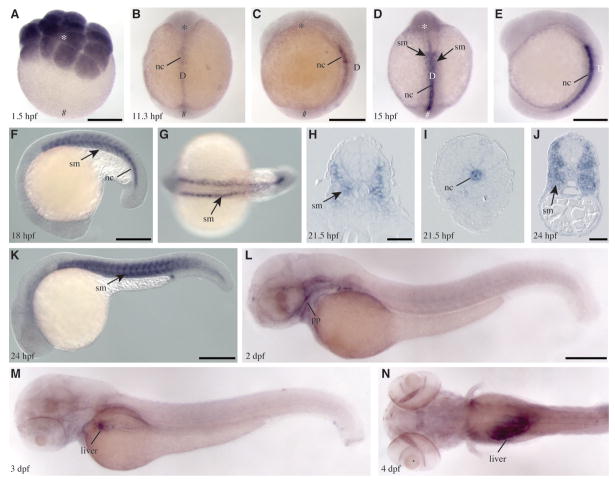
Using RT-PCR, we detected pld1 transcripts in 4-cell stage (1 hpf) embryos (Supplementary Fig. 2), before the onset of the zygotic transcription, thus indicating a maternal RNA deposition. The abundance of pld1 RNA decreased by 5 hpf and started to increase at the end of the gastrula period (9 hpf; Supplementary Fig. 2 and data not shown). A strong pld1 expression was detected in 26 hpf embryos (Supplementary Fig. 2). The spatio-temporal expression pattern of pld1 during zebrafish development was analyzed by whole mount in situ hybridization (Thisse et al., 1993). In a 16-cell stage zebrafish blastula (1.5 hpf) we observed ubiquitous distribution of pld1 transcripts (Fig. 2A). At the early gastrulation (shield stage; 6 hpf), pld1 was ubiquitously but very weakly expressed (data not shown). However, at the 4 somite stage (11.3 hpf), pld1 expression became clearly detectable again and was restricted to the anterior notochord and the adjacent forming somites (Figs. 2B, C). At the 12 somite stage (15 hpf), pld1 expression remained largely confined to the differentiating notochord (Figs. 2D, E) with uneven expression along the anteroposterior axis in the somites flanking the notochord (Fig. 2D, arrows). The expression of pld1 in somites became more pronounced during late segmentation stages (Fig. 2F, G, arrows). The cross-sections through the trunk region further confirmed that pld1 was expressed strongly in the somites in the anterior trunk (Fig. 2H) but remained restricted to the notochord at the posterior trunk (Fig. 2I). After 1 dpf, pld1 was primarily expressed in the somites (Fig. 2K, arrows). At 2 dpf, the expression of pld1 became strongly reduced in the somites (Fig. 2L), and appeared in the pharyngeal pouch tissues (Fig. 2L). At early larval stages (3 and 4 dpf), pld1 RNA was detected mainly in the liver (Figs. 2M, N).
Fig. 2. Spatio-temporal expression of pld1 during zebrafish embryogenesis.
(A) pld1 transcripts are ubiquitously distributed at 16-cell stage (1.5 hpf). At 11.3 hpf (B,C) and 15 hpf (D,E), pld1 expression is restricted to the dorsal notochord (nc) and the adjacent forming somites (sm). (F-I) pld1 expression in somites (sm) becomes more pronounced during late segmentation stages. (H,I) Cross section of an embryo at 21.5 hpf shows pld1 expression in the somites (sm) in the anterior trunk region (H) but in notochord (nc) in the posterior trunk (I). (J, K) At 24 hpf, pld1 is primarily expressed in the somites (sm). (L) At 2 dpf, the expression of pld1 is strongly reduced in the somites and appears in the pharyngeal pouch (pp). (M,N) At 3 dpf and 4 dpf, pld1 is detected mainly in the liver. (A) Animal view: (B,D) dorsal views, animal pole towards the top; (C,E,F,K,L,M) lateral views, anterior to the left; (G,N) dorsal view, anterior to the left. Animal pole ( , ). Vegetal pole (#). Dorsal (D). Scale bar represents 100 μm (H,J) and 250 μm (A,C,E,F,K,L).
Inhibition of Pld1 expression and activity in zebrafish by antisense morpholino oligonucleotides
To determine the role of Pld1 in zebrafish development, we employed two strategies to inhibit its function. First, we designed three antisense morpholino oligonucleotides (MOs) (Fig. 3A) to specifically either block pld1 RNA translation or interfere with its splicing. MO1-pld1 was designed to target the ATG translation initiation site, whereas MO2-pld1 and MO3-pld1 targeted the intron-exon boundary and the exon-intron boundary of the exon encoding the first HKD motif, respectively. A five-nucleotide mismatch morpholino (5mismatch MO2-pld1) corresponding to MO2-pld1 was also designed to serve as a negative control. The effectiveness of MO1-pld1 in blocking pld1 translation was evaluated by using an in vitro translation assay, in which various amounts of MO1-pld1 were added to block the translation of a flag–tagged partial pld1 construct. Subsequently the protein production was analyzed by western blot with the anti-flag antibody. We observed that 10 nM, 0.5 μM and 5 μM MO1-pld1 caused a 34%, 60% and 82% decrease in flag-Pld1 fusion protein expression, respectively (Fig. 3B). The effectiveness of MO2-pld1 and MO3-pld1 in interfering with pld1 splicing was evaluated by RT-PCR amplification (Fig. 3C) and by sequencing the resulting pld1 cDNAs (data not shown). As expected, MO2-pld1 caused the retention of the intron in front of the HKD1 motif-coding exon and introduced a premature stop codon. Comparing the intensity of the RT-PCR band from control embryos, an injection of 3 or 6 ng of MO2-pld1 per embryo caused the production of a incorrectly spliced mRNA (58% and 62%) and a 34% or 50% reduction in native pld1 mRNA. By contrast, injections of 8 ng of 5mismatch MO2-pld1 per embryo did not cause a mis-splicing event. Similar analyses revealed that the injections of MO3-pld1 caused the retention of the intron after the HKD1 motif-coding exon to introduce a stop codon, as corroborated by sequencing of the respective cDNAs. An injection of 3 or 6 ng MO3-pld1 per embryo caused the production of a mis-spliced mRNA and an 88% or 98% reduction in the native pld1 mRNA. Notably, co-injections of MO2-pld1 and MO3-pld1 resulted in a more efficient interference with the pld1 splicing: only 1% of the native pld1 mRNA was detected in the embryos injected with 3 or 6 ng of each MO2-pld1 and MO3-pld1.
Fig. 3. Design and evaluation of effectiveness of antisense oligonucleotides targeting pld1 translation and splicing.
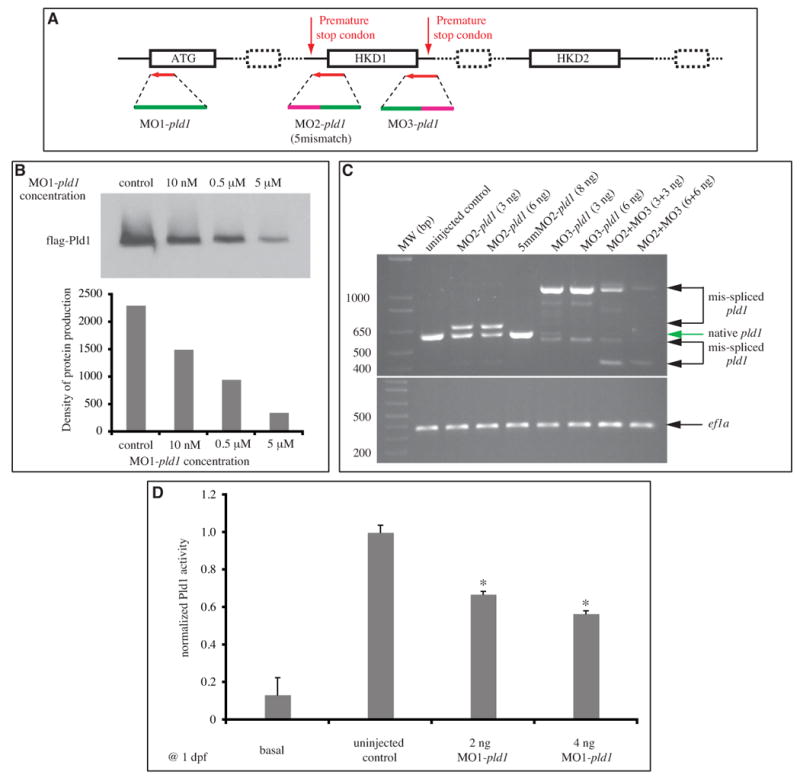
(A) The scheme of morpholinos designed to target pld1. (B) in vitro transcription and translation experiment testing the efficiency of MO1-pld1 to inhibit pld1 translation in a dose-dependent manner. (C) Gel electrophoresis of RT-PCR amplified pld1 fragments to monitor interference by MO2-pld1 and MO3-pld1 compared to the normal splicing of pld1. ef1a is used as a loading control. 5mmMO2-pld1: 5mismatch MO2-pld1. (D) The basal and PMA stimulated PLD activity were measured using a stable isotope method for detecting the formation of PtdBuOH by mass spectrometry. Enzyme activity is significantly reduced in MO1-pld1 injected embryos (*, p<0.01). Data is from three independent experiments. Each condition represents the average measurement from 50 embryos at 1dpf. Error bars depict SEM.
We sought to measure the PLD activity in zebrafish using an advanced stable isotope mass spectrometry (MS)-based assay that allowed assessment of enzyme activity in the whole embryo. We conducted this MS-based lipid analysis to specifically measure the Pld1 activity in homogenates of untreated embryos at 1 dpf using a stable isotope precursor methodology (Brown et al., 2007). In the spectra of PMA + 1-butanol (1-BuOH) treated samples, three peaks increased (Supplementary Fig. 3): 34:1 phosphatidylbutanol (PtdBuOH) at m/z 729, 34:2 PtdBuOH at m/z 727, and 38:6 PtdBuOH at m/z 775. In the PMA + 1-BuOH-d10 treated samples, all of the PtdBuOH peaks shifted 9 Da, confirming their identity as PtdBuOH-d9, the product of PLD activity. The unambiguous identification of PtdBuOH species (resulting from the PMA+1-BuOH treatment) was accomplished by tandem mass spectrometry (MS/MS). The fragmentation spectra of 3 PtdBuOH species are illustrated in Supplementary Fig. 4A-G. Panel A of Fig. S4 shows the MS/MS spectrum of the peak at m/z 729. A detailed description of the product ions used to identify the fatty acyl components is provided in the supplementary materials. This advanced analytical methodology provides a quantitative analysis of changes in Pld product formation and a precise molecular signature for this enzyme activity in zebrafish embryos.
Next we sought to determine whether any systematic changes in Pld activity occurred following the decreased expression resulting from MOs injections. Using mass spectrometry we were able to quantify changes in enzyme activity in the whole embryo. The Pld1 activity in the uninjected control and MO1-pld1 injected zebrafish was also measured by MS (Supplementary Fig. 5). Pld1 activity was significantly impaired by injecting MO1-pld1 (50 embryos were used for 1 sample). In the spectra of PMA+1-BuOH-d10 treated uninjected embryos, three PtdBuOH peaks were observed: 34:1 PtdBuOH-d9 at m/z 738, 34:2 PtdBuOH-d9 at m/z 736, and 38:6 PtdBuOH-d9 at m/z 784. In the PMA+1-BuOH-d10 treated Pld1 morphant samples, the intensity of PtdBuOH peaks decreased. The results of this extensive MS analysis are integrated and presented in Fig. 3D. Together these several lines of evidence demonstrated that injections of MOs targeting pld1 translation or splicing resulted in a significant reduction of its expression and activity.
Loss of Pld1 function results in vascular defects
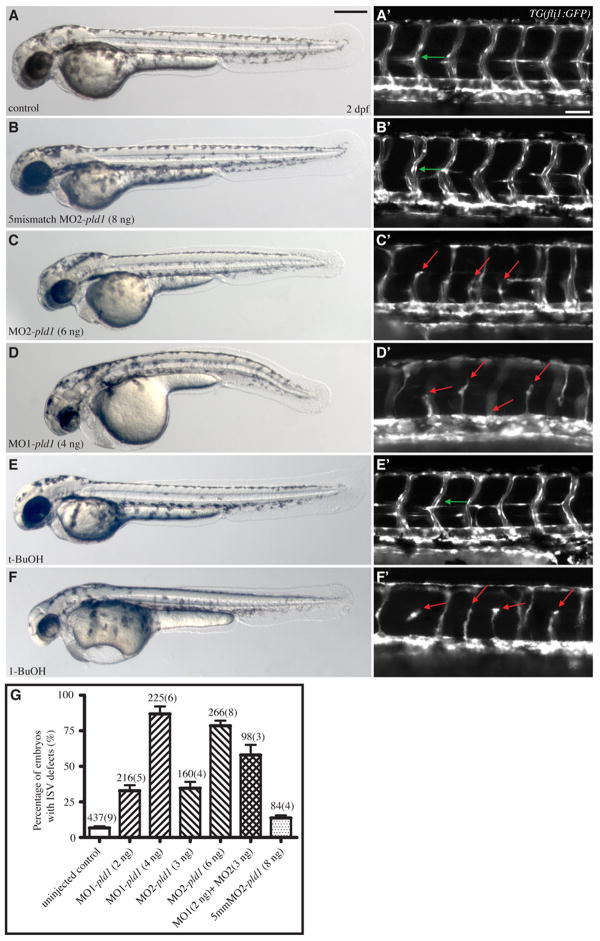
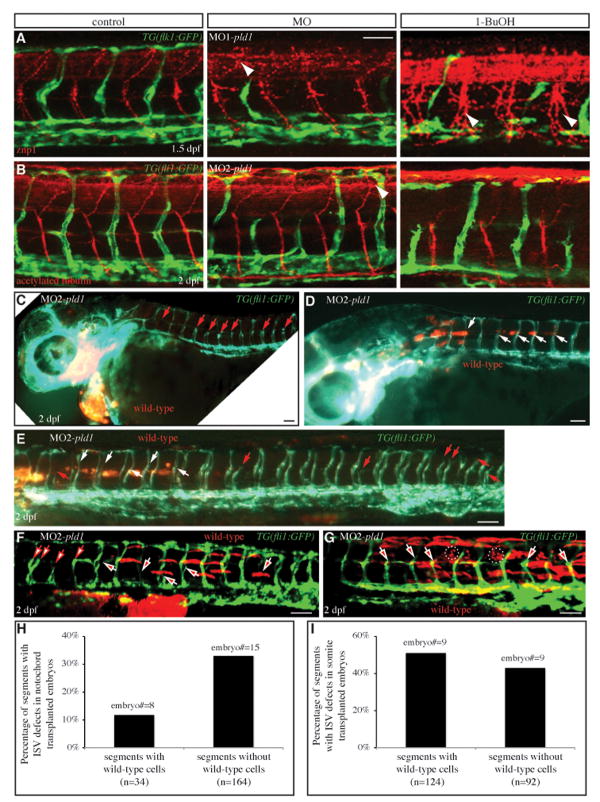
The pld1 morphant embryos underwent the early cleavages during blastula stages and exhibited normal morphogenesis during gastrulation (data not shown). However, at 24 hpf, most of pld1 morphants exhibited little or no circulation and developed pericardiac edema. This phenotype prompted us to investigate the heart and the vascular system development. However, no obvious abnormalities in morphology and heart beat were detected (data not shown). In addition, we monitored the myocardium and endocardium development of pld1 morphants in embryos harboring either TG(flk1:GFP) (Cross et al., 2003) or TG(cmcl2:GFP) transgene (Burns et al., 2005; Huang et al., 2003), and no cardiac defects were observed (data not shown). Compared to wild-type TG(flk1:GFP) embryos (3/3) (Supplementary Fig. 6C), MO2-pld1 (6ng) injected embryos (4/4) (Supplementary Fig. 6D) form the major cranial vessels of the head normally. However, the CCtAs (Cerebellar central arteries) (white arrows) and the retinal IOC vessels (Inner optic circle) (white dotted circle) are disorganized. In the axial trunk vasculature of control TG(flk1:GFP /gata1:DsRed) double transgenic embryos (Supplementary Fig. 6A), we observed normal formation of the caudal vein (CV)— the plexus-like venous network posterior to the yolk extension and normal red blood cell circulation. Similarly, 0.3% 1-BuOH treated embryos (Supplementary Fig. 6B) exhibited a largely normal CV plexus. However, this structure was slightly less complex than that of control embryos and both the axial vessels and the ISV contained fewer red blood cells. We observed similar phenotypes in MO1/MO2-pld1 – injected embryos (Figure 4D′ and data not shown). Importantly, the expression of gata1, a red blood cell marker, was strongly affected in pld1 morphants at an earlier developmental stage (Supplementary Fig. 6E-H). Given that pld1 morphants/1- BuOH treated embryos also exhibited edema and slower heart beat, it is likely that the reduced circulation observed in these animals is due to the combined effects of having a reduced number of red blood cells and a diminished heart output, which likely affects both vascular lumenization and blood cell distribution. Most interestingly, our continued examination of vascular development in pld1 morphants by visualizing an endogenous alkaline phosphatase activity (Habeck et al., 2002) revealed defects in the intersegmental vessels (ISVs) (data not shown). ISVs, which form trunk vascular network by linking the dorsal aorta or the posterior caudal vein (PCV) with the dorsal longitudinal anastomotic vessels, are thought to develop through the process of angiogenesis (Childs et al., 2002; Isogai et al., 2003). To analyze further this ISV associated phenotype, we injected either MO1-pld1, MO2-pld1, MO3-pld1 (data not shown) or 5mismatch control MO2-pld1 into TG(flk1:GFP) (data not shown) or TG(fli1:GFP) zebrafish embryos at 1-cell stage and monitored the development of ISVs (Fig. 4). Compared to control siblings (Fig. 4A), normal ISV development was also observed in 5mismatch MO1-pld1 injected embryos (Fig. 4B) at 1 dpf (data not shown) and 2 dpf (Figs. 4A′, B′, green arrows). By contrast, embryos injected with either 6 ng MO2-pld1 (Fig. 4C) or 4 ng MO1-pld1 (Fig. 4D) exhibited defects of the dorsal outgrowth of ISVs or mispatterning of ISVs with a high frequency (Fig. 4C′, 79%, n=266; Fig. 4D′, 87%, n=225; Fig. 4G), respectively. Both MO1-pld1 and the MO2-pld1 caused ISV defects in a dose-dependent manner. Injections of MO1-pld1 at a 2 ng dosage and of MO2-pld1 at a 3 ng dosage produced ISV defects with a frequency of 33% (n=216) and 35% (n=160), respectively. Whereas co-injections of these amounts of the two MOs resulted in ISV defects at 67% (n=98) (Fig. 4G). As a negative control, the 5mismatch MO2-pld1 at an 8 ng dosage caused only infrequent ISV defects (14%, n=84), in comparison with the spontaneous defects (7%, n=437) observed in uninjected control embryos (Fig. 4G). These results demonstrate that the inhibition of Pld1 function impairs ISV development.
Fig. 4. Disruption of Pld1 signaling impairs development of intersegmental vessels.
(A,B,C,D,E,F) Nomarski and (A′,B′,C′,D′,E′,F′) fluorescent images of trunk region of TG(fli1:GFP) embryos at 2 dpf. (A,A′) uninjected control embryos, (B,B′) embryos injected with 8ng 5mismatch MO2-pld1 and (C,C′) embryos injected with 6ng MO2-pld1, or (D,D′) 4ng MO1-pld1, (E,E′) 0.3% t-butanol treated embryo and (F,F′) to 0.3% 1-butanol treated embryo. (A′,B′,E′) Green arrows point to ISVs with normal morphology, whereas (C′,D′,F′) red arrows point to ISV defects, where ISVs stop in the middle along the ventral-dorsal axis or show completely blocked sprouting from the ventral blood vessels (D′,F′). Scale bar represents 20 μm (A′) and 250 μm (A).
(G) A bar graph shows the percentage of ISV defects in embryos that were injected with different morpholinos. The total number of embryos is indicated above the bar as well as the number of experimental repeats (in parentheses).
1-butanol treatment phenocopies the ISV defects of pld1 morphants
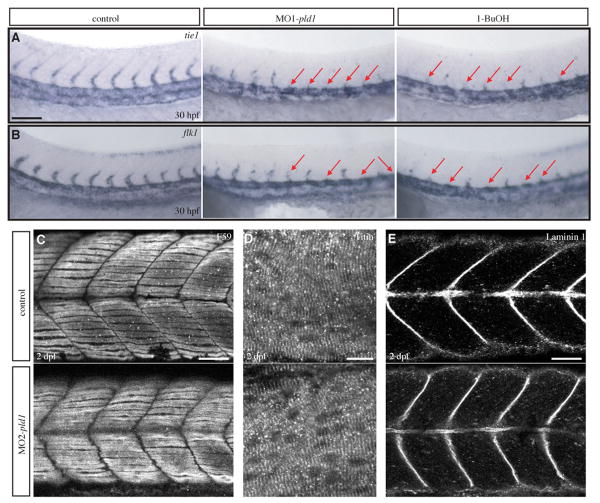
In the presence of low concentrations of primary alcohols such as ethanol, 1-butanol or glycerol, PLD catalyzes a transphosphatidylation reaction to produce the corresponding phosphatidylalcohol at the expense of phosphatidic acid production. In the case of 1-butanol, PtdBuOH, the phosphatidylalcohol generated by the transphosphatidylation reaction, is a lipid not normally present in cells and therefore not readily metabolized. Thus, 1-butanol has been widely used to block PLD signaling by diverting phosphatidic acid production to PtdBuOH, while tertiary(t) butanol, a non-primary alcohol that can not be used in the transphosphatidylation reaction, was used as a control for general toxicity. To test whether blocking PLD signaling with 1-butanol has similar effects on zebrafish vascular development, we incubated zebrafish embryos with 0.3% 1-butanol or t-butanol, as a negative control, between 12 and 24 hpf, which is the window of ISV development (Childs et al., 2002; Isogai et al., 2003). Embryos treated with 1-butanol were slightly shorter, developed pericardiac edema and exhibited blood accumulation in the trunk and around the yolk (Fig. 4F; 98%, n=161). In contrast, no obvious defect was observed in t-butanol treated embryos (Fig. 4E; 5%, n=123) over similar time courses. To determine the effect of Pld signaling blockage on the vascular development, TG(flk1:GFP) (data not shown) or TG(fli1:GFP) (Figs. 4E′, F′) zebrafish embryos were treated with 1-butanol (Fig. 4F′) or t-butanol (Fig. 4E′) between 12 and 24 hpf and the vascular development was examined by fluorescence microscopy. Indeed, 1-butanol treated embryos exhibited very similar ISV defects (Fig. 4F′, red arrows, 96%, n=317) to those described above for pld1 morphants (Figs. 4C′, D′). By contrast, normal ISV development was observed in t-butanol treated embryos (Figure 4E′, green arrows; 95%, n=123). To characterize in more detail the ISV defect in pld1 morphants or 1-butanol treated embryos, we tested the expression of tie1 and flk1, two endothelial marker genes (Fouquet et al., 1997; Lyons et al., 1998). Consistent with the ISV phenotype found in TG(flk1:GFP)/TG(fli1:GFP) zebrafish, we observed either the absence of both tie1 and flk1 positive ISVs or expression of tie1 and flk1 marking shortened sprouting ISVs in either MO1-pld1/ MO2-pld1 injected or 1-butanol treated embryos (Fig. 6A, B, red arrows).
Fig. 6. Effect of Pld1 deficiency on ISV and surrounding tissue development.
(A,B) Loss of Pld1 function impairs ISV development. Whole mount in situ hybridization showing expression of tie1 (A) and flk1 (B) at 30 hpf in the trunk region of uninjected control embryos, and embryos either injected with MO1-pld1 or treated with 1-butanol (1-BuOH). Red arrows indicate stunted ISVs.
(C-E) Somite muscle and notochord development is relatively normal in embryos with disrupted Pld1 signaling. Representative confocal images of whole-mount immunohistochemistry show the morphology of somites, slow muscle and notochord in MO2-pld1 injected embryos at 2 dpf. Lateral view, anterior to the left. (C) F59 antibody was used to stain slow muscle Myosin. (D) Titin antibody was used to label the organization of muscle cell “Z-discs”. (E) Laminin immunoreactivity at the myosepta and the notochord region in control sibling and pld1 morphants embryos. Scale bar represents 10 μm (D), 20 μm (C,E) and 40 μm (A).
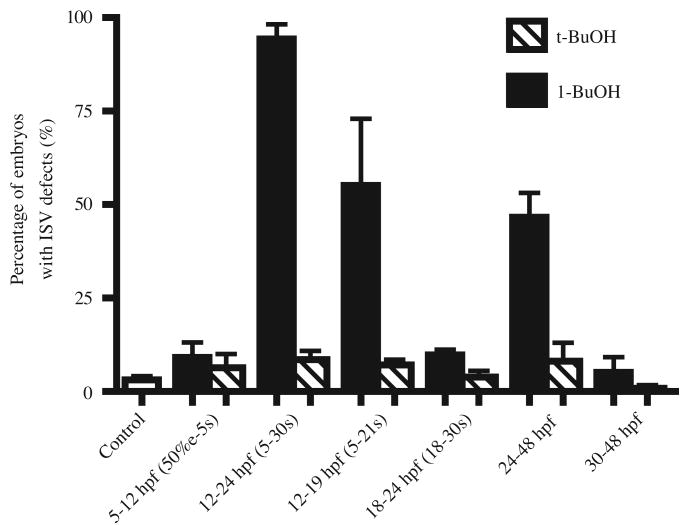
Since the inhibition of PA formation and subsequent lipid signaling by 1-butanol can be released by washing it out in the medium, we applied 1-butanol or t-butanol to TG(flk1:GFP) zebrafish embryos at certain time periods to start to determine the critical time window of Pld signaling in ISV development. When embryos were treated with 1-butanol between 12 and 24 hpf or 12 and 19 hpf, ISV defects were detected in 94% or 55% of total 317 or 153 embryos at 3 dpf, respectively (Fig. 5). Since the primary ISVs do not sprout from dorsal aorta until around 20 hpf (Childs et al., 2002; Isogai et al., 2003), the significant fraction of embryos with ISV defects in the population of embryos treated between 12 and 19 hpf (Fig. 5) suggests that the disruption of Pld1 signaling inhibited the initial migration of ISV endothelial cells along dorsal aorta and/or sprouting from the dorsal aorta. The higher percentage of affected embryos in the population treated between 12 and 24 hpf (Fig. 5) indicates that Pld1 signaling inhibition also interfered with the continued dorsal migration of ISV endothelial cells. When embryos were treated with 1-butanol between 18 and 24 hpf, the growth of ISVs was temporally arrested and then continued when 1-butanol was washed out (Fig. 5 and data not shown). No significant ISV defect was detected when 1-butanol was applied at 3dpf (Fig. 5). However, when embryos were treated with 1-butanol between 24 and 48hpf, 47% of them showed the ISV defect (Fig. 5, n=170), in which the dorsally directed sprouting of ISV was arrested at the level of horizontal myoseptum at 3dpf (data not show). We interpret these results to mean that Pld1 signal is also required for the growth of ISV toward the dorsal halves of the trunk somites. As a control, when embryos were treated with the same concentration of t-butanol in equivalent time windows, no significant ISV defect was observed (Fig. 5, n=1069). These results indicate that Pld1 signaling is required for both the initial sprouting and the continued dorsal growth of ISVs, whereas this time-course experiment analyzes only sensitivity of the end-point of ISV development to different 1-butanol exposure.
Fig. 5. Pld1 signaling is required for ISV development at different developmental stages.
The percentage of embryos showing ISV defects after either 0.3% 1-butanol (1-BuOH, black bars) or 0.3 % t-butanol (t-BuOH, hatched bars) treatment during different time periods of development. Error bars depict standard deviation.
The effect of inhibiting Pld1 signaling on tissues neighboring ISVs
Next we explored the mechanism via which Pld1 deficiency impairs ISV development. Given pld1 expression in developing somites and notochord at the time of ISV morphogenesis (Fig. 2), it is noteworthy that proper segmentation and somite boundary stabilization are required for normal ISV guidance (Shaw et al., 2006). Hence we monitored somite formation and differentiation in pld1 morphants and 1-butanol treated embryos. Similar to control embryos in which muscle fibers were visualized with phalloidin staining at 1 dpf, the somites of pld1 morphants and 1-butanol treated embryos displayed a normal chevron-shape. Moreover, no defect in the somitic boundary formation was found in pld1 morphants and 1-butanol treated embryos (data not shown). The development of slow muscle fibers (F59 antibody) (Fig. 6C, n=12; and data not shown in 1-butanol treatment) and muscle cell differentiation (the sarcomeric organization of Titin) were also relatively normal (Fig. 6D, n=9; and data not shown in 1-butanol treatment). Recent studies have also shown that Laminins, the major constituents of the basement membrane, are involved in notochord differentiation and ISV development (Pollard et al., 2006). Our examination of Laminin1 expression in the trunk region suggested that it is not significantly affected when Pld1 activity is disrupted (Fig. 6E, n=12; and data not shown in 1-butanol treatment). Thus, the ISV defect in Pld1 signaling deficient embryos is unlikely to be related to abnormal segmentation, or somite development. According to a previous report, in some mutants, ISV defects are also associated with impaired notochord differentiation, manifest by persistent expression of the early chordamesoderm markers at 1 dpf (Pollard et al., 2006). However, our analysis of the early chordamesoderm marker shh showed that it was normally downregulated in pld1 morphants or 1-butanol treated embryos (data not shown), suggesting notochord differentiation was not affected when Pld1 function was disrupted.
PLD1 has been recently reported to promote myogenic differentiation in tissue culture (Yoon and Chen, 2008). VEGF and semaphorin are two angiogenic factors expressed in differentiating somites (Halloran et al., 1998; Liang et al., 1998; Shoji et al., 2003). However, our whole mount in situ hybridization analyses of the expression of genes encoding VEGF and semaphorins 3a1, 3a2, and 3E in pld1 morphants and 1-butanol treated embryos found no significant changes in their expression pattern or levels (data not shown). This suggests that Pld1 may regulate the ISV development through a parallel pathway controlling angiogenesis or play a role downstream of these angiogenic pathways.
Inhibition of Pld1 function does not strongly impair the dorsal-ventral guidance of primary/secondary motor axons
Current studies have shown that blood vessel and axon guidance share common molecular mechanisms (Bedell et al., 2005; Carmeliet and Tessier-Lavigne, 2005; Lee et al., 2002; Lu et al., 2004; Torres-Vazquez et al., 2004). In zebrafish, each somitic hemisegment is innervated by three primary motoneurons; the caudal primary motoneuron (CaP), the middle primary motoneuron (MiP) and the rostral primary motoneuron (RoP). The axons of all three motoneurons exit from the spinal cord, and extend ventrally along a common pathway and reach a choice point, the horizontal myoseptum region. After pausing at the choice point, the pathways of the axons diverge in the following manner: CaP extends ventrally along the ventromedial myotome; MiP grows dorsally along the dorsalmedial myotome; and RoP extends laterally through the horizontal myoseptum (Beattie et al., 2002; Eisen et al., 1986). To determine whether Pld signaling is also involved in the guidance of motor neuron axons, we performed immunohistochemistry using a Znp1 antibody to monitor axon guidance in TG(flk1:GFP) zebrafish. Although the growth of intersegmental vessels was dramatically impaired in either MO1-pld1 (4 ng) injected or 1-butanol treated embryos, no significant defects in motoneuron axon (CaP) dorsal-ventral outgrowth were observed in these embryos, compared with control embryos (Fig. 7A). Consistent with these results, in anti-acetylated tubulin antibody labeled embryos, no significant motor neuron axons guidance defects were observed in pld1 morphants or 1-butanol treated embryos (TG(fli1:GFP)) by monitoring the original confocal image stacks (Fig. 7B and data not shown). We noted however, that these ventrally extending axons exhibited more branched morphology (Fig. 7A, B, white arrowheads). These results suggest that Pld1 signaling specifically regulates blood vessel growth but not motor neuron axon guidance in the trunk of zebrafish embryos.
Fig. 7.
(A,B) Disruption of Pld1 signaling differentially affects ISV and motor axon dorsal-ventral morphogenesis. (A) Uninjected control TG(flk1:GFP) embryos were stained with znp1 antibody to visualize primary motor neurons (red), which extend from the dorsal to the ventral side along with ISV (green). (B) Anti-acetylated tubulin antibody detects both the primary and secondary motor neuron cell body and axons (red) in the trunk region of uninjected control TG(fli1:GFP) embryos, and ISV (green) is positioned in between. Embryos with MO1-pld1 injection or 1-butanol treatment exhibit impaired ISV morphology compared with control embryos; however, all the motor neurons are able to connect from the ventral side to the dorsal side. Interestingly, the motoneurons are more branched when Pld1 signaling is disrupted (white arrowheads), compared to control embryos.
(C-E,H) Transplanted wild-type cells in the notochord partially rescue the ISV defects in pld1 morphants. (C-E) Fluorescent images of TG(fli:GFP) pld1 morphants containing Rhodamine-dextran labeled cells (red) transplanted from wild-type donor embryos, and localized in the prechordal plate (C) or anterior notochord (D,E). Lateral view, anterior to the left. ISVs are visualized in green by TG(fli:GFP). White arrows point to normal ISVs in the regions of the chimeric embryos possessing transplanted wild-type cells, whereas red arrows indicate defective ISVs in segments lacking wild-type cells. (H) In chimeric embryos harboring transplanted cells in the notochord, the percentage of ISV defects in segments with wild-type cells (12%, segments number=34, 8 embryos) compared to segments without wild-type cells (33%, segments number=164, 15 embryos).
(F,G,I) Transplanted wild-type cells in the somites cannot rescue the ISV defects inpld1 morphants. (F,G) Fluorescent images of TG(fli:GFP) pld1 morphants containing Rhodamine-dextran labeled cells (red) transplanted from wild-type donor embryos, and localized in the somites. Lateral view, anterior to the left. ISVs are visualized in green by TG(fli:GFP). White arrows with red filling point to defective ISVs in the regions of the chimeric embryos possessing transplanted wild-type cells, whereas red arrows with white filling indicate defective ISVs in segments lacking wild-type cells. (I) In the chimeras exhibiting transplanted cells in the somites, the percentage of ISV defects in segments with wild-type cells (51%, segments number=124, 9 embryos) was comparable to segments without wild-type cells (43%, segments number=92, 9 embryos). Scale bar represents 20 μm (A) and 30 μm (C-G).
Non-autonomous role of Pld1 in ISV development
In order to test further the specificity of Pld1 in ISV development, we undertook two strategies to rescue a normal ISV development in pld1 morphants. Firstly, our efforts to rescue the ISV defects in pld1 morphants were hampered by the fact that injections of synthetic pld1 RNA caused gastrulation defects (data not shown). In our second strategy to restore normal Pld1 activity in pld1 morphants and to identify the tissue source of Pld1 relevant to the ISV development, we transplanted wild-type cells into pld1 morphant embryos. In these experiments, we labeled prospective wild-type donor embryos by injecting Rhodamine-dextran dye at the 1-cell stage. We disrupted normal Pld1 activity in prospective host embryos by injecting MO2-pld1 into transgenic fli1:GFP zebrafish embryos (6 ng/embryo). At the beginning of gastrulation when the Spemann-Mangold organizer equivalent structure, the embryonic shield becomes visible as a thickening of the dorsal blastoderm margin, we transplanted a group of wild-type cells containing Rhodomine-dextran dye from the shield of the wild-type donor embryos into the equivalent region of MO2-pld1 injected TG(fli1:GFP) host embryos. Consistent with previous reports (Miyagi et al., 2004; Saude et al., 2000), some of the transplanted wild-type cells, were successfully integrated into the shield, and subsequently contributed to the prechordal plate mesoderm and the chordamesoderm. Altogether, 15 of the 15 generated chimeric embryos exhibited Rhodomine-dextran labeled cells in the prechordal plate derivatives (Fig. 7C), whereas 8 of them had Rhodomine-dextran labeled cells in both the prechordal plate mesoderm and notochord (Figs. 7D, E and data not shown). In these chimeric embryos, we observed a total of 34 segments containing the transplanted wild-type cells. Using TG(fli1:GFP) to analyze ISV morphogenesis we observed that only 4 (<12%) of these chimeric segments were associated with ISV defects (embryo number =8, Fig. 7E, red arrows; data not shown; Fig. 7H), while the remaining wild-type cell containing segments exhibited relatively normal ISVs (Fig. 7D, E, white arrows). By contrast, 33% of the nonchimeric segments exhibited ISV defects (embryo number=15, segment number=164; Figs. 7D, E, red arrows; and Fig. 7H). Given that pld1 is also expressed in the paraxial mesoderm (Fig. 2), we also carried out experiments in which paraxial mesoderm cells were transplanted into equivalent position of morphant embryos. As expected, these transplanted paraxial cells contributed largely to the somites of the host embryos (Fig. F,G) with a few of them contributing to the ventral spinal cord (Fig. 7G, white dotted circle) due to proximity of prospective neuroectodermal cells to the mesoendoderm cells at early gastrulation (Kimmel et al., 1990). Altogether, 9 generated chimeric embryos exhibited Rhodomine-dextran labeled cells in the paraxial mesoderm derivatives (Fig. 7F,G). In these chimeric embryos, we observed a total of 124 segments containing the transplanted wild-type cells. Using TG(fli1:GFP) to analyze ISV morphogenesis we observed that 64/124 (52%) of these chimeric segments were still associated with ISV defects (embryo number =9, Fig. 7F,G, white arrows with red filling; Fig. 7I), comparable to 43% of the remaining nonchimeric segments with ISV defects (embryo number=9, segment number=92; Figs. 7F, white arrows with red filling; and Fig. 7I). Our transplantation results demonstrated that by providing Pld1 at the endogenous locations in morphants could partially rescue the affected ISV development, thus supporting the specificity of the observed ISV phenotype. Moreover, these experiments suggest that Pld1 influences ISV development in non-autonomous manner by acting in the notochord tissues.
Discussion
Whereas PLD has been implicated in many cellular processes, its in vivo function in vertebrates is not clear due to the lack of loss-of-function studies in the context of vertebrate animal models. Here we cloned the zebrafish ortholog of PLD1, examined its expression and function during zebrafish development, and provided evidence that Pld1 is required for ISV development.
Zebrafish Pld1 shares higher similarity with mammalian PLD1 than with PLD2 (Supplementary Fig. 1), and our biochemical data shows that it is regulated by PKCα and Arf (Fig. 1). These observations support the notion of Pld1 being the ortholog of mammalian PLD1. In fact, an EST encoding a fragment of the zebrafish Pld2 has been recently deposited in Genbank, and cloning of the pld2 gene is in progress.
The pld1 gene product displays a dynamic expression pattern during zebrafish development. Following ubiquitous expression until late gastrulation, the earliest regionalized expression of pld1 was detected in notochord of 4 somite-stage embryos (Figs. 2B, C). Strong pld1 expression in notochord was observed until 18 somite stage (Figs. 2F, G), when it was downregulated in the anterior region and maintained in the posterior nascent notochord when the expression in the somites commences.
Using two different loss-of-function approaches we found that Pld1 is required for proper ISV sprouting and migration. First, we observed similar defects in ISV development, absent or stunted ISVs, in embryos in which Pld1 function was reduced using one MO designed to interfere with pld1 RNA translation and two MOs interfering with its splicing (Figs. 3, 4). Second, these defects occurred in a dose dependent manner (Fig. 4G). Third, ISV defects were not observed when a control morpholino containing 5 mismatches with the MO2-pld1 was used (Figs. 4B′, G). The role of Pld1 in ISV development is further supported by experiments in which Pld1 signaling was blocked using 1-butanol in flk1:GFP/fli1:GFP transgenic zebrafish and monitoring endothelial marker genes, tie1 and flk1, expression in morpholino-injected or chemical treated wild-type embryos (Figs. 4F′, and 6A, B). Importantly, ISV defects were observed in embryos treated with 1-butanol but not with t-butanol (Fig. 5), a non-primary alcohol that cannot be used by Pld1 in the transphosphatidylation reaction. Partial suppression of the ISV morphogenetic defects observed in chimeric pld1morphant embryos containing transplanted wild-type cells (Figs. 7C-E, H) provides another strong evidence that the ISV defects observed in pld1 morphants and 1-butanol treated embryos reflect a specific role of Pld1 in ISV development. These several lines of evidence support the notion that Pld1 plays an important role in regulating ISV morphogenesis.
An important question is to what degree Pld1 function was reduced in these experiments. We have demonstrated the efficiency of both the translation MO and splicing MOs. The translation MO significantly reduced translation of the Pld1 protein in an in vitro translation assay (Fig. 3B). Furthermore, the levels of endogenous transcripts were highly downregulated in embryos injected with MOs targeting pld1 splicing (Fig. 3C). Importantly, we observed significant downregulation of Pld activity in pld1 morphants and embryos treated with 1-butanol, using mass spectrum analyses (Fig. 3D and Supplementary Figs. 3-5). These several lines of evidence indicate that the observed phenotypic defects reflect strong partial loss of Pld1 function. However, it is possible that stronger and/or new defects will be observed when Pld1 function is completely abolished. Possible functional redundancies between Pld1 and Pld2 also await future investigation.
The zebrafish ISV network is formed via a two-step angiogenic mechanism (Isogai et al., 2003). The primary ISV sprouts emerge exclusively from the dorsal aorta at about 20 hpf, reach the dorsolateral roof of the neural tube at about 28 hpf, then branch and form dorsal longitudinal anastomotis vessels (DLAL) by 1.5 dpf. The secondary ISVs sprout exclusively from the posterior cardinal vein in a less synchronous manner after the formation of the primary ISV network is completed. Some of the secondary sprouts connect to the base of primary sprouts and become intersegmental veins. Lineage analyses demonstrated that the endothelial cells forming ISVs are derived from the lateral posterior mesoderm, migrate along with axial vessels before ISV sprouting and then dorsally between somites to their final position (Childs et al., 2002). Pathfinding for the ISV ventral-dorsal migration likely involves multiple repulsive and attractive cues (Bedell et al., 2005; Lee et al., 2002; Lu et al., 2004; Torres-Vazquez et al., 2004). As ISVs in Pld1-deficient embryos were either absent or severely shortened, Pld1 is likely required for ISV sprouting and migration (Figs. 4, 5).
When does Pld1 act to influence ISV development? Transient treatments with 1-butanol established two phases of Pld1 requirement for ISV morphogenesis. The first one during segmentation stages (12-19hpf) and the second at 24-48 hpf, but is no longer required after 30hpf (Fig. 5). Despite our 1-butonal time-course experiment aimed to discover a requirement window of Pld in ISV development, it will be important in the future to ask whether different exposure regimens trigger different mechanisms of ISV interruption, e.g. with earlier exposures affecting the emergence of early arterial sprouts and later exposure impairing the later-emerging venous sprouts. The fact that during the shorter treatments (12-19hpf and 18-24hpf) ISVs halted their outgrowth but resumed it after removal of 1-butanol could be interpreted to mean that pld1 acts in ISV development from 12-30hpf. Interestingly, during segmentation stages to 1 dpf, pld1 is strongly expressed in mature somites and transiently in the differentiating notochord. It is known that somites are the source of a variety of molecules implicated in guidance of blood vessel morphogenesis, including VEGF, Semaphorins, Netrin and Slits (Lu et al., 2004; Shoji et al., 2003; Torres-Vazquez et al., 2004). Many recent publications indicate that axons and blood vessels share similar guidance mechanisms (Bedell et al., 2005; Carmeliet and Tessier-Lavigne, 2005; Lee et al., 2002; Lu et al., 2004; Torres-Vazquez et al., 2004). Although we found an ISV defect in pld1 morphant embryos, we did not observe any striking defect of the motor axon dorsal-ventral guidance in Pld1-deficient embryos using Znp1 (Zn8, Zn12 and anti-acetylated tubulin) antibody staining (Figs. 7A, B and data not shown). However, we noted more branched morphology of these motoneurons. This suggests that the Pld1 activity is specifically required for blood vessel guidance. Whereas Pld1 appears less important for dorsal-ventral motor neuron axon guidance, it may play a role in axonal morphogenesis. In support of this notion, it was recently reported that PLD1 promotes outgrowth of hippocampal dentate granule cell axons by enhancing tissue plasminogen release (Zhang et al., 2005).
The notochord, which produces a variety of secreted signals, plays an essential role in the patterning of surrounding tissues including blood vessels, somites, pancreas, and the neural tube (Stemple, 2005). Hence, it is possible that Pld1 may play some role in tissue patterning and ISV morphogenesis by regulating the notochord differentiation and/or production of notochord-derived signals. Consistent with this notion, some pld1 morphant embryos exhibited curved body axis (Fig. 4D). However, expression of one of the notochord localized proteins, Laminin 1 involved in ISV development (Pollard et al., 2006), was unaffected in pld1 deficient embryos (Fig. 6E). That shh expression becomes downregulated in the notochord in pld1 morphants on schedule (data not shown) further suggests that the notochord differentiation is normal.
A critical role of Pld1 for ISV development in the notochord and not the somites is further supported by the results of our transplantation experiments, in which presence of wild-type notochord cells in pld1 chimeric morphants, correlated with normal ISV development that has not been observed in chimeric embryos harboring transplanted wild-type cells only in the somites. Thus it is tempting to speculate that Pld1 is required in the notochord for a signal that promotes or guides ISV morphogenesis. The fact that treatment with 1-butanol can inhibit ISV formation at a time period (24-48 hpf) when pld1 is not expressed in the notochord could be due to expression of pld2 in this or different tissues of zebrafish embryos at this particular time. Another alternative explanation could be that zebrafish Pld1 protein perdures in the notochord to fulfill its function until this stage of development, even when pld1 transcripts are no longer detectable. Our in situ hybridization analyses revealed that the expression pattern or level of endothelial and blood vessel cells markers (flk1 and tie1) in pld1 morphant or 1-butanol treated embryos were relatively normal, indicating that blood vessel fates are properly specified. Moreover, we have not found any changes in expression of several genes encoding molecules (vegf, angiopoietin, semaphorin 3a1, 3a2 and 3E, slit1 and slit2) implicated in angiogenesis. It is possible that the changes in the expression level of these guidance factors are subtle and in situ hybridization is not sensitive enough to detect the changes. It is also possible that Pld1 may function independently or downstream of these or other guidance signals.
Previous studies of another type of phospholipase, phospholipase C gamma1, revealed its important role in primitive hematopoiesis during zebrafish development (Ma et al., 2007). Zebrafish PLCgamma1 was also implicated in arterial development downstream of VEGF (Lawson et al., 2003). That both phospholipase C gamma1 and Pld1 are required for different aspects of blood vessel development in zebrafish suggests that lipid signaling plays a significant role in the development of the vertebrate vascular system. The distinct requirements for these two phospholipases likely reflect distinct products from each unique enzymatic reaction. PLD has previously been implicated in cell migration. Gomez-Cambronero and colleagues demonstrated that PLD plays a key role in leukocyte adhesion and chemotaxis toward a gradient of chemokines (Gomez-Cambronero et al., 2007). Similarly, expression of active PLD2 is associated with spreading and elongation of EL4 cells in a cultured system (Knoepp et al., 2008). PLD has also been implicated in Dictyostelium discoideum as a gene involved in cell migration (Nagasaki and Uyeda, 2008). PA is the main product of the PLD catalyzed reaction and this bioactive lipid is thought to mediate many of PLD's functions. Delon et al (Delon et al., 2004) demonstrated that PA binds to sphingosine kinase, which phosphorylates sphingosine to generate sphingosine 1-phosphate (S1P), and facilitates its translocation to PA-enriched membrane compartments. S1P, acting through its receptors, was shown to promote blood vessel formation by regulating endothelial cell migration and survival (Kono et al., 2004; Osborne and Stainier, 2003). PA can be further converted to LPA, which activates its G protein coupled receptors. LPA signaling has been implicated in both cancer cell and endothelial cell migration (Mills and Moolenaar, 2003; Osborne and Stainier, 2003). Mazie et al. reported that short-chain analogues of PA induce motility in corneal and other epithelial cell types (Mazie et al., 2006). Subsequent metabolism did not appear to be required and although the downstream targets were not identified the authors noted a striking transactivation of the epidermal growth factor receptor as a possible mechanism. Thus, it is possible that Pld1 plays a role upstream of S1P or LPA to regulate angiogenesis.
In summary, we have cloned a zebrafish homolog of pld1 gene and described its dynamic expression pattern during embryonic development. We characterized the regulatory properties and found that it is a bona fide PLD1 enzyme. Using both loss-of-function and chemical approaches, we identified a novel cell non-autonomous role for Pld1 in the regulation of angiogenesis.
Supplementary Material
Acknowledgments
We thank members of the Brown and LSK group for discussions, Dr. Sarah Childs and Dr. Jesús Torres-Vázquez for advice, Dr. Charles Hong for sharing the fli1:GFP zebrafish transgenic line, Dr. Jesús Torres-Vázquez for sharing the flk1:GFP/gata1:DsRed zebrafish transgenic line and Dr. Sarah Kucenas in B. Appel lab for sharing motoneuron antibodies. We acknowledge excellent fish care by Heidi Beck and other SC Fish Facility Research Assistants. This work was partially supported by grants from the National Institutes of Health (U54 GM69338 to HAB, GM77770 to LSK, and HL073348 to TPZ) as well as support from the Ingram Cancer Professorship endowment to HAB and Martha Rivers Ingram endowment to LSK.
Footnotes
Pld1 acts in zebrafish angiogenesis
References
- Beattie CE, Granato M, Kuwada JY. Cellular, genetic and molecular mechanisms of axonal guidance in the zebrafish. Results Probl Cell Differ. 2002;40:252–69. doi: 10.1007/978-3-540-46041-1_13. [DOI] [PubMed] [Google Scholar]
- Bedell VM, Yeo SY, Park KW, Chung J, Seth P, Shivalingappa V, Zhao J, Obara T, Sukhatme VP, Drummond IA, Li DY, Ramchandran R. roundabout4 is essential for angiogenesis in vivo. Proc Natl Acad Sci U S A. 2005;102:6373–8. doi: 10.1073/pnas.0408318102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HA, Gutowski S, Moomaw CR, Slaughter C, Sternweis PC. ADP-ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell. 1993;75:1137–44. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- Brown HA, Henage LG, Preininger AM, Xiang Y, Exton JH. Biochemical analysis of phospholipase D. Methods Enzymol. 2007;434:49–87. doi: 10.1016/S0076-6879(07)34004-4. [DOI] [PubMed] [Google Scholar]
- Buchanan FG, McReynolds M, Couvillon A, Kam Y, Holla VR, Dubois RN, Exton JH. Requirement of phospholipase D1 activity in H-RasV12-induced transformation. Proc Natl Acad Sci U S A. 2005;102:1638–42. doi: 10.1073/pnas.0406698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CG, Milan DJ, Grande EJ, Rottbauer W, MacRae CA, Fishman MC. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat Chem Biol. 2005;1:263–4. doi: 10.1038/nchembio732. [DOI] [PubMed] [Google Scholar]
- Cai D, Zhong M, Wang R, Netzer WJ, Shields D, Zheng H, Sisodia SS, Foster DA, Gorelick FS, Xu H, Greengard P. Phospholipase D1 corrects impaired betaAPP trafficking and neurite outgrowth in familial Alzheimer's disease-linked presenilin-1 mutant neurons. Proc Natl Acad Sci U S A. 2006;103:1936–40. doi: 10.1073/pnas.0510710103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Exton JH. Determination of interaction sites of phospholipase D1 for RhoA. Biochem J. 2001;355:779–85. doi: 10.1042/bj3550779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature. 2005;436:193–200. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- Childs S, Chen JN, Garrity DM, Fishman MC. Patterning of angiogenesis in the zebrafish embryo. Development. 2002;129:973–82. doi: 10.1242/dev.129.4.973. [DOI] [PubMed] [Google Scholar]
- Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA. Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr Biol. 1997;7:191–201. doi: 10.1016/s0960-9822(97)70090-3. [DOI] [PubMed] [Google Scholar]
- Cross LM, Cook MA, Lin S, Chen JN, Rubinstein AL. Rapid analysis of angiogenesis drugs in a live fluorescent zebrafish assay. Arterioscler Thromb Vasc Biol. 2003;23:911–2. doi: 10.1161/01.ATV.0000068685.72914.7E. [DOI] [PubMed] [Google Scholar]
- Delon C, Manifava M, Wood E, Thompson D, Krugmann S, Pyne S, Ktistakis NT. Sphingosine kinase 1 is an intracellular effector of phosphatidic acid. J Biol Chem. 2004;279:44763–74. doi: 10.1074/jbc.M405771200. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Myers PZ, Westerfield M. Pathway selection by growth cones of identified motoneurones in live zebra fish embryos. Nature. 1986;320:269–71. doi: 10.1038/320269a0. [DOI] [PubMed] [Google Scholar]
- Exton JH. Phospholipase D-structure, regulation and function. Rev Physiol Biochem Pharmacol. 2002;144:1–94. doi: 10.1007/BFb0116585. [DOI] [PubMed] [Google Scholar]
- Fouquet B, Weinstein BM, Serluca FC, Fishman MC. Vessel patterning in the embryo of the zebrafish: guidance by notochord. Dev Biol. 1997;183:37–48. doi: 10.1006/dbio.1996.8495. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Moore S, Bell RM, Dush M. Functional analysis of a phosphatidic acid binding domain in human Raf-1 kinase: mutations in the phosphatidate binding domain lead to tail and trunk abnormalities in developing zebrafish embryos. J Biol Chem. 2003;278:45690–6. doi: 10.1074/jbc.M302933200. [DOI] [PubMed] [Google Scholar]
- Gomez-Cambronero J, Di Fulvio M, Knapek K. Understanding phospholipase D (PLD) using leukocytes: PLD involvement in cell adhesion and chemotaxis. J Leukoc Biol. 2007;82:272–81. doi: 10.1189/jlb.0107033. [DOI] [PubMed] [Google Scholar]
- Habeck H, Odenthal J, Walderich B, Maischein H, Schulte-Merker S. Analysis of a zebrafish VEGF receptor mutant reveals specific disruption of angiogenesis. Curr Biol. 2002;12:1405–12. doi: 10.1016/s0960-9822(02)01044-8. [DOI] [PubMed] [Google Scholar]
- Halloran MC, Severance SM, Yee CS, Gemza DL, Kuwada JY. Molecular cloning and expression of two novel zebrafish semaphorins. Mech Dev. 1998;76:165–8. doi: 10.1016/s0925-4773(98)00124-5. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Altshuller YM, Sung TC, Rudge SA, Rose K, Engebrecht J, Morris AJ, Frohman MA. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J Biol Chem. 1995;270:29640–3. doi: 10.1074/jbc.270.50.29640. [DOI] [PubMed] [Google Scholar]
- Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- Isogai S, Lawson ND, Torrealday S, Horiguchi M, Weinstein BM. Angiogenic network formation in the developing vertebrate trunk. Development. 2003;130:5281–90. doi: 10.1242/dev.00733. [DOI] [PubMed] [Google Scholar]
- Ivanova PT, Milne SB, Byrne MO, Xiang Y, Brown HA. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:21–57. doi: 10.1016/S0076-6879(07)32002-8. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Frohman MA. Phospholipase D: a lipid centric review. Cell Mol Life Sci. 2005;62:2305–16. doi: 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Schilling TF. Origin and organization of the zebrafish fate map. Development. 1990;108:581–94. doi: 10.1242/dev.108.4.581. [DOI] [PubMed] [Google Scholar]
- Knoepp SM, Chahal MS, Xie Y, Zhang Z, Brauner DJ, Hallman MA, Robinson SA, Han S, Imai M, Tomlinson S, Meier KE. Effects of active and inactive phospholipase D2 on signal transduction, adhesion, migration, invasion, and metastasis in EL4 lymphoma cells. Mol Pharmacol. 2008;74:574–84. doi: 10.1124/mol.107.040105. [DOI] [PubMed] [Google Scholar]
- Kono M, Mi Y, Liu Y, Sasaki T, Allende ML, Wu YP, Yamashita T, Proia RL. The sphingosine-1-phosphate receptors S1P1, S1P2, and S1P3 function coordinately during embryonic angiogenesis. J Biol Chem. 2004;279:29367–73. doi: 10.1074/jbc.M403937200. [DOI] [PubMed] [Google Scholar]
- Kook S, Exton JH. Identification of interaction sites of protein kinase Calpha on phospholipase D1. Cell Signal. 2005;17:1423–32. doi: 10.1016/j.cellsig.2005.03.003. [DOI] [PubMed] [Google Scholar]
- LaLonde MM, Janssens H, Rosenbaum E, Choi SY, Gergen JP, Colley NJ, Stark WS, Frohman MA. Regulation of phototransduction responsiveness and retinal degeneration by a phospholipase D-generated signaling lipid. J Cell Biol. 2005;169:471–9. doi: 10.1083/jcb.200502122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Mugford JW, Diamond BA, Weinstein BM. phospholipase C gamma-1 is required downstream of vascular endothelial growth factor during arterial development. Genes Dev. 2003;17:1346–51. doi: 10.1101/gad.1072203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–18. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Lee P, Goishi K, Davidson AJ, Mannix R, Zon L, Klagsbrun M. Neuropilin-1 is required for vascular development and is a mediator of VEGF-dependent angiogenesis in zebrafish. Proc Natl Acad Sci U S A. 2002;99:10470–5. doi: 10.1073/pnas.162366299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Xu X, Chin AJ, Balasubramaniyan NV, Teo MA, Lam TJ, Weinberg ES, Ge R. Cloning and characterization of vascular endothelial growth factor (VEGF) from zebrafish, Danio rerio. Biochim Biophys Acta. 1998;1397:14–20. doi: 10.1016/s0167-4781(97)00233-9. [DOI] [PubMed] [Google Scholar]
- Lu X, Le Noble F, Yuan L, Jiang Q, De Lafarge B, Sugiyama D, Breant C, Claes F, De Smet F, Thomas JL, Autiero M, Carmeliet P, Tessier-Lavigne M, Eichmann A. The netrin receptor UNC5B mediates guidance events controlling morphogenesis of the vascular system. Nature. 2004;432:179–86. doi: 10.1038/nature03080. [DOI] [PubMed] [Google Scholar]
- Lyons MS, Bell B, Stainier D, Peters KG. Isolation of the zebrafish homologues for the tie-1 and tie-2 endothelium-specific receptor tyrosine kinases. Dev Dyn. 1998;212:133–40. doi: 10.1002/(SICI)1097-0177(199805)212:1<133::AID-AJA12>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Ma AC, Liang R, Leung AY. The role of phospholipase C gamma 1 in primitive hematopoiesis during zebrafish development. Exp Hematol. 2007;35:368–73. doi: 10.1016/j.exphem.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Mazie AR, Spix JK, Block ER, Achebe HB, Klarlund JK. Epithelial cell motility is triggered by activation of the EGF receptor through phosphatidic acid signaling. J Cell Sci. 2006;119:1645–54. doi: 10.1242/jcs.02858. [DOI] [PubMed] [Google Scholar]
- McDermott M, Wakelam MJ, Morris AJ. Phospholipase D. Biochem Cell Biol. 2004;82:225–53. doi: 10.1139/o03-079. [DOI] [PubMed] [Google Scholar]
- Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer. 2003;3:582–91. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- Miyagi C, Yamashita S, Ohba Y, Yoshizaki H, Matsuda M, Hirano T. STAT3 noncell-autonomously controls planar cell polarity during zebrafish convergence and extension. J Cell Biol. 2004;166:975–81. doi: 10.1083/jcb.200403110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AJ, Frohman MA, Engebrecht J. Measurement of phospholipase D activity. Anal Biochem. 1997;252:1–9. doi: 10.1006/abio.1997.2299. [DOI] [PubMed] [Google Scholar]
- Nagasaki A, Uyeda TQ. Screening of genes involved in cell migration in Dictyostelium. Exp Cell Res. 2008;314:1136–46. doi: 10.1016/j.yexcr.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Osborne N, Stainier DY. Lipid receptors in cardiovascular development. Annu Rev Physiol. 2003;65:23–43. doi: 10.1146/annurev.physiol.65.092101.142235. [DOI] [PubMed] [Google Scholar]
- Pollard SM, Parsons MJ, Kamei M, Kettleborough RN, Thomas KA, Pham VN, Bae MK, Scott A, Weinstein BM, Stemple DL. Essential and overlapping roles for laminin alpha chains in notochord and blood vessel formation. Dev Biol. 2006;289:64–76. doi: 10.1016/j.ydbio.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Saude L, Woolley K, Martin P, Driever W, Stemple DL. Axis-inducing activities and cell fates of the zebrafish organizer. Development. 2000;127:3407–17. doi: 10.1242/dev.127.16.3407. [DOI] [PubMed] [Google Scholar]
- Shaw KM, Castranova DA, Pham VN, Kamei M, Kidd KR, Lo BD, Torres-Vasquez J, Ruby A, Weinstein BM. fused-somites-like mutants exhibit defects in trunk vessel patterning. Dev Dyn. 2006;235:1753–60. doi: 10.1002/dvdy.20814. [DOI] [PubMed] [Google Scholar]
- Shoji W, Isogai S, Sato-Maeda M, Obinata M, Kuwada JY. Semaphorin3a1 regulates angioblast migration and vascular development in zebrafish embryos. Development. 2003;130:3227–36. doi: 10.1242/dev.00516. [DOI] [PubMed] [Google Scholar]
- Singer WD, Brown HA, Jiang X, Sternweis PC. Regulation of phospholipase D by protein kinase C is synergistic with ADP-ribosylation factor and independent of protein kinase activity. J Biol Chem. 1996;271:4504–10. doi: 10.1074/jbc.271.8.4504. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L, Schier AF, Driever W. Efficient recovery of ENU-induced mutations from the zebrafish germline. Genetics. 1994;136:1401–20. doi: 10.1093/genetics/136.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemple DL. Structure and function of the notochord: an essential organ for chordate development. Development. 2005;132:2503–12. doi: 10.1242/dev.01812. [DOI] [PubMed] [Google Scholar]
- Sung TC, Roper RL, Zhang Y, Rudge SA, Temel R, Hammond SM, Morris AJ, Moss B, Engebrecht J, Frohman MA. Mutagenesis of phospholipase D defines a superfamily including a trans-Golgi viral protein required for poxvirus pathogenicity. Embo J. 1997;16:4519–30. doi: 10.1093/emboj/16.15.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–15. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Topczewska JM, Topczewski J, Shostak A, Kume T, Solnica-Krezel L, Hogan BL. The winged helix transcription factor Foxc1a is essential for somitogenesis in zebrafish. Genes Dev. 2001;15:2483–93. doi: 10.1101/gad.907401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Vazquez J, Gitler AD, Fraser SD, Berk JD, Van NP, Fishman MC, Childs S, Epstein JA, Weinstein BM. Semaphorin-plexin signaling guides patterning of the developing vasculature. Dev Cell. 2004;7:117–23. doi: 10.1016/j.devcel.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Yokozeki T, Yamazaki M, Miyazaki H, Sasaki T, Maehama T, Itoh K, Frohman MA, Kanaho Y. Essential role for phospholipase D2 activation downstream of ERK MAP kinase in nerve growth factor-stimulated neurite outgrowth from PC12 cells. J Biol Chem. 2004;279:37870–7. doi: 10.1074/jbc.M402610200. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Miyagi C, Carmany-Rampey A, Shimizu T, Fujii R, Schier AF, Hirano T. Stat3 Controls Cell Movements during Zebrafish Gastrulation. Dev Cell. 2002;2:363–75. doi: 10.1016/s1534-5807(02)00126-0. [DOI] [PubMed] [Google Scholar]
- Yang SF, Freer S, Benson AA. Transphosphatidylation by phospholipase D. J Biol Chem. 1967;242:477–84. [PubMed] [Google Scholar]
- Yoon MS, Chen J. PLD regulates myoblast differentiation through the mTOR-IGF2 pathway. J Cell Sci. 2008;121:282–9. doi: 10.1242/jcs.022566. [DOI] [PubMed] [Google Scholar]
- Zeng XX, Wilm TP, Sepich DS, Solnica-Krezel L. Apelin and its receptor control heart field formation during zebrafish gastrulation. Dev Cell. 2007;12:391–402. doi: 10.1016/j.devcel.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kanaho Y, Frohman MA, Tsirka SE. Phospholipase D1-promoted release of tissue plasminogen activator facilitates neurite outgrowth. J Neurosci. 2005;25:1797–805. doi: 10.1523/JNEUROSCI.4850-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat Cell Biol. 2007;9:706–12. doi: 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.