Abstract
Morphologically-defined mammalian and molluscan genera (herein “morphogenera”) are significantly more likely to be monophyletic relative to molecular phylogenies than random, under 3 different models of expected monophyly rates: ≈63% of 425 surveyed morphogenera are monophyletic and 19% are polyphyletic, although certain groups appear to be problematic (e.g., nonmarine, unionoid bivalves). Compiled nonmonophyly rates are probably extreme values, because molecular analyses have focused on “problem” taxa, and molecular topologies (treated herein as error-free) contain contradictory groupings across analyses for 10% of molluscan morphogenera and 37% of mammalian morphogenera. Both body size and geographic range, 2 key macroevolutionary and macroecological variables, show significant rank correlations between values for morphogenera and molecularly-defined clades, even when strictly monophyletic morphogenera are excluded from analyses. Thus, although morphogenera can be imperfect reflections of phylogeny, large-scale statistical treatments of diversity dynamics or macroevolutionary variables in time and space are unlikely to be misleading.
Keywords: biogeography, body size, macroecology, macroevolution, systematics
Morphologically-defined genera are the primary analytical units for a wide range of large-scale paleontological and neontological analyses, across topics such as global diversity dynamics (1–8), macroevolutionary trends (9–11), paleoecology (12–14), systematics (15, 16), biogeography (17–19), and conservation biology (20–23). Although assigning rank to sets of species sharing close morphological affinities (usually with 1 or more diagnostic characters) lacks rigorous criteria, analyses of genus-level operational units are less subject to sampling and other biases than species-level analyses (24–26) and generally capture a damped record of species-level patterns (4, 27, 28). However, the proliferation of phylogenetic analyses using molecular data has uncovered an increasing number of paraphyletic or polyphyletic morphogenera (e.g., refs. 29 and 30), calling into question their analytic utility. Yet, despite alarming cases (e.g., ref. 31) and gross generalizations on the value of morphology (e.g., ref. 32), this issue has yet to be addressed quantitatively. Here, we evaluate the phylogenetic status of morphogenera relative to molecular phylogenies in 2 paleobiologically and ecologically important clades, Mammalia and Mollusca, and assess the potential impact of incorrect assumption of generic monophyly on 2 key macroevolutionary and macroecological variables: body size (9, 33–35) and latitudinal range (36–41).
Results
Rates of Paraphyly and Polyphyly.
We coded each of the 425 genera in our database (Table S1) as monophyletic, paraphyletic, or polyphyletic (42) (Fig. 1). The probability of randomly drawing a cladogram consistent with monophyly for a particular morphogenus from the set of all potential cladograms varies with the number of species in each analysis and in each morphogenus, but does not exceed 4.7% for the surveyed morphogenera (see SI Text). All evaluated data partitions significantly exceed this value (Table 1, Fig. 2, Table S2, and Fig. S1), indicating that morphogenera are congruent with molecular phylogenies more often than expected by a random draw of topologies. However, it is not obvious that this is the only model for the expected monophyly rate. Considering several alternatives (see SI Text), we use 25% as a highly conservative upper bound for the expected congruence by chance between taxonomy and molecular cladograms.
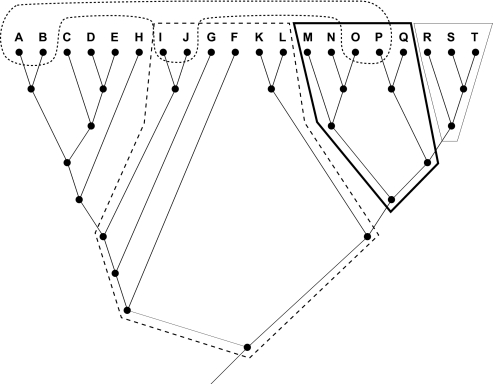
Fig. 1.
Diagrammatic representations of monophyletic, uniparaphyletic, multiparaphyletic, and polyphyletic morphogenera. A named genus is monophyletic with respect to a molecular cladogram, if it is completely congruent with the molecular topology (light polygon: taxa R, S, and T). Nonmonophyletic genera correspond to clades in the molecular cladogram that also contain some members of other genera. A uniparaphyletic morphogenus is one where monophyly is broken by a single branch placed in a different genus (bold polygon: taxa M–Q): this uniparaphyletic group could be made monophyletic by adding the single branch uniting the clade (R, S, T) to the named genus. A multiparaphyletic morphogenus is one where the ancestral state of the basal node is the genus name, but where multiple named branches must be added to the named genus to render the group monophyletic (dashed polygon: taxa I–L, which requires clades (A–H) and (R, S, T) for monophyly). A polyphyletic morphogenus is one where the primitive state of the node defining the last common ancestor is not the genus name (dashed irregular shape: taxa A, B, I, J, O, and P). See ref. 42 and Methods for further details.
Table 1.
Counts and percentages of monophyletic, paraphyletic, and polyphyletic taxa for all taxa in the database
| Taxa type | All Taxa |
Mammalia |
Mollusca |
Mollusca: marine |
Mollusca: nonmarine |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # genera | % | 2SE, % | # genera | % | 2SE, % | # genera | % | 2SE, % | # genera | % | 2SE, % | # genera | % | 2SE, % | |
| Monophyletic | 268 | 63.1 | 4.7 | 150 | 65.8 | 6.2 | 118 | 59.9 | 6.9 | 84 | 68.9 | 8.3 | 34 | 50.7 | 11.9 |
| Nonmonophyletic | 157 | 36.9 | 78 | 34.2 | 79 | 40.1 | 38 | 31.1 | 33 | 49.3 | |||||
| Uniparaphyletic | 55 | 12.9 | 40 | 17.5 | 15 | 7.6 | 9 | 7.4 | 4 | 6.0 | |||||
| Multiparaphyletic | 20 | 4.7 | 9 | 3.9 | 11 | 5.6 | 4 | 3.3 | 6 | 9.0 | |||||
| Nonpolyphyletic | 343 | 80.7 | 199 | 87.3 | 144 | 73.1 | 97 | 79.5 | 44 | 65.7 | |||||
| Polyphyletic | 82 | 19.3 | 3.8 | 29 | 12.7 | 4.5 | 53 | 26.9 | 6.3 | 25 | 20.5 | 7.3 | 23 | 34.4 | 11.3 |
| Total | 425 | 228 | 197 | 122 | 67 | ||||||||||
Bivalve and gastropod mollusks are further divided into marine and nonmarine partitions. Two SE of the observed percentages are given for the rates of monophyly and polyphyly. Rows in italics are the complementary counts and percentages to the cases of monophyly and paraphyly.
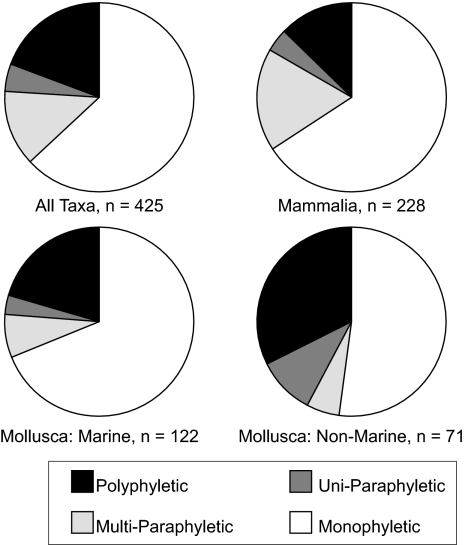
Fig. 2.
Proportion of monophyletic (white), uni-paraphyletic (where 1 branch within a clade is placed in another genus; light gray), multiparaphyletic (where multiple branches within a clade are assigned to different genera; dark gray), and polyphyletic (black) mammalian and molluscan morphogenera (see Fig. 1 and Methods for details on phyletic state definitions); mollusks are partitioned by marine and nonmarine habitat. The majority of morphogenera surveyed were monophyletic, in proportions significantly greater than expected by chance for all partitions (see Table 1).
Overall monophyly, 63 ± 5% (±2 SE), and rates for Mammalia and Mollusca separately, significantly exceed 25% (Table 1 and Fig. 2). Among mollusks, monophyly rates were significant for both marine (69 ± 8%) and nonmarine (51 ± 12%) partitions (Table 1). The lower nonmarine rate appears restricted to nonmarine bivalves (all in the Order Unionoida in our dataset). The monophyly rate for this group does not significantly exceed 25% (35% ± 20%), unlike all other partitions, and the polyphyly rate is 50% (Table S2). Sample sizes are small, but these results are unsurprising, because Unionoida is notoriously variable morphologically, with an unstable and difficult taxonomy (31).
Monophyly rates do not appear to be a sampling artifact. The ability to detect nonmonophyly could be related to the number of taxa in phylogenetic analyses (more exhaustive analyses may uncover hidden morphological convergences) or the number of the taxa in a morphogenus (more complete surveys could reveal paraphyly or polyphyly). Although monophyly rates tend to be higher in smaller analyses for mammals and mollusks as a whole, monophyly rates remain higher than expected by chance with increased numbers of species per morphogenus (Fig. S1). This sampling effect might account for the lower monophyly and higher polyphyly observed among mollusks, where molecular phylogenies average 66.6 species with 6.3 species per morphogenus, compared with 39.9 species per study and 5.3 species per genus for mammals. Molluscan monophyletic and nonmonophyletic morphogenera do not differ significantly in number of species per morphogenus, but do differ in number of species per study (Table 2). However, statistical significance of the latter result hinges on a single analysis (43) of 221 species contributing 23 genera (excluding this study, P = 0.151). For Mammalia, phyletic state does not differ significantly with study size, but does vary weakly with number of species per genus, owing to an overabundance of monophyletic morphogenera with the minimum 3 species: (112 morphogenera with 77 monophyletic). Excluding genera with 3 species removes the significance (P = 0.578), and does not change the main result; monophyly remains significantly >25% (58 ± 9%). This rate is similar to that for mollusks, suggesting that the higher monophyly rate for the complete mammal dataset could reflect sampling.
Table 2.
Kolmogorov-Smirnov tests comparing distributions of number of species in the phylogenetic analyses and number of species sampled in a morphogenus
| Species | Mean species in analysis | Mean species in genus |
|---|---|---|
| Mammalia | ||
| Monophyletic | 41.2 | 5.1 |
| Nonmonophyletic | 37.4 | 5.7 |
| 2-tailed P | 0.524 | 0.029 |
| Mollusca | ||
| Monophyletic | 80.5 | 5.8 |
| Nonmonophyletic | 45.9 | 7.0 |
| 2-tailed P | 0.002 | 0.712 |
Two-tailed P values and mean values for monophyletic and nonmonophyletic genera are given for Mammalia and Mollusca.
If the mammal monophyly rate is inflated by “smaller” studies, then systematic shifts from monophyly to nonmonophyly should be expected with increased sampling. However, for 26 mammalian morphogenera with multiple occurrences, where at least 1 occurrence had 3 species and at least 1 had >3 species, only 3 show this shift, and 1 showed the opposite, becoming monophyletic at greater sampling. Of the remaining cases, 16 did not change phyletic status, and 6 were variable among occurrences with 3 species. Thus, although mammalian phylogenetic analyses tend to be smaller in both species per study and per morphogenus, sampling is not responsible for the higher monophyly rate. Recency and extent of taxonomic revisions within Mammalia (citations in ref. 47) may account for these differences, although detailed tests are needed. Alternatively, multielement mammal skeletons might also provide more potential morphological characters for phylogenetic analysis than do bivalve shells, which should generally increase accuracy, although more slowly than addition of taxa (44–47).
Impact on Body Size and Geographic Range.
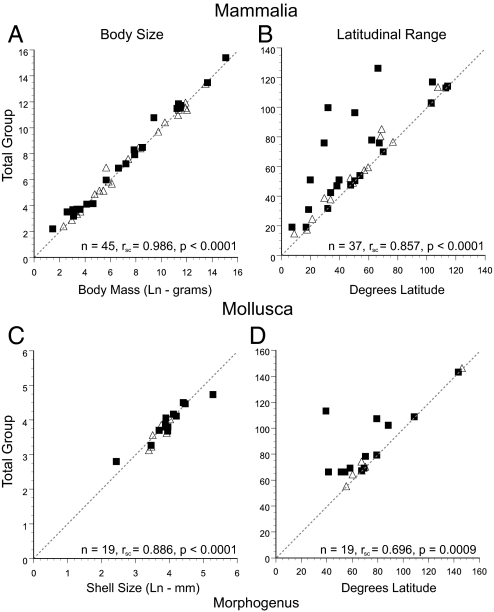
The statistically significant congruence between morphogenera and molecular phylogenies validates the use of morphogenera for many large-scale analytical purposes, but the overall polyphyly rate of 19% (Table 1) suggests that their robustness as proxies for species- or clade-level dynamics should be approached cautiously and evaluated according to specific questions. We assess the potential impact of phylogenetic error for 2 key macroevolutionary and macroecological variables: body size or mass and geographic range (Table S3). For Mammalia and marine Mollusca, highly-significant rank correlations are observed for median values between morphogenera and total groups (groups formed when species are added to render paraphyletic or polyphyletic taxa monophyletic) for both variables (Fig. 3). Because these analyses exclude the monophyletic morphogenera, which by definition correlate perfectly for both variables, they are extremely conservative.
Fig. 3.
Macroecological data for nonmonophyletic morphogenera for Mammalia (A and B) and Mollusca (C and D) are significantly correlated with those for the total group (i.e., all species in the clade defined by the molecular data). Axes give values for the same variables, the x-axes are values for the named morphogenus; y-axes are for the total group. Body size is Ln-g for mammals and Ln-mm (shell size) for mollusks. Latitudinal ranges are in degrees. ■, polyphyletic morphogenera; ▵, paraphyletic morphogenera. Dashed line is y = x, shown for reference. For latitudinal range, polyphyletic or paraphyletic morphogenera cannot have larger ranges than the total group, therefore the triangle under the line y = x cannot be occupied. All Spearman rank correlations shown are significant. These correlations do not include data for the 150 mammalian and 118 molluscan monophyletic morphogenera that would correlate perfectly for both variables.
Discussion
These results likely represent a worst-case scenario for morphogenus monophyly. Much of the compiled molecular work focused on “problem taxa,” those known to be resistant to morphological analysis (e.g., freshwater bivalves, oysters, bovids). Further, our analysis assumes that molecular phylogenies are inerrant, but ≈10% of molluscan and 37% of mammalian morphogenera sampled in multiple analyses shift phyletic status (Table S4). Thus, although most topologies are stable across analyses, some of the mismatches between molecular and morphological data are almost certainly caused by phylogenetic error in molecular trees, not generic assignment. Incomplete molecular sampling of species assigned to morphogenera could artificially depress nonmonophyly rates, but our tests relating monophyly to species number suggest that sampling is not a major factor.
The correlations between morphological and molecular genera in both size and range are remarkably strong, especially in light of the fact that only paraphyletic and polyphyletic genera were included. (The low correlation between molluscan morphogenus and total group latitudinal ranges derives primarily from a single polyphyletic genus: without Ruditapes the Spearman rank correlation increases to 0.92.) These analyses exclude 150 molluscan morphogenera and 118 mammalian morphogenera that were recovered as monophyletic, which would show perfect correspondence between morphogenus and total-group variables. Adding these taxa drives even the lowest observed rank-order correlations to >0.999. The results in Fig. 3 are therefore highly conservative and imply that incorrect assumptions of monophyly should not significantly bias large analyses (see also ref. 27): morphogenera recover biological signal even where only paraphyletic or polyphyletic morphogenera are sampled (highly unlikely, given our results showing monophyly to be far more common than expected by chance). The strength of these correlations probably derives from the significant phylogenetic component recorded for the interspecific evolution of geographic range and body size (33, 37, 47). However, the tighter correlations for body size suggest this character carries a strong phylogenetic signal, such that related species tend to be similar in size, regardless of whether they are direct sister taxa. The larger variance in latitudinal range might reflect allopatric range relationships among closely related taxa, such that paraphyletic and polyphyletic morphogenera are not formed randomly, but tend to exclude species that are spatially disjunct from the rest. If verified by spatially explicit phylogenetic analyses, these results may indicate a mild geographic bias in the formation of morphogenera.
Far from pessimistic views that morphology is of little or no use in recovering phylogenetic relationships (e.g., ref. 32), our results indicate that substantial phylogenetic information is already encoded within traditional morphologic taxonomies, and phylogenetic error is unlikely to obscure biological information, at least for macroecological variables containing strong phylogenetic signal such as body size and geographic range size. The strength of the phylogenetic information in morphology-based taxonomies is encouraging, in that many aspects of macroevolutionary dynamics are best studied by using paleontological data, and morphological data enable joint analysis of extinct and extant taxa, both for resolution of deep nodes (48, 49), and more direct exploration of the physical and biological events that have shaped the modern biota (50, 51). More accurate and precise phylogenetic hypotheses, including rigorous recognition of monophyletic, paraphyletic, and polyphyletic groups, will always improve our understanding of macroevolutionary patterns, but lack of molecular re-evaluation of taxonomies does not preclude robust analyses for many questions, at least for the groups evaluated here. Synoptic datasets will help to circumscribe those organismic variables amenable to comparative analysis using morphology-based taxonomies.
Increasing availability of molecular data can help develop new approaches to morphology-based systematics, by pinpointing characters that reliably capture phylogenetic relationships versus those consistently subject to homoplasy, and will allow testing for among-clade variations in apparent polyphyly, where higher frequencies might derive from, for example, evolutionary exhaustion of character states (52), architectural constraints, fewer adaptive peaks, or plasticity reducing phylogenetic signal. Of course, not all groups will show such strong congruence between morphological taxonomy and molecular phylogeny, scleractinian corals being a troubling example (53), but even there the interplay of modern morphometrics and molecular systematics is beginning to produce more robust mapping of morphology and phylogeny (54, 55). Our results demonstrate the potential for fruitful partnerships between molecular and morphological approaches to biological diversity and add to a growing body of empirical work demonstrating that morphologically defined units can yield meaningful results in a wide variety of biological analyses (e.g., refs. 21 and 39–41).
Methods
Compilation of Morphogenera and Determination of Phyletic Status.
Our literature survey yielded 425 genus occurrences (197 mollusks and 228 nonvolant mammals), representing 315 unique morphogenera (175 mollusks and 140 mammals) from 180 phylogenetic analyses [Table S1; we elevate molluscan subgenera to genus rank, following general paleobiological practice (e.g., ref. 56)]. Total number of genera in surveyed phylogenies ranged from 2 to 109, and total number of species ranged from 6 to 221. The number of species per morphogenus ranged from 3 to 57, with similar values for Mammalia (range 3 to 37) and Mollusca (range 3 to57). For each analysis, we categorized named morphogenera as monophyletic, paraphyletic, or polyphyletic (42) (Fig. 1). Monophyletic morphogenera are those whose species are subtended by the node defining the last common ancestor (LCA) of all species in the morphogenus, with no other taxa subtended by that node. Paraphyly and polyphyly are cases where the species subtended by the LCA are placed in 2 or more morphogenera. Mapping morphogenus name from the tips of the cladogram down to the LCA allows us to determine whether the LCA is included in the morphogenus (42): paraphyletic morphogenera include the LCA in the morphogenus, but not all of its all descendants, whereas polyphyletic morphogenera do not include the LCA (Fig. 1; see ref. 42 for detailed discussion). Some authors (e.g., ref. 57) further partition paraphyly, and we differentiate those whose monophyly is broken by only a single branch, that is, the paraphyletic morphogenus could be made monophyletic by subsuming a single branch into it. We informally term such morphogenera “uniparaphyletic” (Tables S1–S3). This branch could be a single species or a larger clade, so long as subsuming all taxa along that branch renders the morphogenus monophyletic (Fig. 1, bold polygon). In contrast, morphogenera requiring multiple branches to be subsumed to return a monophyletic taxon are termed “multiparaphyletic” (Fig. 1, dashed polygon). Our distinction between uniparaphyly and multiparaphyly is widely, if informally, recognized by the frequent application of “highly paraphyletic” to clades here termed multiparaphyletic (e.g., refs. 58 and 59).
We only tallied ingroup morphogenera in surveyed phylogenetic analyses, because the position of designated outgroup taxa is constrained a priori. In addition, we only sampled morphogenera containing 3 or more species in the phylogenetic analysis and only included studies with at least 2 additional ingroup species not in the morphogenus. The first protocol allows for the multiple potential branch insertions within the morphogenus necessary to recognize polyphyly. The second ensures that there were enough ingroup taxa not included in the named morphogenus to potentially generate that polyphyly.
A “morphogenus” in this analysis represents a recorded occurrence of a morphogenus meeting the above criteria in a surveyed phylogenetic study. As such, a morphogenus can occur multiple times in the database if it has been the subject of multiple phylogenetic analyses; e.g., Mustela (weasels) appears in 9 analyses (Table S1). To ensure that recorded morphogenus occurrences represented independent assessments of phyletic status, if the taxon-by-gene table used in a phylogenetic analysis formed a subset of that in another study, then only the most inclusive analysis was used, and the other was considered subsumed. This occurred most often when researchers expanded on previously published datasets. For studies not wholly subsumed within other analyses, we treated multiple occurrences as separate assessments of the phyletic status, entering each into the dataset.
Only genera with strong support for the defining node were analyzed. Defining “strong” nodal support is a somewhat contentious issue, with disagreement on proper cutoff values across different reconstruction methods (60–63). Following previous authors, we considered bootstrap replicate support ≥70% (61) and Bayesian posterior probabilities ≥95% (64) to be strong. In several instances we noted conflict when different genes or phylogenetic methods recovered different phyletic states for a given morphogenus in the same analysis. In most occurrences 1 topology was weakly supported and therefore excluded from our dataset. One morphogenus (goats Capra) occurred in 2 conflicting phyletic states (monophyletic and uniparaphletic) with strong support for both (highlighted in Table S1). Here, we incorporated only the paraphyletic result into our analyses, conservatively lowering the observed proportion of monophyletic morphogenera.
Frequencies for the 4 phyletic states were tabulated for the total set of taxa and partitioned by taxonomic group. We report surveyed phylogenetic studies, details of the number of taxa included in the analyses and in the morphogenera, including morphogenera and phyletic status, in Table S1.
Macroevolutionary Variables for Morphogenera.
To test for potential biases in measuring macroevolutionary and macroecological variables imparted by incorrectly assumed monophyly, we compiled data on paraphyletic and polyphyletic morphogenera for 2 biologically important variables, body size (9, 33–35) and latitudinal range (36–41). For mammals, body size was measured in grams, drawing on a worldwide database (65). For bivalves, body size was the geometric mean of length and height (40); for gastropods, body size was maximum shell dimension (66). We compared median body mass and total latitudinal range (in degrees) for the named morphogenus to the “total group” (all taxa in the clade defined by the node subtending all species in the morphogenus) by using Spearman rank correlation. We use rank correlation because the variables are not normally distributed, and relative ordering is important in recovering patterns such as within- and among-lineage trends in phenotype evolution (67, 68). Because the species in the morphogenus are part of the total group, these axes are not statistically independent, thus, P values should be taken with some caution. However, the results do demonstrate the ability of nonmonophyletic morphogenera to reproduce the pattern of relative ordering of these variables for the monophyletic clade defined by molecular data. In cases where the same morphogenus was nonmonophyletic in multiple analyses, we used the set of taxa from the phylogenetic analysis where the deviation between morphogenus and total group was greatest, conservatively examining the worst observed fit. Morphogenus and total-group values for macroevolutionary variables are provided in Table S4.
Supplementary Material
Acknowledgments.
We thank P. R. Crane, M. Foote, S. M. Kidwell, K. Roy, J. W. Valentine, P. J. Wagner, and 2 anonymous reviewers for valuable comments and C. S. Hickman, J. H. McLean, and J. D. Taylor for taxonomic advice. This work is supported by a grant from the National Aeronautics and Space Administration and the University of Michigan Society of Fellows.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902973106/DCSupplemental.
References
- 1.Valentine JW. Taxonomic and ecological structure of the shelf benthos during Phanerozoic time. Palaeontology. 1969;12:684–709. [Google Scholar]
- 2.Raup DM, Sepkoski JJ., Jr Periodic extinction of families and genera. Science. 1986;231:833–836. doi: 10.1126/science.11542060. [DOI] [PubMed] [Google Scholar]
- 3.Jablonski D, Raup DM. Selectivity of End-Cretaceous marine bivalve extinctions. Science. 1995;268:389–392. doi: 10.1126/science.11536722. [DOI] [PubMed] [Google Scholar]
- 4.Sepkoski JJ., Jr Rates of speciation in the fossil record. Philos Trans R Soc London Ser B. 1998;353:315–326. doi: 10.1098/rstb.1998.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foote M. Origination and extinction through the Phanerozoic: A new approach. J Geol. 2003;111:125–148. [Google Scholar]
- 6.Stanley SM. An analysis of the history of marine animal diversity. Paleobiology. 2007;33(Suppl):1–55. [Google Scholar]
- 7.Tarver JE, Braddy SJ, Benton MJ. The effects of sampling bias on Palaeozoic faunas and implications for macroevolutionary studies. Palaeontology. 2007;50:177–184. [Google Scholar]
- 8.Alroy J, et al. Phanerozoic trends in the global diversity of marine invertebrates. Science. 2008;321:97–100. doi: 10.1126/science.1156963. [DOI] [PubMed] [Google Scholar]
- 9.Jablonski D. Body-size evolution in Cretaceous molluscs and the status of Cope's rule. Nature. 1997;385:250–252. [Google Scholar]
- 10.Huntley JW, Kowalewski M. Strong coupling of predation intensity and diversity in the Phanerozoic fossil record. Proc Natl Acad Sci USA. 2007;104:15006–15010. doi: 10.1073/pnas.0704960104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novak-Gottshall PM, Lanier MA. Scale dependence of Cope's rule in body size evolution of Paleozoic brachiopods. Proc Natl Acad Sci USA. 2008;105:5430–5434. doi: 10.1073/pnas.0709645105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connolly SR, Miller AI. Global Ordovician faunal transitions in the marine benthos: Ultimate causes. Paleobiology. 2002;28:26–40. [Google Scholar]
- 13.Foote M. Substrate affinity and diversity dynamics of Paleozoic marine animals. Paleobiology. 2006;32:345–366. [Google Scholar]
- 14.Bambach RK, Bush AM, Erwin DH. Autecology and the filling of ecospace: Key metazoan radiations. Palaeontology. 2007;50:1–22. [Google Scholar]
- 15.Wilson J. Sauropod dinosaur phylogeny: Critique and cladistic analysis. Zool J Linn Soc. 2002;136:217–276. [Google Scholar]
- 16.Finarelli JA, Clyde WC. Reassessing hominoid phylogeny: Evaluating congruence in the morphologic and temporal data. Paleobiology. 2004;30:614–651. [Google Scholar]
- 17.Harcourt AH, Coppeto SA, Parks SA. Rarity, specialization, and extinction in primates. J Biogeogr. 2002;29:445–456. [Google Scholar]
- 18.Miller AI. A new look at age and area: The geographic and environmental expansion of genera during the Ordovician radiation. Paleobiology. 1997;23:410–419. doi: 10.1017/s0094837300019813. [DOI] [PubMed] [Google Scholar]
- 19.Jablonski D, Roy K, Valentine JW. Out of the tropics: Evolutionary dynamics of the latitudinal diversity gradient. Science. 2006;314:102–106. doi: 10.1126/science.1130880. [DOI] [PubMed] [Google Scholar]
- 20.Williams PH, Gaston KJ. Measuring more of biodiversity: Can higher-taxon richness predict wholesale species richness? Biol Cons. 1994;67:211–217. [Google Scholar]
- 21.Villasenor JL, Ibarra-Manríquez G, Meave JA, Ortíz E. Higher taxa as surrogates of plant biodiversity in a megadiverse country. Conserv Biol. 2005;19:232–238. [Google Scholar]
- 22.Heino J, Soininen J. Are higher taxa adequate surrogates for species-level assemblage patterns and species richness in stream organisms? Biol Cons. 2007;137:18–89. [Google Scholar]
- 23.Mandelik Y, Dayan T, Chikantunov V, Kravchenko V. Reliability of a higher-taxon approach to richness, rarity, and composition assessments at the local scale. Conserv Biol. 2007;21:1506–1515. doi: 10.1111/j.1523-1739.2007.00823.x. [DOI] [PubMed] [Google Scholar]
- 24.Raup DM. Biases in the fossil record of species and genera. Bull Carnegie Mus Nat Hist. 1979;13:85–91. [Google Scholar]
- 25.Allmon WD. Genera in paleontology: Definition and significance. Hist Biol. 1992;6:149–158. [Google Scholar]
- 26.Sepkoski JJ., Jr Biodiversity: Past, present, and future. J Paleontol. 1997;71:533–539. doi: 10.1017/s0022336000040026. [DOI] [PubMed] [Google Scholar]
- 27.Wagner PJ. Diversity patterns among early gastropods: Contrasting taxonomic and phylogenetic descriptions. Paleobiology. 1995;21:410–439. [Google Scholar]
- 28.Roy K, Valentine JW, Jablonski D, Kidwell SM. Scales of climatic variability and time averaging in Pleistocene biotas: Implications for ecology and evolution. Trends Ecol Evol. 1996;11:458–463. doi: 10.1016/0169-5347(96)10054-9. [DOI] [PubMed] [Google Scholar]
- 29.Lanyon SM. Polyphyly of the blackbird genus Agelaius and the importance of assumptions of monophyly in comparative studies. Evolution (Lawrence, Kans) 1994;48:679–693. doi: 10.1111/j.1558-5646.1994.tb01353.x. [DOI] [PubMed] [Google Scholar]
- 30.de Weerd DRU, Piel WH, Gittenberger E. Widespread polyphyly among Aloplinae snail genera: When phylogeny mirrors biogeography more closely than morphology. Mol Phyl Evol. 2004;33:533–548. doi: 10.1016/j.ympev.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Campbell DC, et al. Phylogeny of North American amblemines (Bivalvia, Unionoida): Prodigious polyphyly proves pervasive across genera. Invert Biol. 2005;124:131–164. [Google Scholar]
- 32.Scotland RW, Olmstead RG, Bennett JR. Phylogeny reconstruction: The role of morphology. Syst Biol. 2003;52:539–548. doi: 10.1080/10635150390223613. [DOI] [PubMed] [Google Scholar]
- 33.Alroy J. Cope's rule and the dynamics of body mass evolution in North American fossil mammals. Science. 1998;280:731–734. doi: 10.1126/science.280.5364.731. [DOI] [PubMed] [Google Scholar]
- 34.Finarelli JA, Flynn JJ. Ancestral state reconstruction of body size in the Caniformia (Carnivora, Mammalia): The effects of incorporating data from the fossil record. Syst Biol. 2006;55:301–313. doi: 10.1080/10635150500541698. [DOI] [PubMed] [Google Scholar]
- 35.Finarelli JA. Mechanisms behind active trends in body size evolution in the Canidae (Carnivora: Mammalia) Am Nat. 2007;170:876–885. doi: 10.1086/522846. [DOI] [PubMed] [Google Scholar]
- 36.Willig MR, Lyons SK. An analytical model of latitudinal gradients of species richness with an empirical test for marsupials and bats in the New World. Oikos. 1998;81:93–98. [Google Scholar]
- 37.Hunt G, Roy K, Jablonski D. Heritability of geographic range sizes revisited. Am Nat. 2005;166:129–135. doi: 10.1086/430722. [DOI] [PubMed] [Google Scholar]
- 38.Lozada T, de Koning GHJ, Kessler M, Klein A-M, Tscharntke T. Geographical range size of tropical plants influences their response to anthropogenic activities. Divers Distrib. 2008;14:59–68. [Google Scholar]
- 39.Roy K, Jablonski D, Valentine JW, Rosenberg G. Marine latitudinal diversity gradients: Tests of causal hypotheses. Proc Natl Acad Sci USA. 1998;95:3699–3702. doi: 10.1073/pnas.95.7.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy K, Jablonski D, Martien KK. Invariant size-frequency distributions along a latitudinal gradient in marine bivalves. Proc Natl Acad Sci USA. 2000;97:13150–13155. doi: 10.1073/pnas.97.24.13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krug AZ, Jablonski D, Valentine JW. Species-genus ratios reflect a global history of diversification and range expansion in marine bivalves. Proc R Soc London Ser B. 2008;275:1117–1123. doi: 10.1098/rspb.2007.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiley EO. Phylogenetics. New York: Wiley; 1981. [Google Scholar]
- 43.Meyer CP. Toward comprehensiveness: Increased molecular sampling within Cypraeidae and its phylogenetic implications. Malacologia. 2004;46:127–156. [Google Scholar]
- 44.Graybeal A. Is it better to add taxa or characters to a difficult phylogenetic problem? Syst Biol. 1998;47:9–17. doi: 10.1080/106351598260996. [DOI] [PubMed] [Google Scholar]
- 45.Wiens JJ. Does adding characters with missing data increase or decrease phylogenetic accuracy? Syst Biol. 1998;47:625–640. doi: 10.1080/106351598260635. [DOI] [PubMed] [Google Scholar]
- 46.Heath TA, Hedtke SM, Hillis DM. Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol. 2008;46:239–257. [Google Scholar]
- 47.Jones KE, Sechrest W, Gittleman JL. In: Phylogeny and Conservation. Purvis A, Gittleman JL, Brooks T, editors. Cambridge, UK: Cambridge Univ Press; 2005. pp. 141–165. [Google Scholar]
- 48.Smith ND, Turner AH. Morphology's role in phylogeny reconstruction: Perspectives from paleontology. Syst Biol. 2005;54:166–173. doi: 10.1080/10635150590906000. [DOI] [PubMed] [Google Scholar]
- 49.Raff RA. Written in stone: Fossils, genes, and evo-devo. Nat Rev Genet. 2007;8:911–920. doi: 10.1038/nrg2225. [DOI] [PubMed] [Google Scholar]
- 50.Jablonski D. Scale and hierarchy in macroevolution. Palaeontology. 2007;50:87–109. [Google Scholar]
- 51.Jablonski D. Biotic interactions and macroevolution: Extensions and mismatches across scales and levels. Evolution (Lawrence, Kans) 2008;62:715–739. doi: 10.1111/j.1558-5646.2008.00317.x. [DOI] [PubMed] [Google Scholar]
- 52.Wagner PJ, Erwin DH. Patterns of convergence in general shell form among Paleozoic gastropods. Paleobiology. 2006;32:316–337. [Google Scholar]
- 53.Fukami H, et al. Conventional taxonomy obscures deep divergence between Pacific and Atlantic corals. Nature. 2004;427:832–835. doi: 10.1038/nature02339. [DOI] [PubMed] [Google Scholar]
- 54.Budd AF, Stolarski J. Searching for new morphological characters in the systematics of scleractinian reef corals: Comparison of septal teeth and granules between Atlantic and Pacific Mussidae. Acta Zool. 2009;90:142–165. [Google Scholar]
- 55.Budd AF, Smith ND. Diversification of a new Atlantic clade of scleractinian reef corals: Insights from phylogenetic analysis of morphologic and molecular data. Paleontol Soc Pap. 2005;11:103–128. [Google Scholar]
- 56.Sepkoski JJ., Jr A compendium of fossil marine animal genera. Bull Am Paleontol. 2002;363:1–563. [Google Scholar]
- 57.Haszprunar G. Die klado-evolutionäre Klassifikation—Versuch einer Synthese. Z Zool Sys Evol. 1986;24:89–109. [Google Scholar]
- 58.Park J-M, Kovaci S, Liber Z, Eddie WMM, Schneeweiss GM. Phylogeny and biogeography of isophyllous species of Campanula (Campanulaceae) in the Mediterranean area. Syst Botany. 2006;31:862–880. [Google Scholar]
- 59.Källersjö M, Bergqvist G, Anderberg AA. Generic realignment in primuloid families of the Ericales s.l.: A phylogenetic analysis based on DNA sequences from three chloroplast genes and morphology. Am J Bot. 2000;87:1325–1341. [PubMed] [Google Scholar]
- 60.Wilcox TP, Zwickl DJ, Heath TA, Hillis DM. Phylogenetic relationships of the dwarf boas and a comparison of Bayesian and bootstrap measures of phylogenetic support. Mol Phyl Evol. 2002;25:361–371. doi: 10.1016/s1055-7903(02)00244-0. [DOI] [PubMed] [Google Scholar]
- 61.Hillis DM, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- 62.Cummings MP, et al. Comparing bootstrap and posterior probability values in the four-taxon case. Syst Biol. 2003;52:477–487. doi: 10.1080/10635150390218213. [DOI] [PubMed] [Google Scholar]
- 63.Alfaro ME, Zoller S, Lutzoni F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol Biol Evol. 2003;20:255–266. doi: 10.1093/molbev/msg028. [DOI] [PubMed] [Google Scholar]
- 64.Huelsenbeck JP, Larget B, Miller RE, Ronquist F. Potential applications and pitfalls of Bayesian inference of phylogeny. Syst Biol. 2002;51:673–688. doi: 10.1080/10635150290102366. [DOI] [PubMed] [Google Scholar]
- 65.Smith FA, et al. Body mass of late Quaternary mammals. Ecology. 2003;84:3403. [Google Scholar]
- 66.Kosnik MA, Jablonski D, Lockwood R, Novak-Gottshall PM. Quantifying molluscan body size in evolutionary and ecological analyses: Maximizing the return on data collection efforts. Palaios. 2006;21:588–597. [Google Scholar]
- 67.Wagner PJ. Contrasting the underlying patterns of active trends in morphologic evolution. Evolution (Lawrence, Kans) 1996;50:990–1007. doi: 10.1111/j.1558-5646.1996.tb02341.x. [DOI] [PubMed] [Google Scholar]
- 68.Finarelli JA, Flynn JJ. The evolution of encephalization in caniform carnivorans. Evolution (Lawrence, Kans) 2007;61:1758–1772. doi: 10.1111/j.1558-5646.2007.00131.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.