Abstract
Amphibians are in decline worldwide. However, their patterns of diversity, especially in the tropics, are not well understood, mainly because of incomplete information on taxonomy and distribution. We assess morphological, bioacoustic, and genetic variation of Madagascar's amphibians, one of the first near-complete taxon samplings from a biodiversity hotspot. Based on DNA sequences of 2,850 specimens sampled from over 170 localities, our analyses reveal an extreme proportion of amphibian diversity, projecting an almost 2-fold increase in species numbers from the currently described 244 species to a minimum of 373 and up to 465. This diversity is widespread geographically and across most major phylogenetic lineages except in a few previously well-studied genera, and is not restricted to morphologically cryptic clades. We classify the genealogical lineages in confirmed and unconfirmed candidate species or deeply divergent conspecific lineages based on concordance of genetic divergences with other characters. This integrative approach may be widely applicable to improve estimates of organismal diversity. Our results suggest that in Madagascar the spatial pattern of amphibian richness and endemism must be revisited, and current habitat destruction may be affecting more species than previously thought, in amphibians as well as in other animal groups. This case study suggests that worldwide tropical amphibian diversity is probably underestimated at an unprecedented level and stresses the need for integrated taxonomic surveys as a basis for prioritizing conservation efforts within biodiversity hotspots.
Keywords: biodiversity estimates, new candidate species, phylogeny, DNA barcoding
The current biodiversity crisis demands the study of broad-scale spatial variation in species richness and endemism to identify areas that merit special conservation attention. Global efforts to minimize biodiversity loss have led to prioritizing biodiversity hotspots (1) which are defined as areas with high concentrations of endemic species and that are undergoing exceptional habitat loss (2–4). A second step is the efficient implementation of conservation measures at a local scale which requires an understanding of spatial patterns of richness and endemism within these hotspots (5). Assessments of such regional priority areas are often hampered by incomplete distributional and taxonomic information. The operational units used to assess conservation priority areas are described species, whereas estimates of undiscovered and undescribed species are usually ignored. Recently, many potential new species have been identified by DNA barcoding, but a taxonomic validation of these species will outdate the short time span left for efficient biodiversity conservation prioritization, and so far it is not clear how this undescribed diversity can nonetheless be considered. This undescribed diversity also bears relevance for understanding the tree of life: The completeness of taxon sampling is one of the major prerequisites for reliable phylogenetic analysis, reconstruction of character evolution, and inference of macroevolutionary processes (6, 7).
Among terrestrial vertebrates, amphibians are characterized by a rapid rate of species discovery (8, 9), with an overall increase in the number of amphibian species globally of 19.4% during the last decade, reaching 6,449 currently recognized species (10). An important acceleration in the rate of new discoveries, mainly from tropical areas, is obvious from many recent studies (11–16). These discoveries are not the result of taxonomic inflation (9, 14, 17), but correspond to real divergent species (18, 19). Although high numbers of undescribed amphibians have been estimated to exist in poorly studied tropical regions (11, 15), these results remain unverified for complete, highly diverse amphibian faunas. In parallel, an increase of threatened amphibian species has been reported worldwide (8, 20, 21). Amphibians are of high conservation concern, with 43% of species being globally threatened (20), most of them in tropical regions with high amphibian diversity.
Madagascar is one of the top priority global hotspots for biodiversity conservation (1), affected by a high rate of habitat destruction (22). Its fauna and flora evolved largely in isolation (23), and many taxa are characterized by a high degree of microendemism within Madagascar (24–27). The native amphibian fauna is constituted by 5 endemic evolutionary lineages of frogs with 100% species-level endemism, 2 of which (the mantellids and the cophyline and scaphiophrynine microhylids) are very species-rich. Large-scale taxonomic inventories conducted since 1991 have led to an increase from 133 to 244 described species, largely due to the exploration of new areas and the application of more efficient techniques. About 46 species were identified during the 1990s mainly based on bioacoustics; the application of combined methods, including molecular genetics, was crucial in the discovery of 51 new species since 2000 as well as in the resurrection of species formerly considered to represent synonyms.
Undescribed diversity may have an important impact on understanding the spatial patterns of endemic radiations on the island, but objective estimates of species numbers are not available so far. We report a comprehensive assessment of morphological, bioacoustic, and genetic variation of the anuran fauna of Madagascar. Our goals are (i) to provide a reliable estimate of the proportion of yet-undescribed amphibian species from Madagascar and their phylogenetic and geographic distribution, (ii) to discuss the impact of our findings for global estimates of amphibian diversity, and (iii) to propose a novel terminology to be better able to assess the increasing number of identified but taxonomically undescribed candidate species of animals.
Results
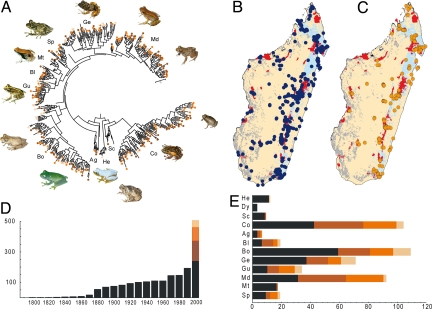
From our integrative analyses of morphological, bioacoustic, and genetic data, we identified many more species of Malagasy frogs than there are names available. The number of described species of Malagasy frogs has slowly increased during the 20th century, reaching 100 described species around 1940, and 133 at the time of the last monographic account in 1991 (28) (Fig. 1). The current number of 244 valid native species indicates an increase of 83.5% since 1991.
Fig. 1.
Phylogenetic, geographic, and historical patterns of undescribed amphibian diversity in Madagascar. (A) Bayesian phylogenetic tree of 236 (out of 244) described species and 258 deeply divergent genealogical lineages of Malagasy frogs (among them 129 CCS and 92 UCS and 37 DCL with >3% genetic divergence to nearest described neighbor) based on a fragment of the mitochondrial 16S rRNA gene. Circles represent CCS (brown), UCS (orange), and DCL (light orange). Inset photos show some of these forms (see SI Appendix). He, Heterixalus (Hyperoliidae); Dy, Dyscophus; Sc, Scaphiophryninae; Co, Cophylinae (Microhylidae); Sp, Spinomantis; Md, Mantidactylus; Mt, Mantella; Gu, Guibemantis; Ge, Gephyromantis; Bo, Boophis; Bl, Blommersia; Ag, Aglyptodactylus (Mantellidae). (B and C) Maps of collecting localities of described species (B) and of CCS and UCS (C) of Malagasy frogs, and remaining primary vegetation (evergreen forests, blue; nonevergreen forests, gray). Current protected area network is shown in red. (D) Cumulative number of species currently considered as valid per 10-year intervals (2001–2008 for the current decade), and the CCS, UCS, and DCL identified in the present paper. (E) Total numbers of described species and CCS, UCS, and DCL in major clades of Malagasy frogs.
We classify frog lineages from Madagascar in 3 categories (Table 1): (i) confirmed candidate species (CCS) are those differing clearly by morphological and bioacoustic characters and usually showing high genetic differentiation that we hypothesize are distinct, undescribed species; (ii) unconfirmed candidate species (UCS) are deep genealogical lineages (15)—bioacoustically and morphologically unstudied and usually derived from geographically distant populations—for which general indications exist that they are distinct, undescribed species; and (iii) deep conspecific lineages (DCL) are deeply divergent genealogical lineages, studied but not having clear morphological or bioacoustic differences with described species. Our data provided an additional 129 CCS, 92 UCS, and 37 DCL. If CCS plus UCS are assumed to represent distinct undescribed species, a 90.6% increase from the current 244 to 465 frog species is projected. This projection would constitute an overall increase of 250% since 1991.
Table 1.
Definitions of proposed categories of candidate species
| Category | General definition | Definition used in Malagasy frogs |
|---|---|---|
| Unconfirmed Candidate Species (UCS) | Default category for deep genealogical lineages of unknown status. The genetic differentiation must be above a threshold value typical for comparisons among closely related species in the group of animals under study. Data deficient for morphology, ecology, and distribution. | Uncorrected pairwise genetic divergences in 16S rRNA gene >3% to all other described species. No data on morphology and bioacoustics due to unavailability of voucher specimens or immature state of vouchers. |
| Confirmed Candidate Species (CCS) | Specimens or populations characterized by a detectable genetic differentiation to all described species, not necessarily above any threshold, but in concordance with at least one of the following criteria:
|
Uncorrected pairwise genetic divergences in 16S rRNA gene to all other described species in most cases >3%, sometimes only 1–2%. Concordance of this molecular differentiation with one of the following:
|
| Deep Conspecific Lineage (DCL) | Deep genealogical lineages above a threshold value typical for comparisons among closely related species in the group of animals under study. One or several of the following must apply:
|
Uncorrected pairwise genetic divergences in 16S rRNA gene > 3% to all other described species in combination with one or several of the following:
|
Undescribed diversity is phylogenetically widespread in Malagasy frogs (Fig. 1A and SI Appendix). We found candidate species in most clades except in a few monospecific or species-poor genera (e.g., Wakea, Dyscophus). Genera such as the colorful Mantella or Heterixalus are rather well studied and consequently contain only a few candidate species, whereas in the 3 most speciose and diverse clades (Mantidactylus, Boophis, and Cophylinae), the number of candidate species is close to or even exceeds the number of described species (Fig. 1E). In general, in most genera the average differentiation in the 16S rRNA gene of CCS and UCS is >4%. Most described species of Malagasy frogs show genetic distances of 6–8% to their closest relatives depending on the period of discovery (9), but these values refer to distances corrected by the Kimura-2-parameter-model (K2P) which are higher than the uncorrected distances we report. The molecular differentiation of CCS and UCS is thus at similar levels as between described species of Malagasy frogs, indicating that they are not the result of taxonomic inflation (e.g., elevating subspecies to specific status).
New discoveries are also geographically ubiquitous. They occur throughout Madagascar, both in poorly explored and in better-studied areas. Described species are known from 451 sites and 87.3% are found in protected areas, whereas CCS and UCS are known from 168 sites and only 66.4% are found in protected areas (Fig. 1 B and C). Even in 2 of the best-studied sites for amphibians in Madagascar, Mantadia/Analamazaotra and Ranomafana National Parks, harboring a total of 94 and 112 species of frogs, we found 10 and 31 CCS and UCS (see SI Appendix). Most described species are known from one or a few localities and have not been found elsewhere. This pattern is consistent among the undescribed species; only 6 of the 219 CCS and UCS are widespread, whereas 63 are currently known from small ranges and 154 from single localities.
Discussion
The Concept of Candidate Species and Their Delimitation.
By integrating molecular, morphological, and bioacoustic data, we have identified a large number of undescribed species of amphibians in Madagascar. The remarkable increase in estimated species numbers is independent of the species concept applied. Almost all described species are well defined as reciprocally monophyletic and strongly differentiated units by molecular datasets and by morphological and/or bioacoustic evidence. The only exceptions are Mantella milotympanum, Mantella nigricans, Dyscophus antongilii, Heterixalus andrakata, Heterixalus variabilis, and Heterixalus tricolor, which are not clearly diagnosable by molecular data and require further study (29). The same combination of character sets defines all CCS (e.g., Fig. 2A), and, based on our ongoing studies, we expect the same for most UCS. Many of these taxa occur in syntopy with their nearest relatives without any signal of admixture, corroborating their species status under biological, evolutionary, and phylogenetic species concepts. In contrast, many DCL correspond to genetically differentiated allopatric populations of widespread species for which the application of a biological species concept is inherently difficult. If criteria such as those for Malagasy primates (30) were applied to these DCL, many of them would be considered evolutionary species as well. Because the process of species formation in amphibians is an active and controversial area of research (31), we currently discourage such conclusions.
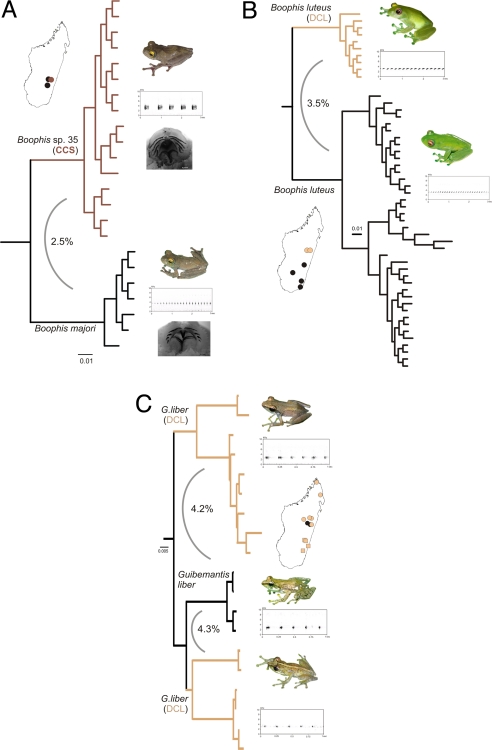
Fig. 2.
Examples of combined phylogeographic, bioacoustic, and morphological evidence used to classify divergent mitochondrial lineages as CCS or DCL. (A) Boophis majori and B. aff. majori are sympatric CCS; despite their only low genetic divergence, they have distinct and constant differences in tadpole morphology and qualitative differences in advertisement calls, without signal of genetic admixture. (B and C) B. luteus (B) and Guibemantis liber (C) consist of deeply divergent genealogical lineages classified as DCL, because the call differences only affect quantitative parameters such as note-repetition rate in B. luteus, and there are no morphological or ecological differences between the populations. Furthermore, genetic admixture of the lineages was detected in G. liber at one locality.
Recent progress in molecular techniques leads to acquiring newly determined DNA sequences at a faster pace than taxonomists are able to follow up with results. In many cases, divergent DNA sequences are observed that probably correspond to distinct, undescribed species, but a taxonomic analysis of these data are impracticable in the short term, especially in morphologically cryptic and highly diverse groups (32, 33). The term “candidate species” is rarely used by zoologists, but has recently been proposed for newly discovered units that probably correspond to undescribed species (31), and we advocate using it in a less formal way than the Candidatus status in microbiology (34–36).
Delimiting species is a resurgence issue in biology for which various explicit procedures have been proposed (37, 38). Most of these procedures require a relatively good state of taxonomic knowledge. For tree-based methods, organisms usually need to be sampled from several populations and to have their phylogenetic relationships well resolved. Many nontree methods require extensive datasets as well. These datasets are usually not available for those genetically divergent individuals for which we propose the category of candidate species. Also, species are known to be often paraphyletic in their mitochondrial haplotypes (39). Even more importantly, phylogenies based on single DNA fragments, such as used in DNA barcoding approaches, are often not sufficiently resolved. We therefore do not recommend explicit tree-based delimitations of candidate species, although such methods are certainly a great improvement for the eventual taxonomic description of species.
Distance-based DNA barcoding methods based on divergence thresholds are prone to 2 kinds of errors. Because there is no fixed time span needed for speciation, there should be a continuum of pairwise genetic divergences of sister species, down to 0% in cases of rapid adaptive speciation. Any threshold will therefore miss a proportion of very young species (false negatives). The second kind of error is to wrongly identify intraspecific genealogical lineages as species (false positives). The accuracy of the method depends on the so-called barcoding gap between intraspecific and interspecific divergences, but previous work on amphibians has shown a wide overlap of these values and absence of a distinct barcoding gap (40). Because DNA barcoding can only be a preliminary tool for a first and rough identification of candidate species, we favor a conservative approach that minimizes the error probability of false positives. This approach will miss species of recent origin, but it will more efficiently help taxonomists to focus on those genealogical lineages likely to be undescribed species. The categories CCS, UCS, and DCL constitute an advance over uncritical approaches to DNA barcoding because they incorporate this useful molecular tool but emphasize the need of complementary data to understand biological reality.
Following our definitions, assignment of a genealogical lineage to the CCS category has the highest reliability of the 3 categories proposed here, and the number of 129 CCS thus provides a minimum estimate of undescribed species in Madagascar. Many DCL are largely based on the lack of evidence for CCS status. Some of these genealogical lineages may thus be upgraded to CCS when more extensive phylogeographic data and integration of nuclear genetic markers are available (e.g., Boophis luteus, Fig. 2B), although in many cases the DCL status is well assessed by comprehensive datasets (e.g., in Guibemantis liber, Fig. 2C). Finally, the UCS status is, by definition, unreliable and is used to accommodate those genealogical lineages with insufficient data.
These category definitions are not restricted to amphibians. In Madagascar and elsewhere, molecular data have been used to detect unexpected levels of cryptic diversity in other animal groups (32, 33, 41–43), and in many cases the biological identity of the previously undescribed genealogical lineages remains largely unassessed. Our definition of CCS (Table 1) relies on genetic divergence combined with a distinct difference in either morphology or in a character that mediates premating isolation (advertisement calls in frogs). Characters and levels of morphological divergence will be different in each animal group, but in general we expect a wide applicability of this approach. More work must focus on understanding conditions under which the concordance of mitochondrial and nuclear gene genealogies can become a defining standard for CCS (15, 44, 45).
Molecular Data in Integrative Taxonomy.
Integrative taxonomic analyses (46) are boosting the discovery and description of amphibians worldwide (17). The global biodiversity crisis requires these kinds of taxonomic studies (21) because they are the prerequisite for understanding diversity patterns and thus for the identification of priority areas for conservation. Despite the fact that recent efforts in amphibian systematics significantly increased the number of species, inventories—especially from tropical areas—are far from complete. These inventories are also needed to complete our knowledge of the tree of life of amphibians.
Amphibians tend to have conservative morphological evolution (47–49), which makes their species-level identification difficult. We here demonstrate the efficiency of an approach that first identifies genealogical lineages with high genetic divergences (40) and in a second step classifies these as CCS, UCS, or DCL by integrating other datasets. This approach does not preclude detailed morphological and bioacoustic studies in cases where biological species show low genetic differentiation. In fact, our approach is close to current taxonomic practice in herpetology: We propose diagnostic character-state differences in morphology or bioacoustics as a central prerequisite for CCS status, agreeing with the fact that most descriptions of new amphibian species are today still based only on morphology, despite the availability of more sophisticated methods (37, 38). To extend current standards in amphibian taxonomy, we strongly recommend the routine inclusion of DNA sequences in descriptions of new species. Such a practice would simplify the subsequent identification of divergent genealogical lineages and their integrative taxonomical study (40), even if this sequence information would not be sufficient to assess phylogenetic relationships of the new species.
Amphibian Diversity Patterns in Madagascar and Conservation Priorities.
Although Madagascar's fauna has been studied extensively during the last century, the increase in species discoveries is making diversity estimates tentative in most cases. Our data suggest that current diversity figures largely underestimate the real diversity of frogs, and probably of other terrestrial vertebrates as well. This underestimation has direct consequences on our understanding of the pattern of biodiversity in Madagascar. Most of the Malagasy amphibian species are known from <10 localities, and many from single localities only (50), making spatial diversity analyses difficult. In many cases, this analysis certainly does not represent species actual distribution ranges, but indicates a real lack of information on species spatial distribution patterns in Madagascar whereas a considerable number of species are probably indeed restricted to very small and isolated ranges.
Important increases in species numbers have also characterized other groups of Malagasy vertebrates during the last 2 decades (30, 51). Numerous undescribed species of reptiles, mammals, and especially freshwater fishes are known to exist but are usually not included in conservation assessments. Probably by far the largest proportion of species of invertebrates remains undescribed as is usual for most tropical regions. For amphibians as well as many other groups of animals, the ongoing discovery of undescribed diversity is being triggered by several factors, including the increase of taxonomic work in Madagascar, incorporating new molecular tools with traditional methods, and an increase in field exploration efforts. Some larger patches of rainforest in Madagascar have not been properly explored yet and probably harbor numerous additional new candidate species. However, many of the amphibians we have identified here are found in some of the best-studied areas of Malagasy rainforests, such as Ranomafana and Analamazaotra/Mantadia National Parks (Fig. 1 B and C; SI Appendix). Taxonomic exploration should thus not be limited to unexplored areas but should include putatively well-known plots as well. The combination of phylogenetic datasets with spatial modeling of species or clades will help to define their potential distribution in space and thereby help to identify target areas for further exploration.
Our findings confirm that spatial patterns of endemism, rarity, and species richness within the Madagascar biodiversity hotspot are poorly known. The protected area network of Madagascar has increased significantly in recent years, and proposals for new protected areas based on multitaxon analyses (52) will cover most of the key biodiversity areas in Madagascar. Almost a quarter (23.3%) of the newly discovered frog candidate species are not found within the currently existing protected areas; many of these are range-restricted and are more likely to disappear given the pace of habitat destruction observed in Madagascar (22). This pattern implies that even a large network of protected areas may not fully protect the current diversity. A conservation strategy should consider protecting additional small rainforest fragments, because they can contain a relatively large proportion of amphibian species (53, 54). The apparent absence from Madagascar of emerging diseases such as the amphibian chytrid fungus, in concert with a strong commitment of national institutions to conserve its biodiversity, characterize Madagascar as a unique opportunity to proactively protect an amphibian fauna so far untouched from catastrophic declines other than those caused by ongoing habitat destruction (55).
Implications for Global Biodiversity Estimates.
This study analyzes the complete amphibian diversity of a tropical biodiversity hotspot by using comprehensive genetic, morphological, and bioacoustic datasets. Previous studies of other hotspots reported high numbers of undescribed amphibian species as well (11, 15), but were more limited in taxonomic and geographic coverage. The estimated 250% increase since 1991 up to possibly 465 species is unparalleled and would make Madagascar one of the top 5 most diverse countries on the planet for amphibians, together with Brazil, Colombia, Ecuador, and Peru. The global importance of Madagascar's amphibian fauna is paramount, especially because of its extreme degree of endemism (100% among the native species). However, it should be taken into account that very few other tropical countries with diverse amphibian faunas have been surveyed as intensively as Madagascar with comparable approaches. The unexpectedly great increase in species numbers that we estimate may therefore not characterize Madagascar as being much more species-rich than other tropical regions. Rather, it exemplifies the power of integrative taxonomic assessments and predicts that applying these to other regions or other groups of organisms may lead to comparably high proportions of novel discoveries.
As a primary conclusion from our study, the number of tropical amphibian species is probably underestimated at an unprecedented level at a global scale. Extrapolating our data to other less-studied tropical regions predicts that the number of amphibian species worldwide could double or possibly even quadruple before saturation in new discoveries can be expected. Amphibians are the vertebrate group with the highest proportion of threatened species (20), and current declines may be affecting more diversity than previously thought. Paradoxically, we are living in an era of simultaneous mass extinction and mass discovery of amphibians (21). Integrative taxonomic inventories, including molecular assessments of diversity as a standard technique, are urgently needed, especially for poorly explored, highly diverse regions.
Materials and Methods
DNA Sequencing and Analysis.
A fragment of the mitochondrial 16S rRNA gene was amplified and sequenced for ca. 2,850 specimens. Based on Neighbor-Joining trees, we identified sequences showing high divergences from reference specimens of described species. Selected sequences were used to compute a tree by using Bayesian inference after determining the appropriate substitution model (see SI Appendix). We used the software TaxI (56) to calculate pairwise distances between sequences, which avoids possible alignment artifacts in the distribution of indels from the global alignment that may affect this computation. Extended methods are available in SI Appendix.
Assessment of Bioacoustic Differentiation.
The calls of many candidate species are documented in a recently compiled sound guide (57). Advertisement calls of anurans are excellent taxonomic indicators, but intraspecific call differences are known (58): Temporal variables and (to a far lesser extent) also frequency (spectral characters) depend on environmental temperature and state of sexual motivation, whereas frequency is mainly influenced by body size. However, these differences are in all cases quantitative and continuous, i.e., note or note interval duration, or fundamental and dominant frequency, become continuously larger or smaller with increasing or decreasing temperature or body size. Similar continuous differences are also known among populations of a species, often because of character displacement or adaptation to environmental factors (59). Most well-documented cases of geographic call variation in anurans refer to such variation in continuous characters (60–62). In contrast, qualitative differences such as the presence/absence of different call types or a melodious vs. unmelodious call structure are rare within species and the few documented cases show the signature of incipient speciation (63). We considered call variation in (i) dominant or fundamental frequency, (ii) note duration, (iii) note interval duration, and (iv) pulse rate as quantitative call differences, insufficient to define a CCS except in situations of sympatry. Differences in (i) number of note types, (ii) general arrangement of note types, and (iii) melodious vs. noisy or pulsed structure of notes were considered to be qualitative call differences, defining a CCS status.
Geographic Analysis.
A georeferenced database was compiled containing amphibian records from literature and our own field inventories. Records were taken into account only if identification was considered reliable, i.e., type localities, records of morphologically distinct species, or records with bioacoustic or molecular data (50). The current protected area network was provided by Conservation International. Distribution maps of all described species and CCS (29) largely form the basis for the geographic analysis. Table S1 in the SI Appendix lists the working names applied herein and in the recently published field guide (29) for the candidate species. The 9 described species for which no genetic data are available (see SI Appendix) are morphologically distinct and have narrow distribution ranges (29), and their confusion with any CCS or UCS is unlikely.
Supplementary Material
Acknowledgments.
We thank Parfait Bora, Neil D'Cruze, Louis du Preez, Liliane Raharivololoniaina, Roger-Daniel Randrianiaina, Jasmin E. Randrianirina, and Che Weldon for their help in the field. We also are grateful to Sandra Nieto Román, Louis Boumans, Angelica Crottini, Susanne Hauswaldt, Meike Kondermann, Gabriele Keunecke, Eva Saxinger, Axel Strauß, Meike Teschke, and Jens Weste who helped to obtain the DNA sequences. David Knox (Conservation International) provided the latest version of the maps of Madagascar's protected area network. The study was made possible by collaboration agreements of the authors' institutions with the Université d'Antananarivo, Département de Biologie Animale, the Association Nationale pour la Gestion des Aires Protegées, and the Parc Botanique et Zoologique de Tsimbazaza, Madagascar. We are grateful to the Malagasy authorities for research and export permits. This work was supported by National Science Foundation Assembling the Tree of Life Program Grant EF-0334939, Consejo Superior de Investigaciones Científicas Intramural Grant 200830I100, grants from the Volkswagen Foundation, Deutsche Forschungsgemeinschaft (VE247/1–1 and VE247/2–1), the European Association of Zoos and Aquaria, and the Wildcare Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Genbank database (accession nos. FJ559069–FJ559372 and FJ217329–FJ217345)
This article contains supporting information online at www.pnas.org/cgi/content/full/0810821106/DCSupplemental.
References
- 1.Mittermeier RA, et al. Hotspots Revisited: Earth's Biologically Richest and Most Endangered Terrestrial Ecoregions. Arlington, VA: Conservation International; 2005. [Google Scholar]
- 2.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 3.Orme CDL, et al. Global hotspots of species richness are not congruent with endemism or threat. Nature. 2005;436:1016–1019. doi: 10.1038/nature03850. [DOI] [PubMed] [Google Scholar]
- 4.Brooks TM, et al. Global biodiversity conservation priorities. Science. 2006;313:58–61. doi: 10.1126/science.1127609. [DOI] [PubMed] [Google Scholar]
- 5.Cowling RM, Pressey RL, Rouget M, Lombard AT. A conservation plan for a global biodiversity hotspot—the Cape Floristic Region, South Africa. Biol Conserv. 2003;112:191–216. [Google Scholar]
- 6.Zwickl DJ, Hillis DM. Increased taxon sampling greatly reduces phylogenetic error. Syst Biol. 2002;51:588–598. doi: 10.1080/10635150290102339. [DOI] [PubMed] [Google Scholar]
- 7.Heath TA, Zwickl DJ, Hillis DM. Taxon sampling affects inferences of macroevolutionary processes from phylogenetic trees. Syst Biol. 2008;57:160–166. doi: 10.1080/10635150701884640. [DOI] [PubMed] [Google Scholar]
- 8.Hanken J. Why are there so many new amphibian species when amphibians are declining? Trends Ecol Evol. 1999;14:7–8. doi: 10.1016/s0169-5347(98)01534-1. [DOI] [PubMed] [Google Scholar]
- 9.Köhler J, et al. New amphibians and global conservation: A boost in species discoveries in a highly endangered vertebrate group. BioScience. 2005;55:693–696. [Google Scholar]
- 10.AmphibiaWeb. Information on amphibian biology and conservation. [Accessed January 14, 2009];2008 Available at www.amphibiaweb.org.
- 11.Meegaskumbura M, et al. Sri Lanka: An amphibian hotspot. Science. 2002;298:379. doi: 10.1126/science.298.5592.379. [DOI] [PubMed] [Google Scholar]
- 12.Faivovich J, et al. Systematic review of the frog family Hylidae, with special reference to Hylinae: Phylogenetic analysis and taxonomic revision. Bull Amer Mus Nat Hist. 2005;294:1–240. [Google Scholar]
- 13.Manamendra-Arachchi K, Pethiyagoda R. The Sri Lankan shrub-frogs of the genus Philautus Gistel, 1848 (Ranidae:Rhacophorinae), with description of 27 new species. Raffles Bull Zool Suppl. 2005;12:163–303. [Google Scholar]
- 14.Stuart BL, Inger RF, Voris HK. High level of cryptic species diversity revealed by sympatric lineages of Southeast Asian forest frogs. Biol Lett. 2006;2:470–474. doi: 10.1098/rsbl.2006.0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouquet A, et al. Underestimation of species richness in Neotropical frogs revealed by mtDNA analyses. PLoS ONE. 2007;2:e1109. doi: 10.1371/journal.pone.0001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De la Riva I. Bolivian frogs of the genus Phrynopus, with descriptions of twelve new species (Anura: Brachycephalidae) Herp Monogr. 2007;21:247–277. [Google Scholar]
- 17.Padial JM, de la Riva I. Taxonomic inflation and the stability of species lists: The perils of ostrich's behavior. Syst Biol. 2006;55:859–867. doi: 10.1080/1063515060081588. [DOI] [PubMed] [Google Scholar]
- 18.Biju SD, Bossuyt F. New frog family from India reveals an ancient biogeographical link with the Seychelles. Nature. 2003;425:711–714. doi: 10.1038/nature02019. [DOI] [PubMed] [Google Scholar]
- 19.Min MS, et al. Discovery of the first Asian plethodontid salamander. Nature. 2005;435:87–90. doi: 10.1038/nature03474. [DOI] [PubMed] [Google Scholar]
- 20.Stuart SN, et al. Status and trends of amphibian declines and extinctions worldwide. Science. 2004;306:1783–1786. doi: 10.1126/science.1103538. [DOI] [PubMed] [Google Scholar]
- 21.Wake DB, Vredenburg VT. Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc Natl Acad Sci USA. 2008;105:11466–11473. doi: 10.1073/pnas.0801921105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper G, Steininger M, Tucker C, Juhn D, Hawkins F. Fifty years of deforestation and forest fragmentation in Madagascar. Env Cons. 2007;34:325–333. [Google Scholar]
- 23.Yoder AD, Nowak MD. Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Ann Rev Ecol Evol Syst. 2006;37:405–431. [Google Scholar]
- 24.Boumans L, Vieites DR, Glaw F, Vences M. Geographical patterns of deep mitochondrial differentiation in widespread Malagasy reptiles. Mol Phylogenet Evol. 2007;45:822–839. doi: 10.1016/j.ympev.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 25.Wilmé L, Goodman SM, Ganzhorn JU. Biogeographic evolution of Madagascar's microendemic biota. Science. 2006;312:1063–1065. doi: 10.1126/science.1122806. [DOI] [PubMed] [Google Scholar]
- 26.Yoder AD, et al. A multidimensional approach for detecting species patterns in Malagasy vertebrates. Proc Natl Acad Sci USA. 2005;102:6587–6594. doi: 10.1073/pnas.0502092102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoder AD, Heckman KL. Mouse lemur phylogeography revises a model of ecographic constraint in Madagascar. In: Fleagle J, Lehman SM, editors. Primate Biogeography: Progress and Prospects. New York: Springer; 2006. pp. 255–268. [Google Scholar]
- 28.Blommers-Schlösser RMA, Blanc CP. Amphibiens (première partie) Faune de Madagascar. 1991;75:1–379. [Google Scholar]
- 29.Glaw F, Vences M. A Field Guide to the Amphibians and Reptiles of Madagascar. 3rd ed. Köln, Germany: Köln, Vences & Glaw; 2007. [Google Scholar]
- 30.Mittermeier RA, et al. Conservation International Tropical Field Guide. Arlington, VA: Conservation International; 2006. Lemurs of Madagascar. [Google Scholar]
- 31.Vences M, Wake DB. Speciation, species boundaries and phylogeography of amphibians. In: Heatwole HH, Tyler M, editors. Amphibian Biology. Vol 6. Chipping Norton, Australia: Surrey Beatty & Sons; 2007. pp. 2613–2669. Systematics. [Google Scholar]
- 32.Blaxter M, Elsworth B, Daub J. DNA taxonomy of a neglected animal phylum: An unexpected diversity of tardigrades. Proc R Soc London B. 2004;271(Suppl 4):S189–S192. doi: 10.1098/rsbl.2003.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blaxter M, et al. Defining operational taxonomic units using DNA barcode data. Philos Trans R Soc London B Biol Sci. 2005;360:1935–1943. doi: 10.1098/rstb.2005.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray RGE, Schleifer KH. Taxonomic notes: A proposal for recording the properties of putative taxa of prokaryotes. Int J Syst Bacteriol. 1994;44:174–176. doi: 10.1099/00207713-44-1-174. [DOI] [PubMed] [Google Scholar]
- 35.Murray RGE, Stackebrandt E. Taxonomic Note: Implementation of the provisional status Candidatus for incompletely described prokaryotes. Int J Syst Bacteriol. 1995;45:186–187. doi: 10.1099/00207713-45-1-186. [DOI] [PubMed] [Google Scholar]
- 36.Stackebrandt E, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- 37.Sites JW, Jr, Marshall JC. Delimiting species: A Renaissance issue in systematic biology. Trends Ecol Evol. 2003;18:462–470. [Google Scholar]
- 38.Sites JW, Jr, Marshall JC. Operational criteria for delimiting species. Annu Rev Ecol Evol Syst. 2004;35:199–227. [Google Scholar]
- 39.Funk DJ, Omland KE. Species-level paraphyly and polyphyly: Frequency, causes and consequences, with insights from animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 2003;34:397–423. [Google Scholar]
- 40.Vences M, Thomas M, Bonett RM, Vieites DR. Deciphering amphibian diversity through DNA barcoding: Chances and challenges. Philos Trans R Soc London Ser B. 2005;360:1859–1868. doi: 10.1098/rstb.2005.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson LE, Goodman SM, Yoder AD. Illumination of cryptic species boundaries in long-tailed shrew tenrecs (Mammalia: Tenrecidae; Microgale) provides insights into geographic variation and distributional constraints. Biol J Linn Soc. 2004;83:1–22. [Google Scholar]
- 42.Yoder AD, et al. Remarkable species diversity in Malagasy mouse lemurs (Primates, Microcebus) Proc Natl Acad Sci USA. 2000;97:11325–11330. doi: 10.1073/pnas.200121897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith MA, Fisher BL, Hebert PDN. DNA barcoding for effective biodiversity assessment of a hyperdiverse arthropod group: The ants of Madagascar. Philos Trans R Soc London Ser B. 2005;360:1825–1834. doi: 10.1098/rstb.2005.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimada T, Matsui M, Yambun P, Lakim M, Mohamed M. Detection of two cryptic taxa in Meristogenys amoropalamus (Amphibia, Ranidae) through nuclear and mitochondrial DNA analyses. Zootaxa. 2008;1843:24–34. [Google Scholar]
- 45.Ahrens D, Monaghan MT, Vogler AP. DNA-based taxonomy for associating adults and larvae in multi-species assemblages of chafers (Coleoptera: Scarabaeidae) Mol Phylogenet Evol. 2007;44:436–449. doi: 10.1016/j.ympev.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 46.Dayrat B. Toward integrative taxonomy. Biol J Linn Soc. 2005;85:407–415. [Google Scholar]
- 47.Cherry LM, Case SM, Wilson AC. Relative rates of morphological evolution in primates, carnivores and frogs. Am Zool. 1977;17:910. [Google Scholar]
- 48.Emerson SB. Convergence and morphological constraint in frogs: Variation in postcranial morphology. Fieldiana Zool. 1986;43:1–19. [Google Scholar]
- 49.Wake DB. Homoplasy—the result of natural-selection, or evidence of design limitations. Am Nat. 1991;138:543–567. [Google Scholar]
- 50.Vieites DR, Nieto-Román S, Vences M. Andreone F, editor. Towards understanding the spatial pattern of amphibian diversity in Madagascar. A Conservation Strategy for the Amphibians of Madagascar. 2008;45:397–340. Mon Mus Reg Sci Nat Torino. [Google Scholar]
- 51.Raxworthy CJ. Introduction to reptiles. In: Goodman SM, Benstead JP, editors. The Natural History of Madagascar. Chicago: Chicago Univ Press; 2003. pp. 946–949. [Google Scholar]
- 52.Kremen C, et al. Aligning conservation priorities across taxa in Madagascar with high-resolution planning tools. Science. 2008;320:222–226. doi: 10.1126/science.1155193. [DOI] [PubMed] [Google Scholar]
- 53.Vallan D. Influence of forest fragmentation on amphibian diversity in the nature reserve of Ambohitantely, highland Madagascar. Biol Conserv. 2000;96:31–43. [Google Scholar]
- 54.Vallan D. Effects of anthropogenic environmental changes on amphibian diversity in the rainforests of eastern Madagascar. J Trop Ecol. 2002;18:725–742. [Google Scholar]
- 55.Andreone F, et al. The challenge of conserving amphibian megadiversity in Madagascar. PLoS Biol. 2008;6:e118. doi: 10.1371/journal.pbio.0060118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steinke D, Salzburger W, Vences M, Meyer A. TaxI—A software tool for DNA barcoding using distance methods. Philo Trans R Soc London Ser B. 2005;360:1975–1980. doi: 10.1098/rstb.2005.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vences M, Glaw F, Marquez R. The Calls of the Frogs of Madagascar. Barcelona: Alosa; 2006. 3 Audio CDs (total duration 220 min) and booklet. [Google Scholar]
- 58.Schneider H, Sinsch U. Contributions of bioacoustics to the taxonomy of Anura. In: Heatwole H, editor. Amphibian Biology. Vol 7. Chipping Norton, Australia: Surrey Beatty & Sons; 2007. pp. 2892–2931. Phylogeny and Systematics. [Google Scholar]
- 59.Gerhardt HC, Huber F. Acoustic Communication in Insects and Anurans: Common Problems and Diverse Solutions. Chicago: Univ Chicago Press; 2002. [Google Scholar]
- 60.Ryan MJ, Rand AS, Weigt LA. Allozyme and advertisement call variation in the túngara frog, Physalaemus pustulosus. Evolution. 1996;50:2435–2453. doi: 10.1111/j.1558-5646.1996.tb03630.x. [DOI] [PubMed] [Google Scholar]
- 61.Hoskin CJ, Higgie H, McDonald KR, Moritz C. Reinforcement drives rapid allopatric speciation. Nature. 2005;437:1353–1356. doi: 10.1038/nature04004. [DOI] [PubMed] [Google Scholar]
- 62.Ryan MJ, Wilczinski W. Evolution of intraspecific variation in the advertisement call of a cricket frog (Acris crepitans, Hylidae) Biol J Linn Soc. 2008;44:249–271. [Google Scholar]
- 63.Boul KE, Funk WC, Darst CR, Cannatella DC, Ryan MJ. Sexual selection drives speciation in an Amazonian frog. Proc R Soc London B. 2007;274:399–406. doi: 10.1098/rspb.2006.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.