Abstract
Spherical capsids are shells of protein subunits that protect the genomes of many viral strains. Although nature displays a range of spherical capsid sizes (reflected by the number of subunits in the formation), specific strains display stringent requirements for forming capsids of specific sizes, a requirement that appears crucial to infectivity. Despite its importance in pathogenicity, little is known regarding the determinants of capsid size. Still less is known about exactly which capsids can undergo maturation events such as buckling transitions—postcapsid-assembly events that are crucial to some virus strains. We show that the exclusive determinant of capsid size is hexamer shape, as defined by subunit–subunit dihedral angles. This conclusion arises from considering the dihedral angle patterns within hexamers belonging to natural canonical capsids and geometric capsid models (deltahedra). From simple geometric models and an understanding of endo angle propagation discussed here, we then suggest that buckling transitions may be available only to capsids of certain size (specifically, T < 7 capsids are precluded from such transformations) and that T > 7 capsids require the help of auxiliary mechanisms for proper capsid formation. These predictions, arising from simple geometry and modeling, are backed by a body of empirical evidence, further reinforcing the extent to which the evolution of the atomistically complex virus capsid may be principled around simple geometric design/requirements.
Keywords: auxiliary proteins, capsid buckling, deltahedra, endo angle constraint
A large number of human- and crop-infecting viruses are protected by spherical capsids (shells) of various sizes that are primarily made up of self-organizing protein subunits (1, 2). Caspar and Klug's (3) seminal article on quasi-equivalence explained how an infinite range of capsid sizes can be “constructed” by combining 60T subunits or 12 pentamers (5-valent subunit clusters) and a variable number of hexamers [10 × (T − 1)] into a closed spherical shell (T ∈ {1,3,4,7, …} and is the triangulation number described in ref. 3).
From the range of possible sizes, generally, subunits from specific viral strains assemble into capsids of specific sizes; the inability to form those native sizes is believed to result in the loss of infectivity. For example, the sobemovirus and birnavirus capsids (4, 5) shown in Fig. 1A are known to be pathogenic primarily in their native T = 3 and T = 13 capsid forms, respectively. Despite its importance in pathogenicity, our picture of capsid size specificity is incomplete. In the present report, we are interested in the structural features (constraints), if any exist, that differentiate between capsids of different sizes (capsid design criteria). An appreciation of these concepts is pressing from a nanotechnological perspective [for the rational design of artificial scalable assemblies that build on current practices, such as in the use of protein fusion and symmetry properties by Padilla et al. (6)] and a therapeutic perspective (to impede the formation of infective native capsids).
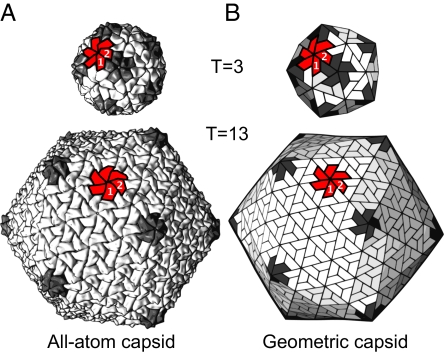
Fig. 1.
Spherical canonical capsids are scalable. (A) Two natural canonical capsids (the T = 3 sobemovirus and T = 13 birnavirus capsids with PDB IDs codes 1smv and 1wce, respectively) are shown to emphasize that spherical capsids come in many sizes that are composed of 12 pentamers (dark gray) and 10 × (T − 1) hexamers (3). (B) We use geometric models as platonic capsid representations for the characterization of structure and function. In each capsid, a single hexamer (colored red) along with 2 subunits (“1” and “2”) are marked to emphasize the structural correspondence between all-atom and geometric capsids.
The size-specificity puzzle gets more interesting given the theoretical evidence that a single subunit shape (the trapezoidal prototile) possesses the ability to tile all of the allowed canonical capsid sizes (T = 1, 3, 4, 7 …) (7), which is backed by evidence of a ubiquitous trapezoidal subunit shape seen in nature (discussed in ref. 8). In these situations, the differences between capsids of different sizes will be seen within the capsid's subunit–subunit dihedral angles,* i.e., size-specificity within canonical capsids (7) may be manifested in the angles at which the generally rigid subunits interact within the capsid.
In the following sections, we attempt to show that the exclusive determinant of canonical capsid size is hexamer shape as defined by the internal subunit–subunit dihedral angles that comprise the hexameric capsomers. We then use knowledge of “endo angle constraints” to predict that only capsids of specific sizes (T ≥ 7) possess the potential to undergo true buckling transitions. Interesting inferences on the requirement of auxiliary proteins in large capsids are also discussed in the context of hexameric flexibility.
Results and Discussion
Hexamer Shapes Encode for Capsid Size.
To understand capsid size specificity, we chose to focus on hexamers and the effect of neighboring pentamers on their shapes for the following reasons: From a geometric perspective, capsids of different sizes (but formed from similarly shaped subunits) must possess identical pentamers [see supporting information (SI) Text], which has also been shown to be true for both cryo-EM (9) and X-ray structures.† Second, hexamer structure in some capsids is known to be influenced by the presence of neighboring pentamers (9, 10), indicating that the arrangement of pentamers may be important for defining hexamer shape.
Comparing dihedral angles between subunits involves defining subunit planes (and then comparing the angle between adjacent planes), which is an imprecise endeavor because the subunit is a 3-dimensional molecule with a rough and complicated atomic surface.‡ Instead, we looked at how similar the pentameric (defined “endo”) dihedral angle is to each of the hexameric angles within a capsid. For any hexamer within capsids possessing highly uniform subunit structures [i.e., strictly canonical capsids (7)], this can be done simply by structurally aligning a pair of adjacent pentameric subunits to each of the 6 pairs of adjacent hexameric subunits (more in Materials and Methods). Each pair–pair structural alignment results in 1 RMSD value, which is low if the angles associated with the pairs are similar (and 0 if the pairs possess identical angles).
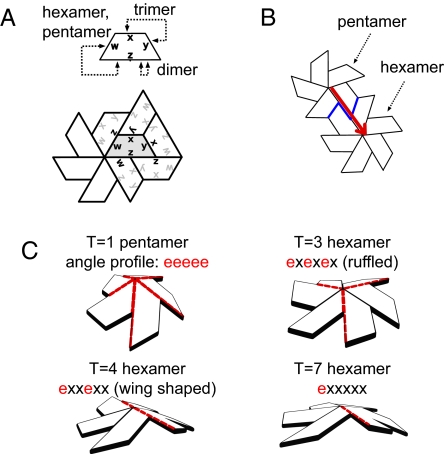
In each capsid studied, for every unique hexamer in a distinct environment (T = 13 capsids have 2 unique hexamer environments, whereas T = 3, 4 and 7 possess just 1), we obtained 6 RMSD values (numbered 1 through 6 in counterclockwise fashion starting with an angle closest to the pentamer) represented as lines (1 for each unique hexamer) and grouped by T number in Fig. 2A (shown separately for each capsid in Fig. S1). Excepting the T = 13 capsid, which possesses 2 unique hexamers (labeled as “hexamer 1” and “hexamer 2”), each line in Fig. 2A is obtained from distinct natural canonical capsid structures (described in Materials and Methods). The qualitative groupings of the lines in this figure suggest that hexamers exist in variously puckered hexamer shapes that are size- or T-specific. For example, all hexamers from T = 3 and T = 4 canonical capsids appear to display characteristic “ruffled” and “wing” shapes, respectively, displayed geometrically in Fig. 3C (that correspond to previously described trimer of dimers and dimer of trimers (11, 12), respectively). Also, all hexameric angles (circled in the x axis in Fig. 2A adjacent to pentameric “endo” angles) are also endo-like in nature, as indicated by the low RMSD values, which is an important outcome of the pentameric endo angle constraint on hexamer shape discussed further on.
Fig. 2.
Hexamer shape is specific to capsid size. (A) The extent to which hexameric dihedral angles (numbered 1 through 6 on the x axis in all graphs) found within natural capsids resemble the endo angle found within the pentamer (angle similarity is proportional to the RMSD). It is evident that hexameric angles adjacent to pentamers (with numbers circled on the x axis) are consistently endo-like, which gives rise to the concept of endo angle propagation (see Endo Angle Constraints). (B) Furthermore, ab initio (geometric) models were used to obtain accurate hexameric dihedral angle values, which reflect the patterns seen in A. Both sources (A and B) indicate that the shapes available to the hexamer is strongly constrained by the size of the capsid.
Fig. 3.
simple geometry describes hexamer shape. (A) A canonical capsid subunit is shown with its bonding rules and 1 local environment. (B) The pentamer–hexamer interface shown in blue possesses a curious effect where the hexameric angle adjacent to a pentamer must also be endo like (or pentameric) in nature. This effect—the endo angle constraint (shown as an arrow from the pentameric angle to that in the hexamer)—can be seen in natural canonical capsids as evidenced by the dihedral angle profiles in Fig. 2A. (C) The result is that hexamers belonging to different capsid sizes (T numbers) possess varying number of endo angles (red dashes) ad may possesses different hexamer shapes.
Fig. 2A, however, useful, cannot be used in making quantitative observations on hexameric geometries that would be required from a capsid design/nanotechnology perspective (because an RMSD does not provide us with angle values, it is only an angle similarity metric). For that, computational models of canonical capsids (deltahedra, described in Materials and Methods) were built for T numbers 1, 3, 4, 7, and 13.§ The dihedral angles present within model hexamers are plotted in Fig. 2B, also arranged per-capsid size. It is clear that these dihedral angle patterns closely resemble those seen in nature for each available capsid size (Fig. 2A). This on its own is interesting because these models were obtained from independent ab initio methods (obtained from nondimensional deltahedron graphs embedded in 3-dimensional space with no application of icosahedral symmetry) but still display natural canonical capsid properties (hexamer shape), reinforcing the natural capsid's geometric/mathematical nature. More relevant, however, is that geometric constructions independently reiterate hexamer size specificity. Second, even in the models, it is evident (partly because of the geometric construction itself) that those hexameric angles shared with pentamers are pentameric (or endo-like) at ≈138.19°, independent of size (or T). SI Text shows that this is a mathematical result of monohedral tilability of the capsid.
Endo Angle Constraints.
From our analysis of both the all-atom capsid structures and geometric models, we can surmise that hexameric dihedral angles are affected by the presence of adjacent pentamers. Fig. 3A represents a canonical capsid subunit (described in ref. 7) with its interaction types that give rise to all possible capsid sizes, and Fig. 3B represents a pentamer–hexamer cluster present in T > 1 capsids. It is evident, if all subunits within a capsid retain similar shape and size, that the angle within the pentamer will be propagated into the adjacent hexamer (indicated by the arrow). This we call the endo angle constraint. From this, it becomes evident that hexameric shapes (Fig. 3C) must be specific to capsid size. This is a natural progression of the endo angle constraint on account of shifting positions (and numbers) of neighboring pentamers around the hexamer [an effect that is corroborated by discussions on empirical (9) and theoretical (10) bacteriophage models].¶
We expect that the dihedral angles within the remainder of the hexamer [the “unconstrained” angles we call exo (or “x”)] must also be indirectly constrained by endo angle propagation, because they must accommodate values suitable to the distribution of the preset endo angles, i.e., the number of endo angles present within a hexamer will be important in determining the possible shapes available to the hexamer.
Considering Larger Capsid Sizes.
As we approach larger capsid sizes (T > 7), the number of hexamers in unique environments will increase. We propose that capsids of all sizes may be created from a small repertoire of hexamer shapes. Early work showed that capsids may be separated into 3 classes distinguishable by distinct size-specific capsid morphologies (obtained from symmetry considerations in ref. 13 and paper models in ref. 3), e.g., capsids with k = 0 (where T = 1, 4, 9, 16, 25 …) belong to class A (in ref. 13) and are most icosahedral in morphology. Extending this class system, we hypothesize that capsids belonging to the same class will possess conserved/common hexamer shapes, and so, the rational modification of a capsid's size within a class will be easier than intraclass size conversions. This explains why T = 4 capsid subunits, which form “wing” shaped hexamers (Fig. 3C), once mutated, are able to assemble exclusively into other sizes within the same morphological class (T = 1, 4, 9, 16, 25, and 36) (14). These interchangeability rules explain how capsids of various sizes may have been sampled from a relatively simple set of capsomer building blocks, leading to a range of capsids seen today.
T > 1 to T = 1 Capsid Transformations.
We find that capsid models of any size possess pentameric angles equaling ≈138.19°, the same as internal angles within an icosahedron and that seen within T = 1 pentamers (see Fig. S2). Considering that no explicit icosahedral symmetry was enforced onto the building of the models, this is not an expected result, because a pentamer (collection of 5 valent plates) possesses a configurational degree of freedom (and could therefore possess nonsimilar angles). This indicates that, even at the most basic geometric level, subunits fated to form T > 1 capsids may possess enough information to assemble into T = 1 capsids, especially if subunit–subunit angles specific to hexamers are prevented from forming, an effect visible in both canonical (5, 15) and noncanonical natural capsids (16, 17). Consequently, all subunits evolved to form T > 1 capsids may possess the potential for T > 1 → T = 1 transformations in specific conditions.
Implications for Anticapsid Therapies.
Our results suggest that rational control of hexamer shape may allow for the redirection of native capsid subunits into nonnative/noninfectious forms, allowing for the development of nonnative but industrially useful assemblies and, more importantly, allowing for rational/combinatorially directed antiviral drug design. An example of such hexamer shape modification is by the binding of organic molecules to specific intrahexamer subunit–subunit interfaces [e.g., the molecule HAP1 that modifies T = 4 capsid assembly in the hepatitis B virus (18)]. The current use of organic molecules in controlling capsid disassembly (discussed in ref. 8) and assembly (e.g., refs. 18–21) provides a possible platform to commence the rational search for such molecules that modulate capsid size via the modification of hexamer shape.
Endo Angles and Buckling Transitions.
Some capsids undergo buckling transitions, where the capsid, once assembled, undergoes a change in morphology from being more spherical to a more faceted (or more “icosahedron-like”) form (22, 23). For capsids that undergo such transitions (distinguished from capsid “swelling events” below), this change in morphology is crucial to the continuation of the virus life cycle. In what follows, we attempt to validate a hypothesis that emerges from our understanding on endo angle constraints. Let us consider hexamers extracted from deltahedra for a range of sizes or T numbers (Fig. 4A). Here, the solid lines represent rigid edges, equilateral triangles represent flat subunits, and “P” marks the dihedral angle that is shared with a pentamer and hence endo constrained at ≈318.19° (the constraint is depicted as dashed lines that prevent specific angles from changing). We hypothesize that buckling transitions should be possible only in T ≥ 7 capsids, where the total number of endo angle constraints per hexamer [given by 6/(T − 1), which is easily derivable]‖ is 1 or lower. From a brief analysis of the graphs in Fig. 4A, it is evident that if the endo angle constraints are “turned on,” i.e., if the dashed lines are treated as solid (locking certain dihedral angles at ≈138.19°), only certain hexamers (possessing 1 or fewer endo angle constraints) will be allowed to sample at least 2 easily obtainable but distinct configurations. Specifically, those “flexible” hexamers must belong to T ≥ 7 capsids, where the average endo angle per hexamer [6/(T − 1)], is ≤1. In this way, although endo angles do not directly constrain all angles in the hexamer (via the arrow depiction in Fig. 3A), in some sizes (T = 3, 4), all hexameric dihedral angles are effectively constrained because of specifically arranged endo angle constraints.
Fig. 4.
Only T ≥ 7 capsid models appear to “buckle.” Hexamer graphs taken from various capsid sizes (where subunits are represented as solid-edged triangles) show that, geometrically, only hexamers from T = 7 canonical capsids (or larger) may undergo changes in shape while maintaining monohedrality (see Endo Angles and Buckling Transitions). (A) This occurs in light of the endo angle constraints (shown effectively as dashed edges) imposed by pentamers (marked by P). (B) Forces applied onto individual dihedral angles within capsid models (see Materials and Methods) indicate that T < 7 capsid models are geometrically rigid upon application of small forces on dihedral angles (indicated by parabolic force-energy profiles and singular minima, shown in Fig. S3), whereas the geometry of T ≥ 7 capsids appear to allow for specific dihedral angles to sample multiple values (shown here for T = 7, 13). (C) The result, especially for T = 7 capsid models, is that hexamers within the capsid are able to sample 2 distinct configurations (blue and red hexamers), a result that parallels buckling transitions in theoretical (26) and experimental studies of the T = 7 capsid (22, 23).
To test this idea, we looked for the availability of accessible conformations to a capsid by physically perturbing (“squeezing and stretching”) dihedral angles within capsid models (deltahedra) of varying sizes (T numbers). The main assumption is that if the simplistic model is not able to sample alternative configurations, then the all-atom capsid that is constrained by simple geometry certainly will not. Here, the trimers are treated as rigid units (forming equilateral triangles, faces of the deltahedron). This is a reasonable assumption if subunit shapes are not greatly changed upon capsid buckling [as is noticed in the bacteriophage HK97, where the morphology change has little effect on the general shape of the subunit (23) while greatly modifying the hexamer pucker state (22)].
For each dihedral angle, we applied stretching and squeezing forces (that try to expand and contract the dihedral angles, discussed in Materials and Methods). The forces were incremented from 0 in small steps (0.00125ε units, with cumulative forces ranging from 0 to ε/8, where ε is the bonded force constant of each bond/edge of the deltahedron), while minimizing the structure at every step. If there is no physical constraint geometrically placed on the specific hexameric angle (on account of the architecture of the model), then the forces will cause a change in the structure, and the recorded energy will remain at zero. If constrained, the capsid will be relatively unyielding to the forces, and the energy will increase harmonically with each step only to fall back into its original state after forces are lifted. Dihedral angle tests showed that all hexameric dihedral angles within the T = 1, 3, and 4 capsids are rigid/constrained within our force regime.
However, analysis of the T = 7 capsid model—where the number of endo angles per hexamer is 1 [i.e., where 6/(T − 1) ≤ 1]—shows that some hexameric angles are able to sample an alternative conformation (indicated by the availability of multiple local minima and hexamer configurations in Fig. 4 A and B, respectively). The change is not instantaneous upon application of infinitesimal force, but depends on overcoming a small energy barrier (akin to going through a transition state). Our results indicate that buckling transitions that require the sampling of 2 distinct conformations may be available only to T ≥ 7 capsids (as evidenced in our T = 7, 13 models). However, we stress that not all large capsids may possess this ability even at a simple geometric level. For example, the T = 9 capsid/deltahedron, which is purely icosahedral in shape (with 20 triangulated facets of 27 subunits), may not possess the ability to easily sample multiple configurations on account of its idealized icosahed-ral shape (which is purely convex and hence geometrically highly stable).
It is noteworthy that buckling of capsids represented by continuum elastic shells have been performed before, where interesting relationships between radius, capsid size, and sphericity were established (24, 25); however, in these studies, the predictions made have yet to be applied to capsids of specific T numbers. The continuum models neglect molecular/geometric features of the capsid (such as hexamer shape), and are therefore not analogous to our investigations, which are centered around subunit-shape-resolved models. It will be interesting to see whether inferences/predictions from continuum and geometric methods converge.
Buckling Transitions Versus Other Maturation Events.
We distinguish between what we call “true” buckling transitions and other maturation events such as capsid swelling (or its inverse: shrinking). Buckling transitions are those transitions that allow a shell to sample 2 morphologies—one being more “spherical” and the other being more “faceted” or icosahedral—without undergoing major changes in subunit–subunit bondedness and subunit shape (23). This excludes the other kind of maturation events—swelling (27–29)—which is theoretically available to any capsid regardless of size. Also, those maturation events requiring gross change in subunit shape [e.g., as seen in Flaviruses (30)] are not considered here.
Swelling is primarily caused by weakening of interfaces (via pH modulation, ion depletion, electrostatic screening, etc.), which causes a radial capsid swell (its converse, “shrinkage,” happens when subunit–subunit interactions are strengthened). These events often accompany the introduction/removal of holes between subunits (commonly found within trimers), which cannot be modeled by simple monohedral tilings/deltahedra (as holes must be considered as additional tiles). Examples of swelling and shrinkage are the T = 3 and unnatural T = 1 plant viruses such as sesbania mosaic virus (that undergo swelling) (27, 31) and T = 4 semliki forest virus and T = 16 herpes virus (that undergo shrinkage from a swollen precursor to a finally more icosahedral-looking capsid) (28, 29), all of which display holes in their expanded or swollen forms. These kinds of swelling/shrinking transformations comprise radial motions that have been given previous theoretical consideration (32, 33) and were not considered here.
Need for Auxiliary Proteins.
We established that the pentamer imposes its endo dihedral angle properties onto adjacent hexameric dihedral angles (Fig. 3B), thereby constraining shapes available to adjacent hexamers. This, along with well-recorded quasiequivalent mechanisms (“switches”) such as order–disorder transitions (reviewed in refs. 34 and 35) are adequate in ensuring the existence of both pentamers and hexamers adjacent to pentamers in small (T ≤ 7) capsids.
However, T > 7 capsids possess >1 hexamer species, where the secondary hexamer type is no longer in contact with any pentamer. Such hexamers may not be directly influenced by the geometric endo angle constraints (and adjacent quasiequivalent mechanisms), and therefore, we argue, may need other (auxiliary) constraints to secure the shape of the isolated hexamer. It is interesting that, so far, all T > 7 spherical capsids have been experimentally found to require auxiliary proteins to form native structures (noted in ref. 34). It is also interesting that, during model construction, all T ≤ 7 capsids did not require any additional constraints to ensure uniform hexamer shapes, whereas the second hexamer that is isolated from the pentamers in the T = 13 model was able to sample at least 2 distinct (and energetically viable) shapes within the capsid, resulting in a nonsymmetric and subunit–subunit bondwise “complicated” capsid structure (“hexamer 2” of the T = 13 capsid model in Fig. 2B is an averaged version of positionally equivalent but architecturally varying hexamers). Based on experimental and geometric studies, we suggest that all T > 7 capsids require auxiliary mechanisms (by means of proteins interaction, etc.) to maintain the shape of secondary hexamers.
Stating that T > 7 capsids must need auxiliary proteins does not preclude the T ≤ 7 capsids from displaying auxiliary proteins—for any capsid size, auxiliary proteins may serve as an excellent mechanism for viral lifecycle control. Our statement implies only that T > 7 capsids may be theoretically/geometrically excluded from forming all required capsomere shapes (to form the final capsid) without auxiliary help in the form of proteins or additional (currently unelucidated) mechanisms to assist in the formation of the secondary hexamers.
Auxiliary Proteins Versus Buckling Availability.
Some T > 7 capsids are known to retain the auxiliary proteins within the final capsid [e.g., the T = 13 birnavirus (5) and reovirus (36, 37)]. This adds an interesting imposition onto T > 7 capsids; because even if they theoretically could buckle, their present morphology may be “locked in” because of contact with the auxiliary proteins. If this is true, the presence of such auxiliary proteins may impede buckling of T > 7 capsids, i.e., buckling transitions may be practically possible only for T = 7 capsids. Currently, empirical data shows direct evidence of buckling transitions exclusively in T = 7 capsids (22, 23, 38), supporting this hypothesis.
Concluding Remarks.
How do capsids form different sizes? The theory of quasiequivalence posits that the coexistence of the pentamer and hexamer allows for capsids of various sizes to exist (3). Here, we have shown, from empirical evidence and ab initio models, that shapes or puckers of the hexamers are strongly indicative of size within all available canonical capsids (opening the possibility of rational design of artificial nanoarrays and hexamer-shape-modifying drugs). After relating canonical capsids to geometrical entities—deltahedra—we were able to use such models and geometric concepts (e.g., endo angle constraints) to arrive at interesting (and empirically supported) general insights and predictions regarding modulation of capsid assembly (auxiliary protein requirements) and postassembly capsid transformations (availability of buckling transitions). Previous work on the capsid subunit (7) and current work on the entire capsid underline the usefulness of simplified but accurate geometric models in elucidating various capsid features, especially those of general import.
Materials and Methods
Natural Capsids Studied.
We studied dihedral angles within X-ray structures of all natural capsids unambiguously denotable as canonical capsids (capsids that are representable by monohedral tilings) (7). The stringency of these qualities is crucial to the dihedral angle comparisons, and so only a portion of those capsids deemed as “canonical” in ref. 7 were studied (those with stricter adherence to monohedrality). The studied virus families, T numbers, and PDB ID codes obtained from the capsid repository VIPERdb (1) are as follows: Nodaviridae (T = 3): fhv [available only in VIPERdb (1)], 1nov, 2bbv, 1f8v; Sobemoviridae (T = 3): 1 smv, 1 × 35, 1f2n, 4sbv, 1 ng0; Tombusviridae (T = 3): 1opo, 1tnv, 1c8n, 2tbv; Tetraviridae (T = 4): 1ohf; Siphoviridae (T = 7l): 2frp, 2ft1, 2fs3, 2fsy, 1ohg; Birnaviridae (T = 13l): 1wce.
Analysis of Angles Within Natural Capsids.
Looking at dihedral angle similarities within 2 quasiequivalent interfaces (say, between adjacent subunit pairs A–B and C–D; an example of a pair “1”–“2” is shown in Fig. 1A) becomes easy when dealing with capsids representable as monohedral tilings (canonical capsids). This is because the subunits within such capsids have little subunit–subunit architectural variability (interface-controlling quasiequivalent switches notwithstanding). Consequently, to check for the similarity between 2 interfaces A–B and C–D, one need only structurally align the Cα traces of AB and CD (both treated as rigid units instead of 2 proteins) and calculate the normalized RMSD. Low RMSD values indicate that the dihedral angles between subunits A and B and subunits C and D are similar (or identical, if the RMSD is 0). From these analyses, we gathered T-specific dihedral angle patterns for hexamers (Fig. 2A).
Creating Capsid Models.
A majority of capsids possess trapezoidal subunits (7), whose interactions are described in Fig. 3A. It is trivial to conclude that subunit trimers caused by “x–y” interactions will remain rigid as a unit if the subunits remain generally rigid. Consequently, it is acceptable to treat each coplanar trimer as a single face, i.e., monohedral capsid models of 60T subunits may be represented as 20T equilateral triangle faced polyhedra otherwise known as deltahedra. The simplest deltahedron, the T = 1 deltahedron, is the 20-faced icosahedron. We created these deltahedra by creating duals of deltahedra in Euclidean space (which, are, interestingly models of buckeyballs) and then obtaining the deltahedra from those duals.
We produced the deltahedral dual (buckeyball) by first generating the graph connectivity by using the spiral code method described by Fowler et al. (39). From this abstract graph description, for each T number, we constructed a planar graph (Schlegel diagram) of the abstract graph by using an algorithm modeled around one described by Bor Plestenjak (40).
This planar graph was then wrapped around a sphere (by using a nonlinear plane to sphere projection). The final minimized structure (minimized so that all edge lengths are equal) will resemble an icosahedral buckeyball. The buckeyballs were transformed into their duals (whose graphs and general shape resemble the required deltahedra). This structure was then minimized to ensure that deltahedra edges are equal (and set arbitrarily to 18 Å). For the capsids studied (T = 1, 3, 4, 7, and 13), the final minimized structures were found to possess the lowest energy possible and were mostly icosahedral (the T = 13 capsid was the exception; see the note on auxiliary proteins in Results and Discussion). These structures were used for the final analysis of (i) hexameric dihedral angle configurations and (ii) availability of buckling transitions.
Assaying Subunit–Subunit Dihedral Angle Constraints.
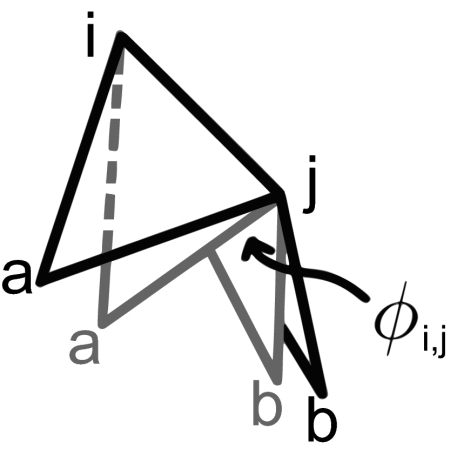
For our capsid structure to possess multiple interchangeable configurations, one would expect a range of allowable values for at least some dihedrals within the capsid (especially within the hexagonal regions). We start with the obtained deltahedra and define the dihedral angles across any edge i,j as φij. Each edge is shared by 2 equilateral triangles (shown in isolation from the entire deltahedron in Fig. 5). The relationship between the dihedral angle φij and the distance between the noncommon point rab is
where m is the height of the equilateral triangles. Therefore, adding a single dihedral restraint across the edge {i,j} is analogous to adding a new bond to the system with potential energy
It is imperative that the force constant kc ≪ krest, where krest is the strength of the bonds making up the deltahedron. This is required because we are studying elastic deformations of dihedral angles (and the dihedral bond) and not the equilateral faces of the deltahedra (although small deviations in shape are acceptable).
Fig. 5.
The dihedral angle φi,[infi]j between 2 equilateral faces sharing edge {i, j} (shown in 2 configurations) depends on the distance between a and b (rab).
We assayed the effect of applying stress onto a dihedral angle with respect to resulting energy change. The study is performed by using the following algorithm:
Initialization step.
(i) Identify the edge {i,j} whose dihedral angle is to be studied. (ii) Assign a restraint energy term Ec as shown above to the appropriate atom pair ({a,b}, in Fig. 5). (iii) Assign rab′ = rab, where rab is the length between atom pair {a,b} in initially obtained (embedded) deltahedron. This ensures that at the first step all energy terms equal zero (because the deltahedron is minimized).
Cycle (until ∣rab′∣ ≤ ∣rmax′∣).
(i) Assign rab′ = rab′ + step_size. This will cause a force to be applied onto a and b because the {a,b} bond length will not be at its equilibrium value. (ii) Allow the structure to relax by energy minimization. At this point, we obtain the total energy of the new structure.
Supplementary Material
Acknowledgments.
This work was supported by grants from the National Institutes of Health Grant RR012255 and National Science Foundation Grant PHY0216576.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
*For example, the average dihedral angle value per capsid will tend towards 180° as we proceed to larger and larger capsid sizes.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811517106/DCSupplemental.
Crystallographically, this is evident when comparing pentamers appearing in capsids of 2 sizes (T > 1 and T = 1) that are formed from chemically identical subunits, e.g., in the birnavirus (PDB ID codes: T = 13: 1wce; T = 1: 1wcd), alfalfa mosaic virus (PDB ID codes: T = 3: 1js9; T = 1: 1yc6), and sesbania mosaic virus (PDB ID codes: T = 3: 1smv, T = 1: 1x36).
Also, many capsid subunits “display” protruding domains (e.g., the P domain of the Tomato Bushy Stunt Virus, PDBID: 2tbv) on the capsid‘s surface, making the choice for a suitable generalized plane even harder.
Note that deltahedra have been previously discussed with respect to spherical capsids, e.g., figure 8 in ref. 3 and figure 3 in ref. 10; however, in both studies, the deltahedra were conceptual tools, and could not be readily related to natural capsid arrangements; only from recent observations of monohedral tilability (7) can we now represent a large number of natural capsids by deltahedra in a structurally meaningful manner.
Ref. 9 dealt with polymorphism within a single capsid, and Moody (10) reasoned that the hexamer shape was modified by the distortion of pentamers due the projection of the pentamer onto the icosahedral insphere (see “hexamer rectification” in (10)). Although a very creative and useful qualitative rationalization of some cryo-EM structures, these rationalizations are clearly not applicable to canonical capsids with uniform subunit shapes.)
From the various definitions of the canonical capsid of triangulation number T (3, 7), we have the number of hexamers per capsid equaling 10(T − 1) and the number of pentamers per capsid equaling 12. Because we have 5 endo angles per pentamer, the average number of endo angles per hexamer that are imposed directly by pentamers must equal 12 × 5/[10(T − 1)] or 6/(T − 1).
References
- 1.Shepherd CM, et al. Viperdb: A relational database for structural virology. Nucleic Acids Res. 2006;34:D386–D389. doi: 10.1093/nar/gkj032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rux J, Burnett R. Spherical viruses. Curr Opin Struct Biol. 2008;8:142–149. doi: 10.1016/s0959-440x(98)80031-6. [DOI] [PubMed] [Google Scholar]
- 3.Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 4.Subramanya H, Gopinath K, Nayudu M, Savithri H, Murthy M. Structure of sesbania mosaic virus at 4.7 a resolution and partial sequence of the coat protein. J Mol Biol. 1993;229:20–25. doi: 10.1006/jmbi.1993.1004. [DOI] [PubMed] [Google Scholar]
- 5.Coulibaly F, et al. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell. 2005;120:761. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Padilla J, Colovos C, Yeates T. Nanohedra: Using symmetry to design self-assembling protein cages, layers, crystals, and filaments. Proc Natl Acad Sci USA. 2001;98:2217–2221. doi: 10.1073/pnas.041614998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mannige R, Brooks C., III Tilable nature of virus capsids and the role of topological constraints in natural capsid design. Phys Rev E. 2008;77 doi: 10.1103/PhysRevE.77.051902. 051902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 9.Choi K, Morais M, Anderson D, Rossmann M. Determinants of bacteriophage φ29 head morphology. Structure (London) 2006;14:1723–1727. doi: 10.1016/j.str.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Moody MF. Geometry of phage head construction. J Mol Biol. 1999;293:401–433. doi: 10.1006/jmbi.1999.3011. [DOI] [PubMed] [Google Scholar]
- 11.Lepault J, et al. Structural polymorphism of the major capsid protein of rotavirus. EMBO J. 2001;20:1498–1507. doi: 10.1093/emboj/20.7.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddy V, Johnson J. Virus Structure and Assembly (Advances in Virus Research) San Diego: Elserver Academic; 2005. pp. 45–468. [Google Scholar]
- 13.Horne R, Wildy P. Symmetry in virus architecture. Virology. 1961;28:348–373. doi: 10.1016/0042-6822(61)90366-x. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira D, Hernandez R, Horton M, Brown D. Morphological variants of sindbis virus produced by a mutation in the capsid protein. Virology. 2003;307:54–66. doi: 10.1016/s0042-6822(02)00034-x. [DOI] [PubMed] [Google Scholar]
- 15.Sangita V, Satheshkumar PS, Savithri HS, Murthy MR. Structure of a mutant t=1 capsid of sesbania mosaic virus: Role of water molecules in capsid architecture and integrity. Acta Crystallogr D. 2005;61:1406–1412. doi: 10.1107/S0907444905024030. [DOI] [PubMed] [Google Scholar]
- 16.Larson S, Lucas R, McPherson A. Crystallographic structure of the t=1 particle of brome mosaic virus. J Mol Biol. 2005;346:815–831. doi: 10.1016/j.jmb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 17.Zubieta C, Schoehn G, Chroboczek J, Cusack S. The structure of the human adenovirus 2 penton. Mol Cell. 2005;17:121–135. doi: 10.1016/j.molcel.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 18.Stray S, et al. A heteroaryldihydropyrimidine enhances and can misdirect assembly of hepatitis B virus capsid. Proc Natl Acad Sci USA. 2005;102:8138–8143. doi: 10.1073/pnas.0409732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourne CR, Finn MG, Zlotnick A. Global structural changes in hepatitis B virus capsids induced by the assembly effector hap1. J Virol. 2006;80:11055–11061. doi: 10.1128/JVI.00933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teschke C, King J, Prevelige PE., Jr Inhibition of viral capsid assembly by 1,1′-bi(4-anilinonaphthalene-5-sulfonic acid) Biochemistry. 1993;32:10658–10665. doi: 10.1021/bi00091a016. [DOI] [PubMed] [Google Scholar]
- 21.Prevelige P., Jr Inhibiting virus-capsid assembly by altering the polymerisation pathway. Proc Natl Acad Sci USA. 1998;16:61–65. doi: 10.1016/s0167-7799(97)01154-2. [DOI] [PubMed] [Google Scholar]
- 22.Conway JF, et al. A thermally induced phase transition in a viral capsid transforms the hexamers, leaving the pentamers unchanged. J Struct Biol. 2007;158:224–232. doi: 10.1016/j.jsb.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wikoff WR, et al. Time-resolved molecular dynamics of bacteriophage hk97 capsid maturation interpreted by electron cryo-microscopy and X-ray crystallography. J Struct Biol. 2006;153:300–306. doi: 10.1016/j.jsb.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Lidmar J, Mirny L, Nelson D. Virus shapes and buckling transitions in spherical shells. Phys Rev E. 2003;68 doi: 10.1103/PhysRevE.68.051910. 051910. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen T, Bruinsma R, Gelbart W. Elasticity theory and shape transitions of viral shells. Phys Rev E. 2005;72 doi: 10.1103/PhysRevE.72.051923. 051923. [DOI] [PubMed] [Google Scholar]
- 26.Tama F, Brooks C., III Diversity and identity of mechanical properties of icosahedral viral capsids studied with elastic network normal mode analysis. J Mol Biol. 2005;345:299–314. doi: 10.1016/j.jmb.2004.10.054. [DOI] [PubMed] [Google Scholar]
- 27.Jacrot B. Studies on the assembly of a spherical plant virus. ii. The mechanism of protein aggregation and virus swelling. J Mol Biol. 1975;95:433–446. doi: 10.1016/0022-2836(75)90201-6. [DOI] [PubMed] [Google Scholar]
- 28.Helenius A, Fries E, Kartenbeck J. Reconstitution of semliki forest virus membrane. J Cell Biol. 1977;75:866–880. doi: 10.1083/jcb.75.3.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trus BL, et al. The herpes simplex virus procapsid: Structure, conformational changes upon maturation, and roles of the triplex proteins vp19c and vp23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 30.Li L, et al. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 31.Sangita V, et al. T=1 capsid structures of sesbania mosaic virus coat protein mutants: Determinants of t=3 and t=1 capsid assembly. J Mol Biol. 2004;342:987–999. doi: 10.1016/j.jmb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Tama F, Brooks C., III The mechanism and pathway of pH induced swelling in cowpea chlorotic mottle virus. J Mol Biol. 2002;318:733–747. doi: 10.1016/S0022-2836(02)00135-3. [DOI] [PubMed] [Google Scholar]
- 33.Zandi R, Reguera D. Mechanical properties of viral capsids. Phys Rev E. 2005;72 doi: 10.1103/PhysRevE.72.021917. 021917. [DOI] [PubMed] [Google Scholar]
- 34.Johnson JE, Speir JA. Quasi-equivalent viruses: A paradigm for protein assemblies. J Mol Biol. 1997;269:665–675. doi: 10.1006/jmbi.1997.1068. [DOI] [PubMed] [Google Scholar]
- 35.Tang J, et al. The role of subunit hinges and molecular “switches” in the control of viral capsid polymorphism. J Struct Biol. 2006;154:59–67. doi: 10.1016/j.jsb.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 36.Grimes J, et al. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 37.Nakagawa A, et al. The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure (London) 2003;11:1227–1238. doi: 10.1016/j.str.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Jiang W, et al. Coat protein fold and maturation transition of bacteriophage p22 seen at subnanometer resolutions. Nat Struct Biol. 2003;10:131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- 39.Fowler PW, Rogers KM. Spiral codes and Goldberg representations of icosahedral fullerenes and octahedral analogues. J Chem Inf Comput Sci. 2001;41:108–111. doi: 10.1021/ci9901486. [DOI] [PubMed] [Google Scholar]
- 40.Plestenjak B. An algorithm for drawing planar graphs. Softw Pract Exper. 1999;29:973–984. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.