Abstract
In bacteria, many “atypical” response regulators (ARRs) lack the conserved residues important for phosphorylation by which typical response regulators switch their output response, suggesting the existence of alternative regulatory mechanisms. However, such mechanisms have not been established. JadR1, an OmpR-type ARR of Streptomyces venezuelae, appears to activate the transcription of jadomycin B (JdB) biosynthetic genes while repressing its own gene. JadR1 activities were inhibited in cells induced to produce JdB, which was found to bind directly to the N-terminal receiver domain of JadR1, causing JadR1 to dissociate from target promoters. The activity of a NarL-type ARR, RedZ, that regulates production of another antibiotic was likewise modulated by the end product (undecylprodigisines), implying that end-product-mediated control of antibiotic pathway-specific ARRs may be widespread. These results could prove relevant to knowledge-based improvements in yield of commercially important antibiotics.
Keywords: JadR1, ligand
Two-component signal transduction systems, consisting of a sensor histidine kinase (HK) and a cognate response regulator (RR), are common in prokaryotes, where they are usually determined by a pair of adjacent genes (1, 2). In response to a signal, the HK autophosphorylates, then transfers the phosphoryl group to an Asp residue of the RR; thus, switching its output response (3). Typically, RRs contain an N-terminal receiver domain (the REC domain) and a C-terminal output domain, in most cases a DNA binding domain (4). In the REC domain, 5 conserved amino acid residues are believed to be vital for phosphorylation, an Asp at the exposed C terminus of the central β-strand c being the phosphoryl acceptor (5). However, some RRs lack 2 or more of the 5 important conserved residues in the REC domain, despite their high predicted structural resemblance to typical RRs (6–11). Such “atypical” (A)RRs are encoded in many bacterial genomes, and their determinants are usually located away from any likely HK determinant (12, 13). Some ARRs are involved in regulation of bacterial growth and development, secondary metabolite biosynthesis, iron transport, cell movement, or virulence (8–10, 14–18). No ARR has been proved to accept phosphoryl groups in vitro, raising the possibility that they use a different signal-responding mechanism, such as a ligand-based mechanism (8, 9, 19, 20); but so far, such a mechanism has not been demonstrated.
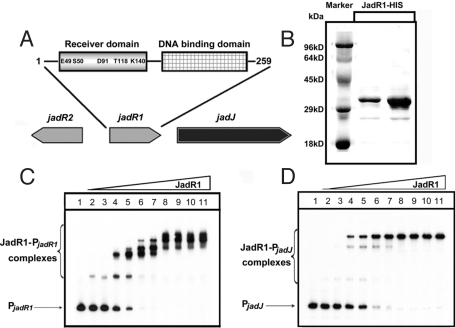
In antibiotic-producing streptomycetes, production of each antibiotic is typically determined by a large gene cluster that usually includes at least one pathway-specific regulatory gene. This is the case for the jadomycins, broad-spectrum cytotoxic polyketide-derived angucycline antibiotics (21, 22) that are produced by Streptomyces venezuelae ISP5230. The jadomycin biosynthetic gene cluster includes 5 regulatory genes (jadW1, -W2, -W3, -R2, and -R1) (23, 24), among which jadR1 is indispensable for jadomycin production (25). The jadR1 is located upstream of, and in the same orientation as, the first structural gene of the jadomycin biosynthetic cluster, jadJ (Fig. 1A) (26). JadR1 is a predicted OmpR type RR, with a winged HTH motif in the C-terminal output domain. In its N-terminal REC domain, the 2 important aspartic acid residues closest to the N terminus are replaced by Glu49 and Ser50. Also, jadR1 is not located near a cognate HK gene; thus, it is an ARR.
Fig. 1.
JadR1 binds to the promoter regions of jadJ and jadR1. (A) The domain organization of JadR1. The amino acids corresponding to the conserved residues of typical RRs are marked. (B) Purification of JadR1-His (C-terminal His-tag) expressed in E. coli. (C and D) Band-shift assays of 2 promoter regions with purified JadR1. Each lane contains 10 ng of γ-32P-labeled PjadR1 (C) or PjadJ (D) probe. Lanes 1–11 contain 0, 1.5, 4.5, 9, 12, 15, 50, 90, 120, 150, and 400 nM JadR1, respectively.
In this study, we show that ligands do indeed bind to and change the activity of an ARR. The ARR is JadR1, and the ligands are the late biosynthetic products of the genes controlled by JadR1, notably jadomycin B (JdB) and its aglycone JdA. We also present evidence that such autoregulation may be a widespread regulatory mechanism for antibiotic biosynthesis in streptomycetes.
Results
JadR1 Binds the Promoter Regions of jadJ and Its Own Gene.
Previous genetic evidence indicated that JadR1 was necessary for the transcription of jadomycin biosynthetic genes (25). To examine the molecular mechanism involved, JadR1-HIS expressed in Escherichia coli was purified to ≈90% purity (Fig. 1B), and used in band-shift assays with 2 potential promoter regions designated PjadR1 (promoter region of jadR1) and PjadJ (promoter region of jadJ). JadR1 showed distinct patterns of binding to these regulatory regions at protein concentrations ranging from 1.5 to 400 nM (Fig. 1 C and D).
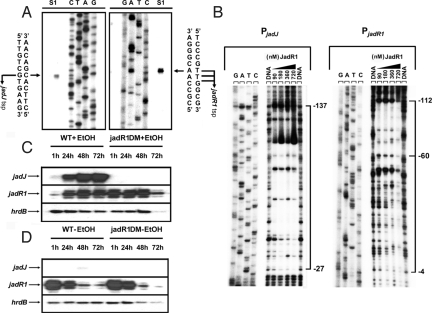
To further understand the regulatory role of JadR1 on jadJ and jadR1 expression, the promoter structures of jadJ and jadR1 and the DNA binding sites were identified (Fig. 2B; Fig. S1 A and B). The transcription start point (tsp) of jadJ was located by high-resolution S1 nuclease protection analysis to a G, 88 nt upstream of the putative jadJ start codon (ATG) (Fig. 2A). The tsp of jadR1 was localized to a T or G, 81, 82, or 83 nt upstream of the putative jadR1 start codon (ATG) (Fig. 2A). Subsequent DNase I footprinting experiments (Fig. 2B; Fig. S1A) revealed that JadR1 protected a large region from −27 to −137 nt relative to the jadJ tsp. The footprint on the jadR1 promoter region covered a region from −4 to −112 nt relative to the jadR1 tsp (Fig. S1B), although at the lowest concentrations of JadR1, only the more upstream part of the promoter region (−112 to −60) was protected. This result may imply that JadR1 binds first to the more upstream binding site, nucleating the occupation of the entire promoter at higher JadR1 levels, leading to obstruction of RNA polymerase binding.
Fig. 2.
The transcriptional level of jadJ and jadR1 with or without ethanol and the JadR1-protected regions upstream of jadJ and jadR1. (A) Determination of the tsps of jadJ and jadR1 by high-resolution S1 mapping of WT RNA (isolated at 48 h after ethanol treatment). (B) DNase I footprinting assays of JadR1 on the jadJ and jadR1 promoter regions. The concentrations (nM) of JadR1 are shown above the lanes. The lanes marked DNA are controls without protein. Each lane contains 200 ng of γ-32P-labeled DNA. The brackets denote the regions protected by JadR1, and the numbers on the right side indicate the distances relative to the respective tsps of jadJ and jadR1. (C and D) The transcriptional level of jadJ and jadR1 with or without ethanol induction was detected by low-resolution S1 analysis. Total RNAs from WT and jadR1DM in the presence (C) or absence (D) of ethanol were hybridized with jadJ and jadR1 probes. hrdB transcription was assayed as a control.
Analysis of the DNA regions interacting with JadR1 did not reveal a highly conserved recognition sequence, although it was possible to identify a repeated but poorly conserved 6-bp candidate consensus sequence. Further analysis will be necessary.
Transcriptional Analysis of jadJ and jadR1.
To evaluate the regulatory significance of the observed patterns of DNA binding, the transcription of jadJ and jadR1 in the WT and a jadR1 disruption mutant (jadR1DM) was analyzed by S1 nuclease protection. In these experiments, we took advantage of the fact that, unlike most secondary metabolites, JdB is produced only when nutritionally unbalanced cultures are subjected to additional stress such as heat shock or ethanol toxicity (22). Total mRNAs were isolated from cultures grown with or without ethanol treatment at various time points (1, 24, 48, and 72 h after ethanol addition) (Fig. 2 C and D).
When ethanol was added to the culture, jadJ mRNA was detectable at 1 h in the WT, and reached its highest abundance at 48–72 h (Fig. 2C), whereas no transcript of jadJ was detected at any time point in jadR1DM. This result indicated that JadR1 positively controls jadJ transcription. However, in the absence of ethanol, little or no jadJ transcript was detectable in either WT or jadR1DM (Fig. 2D), whereas jadR1 mRNA was relatively abundant in both strains (note that the 5′-end of the jadR1 gene is present in jadR1DM, making it possible to assess jadR1 transcription in the deletion mutant). Thus, some additional factors besides JadR1 may be required for jadJ expression; and at least under the conditions used here, the in vivo transcription of jadR1 was not observably dependent on JadR1.
Transcription of jadR1 was inhibited in the WT at 1 h after ethanol, but recovered to the preinduction level by 24 h (Fig. 2C). This inhibition was not observed in jadR1DM, implying that JadR1 is responsible for the inhibition in the WT (i.e., that JadR1 acts as a repressor of its own gene), but that the initial repression could be relieved by some undefined effect at later time points.
Effect of Culture Extract from S. venezuelae on the Activity of JadR1.
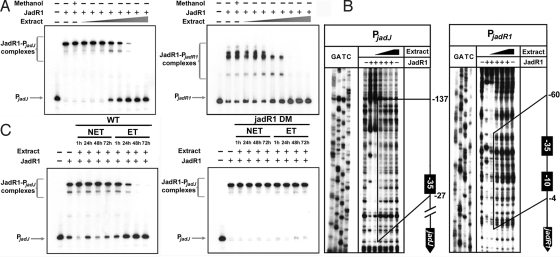
To explain why the initial repression of jadR1 transcription in vivo lost strength with time, we hypothesized that some substance(s) produced by S. venezuelae after ethanol treatment might inhibit DNA binding by JadR1. To explore this hypothesis, a culture supernatant of S. venezuelae grown for 48 h with ethanol treatment was extracted with organic solvent, dried, and redissolved in methanol. Then, different amounts of extract were assayed for their ability to influence binding of JadR1 to PjadR1 and PjadJ (Fig. 3A). Indeed, a high concentration of extract could dissociate JadR1 from both promoter regions. Footprinting experiments showed that the extract influenced the binding of JadR1 to PjadR1 in a concentration-dependent manner (Fig. 3B): JadR1 dissociated from the region stretching from position −4 to −60 at a low concentration of extract, and from the entire protected region at high concentration (Fig. 3B). The results with the jadJ promoter region were more complex. Unexpectedly, at a low concentration of extract, a clearer protected region from −27 to −138 nt indicated enhanced binding of JadR1 to the DNA target (Fig. 3B). However, the protected region gradually disappeared with increasing concentration of extract, indicating progressive dissociation of JadR1 from its DNA target. This result portrayed a subtle regulatory pattern, in which low concentrations of extract stimulated the binding of JadR1 to the jadJ promoter region, whereas the binding was reversed to dissociation at higher concentrations. In vivo, the situation is likely to be more complex, as illustrated by the continued high expression of jadJ in WT cultures at late time points, when jadomycin would have been produced for many hours (Fig. 2C). Investigation of the other jad regulatory genes will be needed to clarify this apparent anomaly.
Fig. 3.
Effect of ligand(s) on the binding activity of JadR1. (A) Band-shift assays of JadR1 (50 nM) with culture extracts in a series of 2-fold dilution steps. Methanol was used as a solvent control. (B) Effect of extract on DNase I footprinting of JadR1 (360 nM) on its DNA targets. The protected regions in the presence of low concentrations of extract are indicated by vertical solid lines. The protected regions relative to the respective tsps of jadJ or jadR1 are numbered on the right side. (C) Band-shift assays of JadR1 (50 nM) with different extracts. NET, no ethanol added; ET, ethanol added.
JdB Modulates the DNA Binding Activity of JadR1 by Interacting with JadR1.
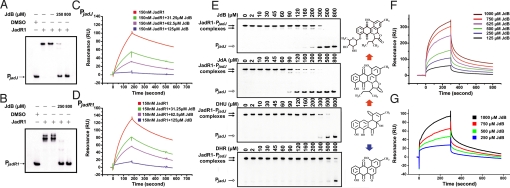
In a more detailed analysis, using extracts from culture supernatants of S. venezuelae WT and jadR1DM, harvested at various time points (1, 24, 48, and 72 h) with or without ethanol treatment, we found that only extracts from WT grown with ethanol markedly inhibited the DNA binding activity of JadR1 (Fig. 3C), inhibitory activity increasing with culture age. This result matched the pattern of abundance of JdB itself, raising the possibility that it might directly modulate JadR1 activity. Therefore, we further purified JdB by HPLC, and assessed its effects in band-shift assays (Fig. 4 A and B). JdB inhibited band-shifting of PjadR1 and PjadJ by JadR1 in a concentration-dependent manner (Fig. 4 A and B). These results were further confirmed by surface plasmon resonance (SPR) analysis of the effect of increasing concentrations of JdB on binding of JadR1 to immobilized PjadJ and PjadR1 (Fig. 4 C and D). In band-shift assays (Fig. 4E), 2,3-dehydro-4-hydroxy-12bH-12-deoxyrabelomycin (DHU) and JdA, both intermediates of JdB biosynthesis, and dehydrorabelomycin (DHR), a shunt product, also showed different degrees of modulation of JadR1 DNA binding activity, whereas several antibiotics with diverse structures had no effect (Fig. S2). This result indicated that the ligand-mediated regulation for JadR1 is pathway-specific. Interestingly, JdA was effective at somewhat lower concentrations than JdB.
Fig. 4.
Effect of JdB and related molecules on the binding of JadR1 to its DNA targets. (A and B) Effect of JdB on the band-shift assay of JadR1 (50 nM) binding to PjadJ (A) and PjadR1 (B). (C and D) Dose-response curves of the inhibition by JdB of the binding of JadR1 (150 nM) to PjadJ (C) or PjadR1 (D). (E) Effect of JdB, JdA, DHU, and DHR on the DNA binding activity of JadR1. Band-shift assays were performed with 50 nM JadR1 and a range of ligand concentrations as indicated. The structures of compounds are present on the right side. Red arrows indicate the direction of biosynthesis, but the blue arrow points to a shunt product. (F) SPR analysis of the binding of JdB to immobilized JadR1. (G) SPR analysis of the binding of JdB to immobilized JadR1R (REC domain of JadR1).
Because DNase I footprinting (Fig. 3B) had revealed no new protected region in the presence of extract containing a high concentration of JdB, the inhibitory effect of JdB was not due to competition with JadR1 for similar binding sites in DNA, as in the case of daunorubicin and its pathway-specific regulator, DnrN (27). This indication that JdB modulates the DNA binding activity of JadR1 by directly interacting with JadR1 was confirmed by SPR between JdB and immobilized JadR1 (Fig. 4F): JdB interacted with JadR1 directly in a dose-dependent manner. A similar result was obtained with just the N-terminal domain of JadR1 (JadR1R) (Fig. 4G). A comparable experiment with the C-terminal domain failed because of the instability of the isolated domain under these conditions.
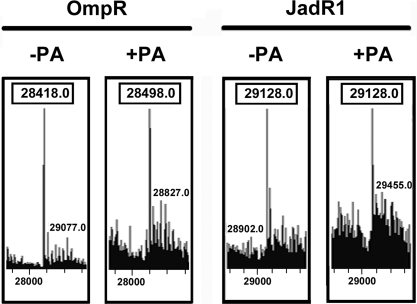
Most typical RRs, including OmpR, that are regulated by phosphorylation can autophosphorylate in vitro in the presence of small molecule phosphodonors such as phosphoramidate (PA), acetyl phosphate, and carbamoyl phosphate. Previous studies had shown that some ARRs could not be phosphorylated in this way (6, 28). Indeed, ARR-JadR1 did not autophosphorylate in the presence of 25 mM PA (Fig. 5).
Fig. 5.
MS analysis of in vitro phosphorylation of OmpR and JadR1 in the presence or absence of PA. −PA indicates that the OmpR and JadR1 proteins were untreated by PA, whereas +PA indicates that the proteins were treated by PA.
These results demonstrated that the activity of an ARR could be directly modulated by a small ligand, and indicated that this was the principal mode of regulation, because JadR1 was not susceptible to in vitro phosphorylation. Interestingly, homologues of jadR1 are present in the biosynthetic clusters for several angucycline antibiotics (29, 30) and for antibiotics that contain an angucycline-like structure (31, 32). The amino acid sequence conservation of such JadR1 homologues may indicate that they have a similar ligand-interactive specificity.
Modulation of DNA Binding Activity of RedZ by Undecylprodigiosin (Red).
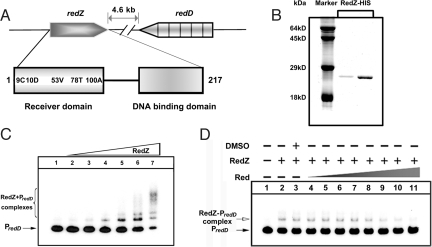
In streptomycetes, pathway-specific ARRs responsible for regulation of secondary metabolite biosynthesis can be categorized into 2 types according to DNA binding motif: the OmpR type represented by JadR1; and the NarL type, which includes RedZ of Streptomyces coelicolor, the most studied streptomycete. RedZ has been deduced to positively regulate Red biosynthesis by activating the transcription of a second pathway-specific activator gene, redD (9, 33). RedZ lacks a pair of Asp residues and a Lys residue in the conserved phosphorylation pocket (Fig. 6A). Therefore, it was proposed that RedZ might use a phosphorylation-independent mechanism of regulation (9). We chose this case to test the possibility that end-product mediated regulation of ARR might be widespread among antibiotic pathways in streptomycetes.
Fig. 6.
Effect of undecylprodigiosin on the DNA binding activity of RedZ. (A) Schematic representation of the relative positions of redZ and redD in the S. coelicolor A3(2) genome. The domain organization of RedZ is indicated by boxes, and the amino acids in RedZ that correspond to the highly conserved residues of typical RRs are marked. (B) Purification of RedZ-HIS expressed in E. coli. (C) Band-shift assays of the interaction of the redD promoter region (PredD) with purified RedZ. Each lane contains 10 ng of γ-32P-labeled PredD. Lanes 1–7 contain 0, 5, 15, 30, 50, 70, and 150 nM purified protein, respectively. (D) Effect of Red on the DNA binding activity of RedZ. Each lane contains 10 ng of γ-32P-labeled PredD. Lanes 2–11 contain 50 nM RedZ and 0, 0, 7.8, 15.6, 31.2, 62.5, 125, 250, 500, and 800 μM undecylprodigiosin, respectively. DMSO was used as a solvent control.
RedZ expressed in E. coli was purified to ≈90% purity (Fig. 6B). Band-shift assays demonstrated that RedZ binds the promoter region of redD in a complex binding pattern (Fig. 6C), strongly supporting the proposal that RedZ regulates redD transcription directly (33). The addition of Red antibiotic (800 μM) inhibited the band shifts (Fig. 6D; note that the very limited solubility of Red in aqueous solvents means that the effective concentration was probably considerably lower). This effect was specific in that adding the same amount of Red to the JadR1-PjadJ complex did not cause any dissociation (Fig. S2). Therefore, as with JadR1, the activity of RedZ is modulated by the end product. Further attempts to study the interaction between RedZ and Red by SPR failed due to the instability of RedZ. However, we note that, unlike the anthracycline daunorubicin (34), the tripyrrole antibiotic Red is not known to bind DNA, and no Red-DNA complex was detected by band-shift assay even in the presence of 800 μM Red (Fig. 6D). Consequently, we believe that Red modulates the activity of RedZ, not by competing for the DNA binding sites, but via interaction with RedZ.
Discussion
Typical RRs are regulated by phosphorylation. However, the fact that there are many ARR genes in bacterial genomes suggests that other switches may control their output responses. In fact, several ARRs appear to exert their function independently of phosphorylation. For example, HP1043 and HP1021 in Helicobacter pylori could not be phosphorylated by low-molecular-weight phosphate donors in vitro, and derivatives of HP1043 carrying either the S51N and D52N single-amino acid substitutions or the S51D52AN double mutation could functionally substitute for the WT protein in vivo (6). In streptomycetes, several ARRs have been studied. Among them, WhiI, which is essential for sporulation in S. coelicolor A3 (2), lacks at least 2 of the conserved residues in typical phosphorylation pockets, and mutation of the residues in conserved positions resulted in modulation, rather than loss, of WhiI function (8, 20). This result led to the proposal that WhiI might be regulated by a small molecule ligand (20). The possibility of posttranscriptional regulation mediated by a coregulator interacting with ARRs has been also suggested for BldM and RedZ (13). Here, we have demonstrated that an ARR (JadR1), instead of being phosphorylated, does indeed interact with small molecules (JdB and its late precursors), causing changes in its own binding to target promoters. Also, the N-terminal Rec domain on its own could bind JdB, suggesting that ligand binding to this domain was analogous to phosphorylation of the Rec domains of typical RRs. We also found that the ability of an ARR of a second subclass (RedZ) to bind to a target promoter was inhibited by addition of Red antibiotic, strengthening the possibility that interactions with ligands may be a widespread mode of modulation of the activity of ARRs.
The structures of JadR1 and RedZ predicted by the SWISS-MODELLING program are strongly similar to those of the templates, which are typical RRs from 2-component systems. Therefore, it is possible that a ligand-modulated strategy might be adopted by some typical RRs, perhaps explaining why there are cases in which RRs with typical phosphorylation pockets exert function independently of phosphorylation (19, 27, 35).
The physiological context of the interaction between ARR-JadR1 and its ligand is intriguing; it is involved in the pathway-specific regulation of genes for the biosynthesis of an antibiotic, and the ligands are the product (JdB), or late intermediates (notably JdA), of the pathway. The strongest interaction was with JdA, suggesting that, after induction by ethanol, an initial low level of expression of the biosynthetic genes yields enough JdA to bind to JadR1; thus, increasing expression. We suppose that high expression of the biosynthetic genes leads to the accumulation of JdB product to a concentration sufficient to inhibit the activation exerted by JadR1. In other words, jadomycins act as autoregulators of their own synthesis.
In streptomycetes, specialized antibiotically-inactive “autoregulators” of antibiotic biosynthesis are well known, the most studied being the gamma-butyrolactone A-factor, which induces streptomycin biosynthesis in Streptomyces griseus (36). Our finding that jadomycin is truly autoregulating puts the use of this term in a new light. Remarkably, the jad gene cluster also contains genes homologous with those that are responsible for the production of gamma-butyrolactones and for implementing their regulatory effects (jadW1, -W2, and -W3) in other streptomycetes. The jadW1 has been shown to have significant roles in regulating jadomycin (24). Future work will address the interplay between these 2 distinct autoregulatory systems, which may in part be responsible for the complex pattern of binding of JadR1 to the promoters of jadR1 and jadJ apparent from Figs. 1 and 2.
It was recently discovered that export of the isochromanequinone polyketide actinorhodin (Act) in S. coelicolor was induced by the binding of a late intermediate in Act biosynthesis to a TetR-like repressor that specifically regulates the gene for the Act transporter (37). Autoregulatory phenomena were also reported in donorubicin and tylosin biosynthesis (38, 39). However, JadR1 is the first example of a primary pathway-specific activator whose activity is controlled by the end product. Also, band-shift assays revealed that DHU, JdA, and DHR also possessed the ability to inhibit the DNA binding activity of JadR1 (Fig. 4E). This raises the possibility of a stepwise autoregulation, in which the first jadomycin-related metabolites to accumulate intracellularly are biosynthetic intermediates/shunt products, which may strongly induce the pathway genes, leading to rapid generation of the JdB end-product to levels high enough for it to serve as an autoregulator for precise modulation of its own synthesis.
It is evident that binding of end-products or late intermediates of antibiotic biosynthetic pathways to pathway-specific regulatory proteins is not confined to any one type of antibiotic, nor to any one type of regulator, nor to any one Streptomyces species. The indication that antibiotic autoregulation may be widespread could prove relevant to knowledge-based improvements in yield of some commercially important secondary metabolites.
Materials and Methods
Bacterial Strains and Growth Conditions.
S. venezuelae ISP5230 and the jadR1 disruption mutant (jadR1DM) were grown on MYM agar (25). The medium and culture conditions for JdB production were as described previously (22). S. coelicolor M145 was grown on minimal medium (MM) containing mannitol as sole carbon source (40); ompR, jadR1, and redZ were expressed in E. coli BL21 (DE3) to obtain large amounts of C-terminally His-tagged proteins.
Construction of jadR1 Disruption Mutant.
A jadR1 disruption mutant (jadR1DM) was constructed as described previously (25), but the apramycin resistance gene used for gene disruption was replaced by a kanamycin resistance gene.
RNA Isolation and S1 Nuclease Protection Analysis.
RNAs as template for S1 nuclease protection analysis were isolated from cultures of WT and jadR1DM grown at 28 °C at various times (1, 24, 48, and 72 h) after ethanol addition or without ethanol. Ethanol was added to 6.5-h cultures, to a final concentration of 6% (vol/vol), before further incubation. The culture conditions for extracting RNAs from cells (22) and S1 nuclease protection analysis were as described previously (41). The hrdB (FJ387221) probe used as a control was prepared by PCR using the unlabeled primer 5′-CGGGAGTGCGGAGTCGGGGG-3′ and the 5′-end [32P]ATP-labeled primer 5′-TGCCCATCAGCCTTTCCCCGC-3′. The jadJ probe was prepared by PCR using the unlabeled primer 5′-AGGCGTGGGTTTCCGCTTCGGC-3′ and the radiolabeled primer 5′-CACGGCCACGCTGCCGATACCC-3′. A DNA sequencing ladder was generated using the radiolabeled primer with an fmol DNA cycle sequencing kit (Promega). The jadR1 probe was prepared by PCR using the radiolabeled primer 5′-CCACGCTCACGCCCGACAGAC-3′ and the unlabeled primer 5′-CGTAGCCGTGCTCGTCGAATTCTC-3′. The protected fragments were analyzed on a 6.0% polyacrylamide gel containing 7 M urea.
Band-Shift and DNase I Footprinting Assays.
Band-shift assays were performed as described previously (41). Briefly, 32P-labeled DNA probes (1,000 cpm) were incubated individually with various quantities of JadR1 at 25 °C for 20 min or RedZ at 0 °C for 1 h, in 20 μL of reaction mixture. After incubation, the binding reactions were separated on a 6% polyacrylamide gel, then the gel was dried and exposed to Biomax radiographic film (Kodak). Purified JdB and Red were dissolved in DMSO, whereas the extracts were dissolved in methanol; 400 mL of culture supernatant of WT at 48 h after ethanol treatment was extracted twice with 100 mL of ethyl acetate, and the extracts were pooled, dried on a rotary evaporator, and redissolved in 1.5 mL of methanol. We used this material undiluted and in a series of 2-fold dilution steps (diluted with methanol) for the band-shift assays shown in Fig. 3A. Dissolved compounds and solvent control were added at 4% (vol/vol) in the reaction mixtures.
For DNase I footprinting assays, binding and DNase I reactions were carried out as described previously (41). The sequence ladder was prepared using an fmol DNA cycle sequencing kit (Promega) with the labeled primers for PjadJ or PjadR1. The primer pairs used to generate the various DNA probes were as follows: for jadJ (361 bp), the unlabeled primer 5′-ACATTCCCGTCCTGTGATCCACC-3′ and the labeled primer 5′-CGCCTTCTCCGTACCCGTTCC-3′; for jadR1 (296 bp), the unlabeled primer 5′- GAAGTGGTCAAGAGTGCCCGTGGTC-3′ and the labeled primer 5′-CGGTTCCCCCTAGCACCTATGTCAC-3′; for redD (423 bp), the unlabeled primer 5′- GAACCGAGGCGACGGAAGGAGG-3′ and the labeled primer 5′- GGTCCATCGTGGCAAGCACTCCC-3′. Cell extracts were added at 4% (vol/vol) in the reaction mixtures for analysis.
Overexpression and Purification of OmpR, JadR1, JadR1 REC Domain, and RedZ.
ompR, jadR1 (and its part encoding REC domain), and redZ were amplified by PCR from DNA of E. coli DH5α, S. venezuelae ISP5230, and S. coelicolor M145 respectively. The jadR1 portion was resequenced; its revised sequence was updated in GenBank (U24659.3). For ompR, primers 5′-GGAATTCCATATGCAAGAGAACTACAAGATTC-3′ and 5′-CCGCTCGAGTGCTTTAGAGCCGTCCGGTAC-3′ were used; for jadR1, primers 5′-ACATATGAGCCTGACGTCCGTAGAAGTGAAG-3′ and 5′-ACTCGAGGCCGCGGCCGAAGCGGAAAC-3′ were used; for the REC domain-encoding part of jadR1, primers 5′-AAACATATGAGCCTGACGTCCGTAGAAGTGAAGG-3′ and 5′- AAACTCGAGTCCGCGGGTGATCGTGCGGG-3′ were used; for redZ, 5′-AAACATATGACGACCCGTGTCCTGGTGTGCTG-3′ and 5′-AAACTCGAGGCGGGCGGGAGTGCCGTAACCC-3′ were used. The amplified fragments were digested with NdeI and XhoI, and then inserted into pET-23b to obtain expression plasmids pET23b::ompR, pET23b::jadR1, pET23b::jadR1R, and pET23b::redZ, respectively. The plasmids were introduced into E. coli BL21 (DE3) for protein overexpression. Purification and concentration of proteins were carried out as described previously (41).
SPR Experiments.
All experiments were performed in HBS buffer (10 mM Hepes, pH 7.4/150 mM NaCl/3 mM EDTA/0.05% Tween 20) on a BIAcore 3000 System (BIAcore) at a flow rate of 30 μL/min and 25 °C. Biotinylated jadR1 and jadJ probes were immobilized on the flow cells of an SA sensor chip at densities of ≈200 response units (RU), whereas JadR1 and JadR1R were immobilized on a CM5 sensor chip at ≈10,000 and ≈5,100 RU, respectively.
HPLC Analysis.
JdB was purified by HPLC on a Shimadzu prominence system with a SPD-20A detector and an YMC-Pack ODS-A reverse-phase column (250 × 10 mm) using a gradient of H2O–acetonitrile (15:85) to 100% acetonitrile in 12 min at a flow rate of 2.5 mL/min. Absorbances were monitored at 313 and 260 nm.
In Vitro Phosphorylation by Liquid-Chromatography-Coupled MS.
Purified OmpR and JadR1 were incubated in kinase buffer (50 mM Tris·HCl, pH 7.5/50 mM KCl/20 mM MgCl2) in the presence or absence of 25 mM ammonium hydrogen PA, which was synthesized according to ref. 42, for 30 min at room temperature, then analyzed using a Thermo Electron LCA Deca XP ion-trap LC-MS system. Masses of the constituents of individual peaks were determined by mass deconvolution.
Supplementary Material
Acknowledgments.
We thank Prof. Keith Chater (John Innes Centre, Norwich, U.K.) for critical reading in preparation of this manuscript, and Prof. Qian-Qun Gu and Mr. Guo-Jian Zhang (Ocean University of China, Qingdao, People's Republic of China) for supplying us with undecylprodigiosin. This work was supported by National Natural Science Foundation of China Grants 30870041 and 30670017 and by the Ministry of Science and Technology of China Grant 2009CB118905.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ387221 and U24659).
This article contains supporting information online at www.pnas.org/cgi/content/full/0900592106/DCSupplemental.
References
- 1.West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 2.Skerker JM, et al. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koretke KK, Lupas AN, Warren PV, Rosenberg M, Brown JR. Evolution of two-component signal transduction. Mol Biol Evol. 2000;17:1956–1970. doi: 10.1093/oxfordjournals.molbev.a026297. [DOI] [PubMed] [Google Scholar]
- 4.Galperin MY. Structural classification of bacterial response regulators: Diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders DA, Gillece-Castro BL, Stock AM, Burlingame AL, Koshland DE., Jr Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem. 1989;264:21770–21778. [PubMed] [Google Scholar]
- 6.Schar J, Sickmann A, Beier D. Phosphorylation-independent activity of atypical response regulators of Helicobacter pylori. J Bacteriol. 2005;187:3100–3109. doi: 10.1128/JB.187.9.3100-3109.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Hara BP, et al. Crystal structure and induction mechanism of AmiC-AmiR: A ligand-regulated transcription antitermination complex. EMBO J. 1999;18:5175–5186. doi: 10.1093/emboj/18.19.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian Y, Fowler K, Findlay K, Tan H, Chater KF. An unusual response regulator influences sporulation at early and late stages in Streptomyces coelicolor. J Bacteriol. 2007;189:2873–2885. doi: 10.1128/JB.01615-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guthrie EP, et al. A response-regulator-like activator of antibiotic synthesis from Streptomyces coelicolor A3(2) with an amino-terminal domain that lacks a phosphorylation pocket. Microbiology. 1998;144:727–738. doi: 10.1099/00221287-144-3-727. [DOI] [PubMed] [Google Scholar]
- 10.Pflock M, et al. The orphan response regulator HP1021 of Helicobacter pylori regulates transcription of a gene cluster presumably involved in acetone metabolism. J Bacteriol. 2007;189:2339–2349. doi: 10.1128/JB.01827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser JS, et al. An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS. Mol Microbiol. 2007;65:319–332. doi: 10.1111/j.1365-2958.2007.05785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutchings MI. Unusual two-component signal transduction pathways in the actinobacteria. Adv Appl Microbiol. 2007;61:1–26. doi: 10.1016/S0065-2164(06)61001-0. [DOI] [PubMed] [Google Scholar]
- 13.Hutchings MI, Hoskisson PA, Chandra G, Buttner MJ. Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicolor A3(2) Microbiology. 2004;150:2795–2806. doi: 10.1099/mic.0.27181-0. [DOI] [PubMed] [Google Scholar]
- 14.Ulijasz AT, Andes DR, Glasner JD, Weisblum B. Regulation of iron transport in Streptococcus pneumoniae by RitR, an orphan response regulator. J Bacteriol. 2004;186:8123–8136. doi: 10.1128/JB.186.23.8123-8136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delany I, Spohn G, Rappuoli R, Scarlato V. Growth phase-dependent regulation of target gene promoters for binding of the essential orphan response regulator HP1043 of Helicobacter pylori. J Bacteriol. 2002;184:4800–4810. doi: 10.1128/JB.184.17.4800-4810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz R, Grossman AR. A response regulator of cyanobacteria integrates diverse environmental signals and is critical for survival under extreme conditions. Proc Natl Acad Sci USA. 1998;95:11008–11013. doi: 10.1073/pnas.95.18.11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonardy S, Freymark G, Hebener S, Ellehauge E, Sogaard-Andersen L. Coupling of protein localization and cell movements by a dynamically localized response regulator in Myxococcus xanthus. EMBO J. 2007;26:4433–4444. doi: 10.1038/sj.emboj.7601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang R, et al. AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus. J Bacteriol. 2004;186:6168–6178. doi: 10.1128/JB.186.18.6168-6178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molle V, Buttner MJ. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol Microbiol. 2000;36:1265–1278. doi: 10.1046/j.1365-2958.2000.01977.x. [DOI] [PubMed] [Google Scholar]
- 20.Ainsa JA, Parry HD, Chater KF. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2) Mol Microbiol. 1999;34:607–619. doi: 10.1046/j.1365-2958.1999.01630.x. [DOI] [PubMed] [Google Scholar]
- 21.Doull JL, Ayer SW, Singh AK, Thibault P. Production of a novel polyketide antibiotic, jadomycin B, by Streptomyces venezuelae following heat shock. J Antibiot. 1993;46:869–871. doi: 10.7164/antibiotics.46.869. [DOI] [PubMed] [Google Scholar]
- 22.Doull JL, Singh AK, Hoare M, Ayer SW. Conditions for the production of jadomycin B by Streptomyces venezuelae ISP5230: Effects of heat shock, ethanol treatment and phage infection. J Ind Microbiol. 1994;13:120–125. doi: 10.1007/BF01584109. [DOI] [PubMed] [Google Scholar]
- 23.Yang K, Han L, Vining LC. Regulation of jadomycin B production in Streptomyces venezuelae ISP5230: Involvement of a repressor gene, jadR2. J Bacteriol. 1995;177:6111–6117. doi: 10.1128/jb.177.21.6111-6117.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Vining LC. Control of growth, secondary metabolism and sporulation in Streptomyces venezuelae ISP5230 by jadW(1), a member of the afsA family of gamma-butyrolactone regulatory genes. Microbiology. 2003;149:1991–2004. doi: 10.1099/mic.0.26209-0. [DOI] [PubMed] [Google Scholar]
- 25.Yang K, Han L, He J, Wang L, Vining LC. A repressor-response regulator gene pair controlling jadomycin B production in Streptomyces venezuelae ISP5230. Gene. 2001;279:165–173. doi: 10.1016/s0378-1119(01)00723-5. [DOI] [PubMed] [Google Scholar]
- 26.Han L, et al. An acyl-coenzyme A carboxylase encoding gene associated with jadomycin biosynthesis in Streptomyces venezuelae ISP5230. Microbiology. 2000;146:903–910. doi: 10.1099/00221287-146-4-903. [DOI] [PubMed] [Google Scholar]
- 27.Furuya K, Hutchinson CR. The DnrN protein of Streptomyces peucetius, a pseudo-response regulator, is a DNA-binding protein involved in the regulation of daunorubicin biosynthesis. J Bacteriol. 1996;178:6310–6318. doi: 10.1128/jb.178.21.6310-6318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruiz D, et al. Phosphorylation-independent activation of the atypical response regulator NblR. Microbiology. 2008;154:3002–3015. doi: 10.1099/mic.0.2008/020677-0. [DOI] [PubMed] [Google Scholar]
- 29.Basnet DB, et al. Angucyclines Sch 47554 and Sch 47555 from Streptomyces sp. SCC-2136: Cloning, sequencing, and characterization. Mol Cells. 2006;22:154–162. [PubMed] [Google Scholar]
- 30.Rebets Y, et al. Function of lanI in regulation of landomycin A biosynthesis in Streptomyces cyanogenus S136 and cross-complementation studies with Streptomyces antibiotic regulatory proteins encoding genes. Arch Microbiol. 2008;189:111–120. doi: 10.1007/s00203-007-0299-5. [DOI] [PubMed] [Google Scholar]
- 31.Galm U, et al. Cloning and analysis of the simocyclinone biosynthetic gene cluster of Streptomyces antibioticus Tu 6040. Arch Microbiol. 2002;178:102–114. doi: 10.1007/s00203-002-0429-z. [DOI] [PubMed] [Google Scholar]
- 32.Ichinose K, Ozawa M, Itou K, Kunieda K, Ebizuka Y. Cloning, sequencing and heterologous expression of the medermycin biosynthetic gene cluster of Streptomyces sp. AM-7161: Towards comparative analysis of the benzoisochromanequinone gene clusters. Microbiology. 2003;149:1633–1645. doi: 10.1099/mic.0.26310-0. [DOI] [PubMed] [Google Scholar]
- 33.White J, Bibb M. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J Bacteriol. 1997;179:627–633. doi: 10.1128/jb.179.3.627-633.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cashman DJ, Scarsdale JN, Kellogg GE. Hydropathic analysis of the free energy differences in anthracycline antibiotic binding to DNA. Nucleic Acids Res. 2003;31:4410–4416. doi: 10.1093/nar/gkg645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connor TJ, Nodwell JR. Pivotal roles for the receiver domain in the mechanism of action of the response regulator RamR of Streptomyces coelicolor. J Mol Biol. 2005;351:1030–1047. doi: 10.1016/j.jmb.2005.06.053. [DOI] [PubMed] [Google Scholar]
- 36.Kato JY, Funa N, Watanabe H, Ohnishi Y, Horinouchi S. Biosynthesis of gamma-butyrolactone autoregulators that switch on secondary metabolism and morphological development in Streptomyces. Proc Natl Acad Sci USA. 2007;104:2378–2383. doi: 10.1073/pnas.0607472104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tahlan K, et al. Initiation of actinorhodin export in Streptomyces coelicolor. Mol Microbiol. 2007;63:951–961. doi: 10.1111/j.1365-2958.2006.05559.x. [DOI] [PubMed] [Google Scholar]
- 38.Jiang H, Hutchinson CR. Feedback regulation of doxorubicin biosynthesis in Streptomyces peucetius. Res Microbiol. 2006;157:666–674. doi: 10.1016/j.resmic.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Fish SA, Cundliffe E. Stimulation of polyketide metabolism in Streptomyces fradiae by tylosin and its glycosylated precursors. Microbiology. 1997;143:3871–3876. doi: 10.1099/00221287-143-12-3871. [DOI] [PubMed] [Google Scholar]
- 40.Kieser T, et al. Practical Streptomyces Genetics. Norwich, U.K.: John Innes Foundation; 2000. pp. 406–418. [Google Scholar]
- 41.Yang H, et al. The tyrosine degradation gene hppD is transcriptionally activated by HpdA and repressed by HpdR in Streptomyces coelicolor, while hpdA is negatively autoregulated and repressed by HpdR. Mol Microbiol. 2007;65:1064–1077. doi: 10.1111/j.1365-2958.2007.05848.x. [DOI] [PubMed] [Google Scholar]
- 42.Sheridan RC, McCullough JF, Wakefield ZT. Phosphoramidic acid and its salts. Inorg Synth. 1971;13:23–26. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.