Abstract
Ig and T cell receptor (TCR) genes consist of separate genomic elements, which must undergo rearrangement and joining before a functional protein can be expressed. Considerable plasticity in the genomic arrangement of these elements has occurred during the evolution of the immune system. In tetrapods, all Ig and TCR chain elements are arranged as translocons. In teleosts, the Ig heavy and TCR chains are translocons, but light chain genes may occur as clusters. However, in chondrichthyes, all of the Ig light and heavy chain genes are arranged as clusters. These clusters vary in number from <10 to several hundred, depending on isotype and species. Here, we report that the germ-line gene for the TCR γ chain in a chondrichthyan, the sandbar shark (Carcharhinus plumbeus), is present as a single locus arranged in a classic translocon pattern. Thus, the shark utilizes 2 types of genomic arrangements, the unique cluster organization for Ig genes and the “conventional” translocon organization for TCR genes. The TCR γ translocon contains at least 5 V region genes, 3 J segment genes, and 1 C segment. As expected, the third hypervariable segment (CDR3), formed by the rearrangement of the Vγ and Jγ segments, contributed the major variability in the intact V region structure. Our data also suggest that diversity may be generated by mutation in the V regions.
Keywords: diversity, translocon
Models proposed for the molecular evolution of the immune system are based on comparative studies of living species. Sharks are critical in this regard, because they belong to an ancient clade (chondrichthyans) that can be traced in the fossil record to the time of the earliest vertebrates. Approximately 450 million years ago, the gnathostomes diverged into 2 groups, the chondrichthyans and the line leading to modern teleosts and tetrapods. It can be concluded that the molecular components of the immune system are ancient and arose before this divergence. This conclusion follows from studies showing that all of the defining elements of the immune system, antibodies, T cell receptors (TCRs), MHC products, and recombination activator genes (RAG), are present in chondrichthyes (1, 2). Thus, continued studies of sharks and rays, the most distant living relatives of mammals with a vertebrate type (VDJ-C recombination) immune system, should provide insights into the molecular origins and evolution of the immune system.
A surprising feature of the organization of shark Ig genes was that the V-J-C of light chains (3–5) and V-D-D-J-C of heavy chains (6–8) are arranged in individual clusters. This feature is in contrast to the translocons of higher vertebrates that consist of large arrays of multiple V segments distantly linked to several J and D segments, as well as C domains (9). Although the genes for the TCR chains α/β and γ/δ have been shown to be present in the skate (10) and horned shark (11, 12), the germ-line loci for these molecules have been only partially characterized. Analyses of cDNA sequences in the skate revealed multiple V region families, and Southern blot analysis of genomic DNA strongly indicated that the skate TCR genes were arranged as translocons (10). This finding was surprising in light of the cluster organization of the Ig genes. Here, we report the sequence of the sandbar shark (Carcharhinus plumbeus) TCR γ chain genomic locus and confirm that it has a prototypical translocon arrangement. Our data show that diversity is generated by mutations in V regions. This finding is significant, because it is generally accepted that hypermutation does not occur, or is very rare (13–15), in TCR V regions. This finding certainly has functional implications for γ/δ T cells in sharks, and it may have phylogenetic significance for understanding the origins of diversity in the immune system.
Results
Cloning of Shark TCR γ Chain.
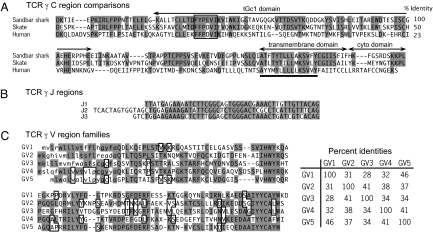
We performed 5′ RACE using degenerate primers based on a conserved C domain amino acid sequence to isolate putative sandbar shark TCR γ chain cDNA clones. A total of 25 cDNA clones were selected from the cDNA library with all of the selected clones sharing identical C region sequences. Searches of the National Center for Biotechnology Information databases by using BLAST showed that the clones had clear identity to known TCR γ chains, the best match being to skate TCR γ chain. The complete sequence of the C region was assembled by using sequences from 5′ and 3′ RACE. Comparison of the derived amino acid sequence of this C region with skate and human sequences, as well as the location of the IgC1 domain, transmembrane (Tm) domain, and cytoplasmic domain are shown in Fig. 1A. The identity with human γ is only 23%, a value much lower than comparable comparisons of Ig and other TCR sequences. This value is too low to indicate specific relatedness to higher vertebrate TCR γ chains and shows that the TCR γ chains have diverged significantly in evolution. The identification of the shark sequence as TCR γ chain is based on homologies with γ chains in BLAST searches, the high identity with the skate sequence (50%), and the conservation of a γ-chain-specific motif in the Tm domain (Fig. 1A).
Fig. 1.
Sequence comparisons of the TCR γ C, J, and V regions identified in cDNA clones. (A) Derived amino acid sequence of the C region and comparison with skate (Raja eglanteria) and human sequences. Only one C region was found in the shark TCR γ cDNA. Identities with the shark sequence are shaded. Domains are indicated by arrows above the diagram, and were identified by comparison with the human sequence. Conserved residues in the TM domain are underlined. Percentage identities are listed on the right. (B) Three J families were identified by cDNA sequence analysis. (C) Five V families, designated GV1, GV2, GV3, GV4, and GV5, were identified by cDNA sequence analysis. Derived amino acid sequences are shown. Putative leader segments are shown in lowercase and were identified by using the SignalP 3.0 program (49). Identical residues are indicated by using both shaded and open boxes. The percentage identities are shown in the matrix on the right.
Southern Blot Analysis.
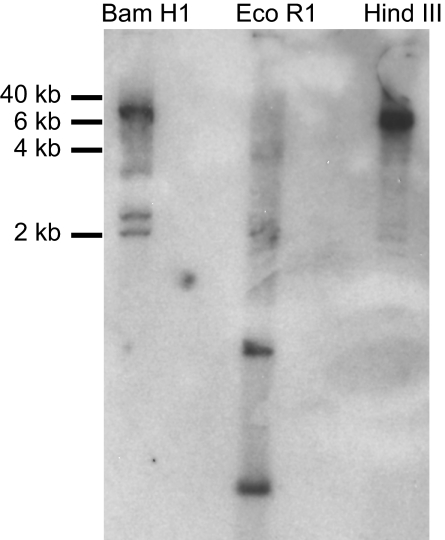
The finding that the cDNA clones isolated above shared identical C region sequences (>99% identity) indicated the presence of only a single copy of the C region gene. To confirm this conclusion, Southern blot analyses were performed with spleen DNA by using the full-length C segment as the probe. As shown in Fig. 2, the BamHI and HindIII DNA preparations each gave rise to one strong hybridizing band, indicating the presence only 1 TCR γ locus in the sandbar shark genome. This conclusion is buttressed by the finding that there are 2 equally dense hybridizing bands in the EcoRI-digested DNA preparation, which is explained by the fact that there is 1 EcoRI restriction site in the C region. Interestingly, several weak hybridizing bands are also seen in each restriction endonuclease DNA preparation (Fig. 2). Because the genomic PCR and sequencing results indicated that lymphocytes in the spleen have undergone gene rearrangement, the multiple weak hybridizing bands are most likely the result of gene rearrangement.
Fig. 2.
Southern blottings of the shark TCR γ locus. Sandbar shark spleen DNA was digested by BamHI (lane 1), EcoRI (lane 2), and HindIII (lane 3) independently. The probe was full-length C region isolated by PCR of cDNA.
Sequencing of the Sandbar Shark TCR γ Locus.
Sandbar shark spleen DNA was purified to perform genomic sequencing of the TCR γ loci. The use of spleen DNA for this purpose has advantages and disadvantages. It is expected that the T cells and B cells in the spleen would be mature cells and have undergone gene rearrangement. Therefore, during the sequencing procedure, some of the clones we obtained contained rearranged V-J segments. However, other cell types in the spleen (e.g., epithelial cells) yielded clones containing unrearranged DNA. Multiple rearranged and unrearranged segments were identified, and careful analysis of the sequences was performed to obtain the complete germ-line DNA sequence. However, spleen DNA is a good target to study the rearrangement events that occur at the genome level of the sandbar shark. Both genomic PCR and Genome Walker DNA walking strategies have been used here. For each genomic PCR, at least 2 pairs of gene-specific primers and multiple high-fidelity polymerases were used to minimize any possible PCR bias and/or PCR error.
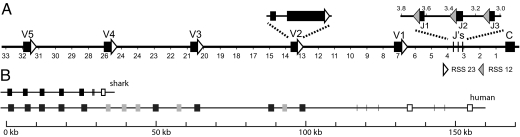
An overview of the TCR γ locus is shown in Fig. 3. The sequence was assembled from 6 PCR products and 10 chromosome walking fragments (Fig. S1). The final sequence was derived from at least 2 independent products in most cases. A few sequence mismatches between different PCR products are present that probably represent allelic differences or hypermutation events, although a small fraction may be due to PCR error. As shown in Fig. 3A, at least 5 V regions, 3 J regions, and 1 C region are arranged in a typical translocon pattern. Five is probably the total number of V genes present, because only 5 V region families were discerned in the 73 cDNA sequences analyzed, and these sequences can be unequivocally matched to the genomic V gene sequences. However, there are possibly more V genes upstream of GV5 that, although rarely expressed in spleen, may be expressed in other tissues. Use of the word “families” to label the different V regions is strictly speaking incorrect here, because each family has only 1 gene member. However, we adopted this terminology, because it is consistent with its use in immunology to describe groups of V regions with <75% amino acid sequence identity.
Fig. 3.
Comparison of shark and human TCR γ loci. (A) Genomic map of the shark TCR γ locus. V, J, and C segments are arranged in a translocon. RSS sequences at the 3′ end of V segments and the 5′ end of the J regions are highlighted by white (RSS 23) and black (RSS 12) triangles. The RSS sequence at the 5′ end of J1 region is shown in gray, because it has not been possible to derive an accurate sequence for an ≈15-bp segment in this region because multiple PCR products show variation in length and sequence. We conclude that this variation represents PCR error. The genomic structure showing the intron present in all of the V segments is illustrated by using the GV2 segment. (B) Diagram comparing the layout of the shark and human TCR γ chain loci. Dark boxes are functional V regions, gray boxes are pseudogenes, vertical lines are J segments, and open boxes are constant regions. The depiction of the human gene is adapted from the Gene View page at the ImMunoGeneTics website (http://imgt.cines.fr).
Consistent with all of the known Ig and TCR V regions, sandbar shark TCR γ V regions also have leader sequences that contain an intron. As mentioned above, several genomic PCR products contained V genes joined to J segments. Because the J to C intron sequences of these clones were identical, we concluded that they were the result of rearrangement events from a single locus, and were not indicative of multiple loci.
Recombination signal sequence (RSS) 23 sequences were identified at the 3′ end of each V region, and RSS 12 sequences at the 5′ end of each J region. Thus, it appears that rearrangement in the shark γ loci follows the “23/12” rule (16) defined in primates and rodents. The RSS segments were analyzed by calculating “RSS information content” (RIC) scores using models developed by Cowell et al. (17). The statistical models used for calculating informational content were learned using mouse Ig and TCR sequences (17), yet the RIC scores for the shark TCR γ locus clearly discriminate the physiologic RSSs. Thus, the informational content of RSS segments is yet another feature of the immune system that has been conserved during vertebrate evolution.
Generation of Diversity.
Based on TCR γ chain C region sequence, 3 different specific primers were designed and used in 5′ RACE to examine the V region repertoire. A total of 73 different clones were obtained and sequenced (Fig. S2). All sequences appeared functional, because there were no frameshift or stop codon mutations. Analysis of the sequences showed that all 3 J region sequences and all 5 V region families identified in the genomic sequence were expressed. The cDNA sequences of the J regions (Fig. 1B) and amino acid sequence of each the V regions are shown in Fig. 1C. Although the J segment sequences are similar, they have different lengths and are clearly distinguishable. For each V region family, examples of all J region sequences were found, and conversely, each J region was found associated with all V regions. There are approximately equal numbers of clones containing GV1 (18), GV2 (19), GV3 (12), and GV4 (17), indicating that there is no bias in the rearrangement of these V segments. Similarly, no bias is apparent in the expression of the 3 J segments. However, we found only 4 clones expressing GV5, suggesting that there may be significantly less rearrangement of this most 5′ distal V region segment.
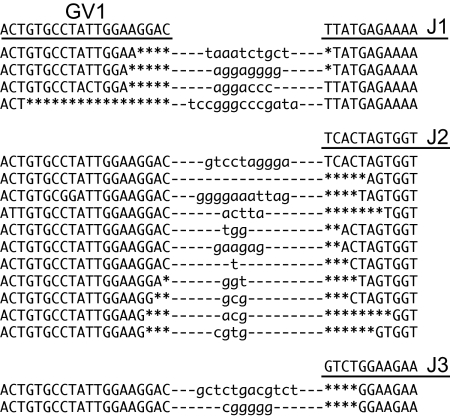
Some CDR3 region sequences of cDNA clones containing GV1 are shown in Fig. 4. The CDR3 region is formed by the joining of V and J regions, and extensive sequence variability is generated at the junction during this process. This diversity is similar to that of higher vertebrates, indicating that the sharks use similar mechanisms of deletion (by exonuclease) and addition [terminal deoxynucleotidyltransferase (TdT)] to generate TCR γ diversity. The length of CDR3 loops varied from 6 to 14 aa, a range similar to that in mice and humans (19). CDR3 loops are usually longer when a V region is joined to the J2 segment, because this segment is larger than the J1 and J3 segments. The same CDR3 sequence variability was seen in genomic PCR fragments generated by using a GV1-specific primer and a C-region-specific primer. Four different-sized PCR products were obtained corresponding to unrearranged TCR γ, and 3 rearrangement events in which the GV1 segment was joined to any of the 3 J region segments. Each of the 3 PCR products containing rearranged DNA was separately cloned, and a total of 18 were sequenced for analysis of CDR3 diversity (Fig. S3). Deletion and addition patterns in this region were identical to those of cDNA sequences, except that ≈50% of the genomic sequences show nonproductive rearrangements, whereas 100% of cDNA products are productive. The number of nucleotide additions ranged from 0 to 13, which could theoretically result in a diversity of nearly 108 sequences. The actual number is probably significantly lower, because the addition of G residues is ≈2-fold higher (37%) compared with A, T, and C residues (≈20% each).
Fig. 4.
Junctional diversity at the CDR3 segments of GV1-containing V regions. Sequences of shark TCR γ cDNA at the V/J junction of GV1 joined to all 3 J segments are shown. The parent genomic sequences are underlined above each group. Deletions (by endonuclease) are shown by asterisks, and nucleotides in lowercase indicate additions (by terminal deoxynucleotidyltransferase).
Surprisingly, comparisons of the cDNA sequences with the parent genomic sequences reveal a high degree of base changes in the cDNA (Fig. S2). Thirty-three of the 73 cDNA sequences had mutations, and 22 of these had 2 or more mutations. Because the mRNA and DNA samples were prepared by using spleen tissue from a single shark, the observed sequence diversity is not due to allelic variation. And it is not due to PCR error, because we used a high-fidelity polymerase. The rare occurrence of changes in C region sequences, and the fact that 33 of the V region sequences had no changes, confirms the fidelity of the polymerase. The average mutation rate of the V region (0.017 per base pair) is much higher than that of the C region (0.7 × 10−4 per base pair) and the expected PCR error rate (≈4.4 × 10−7 per base pair per cycle; in this research all of the PCRs were done with 25–30 cycles), but is comparable with mouse and shark Ig light chain V regions (20). Thus, we conclude that these data demonstrate that somatic mutation occurs in the TCR γ V region. A striking feature is the presence of tandem mutations containing 2–5 nucleotides. Tandem mutations appear to be a unique feature of somatic hypermutation in sharks, because they do not occur in higher vertebrates, and have been documented and characterized only for light and heavy chain V regions in the nurse shark (18, 20–22). Also, there are other similarities between the somatic mutations observed here and those described for the nurse shark.
Phylogeny of TCR γ.
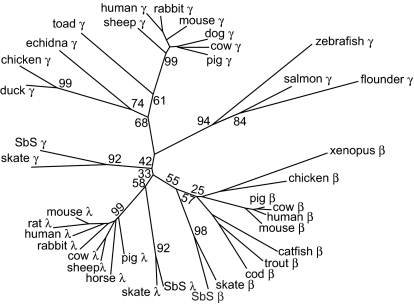
Although the identity of sandbar shark TCR γ chain with those of higher vertebrates is low, we were surprised that the best matches in GenBank searches are mostly with light chains of other species. To further ascertain the relationship of sandbar shark γ with other γ chains, β chains, and λ light chains, we constructed an unrooted phylogenetic tree (Fig. 5) based on C region sequences. When the neighbor-joining method is used, the skate and shark TCR γ chains do not group with the γ chains of higher vertebrates, but rather are in the β/λ clade. We also used unweighted pair group method with arithmetic mean (UPGMA) and maximum parsimony methods with very similar results (Fig. S4). Our interpretation is that ancestral γ chains were closely related to λ and β chains, and diverged in higher vertebrates because of a change in function of γ/δ T cells.
Fig. 5.
Phylogenetic tree, derived from C region comparisons, showing the relationship between TCR γ, Ig λ light chains, and TCR β chains. Sequences were aligned by using MAFFT (44, 45), and the tree was constructed with MEGA4 (48) using the neighbor-joining method (50). Percentage bootstrap values (1,000 replicates) are shown at the major interior branch points (for all of the bootstrap values, see Fig. S4). The horizontal length is proportional to the distance score generated by the computer program. The IgC1 domain amino acid sequences, corresponding to residues 1 to ≈125 using the IMGT numbering scheme (51), of λ light chains and TCR γ and β chains from different species were used to build this tree. The shark TCR γ sequence is from this work; all other sequences were obtained from GenBank.
Similar analysis of the V regions shows that they do not group with any V region family of higher vertebrates. However, skate and shark sequences do cluster, with the 5 shark and 6 skate families forming a tree with 4 groups (Fig. S5). Thus, there has been conservation of Vγ families in elasmobranchs since the divergence of skates and sharks 250 million years ago. The tree indicates that the TCR γ chain locus was well established before this divergence and contained 4 Vγ genes. The repertoire was subsequently expanded by duplication of these genes. For example, the number of sandbar shark V regions was expanded to 5 by an early duplication, giving rise to GV1 and GV5.
Discussion
A surprising feature of the organization of shark Ig genes (23) and the teleost Ig light chain genes (24) is that they are arranged in individual clusters, rather than the translocon arrangement found in higher vertebrates. Here, we show that the sandbar shark TCR γ gene is arranged in a “classic” translocon pattern consisting of at least 5 V regions, 3 J segments, and a single constant region. In higher vertebrate species, the translocon arrangements are not as “pure” as that of shark TCR γ. They are usually more complicated, and contain numerous V, J, and C regions, as illustrated in Fig. 3B. Also, most contain certain cluster characteristics, such as that of the C λ region in mammals, where there are at least 4 separate copies of the J-C λ gene cluster in mice (25), and at least 9 such clusters in human (26). A similar arrangement is also found in the mammalian TCR γ and β gene loci (27–29) and teleost fish light chain genes (24).
The RSSs in the γ locus are typical. As is true for all vertebrates, recombination follows the 12/23 rule, because the V regions have RSS 23s and the J segments have RSS 12s. Analysis of the RIC of shark RSSs and comparison with human and mouse show that the RSSs have been highly conserved. This result is not surprising, because RAG1 and RAG2, which mediate recombination, are also very highly conserved between shark and human. It is apparent that the core mechanisms of rearrangement, the recognition of RSSs and precise cleavage of the DNA by the RAG complex, are highly conserved features of the adaptive immune system.
The presence of both the cluster and translocon type arrangements in sharks and rays raises questions as to the ancestral arrangement pattern. Given the indications that TCR γ chains are more ancient than Ig chains (see below), the most parsimonious explanation is that a simple translocon, as described here, was the ancestral pattern for both TCRs and Igs. Alternately, both patterns may be ancestral and evolved independently from a common precursor after the split of lymphoid cells into separate B and T cell lineages.
In this study, we provide direct evidence for the presence of gene mechanisms used by sharks to generate TCR γ diversity that are similar to human Ig and TCR genes. The sandbar shark TCR γ gene has the capacity to generate enormous diversity by using 3 basic mechanisms. These mechanisms are combinatorial diversity, the introduction of N region diversity during rearrangements, and hypermutation in the V regions. The first 2 are common for TCR and Ig in all species, but the third is unique to immunoglobulins in higher vertebrates. The basic starting point is the 15 different arrangements possible through the 5 V and 3 J segments. With the exception of the decreased representation of TCR GV5, there was no bias in the utilization of these elements. Because the sequence identity of the V regions ranged from 31% to 46%, the rearrangement products represent quite different and distinct backbones on which to build additional diversity. A major component of shark TCR γ variability is contributed by the CDR3 segment formed by the junction of the V/J region. Extensive deletion and addition of nucleotides occur at the V/J junction at the genome level. Therefore, we conclude that the rearrangement mechanisms for generating CDR3 combinational diversity in sharks are essentially no different from those of higher vertebrates. It also seems clear that the incorporation of combinatorial and junctional mechanisms for the generation of diversity in the binding repertoire occurred very early in the evolution of the vertebrate immune system.
Analysis of cDNA sequences clearly shows that somatic mutation may be a third mechanism used to generate diversity in the expressed TCR γ repertoire. This finding is remarkable, because hypermutation does not occur in TCR V regions, or is rare (13–15). It is generally accepted that mutation in the CDR 1 and 2 regions of the α/β receptor would disrupt recognition of class I and II antigen-presenting molecules. This reasoning may not apply to the γ/δ receptors, because these receptors do not require MHC for the recognition of proteins and nonproteins (30, 31). Even so, hypermutation has not been reported to occur in these receptors either. We thought it possible that it may occur in the so-called γ/δ high species such as sheep, pigs, and cows, because γ/δ T cells are 60% of peripheral cells. Despite numerous recent publications characterizing in detail genomic and cDNA sequences in these species (32–34), we could find no reference to hypermutation in TCR γ chains. A recent article noted point mutations in salmon TCR γ chains (35). These results are difficult to interpret, because many mutations were insertions or deletions resulting in frameshifts, with many of these occuring in the C regions. We know of no other report documenting hypermutation in TCR γ V regions. Interestingly, analysis of the mutation patterns shows that they are very similar to hypermutation in the Ig V regions of the nurse shark. Clear distinguishing features are the tandem mutations, which apparently occur only in sharks (18, 20). Thus, it appears that the mechanisms for hypermutation in shark B cells and γ/δ T cells are similar. The high level of mutation in γ V regions emphasizes to us that many γ/δ T cells in the shark recognize and respond to antigen independently of antigen processing and MHC presentation. Considering this finding in conjunction with the expression of a highly diverse repertoire, it is clear that γ/δ cells have a major role in the shark immune system.
The multicluster arrangement raised the specter that Ig genes may not be clonally expressed, because it was difficult to envisage how rearrangement and transcription were regulated in light of what was known of mechanisms in higher vertebrates. However, it has now been unequivocally (18, 36) demonstrated that clonality, an essential requirement of the adaptive immune system, is a feature of Ig expression in B cells of the nurse shark. Hsu and coworkers (18, 36) have proposed a stochastic model for isotype and allelic exclusion, in which limiting amounts of nuclear factors allow the formation of very few complexes able to catalyze transcription and rearrangement. Thus, only 1–3 clusters can be targeted for rearrangement in a single B cell. In contrast to the Ig clusters, the sandbar shark TCR γ locus is a single copy, simple translocon. In common with the Ig clusters, but in marked contrast to the translocons of higher vertebrates, it is small, encompassing ≈30 kb. Thus, for both the Ig clusters and the TCR translocons, part of the processes that are considered to be important for allelic exclusion in higher vertebrates, namely chromatin contraction and rearrangement over long distances, are not a factor here (36). However, our data suggest that allelic exclusion is occurring at the TCR γ locus. All of the cDNA sequences we isolated were functional, i.e., they contained no frameshifts or stop codons. In contrast, approximately half of the genomic rearrangements we examined were nonproductive, i.e., frameshifts were present in the joining regions. These results indicate that nonproductively rearranged and nonrearranged alleles are not expressed, as would be expected if allelic exclusion were occurring. Although these results are not conclusive, we believe that the TCR γ genes are allelically excluded and clonally expressed. This finding suggests that the exclusion model proposed by Hsu et al. (18, 36) may also apply to the TCR γ locus. Another possible explanation of this phenomenon is nonsense-mediated decay of untranslatable mRNA (37). It is possible that we did not identify the mRNAs of nonproductively rearranged DNA because of the decay of these mRNAs.
The nature of the primordial immune receptor is subject to considerable speculation. In its simplest form, we would expect that this protein was a cell surface receptor similar to TCR, but also similar to antibody in its ability to directly bind antigen. Because (i) these features are properties of γ/δ receptors, (ii) γ chains appear phylogenetically primitive (38), and (iii) γ/δ receptors have structural features more in common with Ig Fab than TCR α/β (33, 39, 40), some have speculated that the primordial receptor was a primitive γ/δ molecule (38). Another primitive feature must have been the ability to generate a diverse binding repertoire. Was hypermutation one of these mechanisms? We believe that several considerations indicate it was. Activation-induced cytidine deaminase (AID), the enzyme responsible for hypermutation and isotype switch rearrangement, is conserved in all vertebrates. AID has a role in the generation of diversity in agnathans, even though these animals have a completely different genetic system as the basis of their immune system (41–43). Thus, AID is ancient and was present at the evolutionary origins of the immune system. Our results for sandbar shark γ chain V regions supports this model, because they strongly suggests that hypermutation occurred in the ancestral γ/δ receptor. If so, then it almost certainly occurred in the proposed γ/δ-like primordial receptor. Lee et al. (20) have also proposed that hypermutation is an ancient mechanism for generating diversity, perhaps preceding somatic recombination.
The characterization of the shark immune system continues to yield surprises. The finding that the TCR γ locus in the sandbar shark is a translocon was expected (10), but it is interesting that it is relatively simple and small. It is a major surprise that the V regions undergo hypermutation. This result suggests to us an expanded role for γ/δ T cells in the shark immune system.
Materials and Methods
Preparation of DNA and mRNA.
Sandbar shark spleen was provided by Carl Luer (Mote Marine Laboratories, Sarasota, FL). Sandbar shark spleen genomic DNA was prepared by using the QIAGEN Blood and Cell Culture DNA Kit. Sandbar shark spleen mRNA was prepared by using the Invitrogen MicroFastTrack 2.0 mRNA Isolation Kit.
Degenerate Primer Design.
Based on sequence analysis of the C region and Tm region from multiple species, including skate, chicken, and mammals, a conserved stretch of amino acid sequence (FFPDVI) was identified in the TCR γ chains. Based on this conserved amino acid sequence, gene-specific degenerate primers (GPSs) were designed.
RACE.
The 5′ RACE was performed using the SMART RACE cDNA Amplification Kit (Clontech) according to the manufacturer's instructions. Degenerate primers as well as adaptor-specific primers were used for PCRs using the following conditions: 50 μL reaction total, 34.5 μL of sterile H2O, 5 μL of 10× Advantage 2 PCR buffer, 1 μL of dNTP mix, 1 μL of 50× Advantage 2 polymerase mix, 2.5 μL of 5′ RACE-ready cDNA (1), 5 μL of UPM (universal primer), and 1 μL of GSP. PCR settings: 5 cycles of 94 °C for 30 s, 72 °C for 3 min and 5 cycles of 94 °C for 30 s, 70 °C for 30 s, and 72 °C for 3 min.
Agarose gel electrophoresis of PCRs was performed on a 1% agarose gel, and the band of proper size was carefully excised. The PCR products were purified by using the QIA quick gel purification kit (QIAGEN) and cloned by using the TOPO4 TA vector cloning system (Invitrogen). Mach1-T1 chemically competent cells (Invitrogen) and QIA prep miniprep kit (QIAGEN) were used for transformation and plasmid purification.
Computer Analysis.
The following software packages were used: MAFFT (44, 45) for sequence alignment, MacVector 10.5 (MacVector) for assembly of sequencing projects, sequence alignment and phylogenetic analyses, ClustalW2 (46, 47) for sequence and phylogenic analyses, and MEGA4 (48) for phylogenetic analysis. RSSs were identified by using methods of Cowell et al. (17).
Genomic PCR.
Multiple DNA polymerase kits, including Expand High Fidelity Plus PCR system (Roche), Expand 20 kb Plus PCR system (Roche), and iProof High Fidelity DNA polymerase (Bio-Rad), were used to eliminate possible PCR errors and to increase the chances of getting the right DNA fragment. Multiple pairs of primers were designed based on the cDNA sequence of every V and C region. To rule out the possibility of PCR bias, at least 2 primers were designed for each V region. Normal as well as “touchdown” PCR procedures were performed to get the DNA fragment of interest. Every PCR was performed following the manufacturer's instructions for each DNA polymerase.
Chromosome Walking.
The Universal Genome Walker kit (Clontech) was used in this study. For each library construction, purified genomic DNA was digested with blunt-end restriction enzymes (DraI, EcoRV, PvuII, ScaI, and StuI) independently. An adaptor was added to the end of the digested DNA fragments. Two gene-specific primers (GSP1 and GSP2) were designed based on the cDNA sequence. The nested PCR primer (GSP2) annealed to sequences beyond the 3′ end of the primary primer (GSP1) in 5′ walk (5′ end of GSP in 3′ walk); iProof high fidelity DNA polymerase touchdown PCR procedure was performed. In the primary reaction GSP1 and adaptor primer (AP)1 were used as primers. In the secondary reaction, 1 μL of primary reaction product was used as template, and GSP2 and AP2 were used as primers to perform nested PCR.
Shotgun Sequencing of Large DNA Fragments.
For genome sequencing, shotgun sequencing strategy was applied. The GPS-1 genome primer system (New England Biolabs) was used for this purpose. Individual sequences were assembled by using MacVector.
Southern Blot Analyses.
Sandbar shark spleen DNA was digested by BamHI, EcoRI, and HindIII independently. Digested DNA was transferred to Immobilon-Ny+ transfer membrane (Millipore) by capillary flow under alkaline conditions (1.5 M NaCl/0.5 M NaOH) and fixed to the membrane with UV cross-linking (5,000 μJ). A 32P-labeled full-length C segment was used as the probe. After an overnight hybridization, high-stringency washes were performed in 0.1× SSC/0.1% SDS solution at 65 °C.
Supplementary Material
Acknowledgments.
This work was supported by National Science Foundation Grant MCB-0621853 (to S.F.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. FJ854417–FJ854438 (TCRGV1), FJ854439–FJ854457 (TCRGV2), FJ854458–FJ854470 (TCRGV3), FJ854471–FJ854487 (TCRGV4), FJ854488–FJ854491 (TCRGV5), and FJ854492 (TCR γ genomic locus)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0811283106/DCSupplemental.
References
- 1.Bernstein RM, Schluter SF, Bernstein H, Marchalonis JJ. Primordial emergence of the recombination activating gene 1 (RAG1): Sequence of the complete shark gene indicates homology to microbial integrases. Proc Natl Acad Sci USA. 1996;93:9454–9459. doi: 10.1073/pnas.93.18.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marchalonis JJ, Schluter SF, Bernstein RM, Shen S, Edmundson AB. Phylogenetic emergence and molecular evolution of the immunoglobulin family. Adv Immunol. 1998;70:417–506. doi: 10.1016/s0065-2776(08)60392-2. [DOI] [PubMed] [Google Scholar]
- 3.Hohman VS, Schluter SF, Marchalonis JJ. Diversity of Ig light chain clusters in the sandbar shark (Carcharhinus plumbeus) J Immunol. 1995;155:3922–3928. [PubMed] [Google Scholar]
- 4.Hohman VS, Schuchman DB, Schluter SF, Marchalonis JJ. Genomic clone for sandbar shark lambda light chain: Generation of diversity in the absence of gene rearrangement. Proc Natl Acad Sci USA. 1993;90:9882–9886. doi: 10.1073/pnas.90.21.9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rast JP, et al. Immunoglobulin light chain class multiplicity and alternative organizational forms in early vertebrate phylogeny. Immunogenetics. 1994;40:83–99. doi: 10.1007/BF00188170. [DOI] [PubMed] [Google Scholar]
- 6.Rast JP, Amemiya CT, Litman RT, Strong SJ, Litman GW. Distinct patterns of IgH structure and organization in a divergent lineage of chrondrichthyan fishes. Immunogenetics. 1998;47:234–245. doi: 10.1007/s002510050353. [DOI] [PubMed] [Google Scholar]
- 7.Rumfelt LL, et al. A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc Natl Acad Sci USA. 2001;98:1775–1780. doi: 10.1073/pnas.98.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vazquez M, Mizuki N, Flajnik MF, McKinney EC, Kasahara M. Nucleotide sequence of a nurse shark immunoglobulin heavy chain cDNA clone. Mol Immunol. 1992;29:1157–1158. doi: 10.1016/0161-5890(92)90050-8. [DOI] [PubMed] [Google Scholar]
- 9.Hunkapiller T, Hood L. Diversity of the immunoglobulin gene superfamily. Adv Immunol. 1989;44:1–63. doi: 10.1016/s0065-2776(08)60639-2. [DOI] [PubMed] [Google Scholar]
- 10.Rast JP, et al. alpha, beta, gamma, and delta T cell antigen receptor genes arose early in vertebrate phylogeny. Immunity. 1997;6:1–11. doi: 10.1016/s1074-7613(00)80237-x. [DOI] [PubMed] [Google Scholar]
- 11.Rast JP, Litman GW. T-cell receptor gene homologs are present in the most primitive jawed vertebrates. Proc Natl Acad Sci USA. 1994;91:9248–9252. doi: 10.1073/pnas.91.20.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawke NA, Rast JP, Litman GW. Extensive diversity of transcribed TCR-beta in phylogenetically primitive vertebrate. J Immunol. 1996;156:2458–2464. [PubMed] [Google Scholar]
- 13.Cheynier R, Henrichwark S, Wain-Hobson S. Somatic hypermutation of the T cell receptor V beta gene in microdissected splenic white pulps from HIV-1-positive patients. Eur J Immunol. 1998;28:1604–1610. doi: 10.1002/(SICI)1521-4141(199805)28:05<1604::AID-IMMU1604>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 14.Zheng B, Xue W, Kelsoe G. Locus-specific somatic hypermutation in germinal centre T cells. Nature. 1994;372:556–559. doi: 10.1038/372556a0. [DOI] [PubMed] [Google Scholar]
- 15.Marshall B, Schulz R, Zhou M, Mellor A. Alternative splicing and hypermutation of a nonproductively rearranged TCR alpha-chain in a T cell hybridoma. J Immunol. 1999;162:871–877. [PubMed] [Google Scholar]
- 16.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 17.Cowell LG, Davila M, Kepler TB, Kelsoe G. Identification and utilization of arbitrary correlations in models of recombination signal sequences. Genome Biol. 2002 doi: 10.1186/gb-2002-3-12-research0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee V, et al. The evolution of multiple isotypic IgM heavy chain genes in the shark. J Immunol. 2008;180:7461–7470. doi: 10.4049/jimmunol.180.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rock EP, Sibbald PR, Davis MM, Chien YH. CDR3 length in antigen-specific immune receptors. J Exp Med. 1994;179:323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SS, Tranchina D, Ohta Y, Flajnik MF, Hsu E. Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity. 2002;16:571–582. doi: 10.1016/s1074-7613(02)00300-x. [DOI] [PubMed] [Google Scholar]
- 21.Diaz M, Velez J, Singh M, Cerny J, Flajnik MF. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: The translesion synthesis model of somatic hypermutation. Int Immunol. 1999;11:825–833. doi: 10.1093/intimm/11.5.825. [DOI] [PubMed] [Google Scholar]
- 22.Greenberg AS, et al. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 23.Shamblott MJ, Litman GW. Genomic organization and sequences of immunoglobulin light chain genes in a primitive vertebrate suggest coevolution of immunoglobulin gene organization. EMBO J. 1989;8:3733–3739. doi: 10.1002/j.1460-2075.1989.tb08549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daggfeldt A, Bengten E, Pilstrom L. A cluster type organization of the loci of the immunoglobulin light chain in Atlantic cod (Gadus morhua L.) and rainbow trout (Oncorhynchus mykiss Walbaum) indicated by nucleotide sequences of cDNAs and hybridization analysis. Immunogenetics. 1993;38:199–209. doi: 10.1007/BF00211520. [DOI] [PubMed] [Google Scholar]
- 25.Selsing E, Miller J, Wilson R, Storb U. Evolution of mouse immunoglobulin lambda genes. Proc Natl Acad Sci USA. 1982;79:4681–4685. doi: 10.1073/pnas.79.15.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang H, et al. Identification of three new Ig lambda-like genes in man. J Exp Med. 1986;163:425–435. doi: 10.1084/jem.163.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefranc MP, et al. Molecular mapping of the human T cell receptor gamma (TRG) genes and linkage of the variable and constant regions. Eur J Immunol. 1989;19:989–994. doi: 10.1002/eji.1830190606. [DOI] [PubMed] [Google Scholar]
- 28.Toyonaga B, Yoshikai Y, Vadasz V, Chin B, Mak TW. Organization and sequences of the diversity, joining, and constant region genes of the human T-cell receptor beta chain. Proc Natl Acad Sci USA. 1985;82:8624–8628. doi: 10.1073/pnas.82.24.8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tunnacliffe A, Kefford R, Milstein C, Forster A, Rabbitts TH. Sequence and evolution of the human T-cell antigen receptor beta-chain genes. Proc Natl Acad Sci USA. 1985;82:5068–5072. doi: 10.1073/pnas.82.15.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chien YH, Jores R, Crowley MP. Recognition by gamma/delta T cells. Annu Rev Immunol. 1996;14:511–532. doi: 10.1146/annurev.immunol.14.1.511. [DOI] [PubMed] [Google Scholar]
- 31.Carding SR, Egan PJ. Gammadelta T cells: Functional plasticity and heterogeneity. Nat Rev Immunol. 2002;2:336–345. doi: 10.1038/nri797. [DOI] [PubMed] [Google Scholar]
- 32.Hein WR, Dudler L. TCR gamma delta+ cells are prominent in normal bovine skin and express a diverse repertoire of antigen receptors. Immunology. 1997;91:58–64. doi: 10.1046/j.1365-2567.1997.00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaccarelli G, et al. Genomic organization of the sheep TRG1@ locus and comparative analyses of Bovidae and human variable genes. Gene. 2005;357:103–114. doi: 10.1016/j.gene.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 34.Herzig C, Blumerman S, Lefranc MP, Baldwin C. Bovine T cell receptor gamma variable and constant genes: Combinatorial usage by circulating gammadelta T cells. Immunogenetics. 2006;58:138–151. doi: 10.1007/s00251-006-0097-2. [DOI] [PubMed] [Google Scholar]
- 35.Yazawa R, et al. Functional adaptive diversity of the Atlantic salmon T-cell receptor gamma locus. Mol Immunol. 2008;45:2150–2157. doi: 10.1016/j.molimm.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Malecek K, et al. Immunoglobulin heavy chain exclusion in the shark. PLoS Biol. 2008 doi: 10.1371/journal.pbio.0060157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 38.Richards MH, Nelson JL. The evolution of vertebrate antigen receptors: A phylogenetic approach. Mol Biol Evol. 2000;17:146–155. doi: 10.1093/oxfordjournals.molbev.a026227. [DOI] [PubMed] [Google Scholar]
- 39.Chien YH, Bonneville M. Gamma delta T cell receptors. Cell Mol Life Sci. 2006;63:2089–2094. doi: 10.1007/s00018-006-6020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson IA, Stanfield RL. Unraveling the mysteries of gammadelta T cell recognition. Nat Immunol. 2001;2:579–581. doi: 10.1038/89718. [DOI] [PubMed] [Google Scholar]
- 41.Pancer Z, Cooper MD. The evolution of adaptive immunity. Annu Rev Immunol. 2006;24:497–518. doi: 10.1146/annurev.immunol.24.021605.090542. [DOI] [PubMed] [Google Scholar]
- 42.Alder MN, et al. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310:1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- 43.Pancer Z, et al. Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci USA. 2005;102:9224–9229. doi: 10.1073/pnas.0503792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 48.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 49.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 50.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 51.Lefranc MP, et al. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev Comp Immunol. 2005;29:185–203. doi: 10.1016/j.dci.2004.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.