Abstract
Variation in gene content has been hypothesized to be the primary mode of adaptive evolution in microorganisms; however, very little is known about the spatial and temporal distribution of variable genes. Through population-scale comparative genomics of 7 Sulfolobus islandicus genomes from 3 locations, we demonstrate the biogeographical structure of the pan-genome of this species, with no evidence of gene flow between geographically isolated populations. The evolutionary independence of each population allowed us to assess genome dynamics over very recent evolutionary time, beginning ≈910,000 years ago. On this time scale, genome variation largely consists of recent strain-specific integration of mobile elements. Localized sectors of parallel gene loss are identified; however, the balance between the gain and loss of genetic material suggests that S. islandicus genomes acquire material slowly over time, primarily from closely related Sulfolobus species. Examination of the genome dynamics through population genomics in S. islandicus exposes the process of allopatric speciation in thermophilic Archaea and brings us closer to a generalized framework for understanding microbial genome evolution in a spatial context.
Keywords: archaea, genome evolution, population genomics, horizontal gene transfer, evolutionary rates
A current model of microbial genome dynamics partitions variation into 2 distinct genomic components, core and variable (1), together called the pan-genome (2). The core genome is composed of genes that are common to all strains and held stable through conservation (3). The variable genome is composed of genes not found in all strains, either because genes are gained through horizontal gene transfer or because they are differentially lost. Every unique microbial strain sequenced contains a different suite of variable genes (2, 4–11). Yet, the ecological forces that define the distribution of these variable genes among strains, the sources of genetic material, and the rates at which genes are acquired and lost are unknown. It is largely believed that the variable gene component extends the physiological and ecological capabilities of microbial cells (11–16). Faster than stepwise nucleotide change, the rapid acquisition of genes appears to be responsible for the transfer of a broad range of adaptive functions, most notably antibiotic resistance among divergent bacterial pathogens (8). Although signatures from core housekeeping genes suggest that geographical (17, 18), ecological (19, 20), and biological (21, 22) barriers promote divergence among microbial populations, the mechanisms defining the spatial and temporal distribution of variable genes are not well understood.
Allopatric speciation has been recognized in microorganisms using multilocus sequence analysis of conserved housekeeping genes (23–25). However, it is not known whether the small number of conserved molecular markers used to identify biogeographical patterns obscures patterns of gene flow in other parts of the genome. Two comparative genomic studies have used microarray analysis to assess the biogeographical distribution of genome variation. One, in Helicobacter pylori (6), found a rate of gene content change too rapid to leave a geographical signal. The other, in Sulfolobus islandicus (26), found that the distribution of variable genes differed from the strict geographical structure resolved by previous analysis of core housekeeping genes (18). This study suggests that variable gene content, unlike the core genome, is driven by environmental selection. Using full-genome comparisons, other studies have similarly identified the parallel presence of variable genes in microbial populations that appear differentiated. This has led to the conclusion that the variable gene pool is unbounded, sampled freely by individuals, and incorporated where genes are advantageous (9, 27–30).
To determine the importance of adaptive acquisition of genes to microbial speciation, several studies have mapped variation in gene content onto population structure defined by the core genome, although not in a biogeographical context (9, 11, 29–33, 34). These studies found a rapid flux of genes through genomes, some of which can be associated with parallel adaptive gene gain by divergent strains (5, 9, 10, 28, 35, 36). Gene loss has also been recognized as an important force in genome variation, especially in host-associated microbial populations (37–40). In particular, several derived pathogens, Salmonella enterica Typhi (32, 36) and Shigella (28), and intracellular symbionts, Buchnera (41, 42), have been suggested to undergo significant genome reductions. Gene loss has also been identified in free-living microorganisms (43), and it is often assumed that loss is mechanistically simpler than gain when modeling genome evolution (9, 33, 44, 45), although the relative importance of gene gain and loss to variation in the pan-genome is still the subject of debate. Absolute rates of gene gain and loss provide a basis for comparison among microorganisms and with geological events such as climate and environmental change. Because microorganisms do not leave a fossil record, absolute rates have only been determined in relation to ancient macroevolutionary events (46, 47).
Here, we examine whether there is a biogeographical distribution of variable genes. We use complete genome sequences of the thermoacidophilic crenarchaeon S. islandicus, which has been shown to be geographically isolated through multilocus sequence typing of 7 conserved core genes (18). Our analyses establish geographical isolation in the core and variable genome components of 3 recently diverged and evolutionarily independent S. islandicus populations. Within this well-resolved spatial structure, we examine the rates of gain and loss processes to determine the sources of genome variation as these microbial populations diverge in allopatry.
Results and Discussion
Biogeography of the Core Genome.
Table 1 describes the characteristics of the 7 genomes of S. islandicus sequenced for this study—2 from Yellowstone National Park (YNP), 2 from Lassen National Park (LNP), and 3 from the Mutnovksy Volcano in Kamchatka, Russia. To distinguish between the core and variable genome components, we assigned all genes (coding sequences including pseudogenes) from each of these 7 genomes and the closely related species Sulfolobus solfataricus (48) to homologous gene clusters. We identified 2,169 S. islandicus core gene clusters containing at least 1 sequence from all 7 genomes. Core genes represent, on average, 74% of the total genes in each S. islandicus genome (Table 1). 1,958 Sulfolobus core gene clusters contained at least 1 sequence from the 8 genomes including S. solfataricus.
Table 1.
Summary of S. islandicus genomes and local environments
| Region | Location | Temperature, °C | pH | Strain name | Genome size, MB | No. of genes* | % core |
|---|---|---|---|---|---|---|---|
| LNP | Devil's Kitchen | 78 | 2.7 | L.D.8.5 | 2.71 | 2,988 | 73 |
| LNP | Sulfur Works | 78 | 2.7 | L.S.2.15 | 2.74 | 3,000 | 72 |
| YNP | Geyser Creek | 72 | 3.6 | Y.G.57.14 | 2.70 | 2,966 | 73 |
| YNP | Norris Geyser Valley | 59 | 2.7 | Y.N.15.51 | 2.87 | 3,153 | 69 |
| Kamchatka | Mutnovsky Volcano | 91 | 2.0 | M.14.25 | 2.61 | 2,834 | 77 |
| Kamchatka | Mutnovsky Volcano | 76 | 2.0 | M.16.27 | 2.69 | 2,908 | 75 |
| Kamchatka | Mutnovsky Volcano | 76 | 2.0 | M.16.4 | 2.69 | 2,761 | 79 |
*Gene refers to coding sequence including pseudogenes.
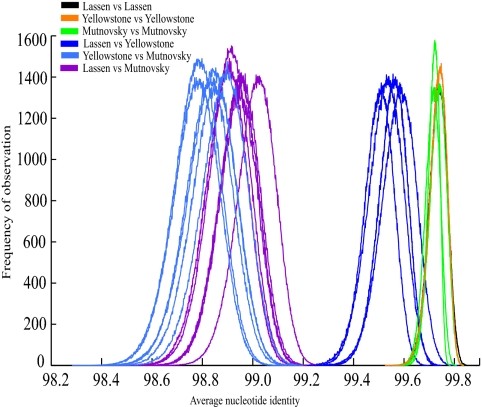
To identify relationships among strains using core genes, the bootstrapped average nucleotide identity (bANI) was calculated from the concatenated alignment of all Sulfolobus core gene clusters for each pair of genomes. Fig. 1 shows that biogeographical patterns are observed using the core genome of our 7 S. islandicus strains with a minimum of 98.78% bANI. The distribution of bANI within each population does not overlap with the distribution between populations. Additionally, the North American populations are genetically more similar than either is to the Mutnovsky population. These results demonstrate that even among very similar strains, a relationship between genetic and geographical distance is resolved that is consistent with isolation by distance in which geographical barriers prevent dispersal among populations.
Fig. 1.
Pairwise bANI for the 7 S. islandicus genomes. Comparisons between LNP (Lassen) strains (black), YNP (Yellowstone) strains (orange), and Mutnovsky strains (green) yield an average identity of 99.73%. Comparisons between North American strains (LNP vs. YNP, dark blue) give an average identity of 99.54%, whereas comparisons between each North American and Mutnovsky strain (magenta and light blue) yield an average identity of 98.89%.
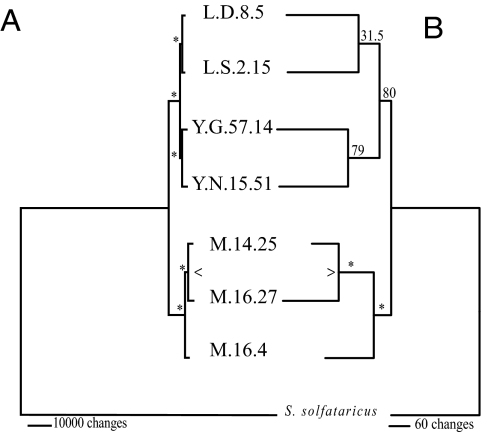
In support of the bANI results, a phylogeny inferred from the concatenated sequence of core gene clusters from the 7 S. islandicus strains and the closely related S. solfataricus (48) resolves a distinct S. islandicus population in each geographical region (Fig. 2A). In addition, a subpopulation within the Mutnovsky region is resolved by the core genome phylogeny. To test for gene flow between populations, we examined the phylogenetic relationships between strains described by each core homologous cluster in comparison to the concatenated core gene phylogeny shown in Fig. 2A, assuming that different tree topologies represent potential gene flow. Seventy-four percent of the individual gene phylogenies support the core gene topology. Ten percent do not have sufficient genetic variation to resolve any pattern significantly. Thirteen percent support an alternate grouping of the 3 Mutnovsky strains, whereas 3% support alternate groupings of the 4 North American strains. These single-gene phylogenetic analyses support isolation by distance among S. islandicus populations, where gene flow is rare in the core genome and occurs primarily within local populations.
Fig. 2.
Maximum parsimony phylogenies inferred from the concatenated core nucleotide genome (A) and presence/absence of gene content matrix (B). Numbers above each branch correspond to bootstrap support for that node in percentage. *, 100%; >, Mutnovsky subpopulation.
Biogeography of the Variable Genome.
A total of 1,060 gene clusters were categorized as variable in S. islandicus because they lacked a representative from at least 1 of the 7 genomes. Fig. 2B shows the relationship among strains based on shared gene content (49) that matches the core gene nucleotide phylogeny (Fig. 2A) with significant bootstrap support. The only node in this phylogeny with less than 50% bootstrap support joins the 2 LNP strains with alternative topologies placing either LNP strain closer to the YNP strain than to the other LNP strain. These data demonstrate that the variable genome has a signal for biogeographical structure consistent with isolation by distance and matching that of the core genome.
These results stand in contrast to previous data described by Grogan et al. (26), where it is suggested that environmental difference rather than geographical isolation drives differences in gene content. Under a model of isolation by distance, populations that are geographically closer to one another are genetically more similar than they are to distant populations because there are a higher number of migrants between them (50). This model is contrasted with one in which environmental selection defines the distribution of genetic diversity (i.e., environments that are more similar have genetically more similar populations) (24). Using comparative genome hybridizations, Grogan et al. (26) showed that their single strain from LNP grouped with Kamchatka strains. In failing to find an association between the geographically proximal YNP and LNP populations, these data suggest the opposite conclusion to those described previously and shown in Fig. 2B (i.e., that environmental selection drives difference in gene content rather than isolation by distance). The difference between our results and those of Grogan et al. (26) reflects our ability to identify the full complement of genes within each genome, and thereby identify variation resulting from gene gain and loss within S. islandicus populations (SI Text). Grogan et al. (26) were only able to identify a subset of lost or highly divergent genes from the genome of S. solfataricus, and therefore missed the unambiguous biogeographical pattern demonstrated here.
Having demonstrated the independence of 3 isolated populations, we determined the history of gene acquisition and loss in each geographical population by mapping gene distribution onto the phylogeny resolved by the core genome (49). The majority of variable gene clusters contained genes that were either present or absent from a single strain, and were thus designated as strain-specific gene gain and loss, respectively. Clusters containing genes present or absent only in all strains from a population were assigned to events earlier in the evolutionary history of the population, before strains diverged as individuals. Although it is possible that these shared genes result from independent gene gain events, the fact that shared genes always map to the same genomic location and are highly similar in sequence makes it more parsimonious to conclude that they result from acquisition by the ancestor to each local population. We examined the annotations of these sets to identify functions that may be locally adaptive in each geographical location (Tables S1 and S2). The majority of shared unique loci have putative annotations as hypothetical or conserved hypothetical proteins. Several population-specific clusters associated with the early divergence between the Mutnovsky and North American populations are 2 gene cassettes with 1 member annotated as putative pilT domain protein. Such 2-gene operons are associated with toxin-antitoxin systems originally described for plasmid maintenance (51). It has recently been hypothesized that these systems originate on plasmids but are co-opted by cells to regulate stress responses to changing environmental conditions (52, 53). In addition, several gene clusters unique to the Mutnovsky subpopulation are associated with the clustered regularly interspaced short palindromic repeat/CRISPR-associated (CRISPR/cas) system. This system is thought to provide Sulfolobus with immunity to foreign genetic elements (54, 55). These data suggest that at least a portion of the biogeographical differentiation in gene content among populations is associated with previous and ongoing interactions between host cells and mobile elements.
Parallel Gain and Loss.
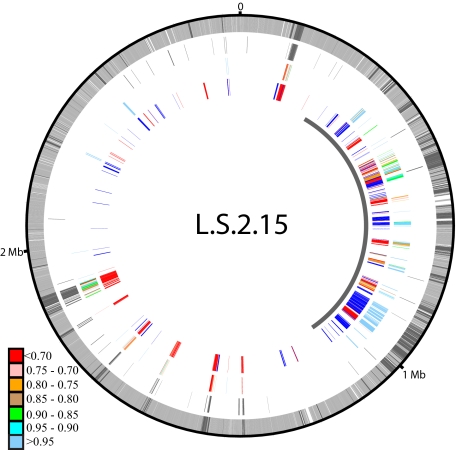
Although the primary pattern shows biogeographical distribution of the variable genome of S. islandicus, 30% of the total variable genome clusters do not map onto the strain phylogeny using parsimony criteria. For example, clusters containing genes found in both strains from LNP and M.16.4 could either result from 2 gene gains (LNP and M.16.4) or 2 gene losses (YNP and M-sub). Because independent gain of the same genes by different populations could indicate their linkage to a common gene pool, these genes were investigated in detail by mapping them onto their genome context, as shown for L.S.2.15 in Fig. 3 and for other genomes in Fig. S1. Two primary patterns were revealed, both consistent with biogeographical differentiation. One pattern is composed of sets of genes that fall into regions of the genome containing virus and plasmid homologs to other mobile elements from other Sulfolobus species (Fig. 3, ring 2). Genes from these clusters are found in different locations in each genome and show no consistent gene order when shared between strains. The average nucleotide identities (ANI) within each of these clusters were significantly more divergent than the core genome (0.18 ± 0.11 and 0.01 ± 0.02, respectively), indicating a different evolutionary history from the core genome. Based on divergence in gene content, order, position in the genome, nucleotide sequence, and association with plasmid and viral genes, the parallel presence of these genes between populations was inferred to result from the independent strain-specific integration of different mobile elements that share a small number of divergent genes. To determine the relationships among integrated genetic elements, we constructed phylogenies for all clusters with 4 or more members with at least 2 from a single population. In the majority of these individual phylogenies, gene sequences are grouped by geographical location (data not shown). In combination with our previous work showing that Sulfolobus viral populations are geographically structured (56), these data provide further evidence for the distribution of mobile elements consistent with biogeography.
Fig. 3.
Physical map of genome dynamics for the L.S.2.15 genome. Rings are numbered beginning with the outer ring. Ring 1 shows the location of S. islandicus core (light gray) and noncore (dark gray) clusters. Ring 2 shows the locations of viral and plasmid homologs. Ring 3 shows the location of clusters whose distribution cannot be determined by parsimony criteria. Colors represent average pairwise nucleotide identity within each cluster, ranging from less than 70% to greater than 95%, as shown in legend to the left. Ring 4 shows designations of gene gain by L.S.2.15 (red), gene loss by any other strain or group of strains (blue), and multiple events (orange) (e.g., gain by North American ancestor followed by loss in L.D.8.5). Innermost arcs denote the location of the variable region.
The other pattern contains sets of variable gene clusters located in the same position relative to conserved core genes in each genome and having conserved gene order that is interrupted only by putative transposases or single-event insertions (strain- or population-specific genes). Nucleotide sequence variation for this set is similar to ANI observed for the core genome (0.02 ± 0.04). Very few genes from this set match virus or plasmid sequences (Fig. 3). These regions are highly conserved in nucleotide sequence, gene content, and gene order, and they also do not match virus and plasmid sequences. Therefore, they are unlikely to result from recent mobile element integration. Instead, we conclude that variation in gene content in these sets represents strain-specific gaps in the genome resulting from differential gene loss from genome regions present in the common ancestor to all S. islandicus strains. The sets of genes contributing to this second pattern are primarily localized to a highly variable portion of each genome (Fig. 3, ring 4) that averages 695 kbp in length beginning ≈300 kbp from the origin in each strain [defined as 1 of 3 replication origins (57, 58)], except in Y.N.15.51, which has had a large inversion (Fig. S1F).
The localization of parallel gene losses to particular shared regions of the genome may represent convergent adaptive loss of function, trimming of genes that are no longer functional within any S. islandicus population [genome streamlining (43)], or regions of the genome that are more prone to gene loss. The majority of the lost genes have no matches to the National Center for Biotechnology Information (NCBI) nonredundant database. This pattern is similar to that recently described in other bacteria in which groups of “orfan” genes (59, 60) that were presumably acquired by horizontal gene transfer are being degraded over time (31).
The remaining clusters whose distributions cannot be determined by parsimony represent small sets of contiguous genes interrupted only by transposable elements that could not be classified into either of the 2 categories described previously because they are not associated with plasmid or virus homologs or linked to core genes. This set is primarily localized to the variable region of the genome described earlier, which contains many small inversions and rearrangements, preventing the use of synteny to identify genome dynamics. A majority of the clusters shared between strains from different populations have different BLAST matches to the NCBI nonredundant database (61), and were therefore designated as differential gain of divergent gene cassettes from different sources. Many of these sets of genes are again associated with the CRISPR/cas system. The fact that divergent CRISPR-associated gene cassettes are gained multiple times by different populations and are found in highly variable regions of the genome suggests the rapid evolution of this putative immune system (62).
Sources of Horizontally Acquired Genes.
All genes from the variable genome identified as gene gains were compared with the NCBI nonredundant database to determine the potential of horizontal gene transfer from divergent species. The majority with significant BLAST (61) hits match genomes of closely related Sulfolobales species or their mobile elements (Table S3), suggesting that horizontal gene transfer from divergent species is a rare event. As in other analyses of microbial species (40, 45, 59, 60, 63), many of the gained genes in S. islandicus have no significant BLAST match to the public databases (Table S3).
Rates of Genome Dynamics.
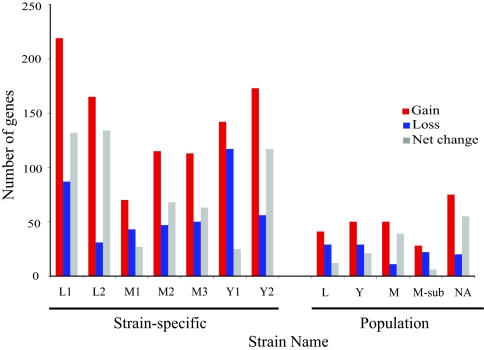
The relatively recent divergence of isolated populations provides a unique opportunity to examine recent mechanisms of genome evolution as populations diverge in allopatry. Summing those clusters designated as uniquely present or absent from each strain or population as gains or losses, respectively, with those assigned as described above, we calculated the total net flux of genes over the evolutionary history of divergence (Fig. 4). As shown in Fig. 4, much of the total gene gain and loss is strain specific, suggesting that it has accelerated since the recent divergence of each strain. However, much of the strain-specific gene gain results from integration of large mobile elements such as viruses and plasmids and may be transient. We are unable to identify the complete rates of strain-specific gene loss because they do not appear within the sequenced genomes but infer from the strain specificity of integrated mobile elements that they are also lost rapidly within these populations. More reliable rates are those shown for each population as they are likely to have occurred earlier in evolutionary history. Fig. 4 shows that gene gain is greater than gene loss at the population level. Unlike many of the host-associated bacteria that have been examined at this scale (32, 36), S. islandicus genomes do not appear to be reducing in size.
Fig. 4.
Tally of genome dynamics by strains and populations. Gene gain (red), gene loss (blue), and net change (gain − loss; gray). Strain names are abbreviated as follows: L1, L.D.8.5; L2, L.S.2.15; M1, M.14.25; M2, M.16.27; M3, M.16.4; Y1, Y.G.57.14; Y2, Y.N.15.51; L, Lassen; Y, Yellowstone; M, Mutnovsky; M-sub, Mutnovsky subpopulation; NA, North America.
The evolutionary independence of each biogeographically isolated S. islandicus population allows us to link the natural history of these populations to geological time scales to calculate absolute rates of these dynamics in years. We estimated the chronology of divergence of each population from the S. islandicus common ancestor using Bayesian divergence dating with the constraint that strain divergence from a regional population could not precede the earliest onset of geothermal activity in that region. Divergence dates for each population are shown on the chronogram in Fig. S2, ranging from 910,000 (±90,000) years ago for the North American and Mutnovsky populations to 140,000 (±25,000) years ago for the M.16.27 and M.14.25 divergence. Based on these dates, the average rate of nucleotide substitution was estimated to be 4.66 × 10−9 (±6.76 × 10−10) substitutions per site per year (Table S4), on the order of the universal rates estimated from comparisons of other species (46, 47, 63) and predicted from laboratory determinations of mutation rates in other Sulfolobus species (64). This unique method for dating microbial populations based on constraining colonization dates with geological events allows estimation of absolute rates of genome dynamics on more recent time scales than have previously been examined.
Using these divergence dates, we estimate an average of 0.26 (±0.03) gene gains and 0.11 (±0.01) gene losses per 1,000 years per lineage (Table S4). The average rate of net gene gain by populations was estimated as 0.08 (±0.03) genes per 1,000 years (Table S4). This is accelerated in comparison to rates of divergence between E. coli and Salmonella (estimated as 0.016 1-kbp length genes per 1,000 years) (63). Without conversions to absolute time in years, the rates of gain and loss per substitution per site suggest that events in S. islandicus are occurring at least twice as fast as those in Streptococcus group B (65). It is possible that S. islandicus has faster rates of genome dynamics than other species; however, it is more likely that the comparisons described here simply reset the lower bounds for these rates because of the recent divergence times estimated for these closely related strains.
Conclusions
Population genomics at this scale reveal the mechanisms of evolution leading to divergence of incipient species (38, 66–69). We have shown that for the thermoacidophilic archaeaon S. islandicus, geographical barriers isolate the core genome and spatially partition the variable gene pool between globally distinct geothermal regions. A geographically structured gene pool promotes local adaptation but dramatically slows gene flow among populations. In this way, the partitioning of the pan-genome pool has great significance for the rate and trajectory of evolution in this species and others that are similarly structured. Thermophilic Archaea represent an ideal system in which to test for biogeographical patterns because they are obligate extremophiles living in geothermal regions that have clear boundaries, are discontinuous, and are geographically distant. Biogeographical isolation has been documented in many microbial species with less restrictive habitat requirements (23, 24) in which the generality of the patterns described here should now be tested. Although populations from these regions are more closely related than the often-used genomic definitions of a species (70), they fit an evolutionary definition of species in which lineages are isolated from one another and evolving independently (71). The geographical structure identified for S. islandicus has allowed us to investigate recent dynamics as populations diverge to become species. Investigations of the dynamics of genome change in a spatial context provide unique insight into mechanisms of local adaptation and gene flow that drive diversity in microorganisms.
Materials and Methods
Genome Sequencing, Annotation, and Clustering.
Seven of the S. islandicus strains, isolated and characterized by multilocus sequence typing (MLST) by Whitaker et al. (18), were chosen to represent the genetic and geographical diversity of each geographical region. At least 2 strains were chosen from each environment so that shared characteristics could be identified. Strains were chosen to represent different geothermal areas within LNP and YNP, as shown in Table 1. Mutnovsky strains were chosen to represent parental and recombinant members of the population, as described by Whitaker et al. (72). Each genome was sequenced by the Department of Energy (DOE) Joint Genome Institute through standard Sanger sequencing methods. Genome assembly and annotation were completed by the Los Alamos National Laboratory (LANL) and Oak Ridge National Laboratory (ORNL) using standard pipeline procedures. ORFs from S. islandicus and S. solfataricus (48) were grouped into homologous clusters by Markov clustering of sequence similarity using MCL v1.006 (73) clustering. Nucleotide sequences for each cluster were aligned with T-Coffee v5.56 (74). Paralogs clustered by MCL were manually split into independent clusters.
bANI and Phylogenetic Analyses.
bANI for the core genome was determined by sampling gene clusters with replacement to construct 3,000 pseudoreplicates the size of the complete core genome. Core cluster alignments were concatenated for each pseudoreplicate and ANI (70) was calculated as the average of the pairwise number of nucleotide identities between strains with gaps treated as missing characters. Phylogenies based on nucleotide alignments were inferred for all clusters of 4 or more members, and the concatenated core genome under both maximum parsimony and maximum likelihood were bootstrapped in PAUP* v4b10 (75). The topologies of 50% consensus phylogenies from 1,000 bootstrap replicates for each individual gene cluster were determined to support the concatenated core gene phylogeny if at least 1 node was shared with the concatenated core topologies and no nodes conflicted.
Integrated Plasmids and Viruses.
Integrated plasmids and viruses were identified by similarity to known plasmids and viruses using BLASTp (61) of all translated ORFs against a database of all published mobile elements associated with Sulfolobus species (76). BLAST results were filtered to include only matches with greater than 40% amino acid identity and greater than 70% coverage of the query from the S. islandicus database.
Genome Dynamics.
Parsimony criteria based on the membership of each homologous gene cluster defined by MCL clustering were used to assign gain and loss dynamics with S. islandicus and in comparison to S. solfataricus. All clusters that mapped to putative transposable elements were excluded from analysis. For these analyses, pseudogene fragments, initially assigned to multiple ORFs, were fused so as not to be counted as multiple events. In addition, the longest representative from each cluster was used as a query for a tBLASTn (61) search against the assembled genome contig. tBLASTn matches of any length with >70% identity that maintained synteny were manually annotated as additional pseudogenes and coded as present for purposes of genome dynamics.
The amino acid sequence of each gene assigned as gain was used in BLASTp (61) queries against the NCBI nonredundant database with an expected score cutoff of 1 × 10−5. Matches were screened such that only hits with 40% or greater identity and 70% or greater coverage were accepted as sources of horizontal gene acquisition. Divergence dates were estimated using Multidivtime v9/25/03 (77). Calibration points represented the onset of geothermal activity for each region and flat priors. Detailed methods and associated references are available in SI Text.
Supplementary Material
ACKNOWLEDGMENTS.
We thank J. Taylor for providing resources for genomic DNA extractions; J. Veysey for developing the methods for bANI analysis; N. Clemons for initial identification of plasmid elements; D. Kirk Nordstrom for discussions of geological dating; G.J. Olsen for discussion on genome analysis methods and tools; and N. Ahlgren, H. Cadillo-Quiroz, I. Cann, K. McMahon, K. Milferstedt, G. Rocap, S. Van Hoy, C. Vanderpool, and S. Wald for helpful comments on the manuscript. Funding for this work was provided by the University of Illinois at Urbana–Champaign and NSF DEB Grant 0816885. S. islandicus genomes were sequenced by the U.S. Department of Energy's Project 0178–051129 under the U.S. Department of Energy's Office of Science Biological and Environmental Research Program and the University of California, Lawrence Livermore National Laboratory (Contract W-7405-Eng-48), Lawrence Berkeley National Laboratory (Contract DE-AC03–76SF00098), and Los Alamos National Laboratory (Contract W-7405-ENG-36).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Genome sequences are available from the Department of Energy Joint Genome Institute, http://www.jgi.doe.gov. The complete genome sequences have been deposited in GenBank (accession nos. CP001399–CP001405). Annotations of genome dynamics reported here are available at www.life.illinois.edu/Sulfolobus_islandicus.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808945106/DCSupplemental.
References
- 1.Lan R, Reeves PR. When does a clone deserve a name? A perspective on bacterial species based on population genetics. Trends Microbiol. 2001;9:419–424. doi: 10.1016/s0966-842x(01)02133-3. [DOI] [PubMed] [Google Scholar]
- 2.Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome.”. Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Coleman ML, et al. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- 5.Chain PSG, et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci USA. 2006;103:15280–15287. doi: 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gressmann H, et al. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 2005;1:e43. doi: 10.1371/journal.pgen.0010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilhelm L, Tripp H, Givan S, Smith D, Giovannoni S. Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data. Biology Direct. 2007;2:27. doi: 10.1186/1745-6150-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–427. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 9.Kettler GC, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lefébure T, Stanhope MJ. Evolution of the core and pan-genome of Streptococcus: Positive selection, recombination, and genome composition. Genome Biol. 2007;8:R71. doi: 10.1186/gb-2007-8-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Retchless AC, Lawrence JG. Temporal fragmentation of speciation in bacteria. Science. 2007;317:1093–1096. doi: 10.1126/science.1144876. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence JG. Gene transfer in bacteria: Speciation without species? Theor Popul Biol. 2002;61:449–460. doi: 10.1006/tpbi.2002.1587. [DOI] [PubMed] [Google Scholar]
- 13.Groisman EA, Ochman H. Pathogenicity islands: Bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 14.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 15.Arnold ML, Sapir Y, Martin NH. Review. Genetic exchange and the origin of adaptations: Prokaryotes to primates. Philos Trans R Soc London B. 2008;363:2813–2820. doi: 10.1098/rstb.2008.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrlich G, Hiller NL, Hu F. What makes pathogens pathogenic. Genome Biol. 2008;9:225. doi: 10.1186/gb-2008-9-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho J-C, Tiedje JM. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl Environ Microbiol. 2000;66:5448–5456. doi: 10.1128/aem.66.12.5448-5456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- 19.Palys T, Berger E, Mitrica I, Nakamura LK, Cohan FM. Protein-coding genes as molecular markers for ecologically distinct populations: The case of two Bacillus species. Int J Syst Evol Microbiol. 2000;50:1021–1028. doi: 10.1099/00207713-50-3-1021. [DOI] [PubMed] [Google Scholar]
- 20.Sikorski J, Nevo E. Adaptation and incipient sympatric speciation of Bacillus simplex under microclimatic contrast at “Evolution Canyons” I and II, Israel. Proc Natl Acad Sci USA. 2005;102:15925–15929. doi: 10.1073/pnas.0507944102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheppard SK, McCarthy ND, Falush D, Maiden MCJ. Convergence of Campylobacter species: Implications for bacterial evolution. Science. 2008;320:237–239. doi: 10.1126/science.1155532. [DOI] [PubMed] [Google Scholar]
- 22.Dykhuizen DE, Green L. Recombination in Escherichia coli and the definition of biological species. J Bacteriol. 1991;173:7257–7268. doi: 10.1128/jb.173.22.7257-7268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramette A, Tiedje JM. Biogeography: An emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb Ecol. 2007;53:197–207. doi: 10.1007/s00248-005-5010-2. [DOI] [PubMed] [Google Scholar]
- 24.Whitaker RJ. Allopatric origins of microbial species. Philos Trans R Soc London B. 2006;361:1975–1984. doi: 10.1098/rstb.2006.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green JL, Bohannan BJM, Whitaker RJ. Microbial biogeography: From taxonomy to traits. Science. 2008;320:1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- 26.Grogan DW, Ozarzak MA, Bernander R. Variation in gene content among geographically diverse Sulfolobus isolates. Environ Microbiol. 2008;10:137–146. doi: 10.1111/j.1462-2920.2007.01439.x. [DOI] [PubMed] [Google Scholar]
- 27.Rocap G, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 28.Yang F, et al. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33:6445–6458. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welch RA, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nubel U, et al. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA. 2008;105:14130–14135. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Passel MWJ, Marri PR, Ochman H. The emergence and fate of horizontally acquired genes in Escherichia coli. PLoS Comput Biol. 2008;4:e1000059. doi: 10.1371/journal.pcbi.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vernikos G, Thomson N, Parkhill J. Genetic flux over time in the Salmonella lineage. Genome Biol. 2007;8:R100. doi: 10.1186/gb-2007-8-6-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makarova K, et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leavis HL, et al. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog. 2007;3:e7. doi: 10.1371/journal.ppat.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holt KE, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;10:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 38.Andersson JO, Andersson SGE. Insights into the evolutionary process of genome degradation. Curr Opin Genet Dev. 1999;9:664–671. doi: 10.1016/s0959-437x(99)00024-6. [DOI] [PubMed] [Google Scholar]
- 39.Ochman H, Moran NA. Genes lost and genes found: Evolution of bacterial pathogenesis and symbiosis. Science. 2001;292:1096–1099. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- 40.Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Ham RCHJ, et al. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci USA. 2003;100:581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moran NA, Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-12-research0054. research0054.0051–0054.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kunin V, Ouzounis CA. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 2003;13:1589–1594. doi: 10.1101/gr.1092603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snel B, Bork P, Huynen MA. Genomes in flux: The evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12:17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- 46.Ochman H, Elwyn S, Moran NA. Calibrating bacterial evolution. Proc Natl Acad Sci USA. 1999;96:12638–12643. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moran NA, Munson MA, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc Lond Ser B. 1993;253:167–171. [Google Scholar]
- 48.She Q, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- 50.Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerdes K, Rasmussen PB, Molin S. Unique type of plasmid maintenance function: Postsegregational killing of plasmid-free cells. Proc Natl Acad Sci USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arcus VL, Rainey PB, Turner SJ. The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 2005;13:360–365. doi: 10.1016/j.tim.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 53.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 55.Lillestøl R, Redder P, Garrett RA, Brügger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Held NL, Whitaker RJ. Viral biogeography revealed by signatures in Sulfolobus islandicus genomes. Environ Microbiol. 2009;11:457–466. doi: 10.1111/j.1462-2920.2008.01784.x. [DOI] [PubMed] [Google Scholar]
- 57.Duggin IG, McCallum SA, Bell SD. Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci USA. 2008;105:16737–16742. doi: 10.1073/pnas.0806414105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lundgren M, et al. Three replication origins in Sulfolobus species: Synchronous initiation of chromosome replication and asynchronous termination. Proc Natl Acad Sci USA. 2004;101:7046–7051. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Daubin V, Ochman H. Bacterial genomes as new gene homes: The genealogy of ORFans in E. coli. Genome Res. 2004;14:1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin Y, Fischer D. On the origin of microbial ORFans: Quantifying the strength of the evidence for viral lateral transfer. BMC Evol Biol. 2006;6:63. doi: 10.1186/1471-2148-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Altschul SF, et al. Gapped BLAST and psi-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simmons SL, et al. Population genomic analysis of strain variation in Leptospirillum group II bacteria involved in acid mine drainage formation. PLoS Biol. 2008;6:e177. doi: 10.1371/journal.pbio.0060177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lawrence JG, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grogan DW, Carver GT, Drake JW. Genetic fidelity under harsh conditions: Analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci USA. 2001;98:7928–7933. doi: 10.1073/pnas.141113098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marri PR, Hao W, Golding GB. Gene gain and gene loss in Streptococcus: Is it driven by habitat? Mol Biol Evol. 2006;23:2379–2391. doi: 10.1093/molbev/msl115. [DOI] [PubMed] [Google Scholar]
- 66.Whitaker RJ, Banfield JF. Population genomics in natural microbial communities. Trends Ecol Evol. 2006;21:508–516. doi: 10.1016/j.tree.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 67.Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:431–440. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- 68.Ward DM, et al. Genomics, environmental genomics and the issue of microbial species. Heredity. 2007;100:207–219. doi: 10.1038/sj.hdy.6801011. [DOI] [PubMed] [Google Scholar]
- 69.Abby S, Daubin V. Comparative genomics and the evolution of prokaryotes. Trends Microbiol. 2007;15:135–141. doi: 10.1016/j.tim.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 70.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiley EO. The evolutionary species concept reconsidered. Syst Zool. 1978;27:17–26. [Google Scholar]
- 72.Whitaker RJ, Grogan DW, Taylor JW. Recombination shapes the natural population structure of the hyperthermophilic archaeon Sulfolobus islandicus. Mol Biol Evol. 2005;22:2354–2361. doi: 10.1093/molbev/msi233. [DOI] [PubMed] [Google Scholar]
- 73.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Notredame C, Higgins DG, Heringa J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 75.Swofford DL. Sunderland, MA: Sinauer Associates; 2003. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods) Version 4. [Google Scholar]
- 76.Brügger K. The Sulfolobus database. Nucleic Acids Res. 2007;35:D413–D415. doi: 10.1093/nar/gkl847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.