Abstract
The striatum, a major component of the brain basal nuclei, is central for planning and executing voluntary movements and undergoes lesions in neurodegenerative disorders such as Huntington disease. To perform highly integrated tasks, the striatum relies on a complex network of communication within and between brain regions with a key role devoted to secreted molecules. To characterize the rat striatum secretome, we combined in vivo microdialysis together with proteomics analysis of trypsin digests and peptidomics studies of native fragments. This versatile approach, carried out using different microdialysis probes and mass spectrometer devices, allowed evidencing with high confidence the expression of 88 proteins and 100 processed peptides. Their secretory pathways were predicted by in silico analysis. Whereas high molecular weight proteins were mainly secreted by the classical mode (94%), low molecular weight proteins equally used classical and non-classical modes (53 and 47%, respectively). In addition, our results suggested alternative secretion mechanisms not predicted by bioinformatics tools. Based on spectrum counting, we performed a relative quantification of secreted proteins and peptides in both basal and neuronal depolarization conditions. This allowed detecting a series of neuropeptide precursors and a 6-fold increase for neurosecretory protein VGF and proenkephalin (PENK) levels. A focused investigation and a long peptide experiment led to the identification of new secreted non-opioid PENK peptides, referred to as PENK 114–133, PENK 239–260, and PENK 143–185. Moreover we showed that injecting synthetic PENK 114–133 and PENK 239–260 into the striatum robustly increased glutamate release in this region. Thus, the combination of microdialysis and versatile proteomics methods shed new light on the secreted protein repertoire and evidenced novel neuropeptide transmitters.
In mammalian brain, the striatum plays a critical role for planning and executing voluntary movements and is also involved in cognitive processes (1). The striatum makes use of a complex network architecture connecting specialized anatomical structures to achieve these highly integrated tasks. It receives projections from primary sensory and motor cortices as well as motor thalamic nuclei and sends projections to downstream basal ganglia structures, thereby influencing the control of cortical and brainstem motor systems (2). In this context, communication within and between brain structures appears as a key element for brain functioning. For cell-to-cell communication, secreted proteins play a pivotal regulatory role. To enter the secretory pathway, it has been long assumed that an N-terminal signal peptide sequence is strictly required. However, recent studies have shown that endoplasmic reticulum- and Golgi-independent or non-classical mechanisms may be responsible for protein secretion (3). The extracellular medium is thus more complex than previously suspected, and its characterization has gained a special interest (4, 5). In silico analyses suggest that mature proteins secreted via classical and non-classical mechanisms share common physicochemical properties (6). In this respect, proteomics is a powerful approach for systematically analyzing proteins present in the extracellular medium (7–9). For neurochemical monitoring of the secretome within the brain, only a few tools provide an appropriate insight into its spatial and temporal dynamics. Microdialysis, in particular, has been shown to be a powerful tool for exploring the extracellular content of the brain in vivo (10–12) and for obtaining vital physiological information that cannot be gleaned from in vitro experiments. The combination of this sampling method with mass spectrometry facilitates investigation of the brain secretome in vivo. However, because of the low concentration of proteins in dialysate, which makes investigations challenging in terms of sensitivity, few studies have combined in vivo brain microdialysis and proteomics/peptidomics analysis (13–16).
In this study, to investigate both proteins and peptides secreted in rat striatum, we performed mass spectrometry analysis of microdialysis fluids. Microdialysis of small and large proteins was carried out using various cutoff probes, and the samples were analyzed through proteomics and peptidomics approaches. In addition, we used spectrum counting (17, 18) to measure the relative abundance of secreted proteins and their processed peptides and to study the modulation of these abundances during neuronal depolarization. This approach allowed us to point out the secretion of new neuropeptides, including neurotransmitters.
EXPERIMENTAL PROCEDURES
Animals
Two-month-old male Wistar rats were studied with procedures strictly in accordance with the recommendations of the European Economic Community (86/609/EEC) for the care and use of laboratory animals.
In Vivo Microdialysis
Samples were collected from 142 rats. Rats were anesthetized with xylazine-ketamine and placed in a stereotaxic frame. Body temperature was kept at 37 °C throughout the experiment with a thermostatically controlled heated blanket. Microdialysis probes (CMA/12, 4-mm active length, 20- or 100-kDa-molecular mass cutoff; CMA Microdialysis, North Chelmsford, MA) were implanted on either side of the brain, in the striatum (coordinates relative to bregma: anteroposterior, +1.0 mm; lateral, ±3.0 mm; dorsoventral, −7.0 mm). Probes were perfused with artificial cerebrospinal fluid (aCSF)1 (145 mm NaCl, 2.68 mm KCl, 1.10 mm MgSO4, 1.22 mm CaCl2 with addition of 3% dextran for 100-kDa probes) at a rate of 1 μl/min for 1 h. Microdialysates were collected on ice over the next hour at a rate of 0.5 μl/min and frozen at −80 °C until MS analysis. We induced neuronal depolarization by increasing the KCl concentration of the dialysis perfusion solution to 145 mm while simultaneously lowering NaCl concentration to keep ionic strength constant. Rats were then killed, and the correct placement of the microdialysis probes was confirmed visually.
Sample Preparation for Mass Spectrometry Analysis
Protein Samples—
For protein analysis, microdialysates were sampled using probes with a 20- or 100-kDa cutoff point. Typically for each analysis, six microdialysate collections from three animals were pooled and digested overnight at 37 °C with 10 μg of trypsin in a 25 mm ammonium bicarbonate solution. Tryptic samples were desalted and concentrated on a C18 ZipTip (Millipore, Bedford, MA) before nano-LC-MS/MS analysis.
Peptide Samples—
For peptide analysis, we used only probes with a 20-kDa cutoff point. For each analysis, six microdialysate collections from three animals were pooled and directly desalted and concentrated on a C18 ZipTip before nano-LC-MS/MS analysis.
Long Peptide Investigation—
This analysis was performed on samples collected in KCl condition using probes with a 20-kDa cutoff point. For each analysis, six microdialysate collections from three animals were pooled, concentrated in a SpeedVac, and directly injected into an LTQ mass spectrometer.
Mass Spectrometry Analysis
For Q-TOF and Orbitrap analysis, the dried peptide extracts were resuspended in 5% ACN and 1% trifluoroacetic acid in water and transferred into vials suitable for nano-LC-MS/MS analysis.
Q-TOF—
Analyses were performed on a nano-LC system coupled to a Q-TOF Ultima mass spectrometer (Waters, Milford, MA). During the chromatography step, peptides were concentrated on a 300-μm × 5-mm PepMap C18 precolumn (LC Packings, Sunnyvale, CA) and separated on a C18 column (75 μm × 150 mm) (LC Packings). Mobile phases consisted of 0.1% formic acid, 97.9% water, 2% ACN (v/v/v) (A) and 0.08% formic acid and 20% water in 79.92% ACN (v/v/v) (B). The nanoflow rate was set to 250 nl/min, and the gradient profile was as follows: from 10 to 40% B in 65 min, from 40 to 90% B in 10 min, constant 90% B for 10 min, and return to 10% B. The system was calibrated every week with [Glu]fibrinopeptide B, and mass shift precision was below 0.4 Da. The mass spectrometer operated in the positive ion electrospray ionization mode with a resolution of 9000–11,000 full-width half-maximum. Data-dependent analysis was used for MS/MS (three most abundant ions in each cycle): 1-s MS (m/z 400–1600) and maximum 5-s MS/MS (m/z 50–1850, continuum mode) with 2-min dynamic exclusion. MS/MS raw data were processed with MassLynx 4.0 software (smooth 3/2 Savitzky Golay) (Waters).
Orbitrap—
Orbitrap analyses were performed on a nano-LC system (Ultimate 3000, Dionex) coupled to an Orbitrap mass spectrometer (ThermoFischer Scientific). During the chromatography step, peptides were concentrated on a 300-μm × 5-mm PepMap C18 precolumn and separated onto a 75-μm × 150-mm C18 column (Gemini C18 phase, Phenomenex, for in-house built columns). Mobile phases consisted of 0.1% formic acid, 97.9% water, 2% ACN (v/v/v) (A) and 0.08% formic acid, 20% water in 79.92% ACN (v/v/v) (B). The nanoflow rate was set at 300 nl/min, and the gradient profile was as follows: constant 4% B for 3 min, from 4 to 55% B in 42 min, from 55 to 90% B in 1 min, constant 90% B for 14 min, and return to 10% B. The system was calibrated weekly with a mixture of caffeine, MRFA peptide, and Ultramark (Sigma), and mass precision was better than 5 ppm. The mass spectrometer operated in the positive ion electrospray ionization mode with a resolution of 30,000 full-width half-maximum. MS and MS/MS data were acquired and processed automatically with Xcalibur 2.0.7 (Thermo Fischer Scientific) and Mascot Distiller 2.1.1.0 (smooth 3/2 Savitzky Golay) (Matrix Science, Boston, MA) softwares.
LTQ—
We investigated long peptides by analyzing samples in an ion trap mass spectrometer (LTQ, Thermo Electron, Bremen, Germany). The instrument was coupled to an Ultimate 3000 (LC Packings) high performance liquid chromatograph. In the chromatography step, microdialysate samples were loaded directly onto an LC Packings monolithic precolumn (polystyrene-divinylbenzene, 200-μm inner diameter, 5 mm long), desalted by running mobile phase A (see below) through the column for 4 min, and separated on an LC Packings monolithic nanocolumn (PepSwift, polystyrene-divinylbenzene, 100-μm inner diameter, 5 cm long). Mobile phases consisted of 0.1% formic acid, 97.9% water, 2% ACN (v/v/v) (A) and 0.08% formic acid and 20% water in 79.92% ACN (v/v/v) (B). The nanoflow rate was set at 300 nl/min, and the gradient profile was as follows: from 0 to 50% B over 120 min, from 60 to 90% B over 10 min, constant 90% B for 10 min, and return to 0% B over 1 min. Mass spectrometry data were acquired in a three-step scanning sequence comprising the full MS scan (450–2000 m/z), the MS data-dependent zoom scan of the most abundant ion, and its MS/MS fragmentation (CID at normalized collision energy of 35%). The equipment was calibrated immediately before analysis by the direct injection of a 20 μm ubiquitin solution (Sigma) via an automated tuning procedure, selecting the 1224 m/z (7+) peak. MS and MS/MS data were acquired and processed automatically with Xcalibur 2.0.7 and Mascot Distiller 2.1.1.0 (smooth 3/2 Savitzky Golay) softwares.
Peptide Sequencing and Protein Precursor Identification
Database searching was performed using the Mascot 2.2.03 program (Matrix Science). Two databases were used: a homemade list of well known contaminants (keratins and trypsin; 21 entries) and an updated compilation of the Swiss-Prot and TrEMBL databases (release 54.8_37.8, 5,678,599 entries in total including 15,495 entries for rat) with Rattus as the selected species. The variable modifications allowed were as follows: N-terminal acetylation, methionine oxidation, and dioxidation, C-terminal amidation, and N-terminal pyroglutamate. For protein samples, “semitrypsin” was selected as enzyme, and two miscleavages were also allowed. For peptidomics studies, “no enzyme” was selected. For Q-TOF analysis, mass accuracy was set to 0.4 Da for both precursor ion (MS mode) and fragments (MS/MS mode). For Orbitrap analysis, these parameters were set to 20 ppm and 1 Da, and for the LTQ they were set to 2 and 0.8 Da, respectively. Mascot data were then transferred to an in-house developed validation software for data filtering according to a significance threshold of <0.05 and the elimination of protein redundancy on the basis of proteins being evidenced by the same set or a subset of peptides. This software is freely available from the authors. Each peptide sequence was checked manually to confirm or contradict the Mascot assignment. Sequences corresponding to irrelevant identifications were discarded. Proteins detected through a single peptide were only considered if this peptide was observed in at least three biological samples, eventually undergoing analytical replicates.
Spectrum Counting
Spectrum counting (SC) was based on Orbitrap analyses with biological and analytical replicates. The spectrum counting is the total number of assigned spectra (redundant plus non-redundant) associated to a given protein. To estimate relative abundance of a given protein, a single value was calculated from individual spectrum counting summed from different analyses, and the resulting spectrum count was normalized by the protein molecular weight (17, 19). To compare protein abundance between basal and KCl samples, spectrum counting was normalized by total spectrum counting of assigned spectra for each analysis and averaged over replicates (20, 21). Peptides shared by multiple proteins were attributed to a single precursor, corresponding to the protein with the best Mascot score. The significance of differences between sets of conditions was assessed by Mann and Whitney test. Differences were considered significant for p values <0.05.
Proteomics Data Mining
Validated hits for each analysis were transferred to a relational database. This database made it possible to carry out specific queries concerning, for example, the number of spectra and/or unique peptides assigned to a given protein, best Mascot score, and best coverage. For each protein of the database, molecular weight was retrieved from UniProt. The SignalP (22) and SecretomeP programs (6) (Center for Biological Sequence Analysis) were used to predict classical and non-classical secretion modes, respectively. For non-secreted precursors, subcellular distribution was retrieved from UniProt and the Ingenuity Pathway Analysis tool (Ingenuity Systems). Associated molecular functions were retrieved from DAVID.
Biological Activity
The non-opioid peptides evidenced in peptidomics experiments, PENK 239–260 and PENK 114–133, were synthesized by Millegen (France). For assessments of biological activity, synthetic PENK 239–260 and PENK 114–133 were delivered into the striatum via microdialysis probes implanted at the coordinates described above. After an equilibration period of 90 min, dialysates were collected at 10-min intervals over a period of 210 min. The first six fractions were collected for basal value determination. The next six fractions were then collected during PENK 239–260 or PENK 114–133 perfusion (10−4 m at 1 μl/min). Nine additional fractions were collected to estimate poststimulation effects over 90 min. Dialysates were collected on ice and stored at −80 °C until analysis. DOPAC, dopamine, HVA, glutamate, and GABA concentrations in dialysates were determined by HPLC with laser-induced fluorescence detection as described previously (23). Amino acid peaks were identified on the basis of retention time, and concentrations were estimated by dividing the peak areas of each amino acid by the value for the corresponding external standard (analytical software class LC10; Shimadzu). Significant differences between conditions were tested using analysis of variance and Fisher's post hoc least significant difference test. Results were deemed significant for a p value <0.05.
RESULTS
Microdialysis Fluid Collection and Analysis—
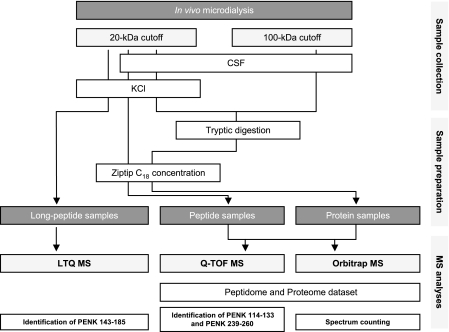
In this study, we combined proteomics and peptidomics analyses of striatal microdialysates to analyze both proteins and endogenous peptides processed from peptide precursors. The overall experimental strategy is summarized in Fig. 1. Samples were collected using probes with a 20- or 100-kDa cutoff point in aCSF or KCl conditions. They underwent trypsin digestion (protein samples, corresponding to what is referred to as proteome analysis) or not (peptide samples, i.e. peptidome analysis) before being concentrated on a ZipTip. Then MS analysis was performed through Q-TOF or Orbitrap MS to characterize the proteome and peptidome (39 and 24 analyses, respectively). Relative quantification (spectrum counting) was performed by Orbitrap MS. For studies devoted to long peptide characterization, microdialysate samples were directly analyzed by LTQ MS.
Fig. 1.
Experimental strategy. Illustration of the various collection and preparative steps for MS analysis of the striatal microdialysate. CSF, cerebrospinal fluid.
Timing of microdialysis sample collection was validated by relative quantification (spectrum counting with normalization based on molecular weight; see below) of blood-specific highly abundant proteins (24) and intracellular proteins (25). Our results show important levels of globin and intracellular proteins only just after probe implantation. These levels subsequently decreased dramatically (p < 0.01) within 1 h (data not shown). Moreover the lack of catalase and carbonic anhydrase in our microdialysate samples suggests that blood contamination is less than 0.005%, which is consistent with previous findings (24).
Striatum Microdialysate Proteome—
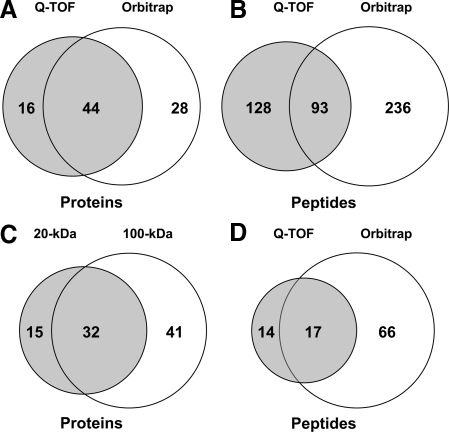
The whole protein data set, i.e. proteins whose expression was evidenced from tryptic fragments and endogenous peptides, is shown in supplemental Table 1. Comparison of the different collection and analytical conditions shows that 60 of the 88 proteins were detected by Q-TOF, and 72 were detected by Orbitrap (Fig. 2A). Only 42% of the peptides evidenced by Q-TOF were detected by Orbitrap (Fig. 2B). Nevertheless 73% of the proteins detected using data from the Q-TOF analyses were confirmed using data from Orbitrap analyses. Indeed despite the high number of MS/MS scans performed by Orbitrap, a certain proportion was not assigned by Mascot because of their low quality. These findings highlight the advantage of combining two MS instruments for deeper insights into proteomes. Elsewhere we detected 47 proteins with the 20-kDa microdialysis probe and 73 with the 100-kDa probe, including 32 common entries (Fig. 2C). Three protein precursors, histone H1.2, excitatory amino acid 2, and brain acid-soluble protein 1, were only detected in our peptidome data set (from one, three, and one unique peptides, respectively). This likely stems the fact that low molecular weight tryptic peptides generated from corresponding endogenous peptides were not resolved by MS (only MH2+ and MH3+ ion peptides with m/z >400 were scanned).
Fig. 2.
Venn diagrams of striatum microdialysate data. The number of proteins (A) and peptides (B) detected by Q-TOF (dark) and Orbitrap (light) analysis is shown. Data obtained with 20- and 100-kDa-cutoff probes were combined for this analysis. C, number of proteins detected in microdialysates sampled with 20-kDa (dark) or 100-kDa (light)-cutoff probes. D, number of endogenous peptides detected in peptidomics analysis by Q-TOF (dark) and Orbitrap (light) analysis of samples collected with 20-kDa-cutoff probes.
To classify the 88 proteins evidenced, we compiled (i) information related to the proteomics data (Mascot score, coverage, and number of unique peptides), (ii) results of secretion mode predictions by bioinformatics tools (SignalP and SecretomeP), and (iii) information (localization and molecular function) retrieved from different protein databases (ExPASy, Ingenuity, and DAVID). Proteins detected in striatal microdialysates could be assigned to 15 major protein categories in terms of molecular function. These include 18 structural molecule proteins, 15 protease inhibitors, nine transfer/carrier proteins, nine neuropeptide hormone proteins, 10 protein-binding proteins, eight calcium-binding proteins, four extracellular matrix proteins, four glycolysis enzymes, three oxygen transporter proteins, three transporters, one DNA-binding protein, one receptor protein, one kinase, and one antioxidant protein. Only one protein (Q5FVI0) was not ascribed a known molecular function. As expected, the nine neuropeptide hormones correspond to nine neuropeptide precursors: proenkephalin (PENK), cholecystokinin (CCKN), secretogranins I and II (SCGI and -II), neurogranin (NEUG), pro-SAAS, neurosecretory protein VGF (VGF), somatostatin, and neuropeptide Y. Furthermore SignalP and SecretomeP analyses predicted 35 proteins to be secreted by classical secretory mechanisms (presence of a signal peptide), 19 were predicted to be secreted by non-classical secretion, and 34 were referred to as “non-secreted.” Information compiled from databases showed that 54 proteins (61.4%) had been reported to be “secreted,” 24 were reported to be located in the “cytoplasm,” nine were reported to be located in the “membrane,” and surprisingly one was reported to be present in the “nucleus” (N-terminal histone H1.2 peptide, detected in peptidomics analysis).
Striatum Microdialysate Peptidome—
From the whole set of peptidomics analysis (including aCSF and KCl sampling and the long peptide experiment), we recorded 100 endogenous peptides: 31 were detected by Q-TOF, 83 were detected by Orbitrap, and three were detected by LTQ with 17% common entries between Q-TOF and Orbitrap (Fig. 2D). The complete peptide data set is available in supplemental Table 2. The 100 peptides originate from 29 protein precursors. Seventy peptides are predicted to be processed from precursors containing a signal peptide sequence and should therefore be able to enter the classical secretory pathway. For neuropeptide precursors, we detected 19 peptides originating from PENK, 10 from NEUG, four from VGF, and one peptide each from the pro-SAAS, SCGI and -II, neuropeptide Y, somatostatin, and CCKN precursors. Several PENK and NEUG peptides correspond to N-terminal or C-terminal truncated peptides processed at dibasic sites. Such peptides are probably produced by endopeptidase cleavage at specific residues followed by carboxy- and aminopeptidase digestion.
Our analysis of possible posttranslational modifications pointed to two peptides (from VGF and CCKN precursors) containing an N-terminal pyroglutamate residue. In addition, four N-terminal peptides, originating from excitatory amino acid transporter 2, thymosin β-4, thymosin β-10, and histone H1.2 precursors, displayed N-terminal acetylation. The N-terminal H1.2 peptide matches with salmon antimicrobial peptide H1, an N-terminal acetylated histone H1-derived peptide previously reported to be secreted in Atlantic salmon skin mucus (26). Thus, this analysis evidences the existence of a wide variety of peptides secreted by the brain and of fragments not previously found in the brain, demonstrating the relevance of our working methods.
Relative Abundance of Proteins and Peptides—
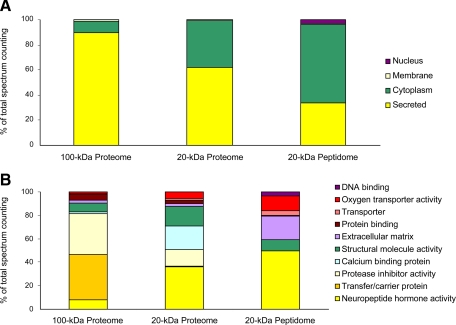
To measure the relative abundance of protein and peptides, we used Orbitrap data to perform spectrum counting and normalized results by molecular weight as described (17, 19). The normalization procedure makes it possible to compare the abundance of proteins of various sizes. Table I shows the most abundant proteins (relative abundance >4%) in various collection and analytical conditions. Albumin, which accounts for about 20% of the total amount of protein present, and NEUG were detected as the major products with the 100- and 20-kDa probes, respectively. Certain high molecular weight transfer/carrier proteins and protease inhibitors were found among the most abundant proteins detected with the 100-kDa probe. Lower molecular weight proteins, such as the protease inhibitor cystatin-C and neuropeptide hormone PENK, also appeared to be abundant in the proteomics analyses carried out with the 20- and 100-kDa probes. Two calcium-binding proteins (Purkinje cell protein 4 and FK506-binding protein 1A) were particularly abundant in analyses performed with the 20-kDa probe. In the peptidomics data, extracellular matrix fibrinogen α and γ were also evidenced as abundant protein precursors. In addition, spectrum counting was calculated for each classification including “molecular function,” “cellular localization,” and “secretory pathway.” Fig. 3 shows relative quantification for different aCSF collection and analytical conditions. The precursors described as “secreted” accounted for 90, 62, and 34% of the 100-kDa proteome, the 20-kDa proteome, and the 20-kDa peptidome, respectively (Fig. 3A). The abundance of neurogranin, classified as cytoplasmic, accounts for the difference in “cellular localization” profile between 20- and 100-kDa samples. SignalP and SecretomeP predictions showed that, whereas most of the proteins of the 100-kDa proteome were secreted by the “classical secretory pathway” (94%), the two secretion modes were equally used (53 and 47%) by the proteins of the 20-kDa proteome. In the 100-kDa proteome, the most abundant molecular functions were “transfer/carrier proteins” (39%) and “protease inhibitor activity” (35%), whereas in the 20-kDa proteome, the most abundant molecular functions were “neuropeptide hormone activity” (36%) and “calcium-binding protein” (20%). The neuropeptide hormone activity function was also the most frequently recorded one in the peptidome (50%) followed by “extracellular matrix” (20%) (Fig. 3B).
Table I.
Abundant proteins evidenced in striatal microdialysate
The table lists proteins for which abundance, as determined by spectrum counting, exceeds 4% of the total amount of protein detected. Microdialysis was performed under basal condition (aCSF perfusion). Proteome analysis was carried out using probes with a 100-kDa (n = 11) or 20-kDa (n = 6) cutoff point. For peptidome analysis (n = 6), we used 20-kDa-cutoff probes. Relative abundance of proteins was deduced from spectrum counting data normalized by molecular mass.
| Description | UniProt ID | Molecular mass | Abundance | Secretory pathway | Molecular function |
|---|---|---|---|---|---|
| Da | % | ||||
| 100-kDa proteome | |||||
| Serum albumin precursor | ALBU_RAT | 68,686 | 20.1 | Classical secretion | Transfer/carrier protein |
| Cystatin-C precursor | CYTC_RAT | 15,427 | 11.0 | Classical secretion | Protease inhibitor activity |
| Apolipoprotein E precursor | APOE_RAT | 35,731 | 8.1 | Classical secretion | Transfer/carrier protein |
| Thymosin β-4 | TYB4_RAT | 5,050 | 6.6 | Non-classical secretion | Structural molecule activity |
| α1-Antiproteinase precursor | A1AT_RAT | 46,107 | 7.0 | Classical secretion | Protease inhibitor activity |
| α1-Inhibitor 3 precursor | A1I3_RAT | 163,670 | 4.9 | Classical secretion | Protease inhibitor activity |
| Proenkephalin A precursor | PENK_RAT | 30,912 | 4.5 | Classical secretion | Neuropeptide hormone activity |
| 20-kDa proteome | |||||
| Neurogranin | NEUG_RAT | 7,492 | 19.8 | Other | Neuropeptide hormone activity |
| Thymosin β-4 | TYB4_RAT | 5,050 | 15.8 | Non-classical secretion | Structural molecule activity |
| Proenkephalin A precursor | PENK_RAT | 30,912 | 13.6 | Classical secretion | Neuropeptide hormone activity |
| Cystatin-C precursor | CYTC_RAT | 15,427 | 13.3 | Classical secretion | Protease inhibitor activity |
| Purkinje cell protein 4 | PCP4_RAT | 107,22 | 9.5 | Non-classical secretion | Calcium-binding protein |
| FK506-binding protein 1A | FKB1A_RAT | 11,915 | 9.5 | Other | Calcium-binding protein |
| β1 globin | Q9QUT6_9MURI | 15,824 | 4.3 | Other | Oxygen transporter activity |
| Peptidome | |||||
| Neurogranin | NEUG_RAT | 7,492 | 42.8 | Other | Neuropeptide hormone activity |
| Fibrinogen α chain precursor | FIBA_RAT | 86,632 | 13.0 | Classical secretion | Extracellular matrix |
| β1 globin | Q9QUT6_9MURI | 15,824 | 12.9 | Other | Oxygen transporter activity |
| Fibrinogen γ chain precursor | FIBG_RAT | 50,600 | 5.7 | Classical secretion | Extracellular matrix |
Fig. 3.
SC of microdialysate sampled with 20- or 100-kDa cutoff points. SC was obtained from proteome and peptidome data. Proteome, samples collected with either 100- or 20-kDa-cutoff membranes and undergoing trypsin digestion before MS analysis. Peptidome, samples collected with 20-kDa-cutoff membranes and submitted to MS analysis without trypsin digestion. For the 100-kDa proteome, six biological samples were analyzed. For the 20-kDa proteome and peptidome, three biological samples were studied. SC was grouped from cellular localization (A) and molecular function (B).
Influence of Depolarization on Protein and Peptide Abundance—
We induced neuronal depolarization using microdialysis probes with a 20-kDa cutoff point perfused with 145 mm KCl. To allow comparisons between samples, spectrum counting was normalized by number of assigned spectra (20, 21). No significant differences were observed for proteomics data, even for neuropeptide precursors. This may be due to the tryptic peptides generated from these very short neuropeptides not being resolved by MS because of their low molecular weight. We therefore focused our analysis on peptidomics data. The total number of spectra assigned through peptidomics analysis differed significantly between basal (aCSF) and KCl conditions (31 ± 11 versus 53 ± 3.6, p < 0.05; n = 6 replicates from three biological samples). This difference mainly rests on an increase for proteins (peptides) of the classical secretory pathway (23 ± 8.5 versus 40 ± 1.5, p < 0.05; n = 6).
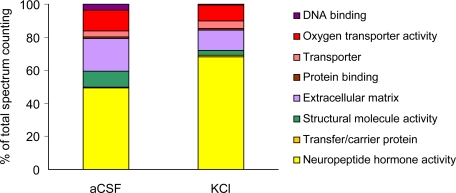
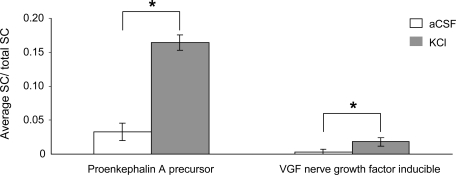
Spectrum counting showed a higher proportion of neuropeptide hormone activity in the KCl than in the aCSF perfusion condition (68.9 versus 49.7% of total spectrum counting) as indicated in Fig. 4. Indeed neuronal depolarization led to the detection of a series of proteins (CCKN, pro-SAAS, SCGI, and SCGII) corresponding to neuropeptide precursors absent in basal conditions. In addition, PENK and VGF, albeit present in the basal condition, displayed a robust increase in the presence of KCl (Fig. 5).
Fig. 4.
Molecular function of endogenous peptides. Spectrum counting was carried out on samples collected in aCSF or KCl condition using 20-kDa-cutoff probes.
Fig. 5.
Spectrum counting of neuropeptide precursors. Samples were collected in aCSF or KCl condition using 20-kDa-cutoff probes. SC was normalized by total SC and averaged from six analyses. Each error bar corresponds to the standard error of the mean. *, p < 0.05 by Mann and Whitney test.
Identification of New PENK Neuropeptides—
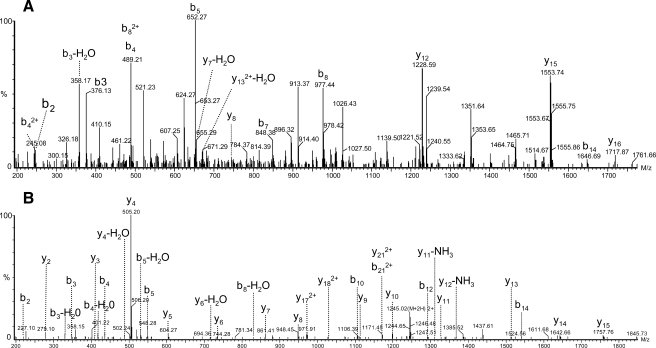
In KCl-infused animals, our peptidome data set revealed the presence of 16 non-opioid peptides originating from PENK (supplemental Table 2), accounting for 18.9% of precursor sequence coverage. Based on this data set, we were able to evidence three subsets of PENK peptides (supplemental Fig. 1). The first subset corresponded to the sequence SPQLEDEAKE (PENK 198–207), which was previously shown to be a striatal processed and secreted peptide (15). Given the general rules governing prohormone processing (cleavage at dibasic sites or before single Arg residues) (27–29), we hypothesized that peptides clustered in the two other subsets corresponded to truncated forms of the non-opioid peptides MDELYPVEPEEEANGGEILA (PENK 114–133) and FAESLPSDEEGESYSKEVPEME (PENK 239–260). For characterizing these two putative peptides, we performed MS/MS focused on doubly charged and triply charged peptides corresponding to MH+ 2205 (PENK 114–133) and 2489 (PENK 239–260). LC-MS/MS analysis performed in the KCl condition allowed detecting PENK 114–133 and PENK 239–260 with Mascot scores of 23 and 42, respectively (Fig. 6). The comparison of MS/MS spectra obtained from synthetic and endogenous peptides definitively confirmed the amino acid sequence (supplemental Fig. 2).
Fig. 6.
Identification of new PENK neuropeptides. Q-TOF MS/MS spectra of MH2+ MDELYPVEPEEEANGGEILA (PENK 114–133) (A) and MH2+ FAESLPSDEEGESYSKEVPEME (PENK 239–260) (B) from microdialysate sampled in KCl condition are shown.
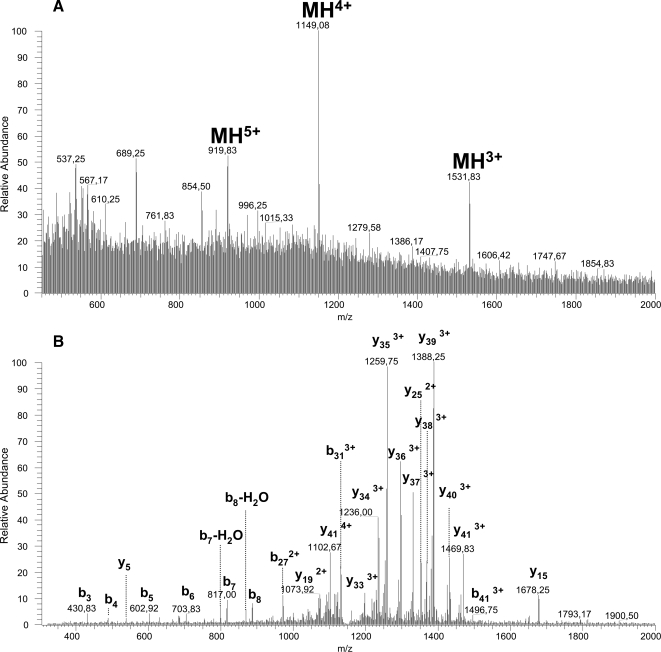
From proteome data, an additional PENK peptide (DADEGDTLANSSDLLK) was detected in trypsin digests. This peptide putatively corresponds to the N-terminal part of the processed non-opioid peptide PENK 143–185. To evidence this 43-amino acid-long peptide, we analyzed samples collected in the KCl condition by LTQ MS. From MS and MS/MS spectra, we focused on multicharged peptides corresponding to MH+ 4593 (PENK 143–185). MH3+, MH4+, and MH5+ ions were detected at m/z 1531.7 (Δ+0.03), 1149.08 (Δ+0.07) and 919.8 (Δ+0.4), respectively (Fig. 7A). Because the MH3+ peptide ion was not fragmented and because Mascot is unable to resolve the MH4+ and MH5+ peptide ion MS/MS spectra, identification was based on the theoretical spectrum generated by MS products (ProteinProspector). Fragment annotation confirmed the peptide sequence DADEGDTLANSSDLLKELLGTGDNRAKDSHQQESTNNDEDSTS and thus the occurrence of processed PENK 143–185 in striatal microdialysates (Fig. 7B). Long peptide experiments also pointed to two additional peptides originating from the VGF precursor. These peptides were evidenced from MH3+ 1157.9 (GGGEDEVGEEDEEAAEAEAEAEEAERARQNALL) and MH4+ 1042.1 (APPGRSDVYPPPLGSEHNGQVAEDAVSRPKDDSVPEVRAA) with Mascot scores of 117 and 72, respectively.
Fig. 7.
Long peptide analysis of a new PENK neuropeptide. MS (A) and MS/MS (B) spectra of the peptide DADEGDTLANSSDLLKELLGTGDNRAKDSHQQESTNNDEDSTS (PENK 143–185) detected in microdialysate sampled in KCl condition are shown.
Biological Activity—
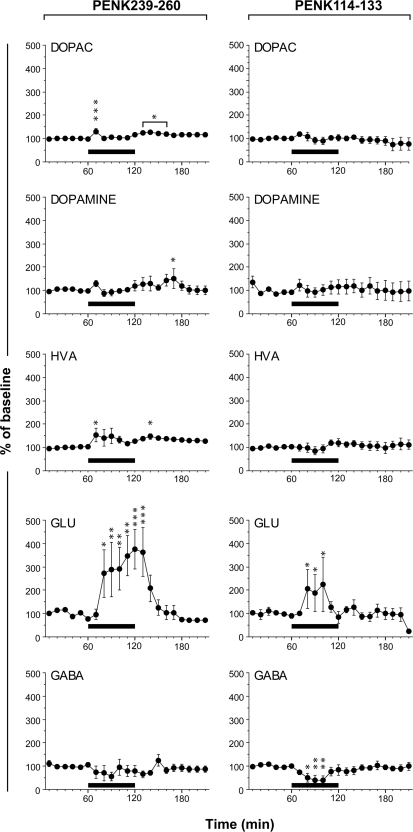
Haskins et al. (15) previously demonstrated the neuroactivity of new PENK peptides detected by MS analysis of the rat striatum microdialysate. Here we screened two new non-opioid peptides derived from PENK (PENK 239–260 and PENK 114–133) for neurochemical activity by infusing the striatum with the synthetic peptides by microdialysis and monitoring the amino acids present in the dialysate. The samples collected were analyzed by HPLC with laser-induced fluorescence detection, making it possible to monitor six amino acids at 10-min intervals (Fig. 8). We found that the release of glutamate in the striatum was rapidly increased by PENK 239–260 and PENK 114–133. Notably PENK 239–260 elicited a highly significant 3-fold increase of glutamate concentration that persisted throughout the duration of peptide infusion. The PENK 114–133 effect on glutamate concentration was much smaller, but this peptide also was shown to decrease GABA concentration. DOPAC, dopamine, and HVA concentrations scattered around basal levels irrespectively of the presence of PENK 239–260 and PENK 114–133.
Fig. 8.
Effect of peptides infusion on amino acid concentrations in the striatum. Changes, recorded at 10-min intervals, in DOPAC, dopamine, HVA, glutamate (GLU), and GABA concentrations during the delivery (—) of 10−4 m peptides PENK 239–260 or PENK 114–133 through the dialysis probe. Results are expressed as percentage of base line. Each error bar corresponds to the standard error of the mean. Asterisks indicate statistically significant changes (*, p < 0.05; **, p < 0.01; ***, p < 0.005) in analysis of variance and Fisher's post hoc least significant difference test for comparisons with the base line. Each point is the mean value obtained from three to four animals.
DISCUSSION
Here we present a thorough characterization of the rat striatal microdialysate using a parallel peptidome and proteome profiling drawn with different MS instruments and microdialysis probes. The data set consists of 88 proteins and 100 endogenous peptides derived from 29 protein precursors. Our assay was flexible enough to detect high molecular weight molecules as well as native peptides that were shown to be biologically active. Bioinformatics tools (SignalP and SecretomeP) and evaluation of brain damage (based on cytoplasmic protein levels) provide strong evidence to suggest that we sampled secreted molecules. Moreover most of the peptides and proteins whose expression was recorded in this study are brain-specific, and several of them (PENK, Purkinje cell protein 4 (PCP4), and RGD1307215 protein) are encoded by genes highly expressed in the striatum (30). In line with previous studies of cerebrospinal fluid or brain secretome (7, 8, 14), we detected proteins (serine protease inhibitors, fibrinogen β, enolase, and complement C3) up-regulated in traumatic brain injury (31) and plasma proteins such as apolipoprotein A-I and α1-antiproteinase. However, kinetics analysis demonstrated that blood contamination and cell damage occurred mostly just after probe implantation and are therefore minimal using our experimental setup (sample collection after 1 h).
Repertoire of Proteins Secreted in the Striatum—
The proteome repertoire characterized in this study consists of 88 proteins. Previous in vitro (8, 9, 32) and in vivo (14, 15) studies of proteins secreted by brain cells listed five to 43 proteins. The larger number of proteins detected in our study highlights the relevance of our approach. Moreover to deeply explore our data set and obtain relative quantification of dialysate proteins, we performed spectrum counting analysis of samples collected using 100- and 20-kDa-cutoff probes. For high molecular weight samples, 90% of the protein population is described as secreted. The classical secretion pathway, which successively transports proteins from endoplasmic reticulum to the Golgi apparatus and finally into secretory vesicles, appears to be the major route (94%). For 20-kDa samples, 62% of the proteins were classified as secreted with a similar proportion being routed by the classical and non-classical secretory pathways. For such samples, the relatively large fraction of proteins referred to as “cytoplasmic” may reflect alternative secretion mechanisms. Indeed metabolic enzymes, cytoskeletal proteins, and chaperones (33) have been shown to be secreted by the endocytic pathway (34, 35).
Concerning molecular functions, in 100-kDa samples the abundance of proteins of the transfer/carrier protein group was high. The level of albumin, a protein recently shown to be secreted by microglial cells (36), largely accounts for the high representation of this function. In addition, several other carrier proteins such as apolipoprotein E, acyl-CoA-binding protein, phosphatidylethanolamine-binding protein A, and ceruloplasmin are also detected as abundant products in dialysates. These proteins are secreted by astrocytes (8). Apolipoprotein E and acyl-CoA-binding protein play an important role in lipid distribution and metabolism (37, 38), and ceruloplasmin may contribute to heme and iron sequestration in the central nervous system (39). The presence of these proteins in microdialysates suggests that the striatal secretome supplies a variety of carrier proteins that are essential for the homeostasis of the central nervous system microenvironment.
In 20-kDa proteome samples, calcium-binding protein is an abundant molecular function. The efficiency of synaptic transmission is critically dependent on cytosolic [Ca2+], and calcium signaling is crucial for several aspects of plasticity at glutamatergic synapses. SecretomeP predicts a “non-classical” secretion pathway for the most abundant calcium-binding protein, PCP4. PCP4 has a conserved IQ motif, which may serve as a binding site for various EF-hand proteins, including calmodulin (CaM) and CaM-like proteins. CaM kinase II δ chain and A-kinase anchor protein 5, both present in microdialysates, also bind calmodulin. A-kinase anchor protein 5 has been shown to be located within the postsynaptic membrane (40), and CaM kinase II δ chain is a calcium-dependent protein kinase that has been implicated in various aspects of synaptic plasticity. Bioinformatics tools predicted the secretion of these proteins through non-classical mechanisms. We hypothesize that detection of a number of calcium- and calmodulin-binding proteins in striatal secretome may highlight neuronal activity and synaptic plasticity. Furthermore these proteins may be involved in the regulation of extracellular [Ca2+].
The “structural molecule activity” function is evidenced in both 20- and 100-kDa samples. Several proteins with such a function (actin, tubulin, neurofilament proteins, α-internexin, profilin, and MAP-1) are potentially associated with the synaptic vesicle membrane (41). Their occurrence, together with that of proteins of the protease inhibitor activity group, points to the control of neuronal architecture. Indeed protease inhibitor activity proteins contribute to the pattern of neuronal connections associated with long lasting forms of synaptic plasticity (42).
Repertoire of Striatal Neuropeptides—
As expected, in the peptidome data set, the most abundant endogenous peptides belong to the neuropeptide hormone activity category. Neuropeptides are required for cell-to-cell communication in neurotransmission and for the regulation of endocrine functions. They are processed from protein precursors by a proteolytic mechanism, packaged into large dense core vesicles, and released to bind receptors on the postsynaptic target neuron. Our peptidome data set shows that these peptides are all processed at dibasic sites or before single Arg residues as previously reported for polypeptides detected in cerebrospinal fluid (43). In our peptidome, we evidenced new processed and secreted neuropeptides: VGF peptide 24–63, SCGI peptide 304–321; and PENK peptides 114–133, 143–185, and 239–260. Moreover the processing of the VGF peptide 375–420 is suggested both by peptidomics (cleavage at the dibasic 373–374 site) and proteomics data (cleavage at the dibasic 421–422 site). Thus, our data add new candidates to prior studies of the rat neuropeptidome (44–46).
We also detected a prohormone-like processed peptide (GPGPGGPGGAGGARGGAGGGPSGD), referred to as NEUG 55–78. To our knowledge, NEUG processing has never been reported. This protein has been described as cytoplasmic, and its secretion is not predicted by bioinformatics tools. Although the presence of cytoplasmic peptides could result from normal degradation by exosomes, peptidomics data suggest a different process for NEUG peptides. NEUG displays an IQ motif involved in calmodulin binding. However, the processed peptide present in microdialysates does not contain this motif. Furthermore the protein displays several dibasic sites, a feature present in neuropeptide precursors, and the C-terminal NEUG peptide 55–78 is specifically processed at one such dibasic site (53–54 site). This prohormone-like processing and the high abundance of NEUG peptides in microdialysates are highly consistent with striatal secretion and suggest a new bioactivity for NEUG. The absence of a signal peptide in NEUG suggests that this peptide is secreted via a non-classical secretory mechanism.
Identification of New Biologically Active Neuropeptides—
As expected, the three non-opioid PENK peptides (PENK 114–133, PENK 143–185, and PENK 239–260) described here are encompassed by dibasic sites, consistent with their production via a prohormone convertase activity. One characteristic feature of these peptides is a very high content of acidic residues, a hallmark for secreted peptides (43, 47, 48). The data obtained in the experiments with KCl, along with previous data (15), definitively demonstrate that all PENK fragments are co-secreted in the striatum. PENK was initially characterized as a precursor of the neurotransmitter opioid peptides Met-enkephalin, Leu-enkephalin, heptapeptide, and octapeptide. There is experimental evidence showing that opioid peptides decrease the excitatory postsynaptic potential (49, 50). Conversely recent results have suggested that the non-opioid PENK peptide 198–209 increases the release of glutamate and aspartate and that PENK 219–229 increases the release of glutamate and GABA (15). We demonstrate here that PENK 239–260 and, to a lesser extent, PENK 114–133 strongly increase the release of glutamate and that PENK 114–133 decreases the release of GABA. As the endopeptidase process ensures the release of all PENK peptides into the extracellular space, it is worth emphasizing that opioid and non-opioid peptides originating from PENK exert opposite effects on neurotransmitters.
In conclusion, the versatile proteomics approach described here enables in-depth characterization of the rat striatal secretome. Furthermore we demonstrate that a combination of microdialysis sampling and peptidomics analysis can be used to identify new biologically active neuropeptides and to explore temporally and spatially their release process in specific brain regions.
Supplementary Material
Acknowledgments
We thank Y. Vandenbrouck and A. Martel for providing bioinformatic tools, M. Ferro for helpful discussions, E. Diguet for the initial screening of PENK 239–260 neuroactivity, M. Bellucci for precious artwork, and P. Legrain for initiating discussions during an early phase of this project.
Footnotes
Published, MCP Papers in Press, January 21, 2009, DOI 10.1074/mcp.M800501-MCP200
The abbreviations used are: aCSF, artificial cerebrospinal fluid; CCKN, cholecystokinin; DOPAC, dihydroxyphenylacetic acid; GABA, γ-aminobutyric acid; HVA, homovanillic acid; NEUG, neurogranin; PENK, proenkephalin A precursor; SC, spectrum counting; SCG, secretogranin; VGF, neurosecretory protein VGF; LTQ, linear trap quadrupole; DAVID, Database for Annotation, Visualization, and Integrated Discovery; PCP4, Purkinje cell protein 4; CaM, calmodulin.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Yin, H. H., and Knowlton, B. J. ( 2006) The role of the basal ganglia in habit formation. Nat. Rev. Neurosci. 7, 464–476 [DOI] [PubMed] [Google Scholar]
- 2.Nauta, W. J. H. ( 1989) Reciprocal links of the corpus striatum with the cerebral cortex and limbic system: a common substrate for movement and thought?, in Neurology and Psychiatry: a Meeting of Minds (Mueller, J., ed) pp. 43–63, S. Karger AG, Basel
- 3.Nickel, W. ( 2005) Unconventional secretory routes: direct protein export across the plasma membrane of mammalian cells. Traffic 6, 607–614 [DOI] [PubMed] [Google Scholar]
- 4.Keller, M., Ruegg, A., Werner, S., and Beer, H. D. ( 2008) Active caspase-1 is a regulator of unconventional protein secretion. Cell 132, 818–831 [DOI] [PubMed] [Google Scholar]
- 5.Chiellini, C., Cochet, O., Negroni, L., Samson, M., Poggi, M., Ailhaud, G., Alessi, M. C., Dani, C., and Amri, E. Z. ( 2008) Characterization of human mesenchymal stem cell secretome at early steps of adipocyte and osteoblast differentiation. BMC Mol. Biol. 9, 26–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bendtsen, J. D., Jensen, L. J., Blom, N., Von Heijne, G., and Brunak, S. ( 2004) Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17, 349–356 [DOI] [PubMed] [Google Scholar]
- 7.Zougman, A., Pilch, B., Podtelejnikov, A., Kiehntopf, M., Schnabel, C., Kumar, C., and Mann, M. ( 2008) Integrated analysis of the cerebrospinal fluid peptidome and proteome. J. Proteome Res. 7, 386–399 [DOI] [PubMed] [Google Scholar]
- 8.Lafon-Cazal, M., Adjali, O., Galeotti, N., Poncet, J., Jouin, P., Homburger, V., Bockaert, J., and Marin, P. ( 2003) Proteomic analysis of astrocytic secretion in the mouse. Comparison with the cerebrospinal fluid proteome. J. Biol. Chem. 278, 24438–24448 [DOI] [PubMed] [Google Scholar]
- 9.Thouvenot, E., Lafon-Cazal, M., Demettre, E., Jouin, P., Bockaert, J., and Marin, P. ( 2006) The proteomic analysis of mouse choroid plexus secretome reveals a high protein secretion capacity of choroidal epithelial cells. Proteomics 6, 5941–5952 [DOI] [PubMed] [Google Scholar]
- 10.Clough, G. F. ( 2005) Microdialysis of large molecules. AAPS J. 7, E686–E692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wotjak, C. T., Landgraf, R., and Engelmann, M. ( 2008) Listening to neuropeptides by microdialysis: echoes and new sounds? Pharmacol. Biochem. Behav. 90, 125–134 [DOI] [PubMed] [Google Scholar]
- 12.Frost, S. I., Keen, K. L., Levine, J. E., and Terasawa, E. ( 2007) Microdialysis methods for in vivo neuropeptide measurement in the Stalk-median eminence in the Rhesus monkey. J. Neurosci. Methods 168, 26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haskins, W. E., Wang, Z., Watson, C. J., Rostand, R. R., Witowski, S. R., Powell, D. H., and Kennedy, R. T. ( 2001) Capillary LC-MS2 at the attomole level for monitoring and discovering endogenous peptides in microdialysis samples collected in vivo. Anal. Chem. 73, 5005–5014 [DOI] [PubMed] [Google Scholar]
- 14.Maurer, M. H., Berger, C., Wolf, M., Futterer, C. D., Feldmann, R. E., Jr., Schwab, S., and Kuschinsky, W. ( 2003) The proteome of human brain microdialysate. Proteome Sci. 1, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haskins, W. E., Watson, C. J., Cellar, N. A., Powell, D. H., and Kennedy, R. T. ( 2004) Discovery and neurochemical screening of peptides in brain extracellular fluid by chemical analysis of in vivo microdialysis samples. Anal. Chem. 76, 5523–5533 [DOI] [PubMed] [Google Scholar]
- 16.Maurer, M. H., Haux, D., Sakowitz, O. W., Unterberg, A. W., and Kuschinsky, W. ( 2007) Identification of early markers for symptomatic vasospasm in human cerebral microdialysate after subarachnoid hemorrhage: preliminary results of a proteome-wide screening. J. Cereb. Blood Flow Metab. 27, 1675–1683 [DOI] [PubMed] [Google Scholar]
- 17.Blondeau, F., Ritter, B., Allaire, P. D., Wasiak, S., Girard, M., Hussain, N. K., Angers, A., Legendre-Guillemin, V., Roy, L., Boismenu, D., Kearney, R. E., Bell, A. W., Bergeron, J. J., and McPherson, P. S. ( 2004) Tandem MS analysis of brain clathrin-coated vesicles reveals their critical involvement in synaptic vesicle recycling. Proc. Natl. Acad. Sci. U. S. A. 101, 3833–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, H., Sadygov, R. G., and Yates, J. R., III ( 2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 19.Bergeron, J. J., and Hallett, M. ( 2007) Peptides you can count on. Nat. Biotechnol. 25, 61–62 [DOI] [PubMed] [Google Scholar]
- 20.Zhang, B., VerBerkmoes, N. C., Langston, M. A., Uberbacher, E., Hettich, R. L., and Samatova, N. F. ( 2006) Detecting differential and correlated protein expression in label-free shotgun proteomics. J. Proteome Res. 5, 2909–2918 [DOI] [PubMed] [Google Scholar]
- 21.Xia, Q., Wang, T., Park, Y., Lamont, R. J., and Hackett, M. ( 2007) Differential quantitative proteomics of Porphyromonas gingivalis by linear ion trap mass spectrometry: non-label methods comparison, q-values and LOWESS curve fitting. Int. J. Mass Spectrom. 259, 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. ( 1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 23.Boulet, S., Lacombe, E., Carcenac, C., Feuerstein, C., Sgambato-Faure, V., Poupard, A., and Savasta, M. ( 2006) Subthalamic stimulation-induced forelimb dyskinesias are linked to an increase in glutamate levels in the substantia nigra pars reticulata. J. Neurosci. 26, 10768–10776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.You, J. S., Gelfanova, V., Knierman, M. D., Witzmann, F. A., Wang, M., and Hale, J. E. ( 2005) The impact of blood contamination on the proteome of cerebrospinal fluid. Proteomics 5, 290–296 [DOI] [PubMed] [Google Scholar]
- 25.Burgess, J. A., Lescuyer, P., Hainard, A., Burkhard, P. R., Turck, N., Michel, P., Rossier, J. S., Reymond, F., Hochstrasser, D. F., and Sanchez, J. C. ( 2006) Identification of brain cell death associated proteins in human post-mortem cerebrospinal fluid. J. Proteome Res. 5, 1674–1681 [DOI] [PubMed] [Google Scholar]
- 26.Luders, T., Birkemo, G. A., Nissen-Meyer, J., Andersen, O., and Nes, I. F. ( 2005) Proline conformation-dependent antimicrobial activity of a proline-rich histone h1 N-terminal Peptide fragment isolated from the skin mucus of Atlantic salmon. Antimicrob. Agents Chemother. 49, 2399–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirabeau, O., Perlas, E., Severini, C., Audero, E., Gascuel, O., Possenti, R., Birney, E., Rosenthal, N., and Gross, C. ( 2007) Identification of novel peptide hormones in the human proteome by hidden Markov model screening. Genome Res. 17, 320–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amare, A., Hummon, A. B., Southey, B. R., Zimmerman, T. A., Rodriguez-Zas, S. L., and Sweedler, J. V. ( 2006) Bridging neuropeptidomics and genomics with bioinformatics: prediction of mammalian neuropeptide prohormone processing. J. Proteome Res. 5, 1162–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Southey, B. R., Amare, A., Zimmerman, T. A., Rodriguez-Zas, S. L., and Sweedler, J. V. ( 2006) NeuroPred: a tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 34, W267–W272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brochier, C., Gaillard, M. C., Diguet, E., Caudy, N., Dossat, C., Segurens, B., Wincker, P., Roze, E., Caboche, J., Hantraye, P., Brouillet, E., Elalouf, J. M., and de Chaldee, M. ( 2008) Quantitative gene expression profiling of mouse brain regions reveals differential transcripts conserved in human and affected in disease models. Physiol. Genomics 33, 170–179 [DOI] [PubMed] [Google Scholar]
- 31.Conti, A., Sanchez-Ruiz, Y., Bachi, A., Beretta, L., Grandi, E., Beltramo, M., and Alessio, M. ( 2004) Proteome study of human cerebrospinal fluid following traumatic brain injury indicates fibrin(ogen) degradation products as trauma-associated markers. J. Neurotrauma 21, 854–863 [DOI] [PubMed] [Google Scholar]
- 32.Thouvenot, E., Urbach, S., Dantec, C., Poncet, J., Seveno, M., Demettre, E., Jouin, P., Touchon, J., Bockaert, J., and Marin, P. ( 2008) Enhanced detection of CNS cell secretome in plasma protein-depleted cerebrospinal fluid. J. Proteome Res. 7, 4409–4421 [DOI] [PubMed] [Google Scholar]
- 33.Pisitkun, T., Shen, R. F., and Knepper, M. A. ( 2004) Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U. S. A. 101, 13368–13373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denzer, K., Kleijmeer, M. J., Heijnen, H. F., Stoorvogel, W., and Geuze, H. J. ( 2000) Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 113, 3365–3374 [DOI] [PubMed] [Google Scholar]
- 35.Chaput, N., Flament, C., Viaud, S., Taieb, J., Roux, S., Spatz, A., André, F., LePecq, J. B., Boussac, M., Garin, J., Amigorena, S., Théry, C., and Zitvogel, L. ( 2006) Dendritic cell derived-exosomes: biology and clinical implementations. J. Leukoc. Biol. 80, 471–478 [DOI] [PubMed] [Google Scholar]
- 36.Ahn, S. M., Byun, K., Cho, K., Kim, J. Y., Yoo, J. S., Kim, D., Paek, S. H., Kim, S. U., Simpson, R. J., and Lee, B. ( 2008) Human microglial cells synthesize albumin in brain. PLoS ONE 3, e2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oikari, S., Ahtialansaari, T., Heinonen, M. V., Mauriala, T., Auriola, S., Kiehne, K., Folsch, U. R., Janne, J., Alhonen, L., and Herzig, K. H. ( 2008) Downregulation of PPARs and SREBP by acyl-CoA-binding protein overexpression in transgenic rats. Pfluegers Arch. Eur. J. Physiol. 456, 369–377 [DOI] [PubMed] [Google Scholar]
- 38.Mahley, R. W. ( 1988) Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science 240, 622–630 [DOI] [PubMed] [Google Scholar]
- 39.Patel, B. N., and David, S. ( 1997) A novel glycosylphosphatidylinositol-anchored form of ceruloplasmin is expressed by mammalian astrocytes. J. Biol. Chem. 272, 20185–20190 [DOI] [PubMed] [Google Scholar]
- 40.Carr, D. W., Stofko-Hahn, R. E., Fraser, I. D., Cone, R. D., and Scott, J. D. ( 1992) Localization of the cAMP-dependent protein kinase to the postsynaptic densities by A-kinase anchoring proteins. Characterization of AKAP 79. J. Biol. Chem. 267, 16816–16823 [PubMed] [Google Scholar]
- 41.Burre, J., and Volknandt, W. ( 2007) The synaptic vesicle proteome. J. Neurochem. 101, 1448–1462 [DOI] [PubMed] [Google Scholar]
- 42.Yong, V. W., Power, C., Forsyth, P., and Edwards, D. R. ( 2001) Metalloproteinases in biology and pathology of the nervous system. Nat. Rev. Neurosci. 2, 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stark, M., Danielsson, O., Griffiths, W. J., Jornvall, H., and Johansson, J. ( 2001) Peptide repertoire of human cerebrospinal fluid: novel proteolytic fragments of neuroendocrine proteins. J. Chromatogr. B Biomed. Sci. Appl. 754, 357–367 [DOI] [PubMed] [Google Scholar]
- 44.Bora, A., Annangudi, S. P., Millet, L. J., Rubakhin, S. S., Forbes, A. J., Kelleher, N. L., Gillette, M. U., and Sweedler, J. V. ( 2008) Neuropeptidomics of the supraoptic rat nucleus. J Proteome Res. 7, 4992–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkin, M. C., Wei, H., O'Callaghan, J. P., and Kennedy, R. T. ( 2005) Sample-dependent effects on the neuropeptidome detected in rat brain tissue preparations by capillary liquid chromatography with tandem mass spectrometry. Anal. Chem. 77, 6331–6338 [DOI] [PubMed] [Google Scholar]
- 46.Dowell, J. A., Heyden, W. V., and Li, L. ( 2006) Rat neuropeptidomics by LC-MS/MS and MALDI-FTMS: enhanced dissection and extraction techniques coupled with 2D RP-RP HPLC. J Proteome Res. 5, 3368–3375 [DOI] [PubMed] [Google Scholar]
- 47.Bernay, B., Baudy-Floc'h, M., Zanuttini, B., Gagnon, J., and Henry, J. ( 2005) Identification of SepCRP analogues in the cuttlefish Sepia officinalis: a novel family of ovarian regulatory peptides. Biochem. Biophys. Res. Commun. 338, 1037–1047 [DOI] [PubMed] [Google Scholar]
- 48.Bernay, B., Baudy-Floc'h, M., Gagnon, J., and Henry, J. ( 2006) Ovarian jelly-peptides (OJPs), a new family of regulatory peptides identified in the cephalopod Sepia officinalis. Peptides 27, 1259–1268 [DOI] [PubMed] [Google Scholar]
- 49.Zhu, W., and Pan, Z. Z. ( 2005) Mu-opioid-mediated inhibition of glutamate synaptic transmission in rat central amygdala neurons. Neuroscience 133, 97–103 [DOI] [PubMed] [Google Scholar]
- 50.Jiang, Z. G., and North, R. A. ( 1992) Pre- and postsynaptic inhibition by opioids in rat striatum. J. Neurosci. 12, 356–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.