Abstract
The circadian clock is an endogenous mechanism that coordinates biological processes with daily and seasonal changes in the environment. Heterodimerization of central clock components is an important way of controlling clock function in several different circadian systems. CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) are Myb-related proteins that function in or close to the central oscillator in Arabidopsis (Arabidopsis thaliana). Single mutants of cca1 and lhy have a phenotype of short-period rhythms. cca1 lhy double mutants show an even shorter period phenotype than the cca1 single mutant, suggesting that CCA1 and LHY are only partially functionally redundant. To determine whether CCA1 and LHY act in parallel or synergistically in the circadian clock, we examined their expression in both light-grown and etiolated seedlings. We have shown that LHY and CCA1 bind to the same region of the promoter of a Light-harvesting chlorophyll a/b protein (Lhcb, also known as CAB). CCA1 and LHY can form homodimers, and they also colocalize in the nucleus and heterodimerize in vitro and in vivo. In Arabidopsis, CCA1 and LHY physically interact in a manner independent of photoperiod. Moreover, results from gel filtration chromatography indicate that CCA1 and LHY are present in the same large complex in plants. Taken together, these results imply that CCA1 and LHY function synergistically in regulating circadian rhythms of Arabidopsis.
Circadian clocks are autoregulatory, endogenous mechanisms that control numerous physiological and molecular processes in organisms ranging from cyanobacteria to humans. In plants, leaf movements, stomata opening, hypocotyl elongation, and the expression of a large number of genes show circadian rhythms (Harmer and Kay, 2000; Edwards et al., 2006; Yakir et al., 2007; Michael et al., 2008). Although circadian clock components are divergent across the kingdoms, most organisms share a conserved mechanism of generating rhythmicity: a central oscillator based on transcriptional negative feedback loops (Dunlap, 1999). In Arabidopsis (Arabidopsis thaliana), three genes have been suggested as core components of the central oscillator: CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and TIMING OF CAB EXPRESSION1 (TOC1). CCA1 and LHY bind directly to the promoter of TOC1, negatively regulating TOC1 expression, and TOC1 participates in the positive regulation of CCA1 and LHY expression through an unknown mechanism (Alabadí et al., 2001). Other genes, such as LUX ARRHYTHMO, also known as PHYTOCLOCK1, GIGANTEA (GI), EARLY FLOWERING3 (ELF3), ELF4, TIME FOR COFFEE, and PSEUDORESPONSE REGULATOR3/5/7/9 (PRR3/5/7/9), have also been suggested to function in or close to the central oscillator (Doyle et al., 2002; Hazen et al., 2005; Locke et al., 2005; Nakamichi et al., 2005; Edwards et al., 2006; Gould et al., 2006; Ding et al., 2007; McWatters et al., 2007).
CCA1 and LHY are closely related transcription factors that contain a single Myb domain (Wang et al., 1997; Schaffer et al., 1998). They both have circadian rhythms of expression peaking soon after dawn. cca1 and lhy single mutants have short-period phenotypes, and overexpression of either gene causes arrhythmicity in expression of clock-regulated genes, leaf movement, and hypocotyl elongation and leads to dramatically reduced levels of endogenous CCA1 and LHY transcripts (Schaffer et al., 1998; Wang and Tobin, 1998; Green and Tobin, 1999; Alabadí et al., 2002; Mizoguchi et al., 2002). Therefore, CCA1 and LHY negatively regulate their own and each other's expression.
However, regulation of clock genes is not restricted to the level of transcription; a multitude of protein interactions are required for the generation and maintenance of rhythmicity within the central oscillator. Perhaps the best-characterized example is the Drosophila clock in which PERIOD (PER) protein interacts with TIMELESS (TIM) and inhibits their own transcription by repressing the activity of dCLOCK (dCLK) and CYCLE (CYC; Gekakis et al., 1995; Meyer et al., 2006). The dCLK/CYC heterodimer not only activates PER and TIM transcription but also the transcription of VRILLE and PAR DOMAIN PROTEIN1, which encode negative and positive regulators of CLK transcription, respectively (for review, see Hardin, 2004). Recent discoveries demonstrate that protein interactions are also important for proper clock function in Arabidopsis. The F-box protein ZEITLUPE (ZTL) binds TOC1 and causes it to be degraded through a CULLIN1-containing SCF (for Skp1-Cullin-F-box protein) complex (Más et al., 2003; Harmon et al., 2008). ZTL also regulate PRR5 degradation through direct binding (Kiba et al., 2007). TOC1/ZTL interaction can be competed by the TOC1/PRR3 interaction and possibly prevents TOC1 degradation in the vascular tissues (Para et al., 2007; Fujiwara et al., 2008). In addition, ZTL protein is stabilized by direct interaction with GI in blue light (Kim et al., 2007).
In this work, we show that CCA1 and LHY have similar expression patterns and that they can form homodimers and heterodimers in vitro and in vivo. CCA1 and LHY also colocalize in the nucleus and physically interact in plants. Furthermore, we find that they are present in the same large complex. These observations suggest that CCA1 and LHY function synergistically in regulating the circadian rhythms of Arabidopsis.
RESULTS
A cca1 lhy Double Mutant Has More Severe Phenotypes Than a cca1 Single Mutant
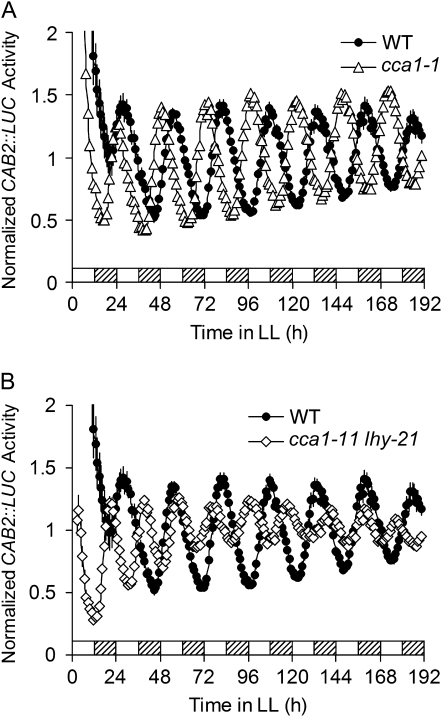
cca1 and lhy single loss-of-function mutants confer short periods (Green and Tobin, 1999; Alabadí et al., 2002; Mizoguchi et al., 2002) and an initial study showed that a cca1 lhy double loss-of-function mutant (cca1-1 lhy-11) is unable to maintain rhythms of clock-controlled RNAs for more than a few cycles under constant light (LL; Alabadí et al., 2002; Mizoguchi et al., 2002). However, the lhy allele used in that study was not truly null, because a truncated LHY protein is still expressed (Supplemental Fig. S1). Therefore, it is possible that the truncated LHY retained partial clock-associated function in the cca1-1 lhy-11 double mutant. We therefore set out to test whether plants that are completely null for both the CCA1 and LHY proteins are able to sustain rhythmicity in LL. The CAB2∷LUC (for LUCIFERASE) circadian reporter (Anderson et al., 1994; Knowles et al., 2008) was introduced into wild-type Wassilewskija (Ws), cca1-1 (a single-null mutant in the Ws background), and cca1-11 lhy-21 (a double-null mutant in the Ws background) that is completely missing both proteins (Supplemental Fig. S1; Locke et al., 2005) seedlings by transformation (Clough and Bent, 1998). Figure 1 demonstrates that the CAB2∷LUC rhythm in cca1-11 lhy-21 seedlings persists for at least 8 d in LL. Although some initial dampening (decreasing amplitude over time) was evident in the double mutant, the rhythm stabilized after three to four cycles. This is in contrast to reports that indicate rhythms dampen out completely in the double mutants (Mizoguchi et al., 2002; Locke et al., 2005). Compared to the cca1-1 single mutant, which displayed a short-period phenotype (24.77 ± 0.07 h versus 26.41 ± 0.16 h in wild type), the cca1-11 lhy-21 double mutant exhibits reduced amplitude as well as a very short period (19.73 ± 0.12 h) of CAB2∷LUC rhythm (Fig. 1). These results indicate that CCA1 and LHY are required for sustaining robust rhythms and proper period in LL; however, in their absence, other presumably closely related proteins can partially replace this function. The short period of the single cca1-1 mutant and the additive period phenotype of the cca1-11 lhy-21 double mutant indicate that CCA1 and LHY are only partially redundant, in which case they might function in parallel and/or work together in regulating circadian rhythms.
Figure 1.
The cca1-11 lhy-21 double mutant has a shorter period than the cca1-1 single mutant. A, Comparison of the CAB2∷LUC activity in wild type (Ws) and cca1-1. B, Comparison of the CAB2∷LUC activity in wild type (Ws) and cca1-11 lhy-21. CAB2∷LUC seedlings in wild-type (Ws), cca1-1, and cca1-11 lhy-21 backgrounds were grown in 12L:12D for 8 d and then transferred to LL. Mean bioluminescence traces ± SEM (n = 38–40) are shown. Each point was normalized to the average luminescence value for the entire run. Subjective day and subjective night are denoted by white and hatched bars, respectively. All of these experiments were done at least twice with similar results. WT, Wild type.
CCA1 and LHY Have Similar Expression Patterns
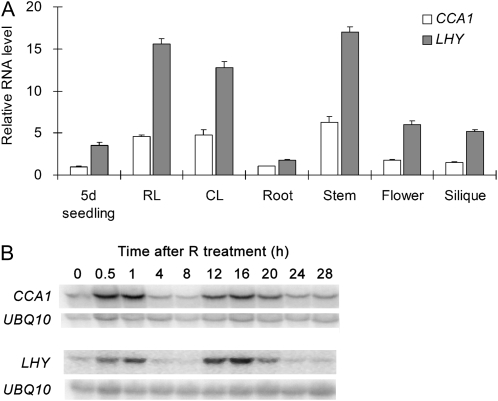
Both CCA1 and LHY are expressed rhythmically with peaks of expression occurring soon after dawn in plants grown under diurnal conditions or transferred to LL (Schaffer et al., 1998; Wang and Tobin, 1998). To determine whether CCA1 and LHY function in parallel or synergistically, their tissue-specific expression was examined. Different Arabidopsis tissues from light-grown plants were harvested at 1 h after dawn (zeitgeber time [ZT]-1) when CCA1 and LHY are most abundant, and quantitative real-time PCR (qRT-PCR) was used to measure their levels of expression. Figure 2A shows that both CCA1 and LHY are expressed at higher levels in leaves and stems compared to roots, inflorescences, and siliques. Although CCA1 and LHY have similar expression patterns in different tissues, LHY is generally more highly expressed than CCA1 (Fig. 2A).
Figure 2.
CCA1 and LHY have similar expression patterns. A, Comparison of the expression of CCA1 and LHY in different tissues (RL, rosette leaf; CL, cauline leaf). Five-day-old (5d) seedlings were grown on plates, and the other tissues were harvested from 4-week-old soil-grown plants. All samples were collected at ZT-1. The RNA was subjected to qRT-PCR analysis to measure levels of CCA1 and LHY relative to RH8 (an internal control). Reactions were performed in triplicate. Error bars denote ± sd. B, CCA1 and LHY are expressed with similar kinetics in etiolated seedlings exposed to R. Time course of CCA1 and LHY expression in 5-d-old etiolated seedlings after exposure to 2 min of R. Gel blots of total RNA were hybridized with RNA probes of CCA1, LHY, and UBQ10 (control). All of these experiments were done at least twice with similar results.
In etiolated seedlings, CCA1 and LHY are expressed at very low levels, but light can induce CCA1 and LHY expression (Wang and Tobin, 1998; Martinez-Garcia et al., 2000). To examine whether LHY could be regulated by light and the circadian clock in a similar manner to CCA1 in etiolated seedlings, we compared the expression of CCA1 and LHY in etiolated seedlings treated with a brief exposure to red light (R). This experimental approach allows the acute response to light and the circadian response to be separated (Millar et al., 1992; Anderson and Kay, 1997). Arabidopsis seedlings were grown for 5 d in the dark, exposed to 2 min of R, and tissue was harvested at various times up to 28 h following R exposure. Consistent with previous reports (Wang and Tobin, 1998; Martinez-Garcia et al., 2000), both CCA1 and LHY mRNA levels increased within 0.5 h and peaked at 1 h after R treatment (Fig. 2B), demonstrating that light induces both LHY and CCA1 expression in etiolated seedlings. Following the acute response to light, the expression of both CCA1 and LHY mRNAs reached trough levels in approximately 8 h and rose transiently to peak at approximately 16 h after R treatment (Fig. 2B). The second peak is most likely due to the action of an endogenous circadian oscillator (Millar and Kay, 1996). This result demonstrates that both light and the circadian clock regulate the expression of CCA1 and LHY in a similar way in etiolated seedlings. Taken together, the results indicate that CCA1 and LHY have similar expression patterns in both light-grown plants and etiolated seedlings.
LHY and CCA1 Bind to the Same Region of the Light-harvesting chlorophyll a/b protein Promoter
CCA1 and LHY show similar regulation by light and the circadian clock. In addition, CCA1 mediates activation of Light-harvesting chlorophyll a/b protein (Lhcb) genes by binding directly to their promoters (Wang et al., 1997). It has previously been shown that the Myb domain of CCA1 interacts with two closely spaced binding sites (−92 to −105 and −111 to −122 relative to the transcription start site) in the Lhcb1*3 promoter (Wang et al., 1997). The extensive similarity (87% identity) between the Myb domains of the CCA1 and LHY proteins prompted us to examine whether LHY can bind to the same regions of the Lhcb1*3 promoter as CCA1. Figure 3 shows the results from a representative electrophoretic mobility shift assay (EMSA) using Escherichia coli-produced CCA1 and LHY proteins and wild-type and mutant promoter fragments of Lhcb1*3 as probes. DNA-binding activities were observed for both proteins using the wild-type A2 fragment of the Lhcb1*3 promoter (Fig. 3A). This A2 fragment was used previously to characterize CCA1 binding ability (Wang et al., 1997). The mutated m1 fragment carries several point mutations spanning both CCA1 binding sites in the Lhcb1*3 promoter (Fig. 3C). These mutations were previously shown to abolish binding of CCA1 to the wild-type A2 fragment (Wang et al., 1997). As shown in Figure 3A, the altered nucleotides of the m1 fragment also completely abolished the binding of LHY.
Figure 3.
LHY binds to the same region of the Lhcb1*3 promoter as CCA1. A, EMSA analysis of CCA1 and LHY binding on the Lhcb1*3 promoter. The m1 probe contains the point mutations shown in C. WT, Wild type. B, Phenanthroline-copper footprinting of LHY on the wild-type A2 fragment of the Lhcb1*3 promoter. An EMSA gel with LHY and the wild-type A2 fragment was treated with phenanthroline-copper to cleave DNA, and the DNA from each band was recovered and separated on a sequencing gel. The lines between or next to the lanes highlight the regions protected by the proteins. F, Free probe. B1 and B2 refer to the DNA-protein complexes containing LHY used in the footprint analysis. C, Nucleotide sequence of the wild-type and m1 probes. WT and m1 denote the sequences of the wild-type A2 fragment of the Lhcb1*3 promoter and of the m1 mutated fragment, respectively. Dashes in the sequence of m1 represent nucleotides identical to those of the wild-type fragment. Solid and dashed lines above the sequence of the wild-type fragment represent the regions protected by LHY and CCA1, respectively. The region protected by CCA1 is from Wang et al. (1997). All of these experiments were done at least twice with similar results.
To characterize the LHY-binding site on the Lhcb1*3 promoter in more detail, we carried out 1,10-phenanthroline-copper footprinting analysis. We have previously used this method to characterize CCA1 binding to the same fragment of the Lhcb1*3 promoter (Wang et al., 1997). After performing an EMSA to resolve the DNA-protein complexes, the gel was treated with 1,10-phenanthroline-copper, and the DNA in the two complexes (B1 and B2; Fig. 3A) was recovered and resolved in a sequencing gel. As shown in Figure 3B, in complex B1, the −96 to −107 region was protected and in complex B2, regions from −92 to −107 and from −111 to −130 were protected. This result suggests that the two complexes of different mobilities are a result of the presence of two separate binding sites on this fragment and that the −96 to −107 region probably is the higher affinity binding site for LHY. A similar observation in which one complex has a larger protected region than the other was also made in CCA1 footprinting analysis (Wang et al., 1997). Figure 3C summarizes the results from the phenanthroline-copper footprinting and compares the regions of the wild-type A2 fragment protected by CCA1 and LHY. These results demonstrate that the two proteins bind to the same region of the Lhcb1*3 promoter.
CCA1 Interacts with LHY in Vitro
In Drosophila, CYC heterodimerizes with dCLK and activates the transcription of PER and TIM through binding to the E-boxes of the PER and TIM promoters (Shirasu et al., 2003). Because CCA1 and LHY bind to the same region of the Lhcb1*3 promoter, we investigated whether CCA1 heterodimerizes with LHY using a reciprocal interaction assay. Glutathione S-transferase (GST) fusions of CCA1 and LHY were immobilized on glutathione-agarose beads and incubated with [35S]Met-labeled CCA1 or LHY synthesized by in vitro transcription-translation. LUC was used as a control. Figure 4A shows that in the presence of either GST-CCA1 or GST-LHY, but not with GST alone, both CCA1 and LHY proteins synthesized in vitro were efficiently bound, while LUC was not bound by either of the two GST fusion proteins. These results demonstrate that CCA1 and LHY not only homodimerize but can also form a heterodimer in vitro.
Figure 4.
CCA1 and LHY can physically interact in vitro, and the region of CCA1 downstream of the Myb domain is important for this interaction. A, CCA1 and LHY can homo- and heterodimerize in vitro. Glutathione-agarose beads containing approximately 1 μg of GST, GST-CCA1, or GST-LHY were mixed with 20 μL of 35S-labeled CCA1, LHY, or LUC. Proteins interacting with GST, GST-CCA1, or GST-LHY were eluted and resolved in SDS-PAGE, followed by autoradiography. Input represents 10% of the 35S-labeled protein used. B, The region between amino acids 136 and 316 of CCA1 is essential for interaction with LHY in vitro. The Myb domain of CCA1 is shown. The region of CCA1 filled with black and denoted with an asterisk indicates the 136- to 316-amino acid region that interacts with LHY. All of these experiments were done at least twice with similar results.
To identify the region of CCA1 that is responsible for interacting with LHY, we carried out interaction assays using [35S]Met-labeled LHY synthesized in vitro and various deletion constructs of CCA1 fused to GST. Figure 4B shows that in the presence of immobilized GST-CCA1 proteins lacking either the 316 or 436 N-terminal amino acids of CCA1 or a 472-amino acid portion from the C terminus, LHY could not be efficiently bound. Taken together, these data suggest that a region of CCA1 between amino acids 136 and 316 is important for interaction with LHY. It is worth mentioning that the LHY-interactive domain of CCA1 lies outside the DNA-binding Myb domain.
CCA1 and LHY Can Form Homodimers in Vivo
To examine the homodimerization of CCA1 in vivo, we carried out in vivo transient expression in Nicotiana benthamiana. Expression constructs of 35S∷YFP (yellow fluorescent protein)-CCA1 and 35S∷MYC-CCA1 were coinfiltrated into N. benthamiana and total protein was extracted from infiltrated leaves. Immunoprecipitation with anti-GFP antibody and subsequent detection of MYC-CCA1 using anti-MYC antibody (Fig. 5A) showed that anti-GFP antibodies not only immunoprecipitate YFP-CCA1, but also coimmunoprecipitate MYC-CCA1. To examine the homodimerization of LHY in vivo, similar immunoprecipitation was performed using anti-GFP antibodies with total protein extracts from N. benthamiana leaves coexpressing YFP-LHY and haemagglutinin (HA)-LHY. Figure 5B shows that HA-LHY is coprecipitated with YFP-LHY. These results indicate that both CCA1 and LHY can homodimerize in vivo.
Figure 5.
CCA1 and LHY can form homodimers and heterodimers in vivo. A, CCA1 homodimerizes in planta. Immunoprecipitation (IP) by α-GFP antibody was performed on total protein extracts from N. benthamiana leaves transiently expressing YFP-CCA1 and MYC-CCA1. B, LHY homodimerize in planta. Immunoprecipitation by α-GFP antibody was performed on total protein extracts from N. benthamiana leaves transiently expressing YFP-LHY and HA-LHY. C, CCA1 and LHY colocalize in the nucleus. Left, CFP fluorescence localization of the transiently expressed LHY-CFP in N. benthamiana leaves; middle, YFP fluorescence localization of the transiently expressed YFP-CCA1 in N. benthamiana leaves; right, merged image of the CFP and YFP fluorescence in N. benthamiana leaves transiently expressing the two constructs. Bars = 10 μm. D, In planta heterodimerization of CCA1 and LHY requires the region between amino acids 136 and 316 of CCA1. Immunoprecipitation by α-LHY antibody was performed on total protein extracts from N. benthamiana leaves transiently expressing HA-LHY with MYC-CCA1 (lanes 1, 8, and 15), MYC-CCA1Δ86N (lanes 2, 9, and 16), MYC-CCA1Δ316N (lane 3, 10, and 17), MYC-CCA1Δ436N (lanes 4, 11, and 18), MYC-CCA1Δ86C (lanes 5, 12, and 19), MYC-CCA1Δ270C (lanes 6, 13, and 20), and MYC-CCA1Δ472C (lanes 7, 14, and 21). Input and supernatant after immunoprecipitation were run next to the immunoprecipitation product as a control. One hundred micrograms of total protein was analyzed as input. All of these experiments were done at least twice with similar results.
CCA1 and LHY Colocalize in the Nucleus and Heterodimerize in Vivo
Because both CCA1 and LHY have been reported in the nucleus in plants (Wang et al., 1997; Carré and Kim, 2002), we first examined whether they colocalize in vivo. Expression constructs of 35S∷YFP-CCA1 and 35S∷LHY-CFP (cyan fluorescent protein)-HA were coinfiltrated into N. benthamiana. Confocal microscopy results (Fig. 5C) show that both YFP-CCA1 and LHY-CFP are localized in the nucleus and their expression overlaps. To verify the heterodimerization between CCA1 and LHY, MYC-CCA1 and HA-LHY were coexpressed in N. benthamiana. Immunoprecipitation with anti-LHY antibody and subsequent detection of MYC-CCA1 (Fig. 5D) showed that CCA1 interacts strongly with LHY in vivo. We extended these trials to test different regions of CCA1 and found that a region of CCA1 between amino acids 136 and 316 is important for interaction with LHY (Fig. 5D) in vivo. Protein product from one of the CCA1 deletion constructs (35S∷MYC-CCA1Δ316N) can only be detected by anti-CCA1 antibody, but not anti-MYC antibody (Fig. 5D), indicating that the MYC tag may be cleaved off from CCA1Δ316N protein. As expected, protein products from CCA1 C-terminal deletion constructs (35S∷MYC-CCA1Δ270C and 35S∷MYC-CCA1Δ472C) are not recognized by anti-CCA1 antibody, which is generated against the C-terminal portion of the CCA1 protein (Fig. 5D). Taken together, the results support the in vitro findings, demonstrating that CCA1 and LHY colocalize in the nucleus, and the region of CCA1 downstream of the Myb domain contains the determinants for LHY interactions in planta.
CCA1 and LHY Physically Interact in Arabidopsis
The physical interaction between CCA1 and LHY was also observed in wild-type (ecotype Columbia [Col]) Arabidopsis (Fig. 6A). Immunoprecipitation was performed using anti-LHY antibodies with total extracts from 2-week-old seedlings grown in 12 h light:12 h darkness (12L:12D) and harvested at ZT-1 and ZT-23 (1 h before dawn). Figure 6A shows that CCA1 coprecipitated with LHY and their interaction is not light dependent in Arabidopsis.
Figure 6.
CCA1 and LHY can interact in plants and their interaction is independent of photoperiod. Western-blot analysis of immunoprecipitation (IP) with α-LHY antibody and detection with antibody to LHY and CCA1. A, CCA1 coimmunoprecipitates with LHY in plant extract. Two-week-old seedlings grown in 12L:12D were harvested at ZT-1 and ZT-23. B, CCA1 coimmunoprecipitates with LHY throughout the diurnal cycle. Two-week-old seedlings grown in 12L:12D were harvested at different times as indicated. One hundred micrograms of total protein was analyzed as input. Light and dark periods are denoted by white and black bars, respectively. All of these experiments were done at least twice with similar results.
To further determine whether CCA1/LHY interaction is affected by photoperiod, wild-type Col Arabidopsis plants grown in 12L:12D were harvested at different times and a similar immunoprecipitation was conducted. The amount of LHY immunoprecipitated and the amount of CCA1 coimmunoprecipitated were correlated with the LHY and CCA1 levels in the total extract, suggesting that CCA1 can interact with LHY whenever they both are present. Similar results were obtained when Arabidopsis plants were grown in LL and harvested at different circadian times (data not shown). These data demonstrate that CCA1 and LHY physically interact in a manner independent of photoperiod in plants (Fig. 6).
CCA1 and LHY Are Found in a Large Complex
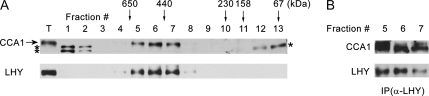
To examine whether the interaction of CCA1 and LHY is stable in plants, gel filtration chromatography was performed using total extract from wild-type (Col) Arabidopsis, which was prepared from 2-week-old seedlings grown in 12L:12D and harvested at ZT-1. We expected to observe heterodimers of about 150 kD. Surprisingly, we were unable to detect any monomers or heterodimers of CCA1 and LHY. CCA1 and LHY monomers are predicted to be 67 and 70 kD, respectively, but migrate at 75 and 80 kD, respectively, when subject to electrophoresis in denaturing conditions (Wang and Tobin, 1998; Daniel et al., 2004). We found that both CCA1 and LHY are present in a large complex (>440 kD; Fig. 7A).To determine whether CCA1 and LHY are present in the same complex, fractions 5, 6, and 7 were subjected to the immunoprecipitation with anti-LHY antibodies. CCA1 coprecipitated with LHY in those fractions, indicating that CCA1 and LHY are present in the same large complex (Fig. 7B). This observation suggests that CCA1 and LHY could function as part of a complex and work cooperatively with other proteins in regulating circadian rhythms.
Figure 7.
CCA1 and LHY are present in the same large complex. A, CCA1 and LHY comigrate in gel filtration chromatography. Shown is western-blot analysis of fractions collected from gel filtration and detection with antibody to LHY and CCA1. The arrow indicates the band of CCA1 and the asterisks indicate cross-reactive bands. Total (T) represents 5% of the protein extract used for gel filtration. B, CCA1 coimmunoprecipitates with LHY in fractions 5, 6, and 7. Protein extracts were prepared from 2-week-old seedlings grown in 12L:12D and harvested at ZT-1. All of these experiments were done at least twice with similar results.
DISCUSSION
It has been shown that heterodimerization of central clock components is an important way of controlling clock function in other circadian systems (Dunlap, 1999). Examples include the interaction of PER and TIM in Drosophila as a means of controlling the nuclear entry and degradation of both proteins (Edery, 1999; Meyer et al., 2006), the interactions among Kai proteins in cyanobacteria (Iwasaki et al., 1999) and heterodimers of the transcription factors mCLOCK and BMAL1, and interactions among three mPER (PER1–PER3) and two CRYPTOCHROME (mCRY1 and mCRY2) proteins in mammals (Reppert and Weaver, 2002; Lowrey and Takahashi, 2004). In this study, we demonstrate that CCA1 and LHY have similar expression patterns, bind to a similar region of the Lhcb1*3 promoter, colocalize in the nucleus, and that they can physically interact both in vitro and in plants. These findings are consistent with the evidence showing the involvement of dimerization between core oscillator components in the mammalian system (Kwon et al., 2006) and in Drosophila (Meyer et al., 2006), suggesting that homodimerization and heterodimerization might be conserved aspects in the regulation of eukaryotic circadian clocks.
The observation that cca1-1 plants show persistent rhythms in LL, albeit with a shorter period length than wild type, has been attributed to the presence of functionally redundant components in the circadian system, particularly to the CCA1-homolog LHY (Green and Tobin, 1999). We have shown that in mutants lacking both CCA1 and LHY, rhythms are still generated and persist in LL, but with a reduced amplitude and shortened period length (Fig. 1). Similar results have been reported elsewhere (Locke et al., 2005). Hence, a compromised, but functioning oscillator continues to operate in the absence of the two proteins. It is known that CCA1 and LHY belong to a 10-member family of Myb-related proteins that have significant homology in their Myb domains (Yanhui et al., 2006). Moreover, some of the single Myb domain proteins in this family have been reported to be under circadian control (Somers, 1999; Strayer and Kay, 1999; Kuno et al., 2003; Zhang et al., 2007). One can postulate that in the absence of CCA1 and LHY, some or all of the eight remaining members can substitute for them to some extent so that some rhythmic expression is maintained.
The phenotype of the single mutants and the enhanced phenotype of cca1 lhy double mutants (Fig. 1) indicate that CCA1 and LHY are not entirely functionally redundant. One possibility is that CCA1 and LHY have distinct roles and work in parallel in the control of circadian rhythms. Alternatively, CCA1 and LHY may have similar functions, but either the dosage of their combined gene products or the interactions between them are crucial for their function. cca1 and lhy single mutants have similar phenotypes (Green and Tobin, 1999; Alabadí et al., 2002; Mizoguchi et al., 2002), and overexpression of each gene has similar effects on circadian rhythms (Schaffer et al., 1998; Wang and Tobin, 1998). Together with the evidence presented here that CCA1 and LHY have similar expression patterns (Fig. 2) and that LHY and CCA1 bind to the same region of the Lhcb1*3 promoter (Fig. 3), this suggests that CCA1 and LHY have closely related functions within the circadian system. It has been shown that proteins containing a single Myb domain could bind DNA as dimers (Jin and Martin, 1999). In Drosophila, the transcription factors CYC and dCLK are localized in the nucleus and activated by dimerization. The complex of CYC and dCLK binds to the E-boxes of the PER and TIM promoters and activates their expression (Shirasu et al., 2003). Our results showed that CCA1 and LHY not only homodimerize but also colocalize in the nucleus and form heterodimers in vitro and in vivo (Figs. 4 and 5). The interaction between CCA1 and LHY was confirmed in plants using coimmunoprecipitation with total plant extract of wild-type Arabidopsis (Fig. 6). In addition, we determined that the region of CCA1 between amino acids 136 and 316 is responsible for interacting with LHY (Figs. 4B and 5D). Although the functional significance of CCA1 and LHY dimerization has yet to be elucidated, one hypothesis is that homodimers and heterodimers may have different effects on nuclear entry, DNA binding, and/or protein degradation. In wild-type plants, heterodimers of CCA1 and LHY could be important for proper feedback inhibition and in single mutants of cca1 and lhy, CCA1 or LHY may form homodimers, which could maintain rhythmic oscillations but cause the feedback inhibition to occur faster and result in a short-period phenotype. This is consistent with the notion that CCA1 and LHY have different biochemical activities (Gould et al., 2006) and they are only partially redundant in the circadian system.
In an effort to detect CCA1/LHY heterodimers in plants, wild-type plant extracts were fractionated by gel filtration chromatography. Interestingly, both CCA1 and LHY are found in a large complex (Fig. 7A). This novel finding will open up new opportunities for future work. Because CCA1 and LHY physically interact (Fig. 7B), it is likely that they are present in the same complex. The calculated molecular mass of heterodimers of CCA1/LHY is about 150 kD, yet the CCA1/LHY-containing complex is >400 kD (Fig. 7). Discovery of other components in the complex will significantly increase our knowledge of the functions of CCA1 and LHY and of the central oscillator of the circadian clock in Arabidopsis. To discover other components of the protein complex, transgenic plants expressing tandem affinity purification-tagged CCA1 have been generated and will be used to purify the complex.
In summary, our study shows that CCA1 and LHY not only homodimerize, but also colocalize in the nucleus and heterodimerize. They are also present in a protein complex, suggesting that CCA1 and LHY function synergistically in the control of circadian rhythms of Arabidopsis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col was used for all experiments described unless stated otherwise. Seeds were stratified for 3 d in the dark at 4°C and then sown onto either solid Murashige and Skoog medium (Murashige and Skoog, 1962) containing 1.5% agarose or soil. Seedlings were grown under a 12-h-fluorescent light (40 μmol m−2 s−1):12-h-dark photoperiod at a constant temperature of 22°C. Growth of etiolated seedlings and brief R treatments of seedlings were carried out as previously described (Wang et al., 1997).
Plasmid Constructs
To create the 35S-YFP-LHY and 35S-HA-LHY constructs, LHY cDNA was generated by PCR using the full-length LHY cDNA in pBluesript SK (±) plasmid (Knowles et al., 2008) as template and the primers LHY-F, 5′-CACCATGGATACTAATACATCTGGAG-3′ and LHY-R(S), 5′-TCATGTAGAAGCTTCTCCTTC-3′. Inserts were cloned into the pEarleyGate 104 (35S-YFP-LHY) and pEarleyGate 201 (35S-HA-LHY) vectors (Earley et al., 2006) using the GATEWAY recombination system (Invitrogen). To eliminate the stop codon of LHY cDNA, the LHY-R(NS) primer 5′-TGTAGAAGCTTCTCCTTCCAA-3′ was used in combination with LHY-F. LHY cDNA without stop codon (LHY-NS) was cloned into the pEarleyGate 102 vector (Earley et al., 2006) to create 35S-LHY-CFP-HA construct.
To create the 35S-YFP-CCA1 and 35S-MYC-CCA1 constructs, CCA1 cDNA was generated by PCR using the full-length CCA1 cDNA in pGEX-3X (Sugano et al., 1998) as template and the primers CCA1-F, 5′-CACCATGGAGACAAATTCGTCTGG-3′ and CCA1-R, 5′-TCATGTGGAAGCTTGAGTTTC-3′. Inserts were cloned into the pEarleyGate 104 (35S-YFP-CCA1) and pEarleyGate 203 (35S-MYC-CCA1) vectors (Earley et al., 2006) using the GATEWAY recombination system (Invitrogen). To generate 35S-MYC-CCA1 deletion constructs, PCR was done using primers CCA1Δ86N-F, 5′-CACCCTAGACATAGCTATTCCTCC-3′; CCA1Δ316N-F, 5′-CACCGCTGCTAGTGCTTGGTGGG-3′; CCA1Δ436N-F, 5′-CACCGAGGCGGATGCATCAGAAAG-3′ in combination with CCA1-R and CCA1Δ86C-R, 5′-TCATTGTTCTTGTTGTTGTTGTTCTTC-3′; CCA1Δ270C-R, 5′-TCAACCACCTGAACTAAGAGGAG-3′; and CCA1Δ472C-R, 5′-TCACTCAGGATGCGACACTTTTTC-3′ in combination with CCA1-F. Inserts were cloned into the pEarleyGate 203 vector (Earley et al., 2006) using the GATEWAY recombination system (Invitrogen).
PCR amplifications were performed using Pfu DNA polymerase (Stratagene) following standard PCR protocol. All vectors prepared were direct sequenced.
Bioluminescence Assays
Bioluminescence assays were performed using 96-well plates as described by Onai et al. (2004) and Knowles et al. (2008).
RNA Extraction and qRT-PCR
RNA extraction and qRT-PCR were carried out as previously described (Knowles et al., 2008). RNA HELICASE8 (RH8; Schaffer et al., 2001) was used as a noncycling reference. All reactions were performed in triplicate. The primers for qRT-PCR were: CCA1_F, 5′-GGGGTGTGAATGATGGAAAAGA-3′; CCA1_R, 5′-CGATCTTCATTGGCCATCTCAG-3′; LHY_F, 5′-GACAACGCGGTTCAAGATGTTC-3′; LHY_R, 5′-CCAAGGGTAGTTTTGCATGCTG-3′; RH8_F, 5′-CAATGGCTTCGAAGAGGTCAGA-3′; and RH8_R, 5′-TGGGTCGATAACTTCGGATTGA-3′. The RNA gel-blot analysis and the production of RNA probes of CCA1, LHY, and UBQ10 were carried out as previously described (Wang and Tobin, 1998).
Recombinant Proteins
GST-CCA1 and GST-LHY fusion proteins were expressed in Escherichia coli and were purified as described previously (Sugano et al., 1998). The GST-CCA1 deletions were generated by deleting the appropriate fragment from pGEX-CCA1 (Sugano et al., 1998) and re-ligating the remaining portion of the construct (e.g. Δ86N represents 86-amino acid deletion from N terminus and Δ86C represents 86-amino acid deletion from C terminus, etc.).
EMSA and Footprinting
The wild-type A2 and mutant m1 probes have been described previously (Sun et al., 1993). CCA1 and LHY proteins were produced in E. coli as GST fusions and released from the GST portion by cleavage with factor Xa as described by Sugano et al. (1998). Each DNA-binding reaction included 0.3 ng of 32P-labeled DNA probe, 1 μg of bovine serum albumin, and 0.5 μg of poly (dI-dC). Labeled probes were incubated in the presence or absence of approximately 5 ng of recombinant CCA1 or LHY proteins for 15 min prior to electrophoresis. The EMSA buffer and electrophoresis conditions have been described previously (Sun et al., 1993). DNA-protein complexes were detected by autoradiography.
Phenanthroline-copper footprinting was carried out as described previously (Wang et al., 1997). The A2 fragment was labeled with 32P at the 3′-end of the antisense strand (Sun et al., 1993). The EMSA reactions contained 1.5 × 106 cpm of end-labeled probe, 120 ng of Factor Xa-digested LHY protein, and 3 μg poly(dI-dC).
In Vitro Interaction Assay
CCA1, LHY, and LUC proteins labeled with [35S]Met were synthesized in vitro in 50 μL of rabbit reticulocyte lysate using a coupled transcription translation system (Promega). In vitro interaction assays were performed as described previously (Sugano et al., 1998).
Nicotiana benthamiana Infiltration
Nicotiana benthamiana leaves were agroinfiltrated as described previously (Voinnet et al., 2003; Kim et al., 2007). Agrobacterium tumefaciens strain C58C1 cells were transformed with the appropriate binary vectors using electroporation. A. tumefaciens positive clones were grown overnight and resuspended in infiltration buffer (10 mm MgCl2, 10 mm MES, and 100 μm acetosyringone) to reach a concentration of 0.8 OD600. This suspension was pressure-infiltrated into tobacco leaves using a syringe. Leaves were analyzed or harvested 2 d after infiltration.
Confocal Microscopy
All imaging was done using a Carl Zeiss 510 Meta laser scanning confocal microscope with a Plan-Apochromat 63×/1.4 oil DIC objective. To image YFP and CFP together, we used multitracking in stack mode and a 488/543 main dichroic. YFP was excited with an argon laser at 25% to 50% of its output that was attenuated to 7% to 9% at 514 nm. The emission was sent through a 530- to 600-nm band-pass filter for detection of YFP. CFP was stimulated by a 458-nm laser line that was attenuated to 7% to 9%. The emission was passed through a 475- to 525-nm band-pass filter for detection of CFP.
Protein Analysis and Coimmunoprecipitation
Proteins were extracted in immunoprecipitation buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 1 mm EDTA, 10% glycerol, 1% Triton X-100, 2 mm phenylmethylsulphonyl fluoride, 50 μm MG115, 50 μm MG132, and protease inhibitor cocktail [Roche]). The immunoprecipitation was performed as described by Shalitin et al. (2002) using either anti-GFP (sc-8334; Santa Cruz Biotechnology) or anti-LHY antibody (Daniel et al., 2004). Immunoblotting was performed as described (Lu and Hrabak, 2002) with the appropriate primary antibody (affinity-purified anti-CCA1 antibody; Wang and Tobin, 1998), affinity-purified anti-LHY antibody (Daniel et al., 2004), anti-GFP (sc-8334; Santa Cruz Biotechnology), anti-MYC (Upstate), or anti-HA (Abcam).
Gel Filtration Chromatography
Gel filtration was performed as described previously (Torii et al., 1998) except 1× gel filtration buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 5 mm MgCl, 10% glycerol, 0.1% NP-40, 2 mm dithiothreitol, 0.7 μg/mL pepstatin, 1 mm phenylmethylsulphonyl fluoride, 10 μm MG115, 10 μm MG132, and protease inhibitor cocktail [Roche]) was used for extraction and gel filtration. The extract was passed through a Superdex 200 (Pharmacia) gel filtration column. After a 7-mL void volume, 13 fractions of 0.5 mL each were collected and analyzed for the presence of CCA1 and LHY by western-blot analysis.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Truncated LHY protein is present in the cca1-1 lhy-11 double loss-of-function mutant.
Supplementary Material
Acknowledgments
We thank Candace Webb for her critical reading of the manuscript.
This work was supported by the National Institutes of Health (grant no. GM23167 to E.M.T.).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Elaine M. Tobin (etobin@ucla.edu).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883 [DOI] [PubMed] [Google Scholar]
- Alabadí D, Yanovsky MJ, Más P, Harmer SL, Kay SA (2002) Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr Biol 12 757–761 [DOI] [PubMed] [Google Scholar]
- Anderson SL, Kay SA (1997) Phototransduction and circadian clock pathways regulating gene transcription in higher plants. Adv Genet 35 1–34 [DOI] [PubMed] [Google Scholar]
- Anderson SL, Teakle GR, Martino-Catt SJ, Kay SA (1994) Circadian clock- and phytochrome-regulated transcription is conferred by a 78 bp cis-acting domain of the Arabidopsis CAB2 promoter. Plant J 6 457–470 [DOI] [PubMed] [Google Scholar]
- Carré IA, Kim JY (2002) MYB transcription factors in the Arabidopsis circadian clock. J Exp Bot 53 1551–1557 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Daniel X, Sugano S, Tobin EM (2004) CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci USA 101 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Millar AJ, Davis AM, Davis SJ (2007) TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19 1522–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognar L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419 74–77 [DOI] [PubMed] [Google Scholar]
- Dunlap JC (1999) Molecular bases for circadian clocks. Cell 96 271–290 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 616–629 [DOI] [PubMed] [Google Scholar]
- Edery I (1999) Role of posttranscriptional regulation in circadian clocks: lessons from Drosophila. Chronobiol Int 16 377–414 [DOI] [PubMed] [Google Scholar]
- Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, Straume M, Smith JQ, Millar AJ (2006) FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salome PA, McClung CR, Somers DE (2008) Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo- response regulator proteins. J Biol Chem 283 23073–23083 [DOI] [PubMed] [Google Scholar]
- Gekakis N, Saez L, Delahaye-Brown AM, Myers MP, Sehgal A, Young MW, Weitz CJ (1995) Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science 270 811–815 [DOI] [PubMed] [Google Scholar]
- Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, et al (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM (1999) Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96 4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE (2004) Transcription regulation within the circadian clock: the E-box and beyond. J Biol Rhythms 19 348–360 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Kay SA (2000) Microarrays: determining the balance of cellular transcription. Plant Cell 12 613–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon F, Imaizumi T, Gray WM (2008) CUL1 regulates TOC1 protein stability in the Arabidopsis circadian clock. Plant J 55 568–579 [DOI] [PMC free article] [PubMed]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Taniguchi Y, Ishiura M, Kondo T (1999) Physical interactions among circadian clock proteins KaiA, KaiB and KaiC in cyanobacteria. EMBO J 18 1137–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41 577–585 [DOI] [PubMed] [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua NH (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449 356–360 [DOI] [PubMed] [Google Scholar]
- Knowles SM, Lu SX, Tobin EM (2008) Testing time: Can ethanol-induced pulses of proposed oscillator components phase shift rhythms in Arabidopsis? J Biol Rhythms 23 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno N, Moller SG, Shinomura T, Xu X, Chua NH, Furuya M (2003) The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell 15 2476–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Lee J, Chang SH, Jung NC, Lee BJ, Son GH, Kim K, Lee KH (2006) BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol Cell Biol 26 7318–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1: 2005.0013 [DOI] [PMC free article] [PubMed]
- Lowrey PL, Takahashi JS (2004) Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet 5 407–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Hrabak EM (2002) An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol 128 1008–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288 859–863 [DOI] [PubMed] [Google Scholar]
- Más P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426 567–570 [DOI] [PubMed] [Google Scholar]
- McWatters HG, Kolmos E, Hall A, Doyle MR, Amasino RM, Gyula P, Nagy F, Millar AJ, Davis SJ (2007) ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol 144 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Saez L, Young MW (2006) PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science 311 226–229 [DOI] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al (2008) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Kay SA (1996) Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci USA 93 15491–15496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Chua NH, Kay SA (1992) A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carré IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2 629–641 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T (2005) PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46 686–698 [DOI] [PubMed] [Google Scholar]
- Onai K, Okamoto K, Nishimoto H, Morioka C, Hirano M, Kami-Ike N, Ishiura M (2004) Large-scale screening of Arabidopsis circadian clock mutants by a high-throughput real-time bioluminescence monitoring system. Plant J 40 1–11 [DOI] [PubMed] [Google Scholar]
- Para A, Farré EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA (2007) PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19 3462–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR (2002) Coordination of circadian timing in mammals. Nature 418 935–941 [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229 [DOI] [PubMed] [Google Scholar]
- Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C (2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417 763–767 [DOI] [PubMed] [Google Scholar]
- Shirasu N, Shimohigashi Y, Tominaga Y, Shimohigashic M (2003) Molecular cogs of the insect circadian clock. Zoolog Sci 20 947–955 [DOI] [PubMed] [Google Scholar]
- Somers DE (1999) The physiology and molecular bases of the plant circadian clock. Plant Physiol 121 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer CA, Kay SA (1999) The ins and outs of circadian regulated gene expression. Curr Opin Plant Biol 2 114–120 [DOI] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Green RM, Wang ZY, Tobin EM (1998) Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci USA 95 11020–11025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Doxsee RA, Harel E, Tobin EM (1993) CA-1, a novel phosphoprotein, interacts with the promoter of the cab140 gene in Arabidopsis and is undetectable in det1 mutant seedlings. Plant Cell 5 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, McNellis TW, Deng XW (1998) Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J 17 5577–5587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33 949–956 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217 [DOI] [PubMed] [Google Scholar]
- Yakir E, Hilman D, Harir Y, Green RM (2007) Regulation of output from the plant circadian clock. FEBS J 274 335–345 [DOI] [PubMed] [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, et al (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60 107–124 [DOI] [PubMed] [Google Scholar]
- Zhang X, Chen Y, Wang ZY, Chen Z, Gu H, Qu LJ (2007) Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J 51 512–525 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.