Abstract
The I subunit of magnesium-chelatase (CHLI) is encoded by two genes in Arabidopsis (Arabidopsis thaliana), CHLI1 and CHLI2. Conflicting results have been reported concerning the functions of the two proteins. We show here that the chli1/chli1 chli2/chli2 double knockout mutant was albino. Comparison with the pale-green phenotype of a chli1/chli1 single knockout mutant indicates that CHLI2 could support some chlorophyll biosynthesis in the complete absence of CHLI1. Real-time quantitative reverse transcription-polymerase chain reaction showed that CHLI2 was expressed at a much lower level than CHLI1. The chli1/chli1 chli2/chli2 double mutant could be fully rescued by expressing a transgene of CHLI2 driven by the CHLI1 promoter. These results suggest that differences between CHLI1 and CHLI2 lie mostly in their expression levels. Furthermore, both the chli1/chli1 and chli2/chli2 single knockout mutants had lower survival rates during de-etiolation than the wild type, suggesting that both genes are required for optimal growth during de-etiolation. In addition, we show that a semidominant chli1 mutant allele and the chli1/chli1 chli2/chli2 double mutant accumulated Lhcb1 transcripts when treated with the herbicide norflurazon, indicating that knocking out the CHLI activity causes the genome-uncoupled phenotype.
Magnesium (Mg)-chelatase catalyzes the first committed step toward chlorophyll synthesis in the tetrapyrrole biosynthesis pathway. The enzyme inserts Mg2+ into protoporphyrin IX and produces Mg-protoporphyrin IX. The enzyme is composed of three subunits: CHLH, CHLD, and CHLI (corresponding to BchH, BchD, and BchI in Rhodobacter and XAN-F, XAN-G, and XAN-H in barley [Hordeum vulgare], respectively). Both in vitro and in vivo evidence has shown that all three subunits are essential for the Mg-chelatase activity (Gibson et al., 1995; Willows et al., 1996; Kannangara et al., 1997). The H subunit binds protoporphyrin IX and may be the catalytic subunit for the metallation reaction. The D and I subunits form an activation complex in an ATP- and Mg2+-dependent manner. In Arabidopsis (Arabidopsis thaliana), both the D and H subunits are encoded by single genes.
CHLI belongs to the AAA+ (ATPases associated with various cellular activities) family of ATPases. It forms a ring-shaped homohexamer. Many semidominant and recessive alleles of chli mutants have been isolated from various species (Kjemtrup et al., 1998; Fitzmaurice et al., 1999; Hansson et al., 2002; Soldatova et al., 2005). The semidominant alleles are all missense mutations that still produce full-length proteins. The mutant proteins can assemble with the wild-type proteins into the hexameric ring, hindering the ATPase activity and therefore resulting in the semidominant phenotype (Hansson et al., 2002).
CHLI isoforms in Arabidopsis are encoded by two genes: CHLI1 (At4g18480) and CHLI2 (At5g45930). CHLI1 seems to be the major functional form, since chlorophyll levels in chli1-null mutants are reduced to 10% to 17% of the wild-type level (Rissler et al., 2002). Some reports have shown that CHLI1 and CHLI2 RNAs are expressed at similar levels (Rissler et al., 2002; Apchelimov et al., 2007). Because CHLI2 protein was not detected even in isolated chli1 mutant plastids, it has been proposed that CHLI2 is subjected to rapid posttranslational turnover and does not accumulate in vivo (Rissler et al., 2002). Alternatively, it has also been suggested that CHLI2 cannot assemble into the hexameric ring structure due to changes in residues in its C terminus (Apchelimov et al., 2007). However, from a miniarray experiment examining all genes involved in tetrapyrrole biosynthesis (Matsumoto et al., 2004) and also from the Genevestigator (https://www.genevestigator.ethz.ch/; Zimmermann et al., 2004) and MPSS (for Massively Parallel Signature Sequencing; http://mpss.udel.edu/at/; Brenner et al., 2000) databases, the expression level of CHLI2 is shown to be much lower than that of CHLI1. It is possible, therefore, that CHLI2 function could not be observed due to its very low expression level. Indeed, it has been shown that recombinant CHLI2 has ATPase activity, although with a lower Vmax and KmATP than CHLI1 (Kobayashi et al., 2008). A double mutant of a partially functional light-green allele of chli1, cs (or ch42-2), with a chli2-knockout mutant, is albino. This result indicates that at least in the presence of a partially functional CHLI1, CHLI2 can contribute to some chlorophyll biosynthesis (Kobayashi et al., 2008).
The tetrapyrrole biosynthesis pathway also seems to be important for retrograde signaling from plastids to the nucleus. One frequently used method for studying the signaling pathway is to treat plants with the herbicide norflurazon (Nf), which inhibits carotenoid biosynthesis and causes photo-oxidative damage to plastids. This treatment results in repressed expression of nucleus-encoded photosynthetic genes like Lhcb1 in response to signals sent by the damaged plastids. Arabidopsis mutants that still express Lhcb1 in the presence of Nf have been identified and are named genome uncoupled (gun) mutants (Susek et al., 1993; Cottage et al., 2008). Noticeably, four of the original five gun mutants are defective in tetrapyrrole biosynthesis. The gun2 and gun3 mutants are defective in heme oxygenase and phytochromobilin synthase, respectively. The gun5 mutant has a missense mutation in CHLH, and GUN4 encodes a regulator of Mg-chelatase (Mochizuki et al., 2001; Larkin et al., 2003). A knockout mutation in CHLD also results in the gun phenotype (Strand et al., 2003). Therefore, it seems that perturbation of Mg-chelatase activity would result in the gun phenotype. However, two alleles of the chli1 mutants, ch42 (ch42-1) and cs (ch42-2), do not accumulate Lhcb1 transcripts when treated with Nf and therefore are not gun mutants (Mochizuki et al., 2001). It has been proposed that the presence of CHLI2 in the chli1 mutants may be sufficient to allow functioning of the retrograde signaling pathway (Nott et al., 2006).
During our search for Arabidopsis mutants defective in protein import into chloroplasts, several pale-green or albino mutants were collected from various sources. One of the mutants from the Arabidopsis Biological Resource Center (ABRC), cs215, although not defective in protein import (data not shown), showed an interesting semidominant phenotype. Homozygous cs215 mutants were albino, while heterozygous plants were pale green. Positional cloning of the cs215 locus revealed that cs215 is a new allele of chli1 mutants. To compare cs215 with other chli mutants, we obtained T-DNA or Ds insertion alleles of chli1 and chli2 mutants and generated various double mutants. Our results indicated that the low functionality of CHLI2 was mostly due to its low expression level compared with that of CHLI1. We further tested the homozygous cs215 mutant and the chli1/chli1 chli2/chli2 double mutant for the gun phenotype. When treated with Nf, the chli1/chli1 chli2/chli2 double mutant accumulated Lhcb1 to a level similar to that observed in a chld-knockout mutant and the cs215 homozygous mutant accumulated an even higher level of Lhcb1, similar to that observed in a chlh-knockout mutant, suggesting that knocking out the CHLI activity also caused the gun phenotype.
RESULTS
Identification of the cs215 Locus
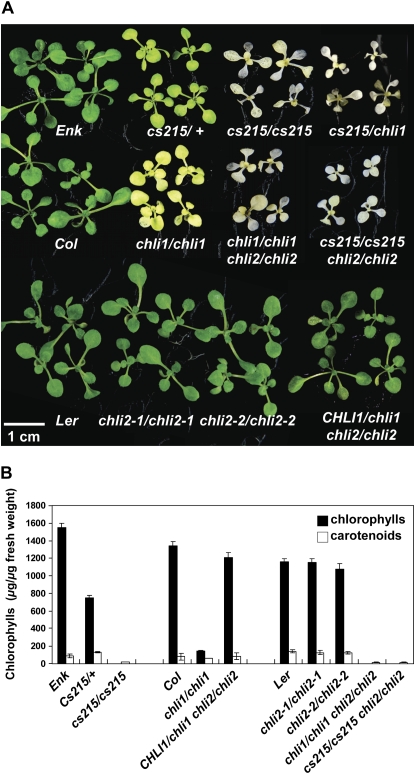
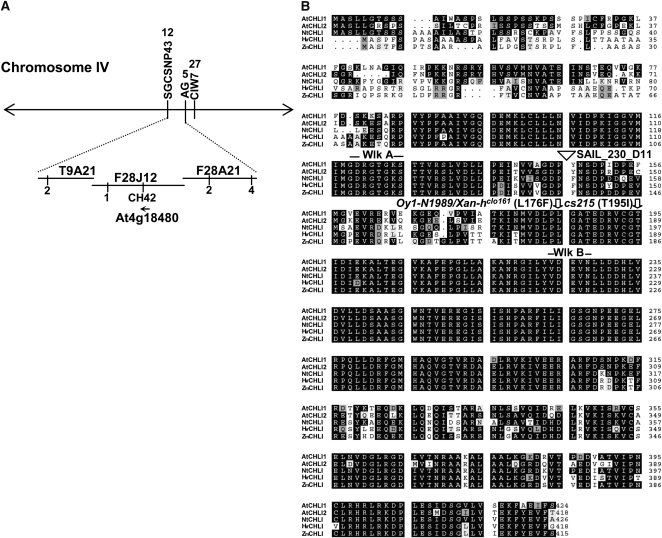
Homozygous cs215 (cs215/cs215) mutants were albino, while heterozygous (cs215/+) plants were pale green (Fig. 1). To identify the cs215 locus, we crossed the cs215/+ mutant with Columbia (Col) and Landsberg erecta (Ler) wild-type plants because the cs215 mutant is in the ecotype Enkheim (Enk). We identified PCR-based polymorphic markers that distinguish between the Enk and Col/Ler ecotypes (see “Materials and Methods”). Initial mapping placed the cs215 locus between markers AG and SGCSNP43 on chromosome IV (Fig. 2A). Data from three recombinant plants delimited the cs215 locus to the region encompassed by bacterial artificial chromosomes F28J12 and F28A21. One of the genes in this region, At4g18480 encoding CHLI1, has been shown to have mutant alleles with semidominant pale-green phenotypes (Kjemtrup et al., 1998; Fitzmaurice et al., 1999; Hansson et al., 2002; Soldatova et al., 2005). Therefore, we sequenced the region of At4g18480 from cs215 and found that cs215 indeed had a C-to-T mutation at nucleotide 584 that converted a Thr at residue 195 to an Ile (Fig. 2B). Pale-green plants were heterozygous at this position. Thr-195 is located between the Walker A and Walker B motifs of the ATPase domain of CHLI and is only 10 residues downstream from the mutations in the barley Xan-hclo161 and maize (Zea mays) Oy1-N1989 mutants (Fig. 2B), both of which are semidominant alleles of the chli mutants. When a cs215/+ plant was crossed with a chli1-knockout mutant (see below), the F1 seedlings had the same phenotype as cs215/cs215 (cs215/chli1; Fig. 1A), further supporting that cs215 was allelic to chli1. We also sequenced the CHLI2 gene from the Enk ecotype, and the result indicated that CHLI2 from Enk has an identical deduced amino acid sequence to that of CHLI2 from Col (data not shown). Therefore, it is unlikely that the severe phenotype of cs215 was caused by a synthetic effect with natural variations in the CHLI2 gene of the Enk ecotype.
Figure 1.
Phenotypes of various mutants and double mutants. A, Plants of the indicated genotypes were grown on MS medium for 12 d. B, Chlorophyll and carotenoid contents in 12-d-old plants as shown in A. Data shown are means ± sd from five independent samples per genotype, each sample containing five to eight plants.
Figure 2.
Identification of the cs215 locus. A, Summary of cs215 mapping. Vertical lines indicate the positions of PCR-based markers. Values beneath the lines indicate the number of recombinant plants. The direction of transcription of the At4g18480 open reading frame is indicated (arrow). B, Alignment of Arabidopsis CHLI1 and related proteins. Alignment of Arabidopsis CHLI1 (AtCHLI1) and CHLI2 (AtCHLI2), Sulfur from tobacco (Nicotiana tabacum; NtCHLI), and CHLI from barley (HvCHLI) and maize (ZmCHLI). Walker A (Wlk A) and Walker B (Wlk B) motifs of the ATP-binding fold are marked above the sequences. Mutation positions of the chli1 (SAIL_230_D11), cs215, and maize Oy1-N1989/barley Xan-hclo161 mutants are also indicated.
CHLI2 Supports Some Chlorophyll Biosynthesis in the Absence of CHLI1
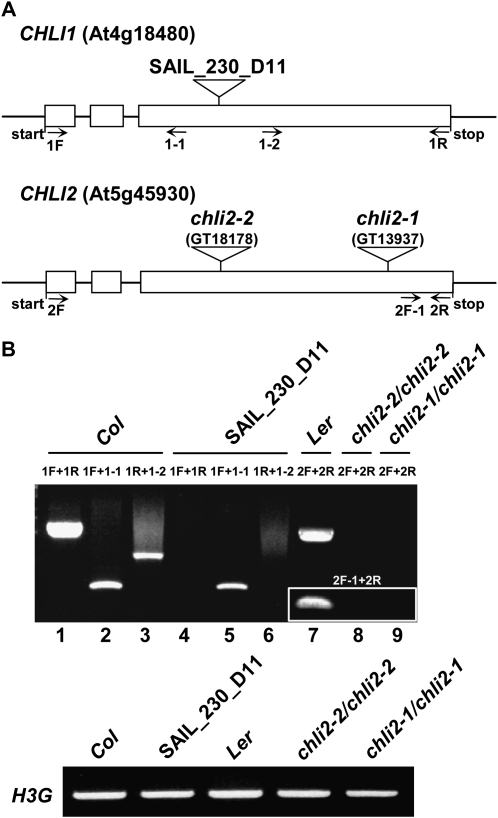
To compare the phenotype of c215 with those of other chli1 mutants, a mutant line with a T-DNA insertion in the third exon of the CHLI1 gene (SAIL_230_D11; Fig. 3A) was obtained from the ABRC. This mutant produced a truncated CHLI1 mRNA but no full-length transcripts or transcripts behind the T-DNA insertion site (Fig. 3B, lanes 4–6). It was yellow in appearance (Fig. 1A) and had less than 10% of the wild-type level of chlorophylls (Fig. 1B), similar to the levels reported for other chli1-null mutants (Rissler et al., 2002). These data suggested that SAIL_230_D11 is most likely also a chli1-null mutant. SAIL_230_D11 will be referred to as the chli1/chli1 mutant herein.
Figure 3.
Transcript analyses of the chli1/chli1 and chli2/chli2 mutants. A, Schematic representation of the structures of the CHLI genes and the location of each insertion site. Exons are represented by white boxes, and introns are represented by lines between boxes. Location and direction of primers used for RT-PCR are also indicated (arrows). B, Analyses of CHLI transcripts in chli1/chli1 and chli2/chli2 mutants. Total RNA extracted from wild-type and mutant plants was analyzed by RT-PCR. Histone 3G (H3G; At4g40040) transcripts were analyzed as a control.
Comparison of chli1/chli1 with cs215/cs215 revealed that chli1/chli1 had more chlorophylls than cs215/cs215 (Fig. 1B). Arabidopsis has two genes encoding CHLI: CHLI1 and CHLI2. It is possible that in the chli1/chli1 mutant, CHLI2 can still support some chlorophyll biosynthesis. In the cs215/cs215 mutant, the presence of the cs215 mutant protein might prevent CHLI2 from functioning, resulting in the phenotype difference between cs215/cs215 and chli1/chli1. To verify this, we obtained two mutant lines, GT13937 (Kobayashi et al., 2008) and GT18178, with Ds transposon insertion in the CHLI2 gene (Fig. 3A). Reverse transcription (RT)-PCR analyses indicated that neither mutant produced any full-length CHLI2 mRNA or any CHLI2 mRNA 3′ to the Ds insertion site (Fig. 3B, lanes 8 and 9 and inset). Both mutants were wild type in appearance (Fig. 1A) and in chlorophyll levels (Fig. 1B). GT13937 and GT18178 are referred to herein as chli2-1 and chli2-2, respectively.
We crossed cs215/+ and CHLI1/chli1 with chli2-1/chli2-1 and chli2-2/chli2-2 to generate various double mutants. Results from crosses to chli2-1/chli2-1 and to chli2-2/chli2-2 were identical, and data from chli2-1 are presented unless specified. Double mutants of chli1/chli1 chli2/chli2 were albino (Fig. 1A) with no detectable chlorophylls (Fig. 1B). Compared with the chli1/chli1 single mutant, the albino phenotype of the double mutant indicated that the CHLI2 protein must be stable enough to provide some Mg-chelatase activity even in the absence of CHLI1. Furthermore, double mutants of cs215/cs215 chli2/chli2 were indistinguishable from the cs215 single (cs215/cs215 CHLI2/CHLI2) mutant (Fig. 1). Therefore, it is likely that the presence of the cs215 mutant protein had prevented CHLI2 from functioning. This may be because the expression level of CHLI1 is much higher than that of CHLI2, as shown previously by Matsumoto et al. (2004). In the cs215 mutant, the high amount of cs215 mutant protein may out-compete the low amount of CHLI2 in assembly with CHLD. It is also possible that the expression levels of CHLI1 and CHLI2 were similar (Rissler et al., 2002; Apchelimov et al., 2007) and that the presence of the cs215 mutant protein inhibited the expression of CHLI2 as suggested previously (Soldatova et al., 2005).
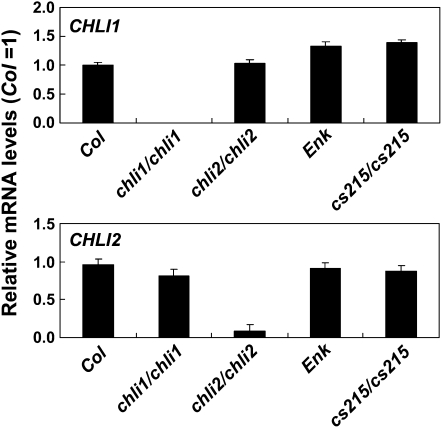
The Expression Level of CHLI2 Is Much Lower Than That of CHLI1 and Is Not Affected by Mutations in CHLI1
To clarify the role of CHLI2, we compared the expression levels of CHLI1 and CHLI2 using real-time quantitative RT-PCR. Amounts of CHLI1 and CHLI2 RNA were determined by comparing with standard curves generated from known quantities of CHLI1 and CHLI2 plasmid DNA. The results showed that in 14-d-old seedlings, the ratio of CHLI1 to CHLI2 RNA was 5.86 ± 1.31 (n = 5). This result agrees with the reported miniarray data (Matsumoto et al., 2004) and is also supported by data found in Genevestigator (expression value of 9,764 versus 2,121 in rosette leaves) and MPSS (928 versus 132 transcripts per million in 21-d-old leaves) databases. The level of CHLI2 was not changed in the chli1/chli1 or cs215/cs215 mutants (Fig. 4, bottom), indicating that mutations in CHLI1 did not affect the expression of CHLI2. The level of CHLI1 was also not changed in the chli2/chli2 or cs215/cs215 mutants (, top).
Figure 4.
CHLI1 and CHLI2 mRNA levels in the wild type and chli mutants. Total RNA was isolated from 12-d-old seedlings. CHLI1 and CHLI2 mRNA levels were determined by real-time quantitative RT-PCR as described in “Materials and Methods.” Data shown are means ± sd from four independent samples per genotype, and each sample contained five plants.
CHLI2 Driven by the CHLI1 Promoter Can Rescue the chli1/chli1 chli2/chli2 Double Mutant
It has been suggested that CHLI2 is defective in hexameric ring assembly due to changes in its C terminus (Apchelimov et al., 2007). However, it has also been shown that CHLI2 is an active ATPase (Kobayashi et al., 2008). To test the function of CHLI2 in vivo, we fused the CHLI2 gene behind a 2.1-kb CHLI1 promoter fragment and used this transgene (referred to herein as pCHLI1∷CHLI2) to complement the chli1/chli1 chli2/chli2 double mutant. Transgenic plants with one copy of the pCHLI1∷CHLI2 transgene were referred to as the pCHLI1∷CHLI2 (chli1/chli1 chli2/chli2) plants. For fair comparison, these plants were compared with plants with only one endogenous copy of CHLI1 (genotype CHLI1/chli1 chli2/chli2 and referred to herein accordingly). Nonetheless, CHLI1/chli1 chli2/chli2 plants were similar to wild-type plants in both appearance (Fig. 1A) and chlorophyll levels (Fig. 1B).
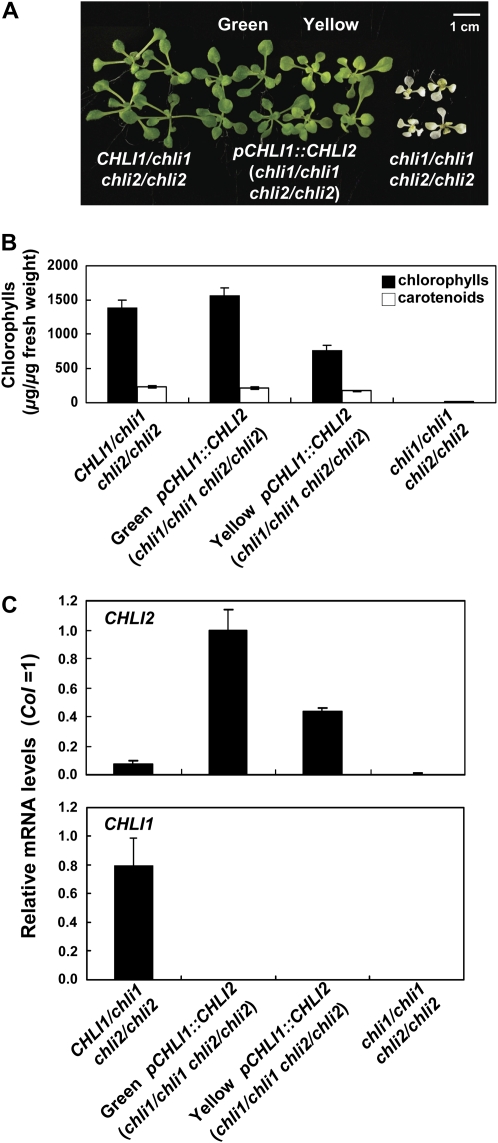
The pCHLI1∷CHLI2 (chli1/chli1 chli2/chli2) transgenic plants had phenotypes that ranged from fully green to slightly yellow in younger leaves (Fig. 5A, Green and Yellow, respectively). The Green transgenic plants had chlorophyll levels similar to those of CHLI1/chli1 chli2/chli2 plants (Fig. 5B). This result indicated that CHLI2 could substitute for CHLI1 if expressed at a sufficient level. The Yellow transgenic plants had chlorophyll and CHLI2 transcript levels about half of those in the Green transgenic plants (Fig. 5, B and C). This result further demonstrated that the expression level of CHLI2 was correlated with the level of chlorophyll biosynthesis activity observed.
Figure 5.
CHLI2 driven by the CHLI1 promoter rescued the chli1/chli1 chli2/chli2 double mutant. A, Plants of the indicated genotypes were grown on MS medium for 12 d and then photographed. The pCHLI1∷CHLI2 (chli1/chli1 chli2/chli2) transgenic plants had phenotypes that ranged from fully green (Green) to slightly yellow in younger leaves (Yellow). Representative transgenic lines were photographed and used for analyses shown in B and C. B, Chlorophyll and carotenoid contents of 12-d-old plants as shown in A. Data shown are means ± sd derived from five independent samples per genotype, each sample containing five to eight plants. C, Comparison of CHLI2 mRNA levels in the wild type and chli mutants. Seedlings were grown as described in A. mRNA levels were determined by real-time quantitative RT-PCR as described in “Materials and Methods.” Data shown are means ± sd derived from five independent samples per genotype, each one containing five to eight plants. CHLI1 transcript levels were also determined as controls.
The chli Mutants Have Lower Survival Rates during De-Etiolation
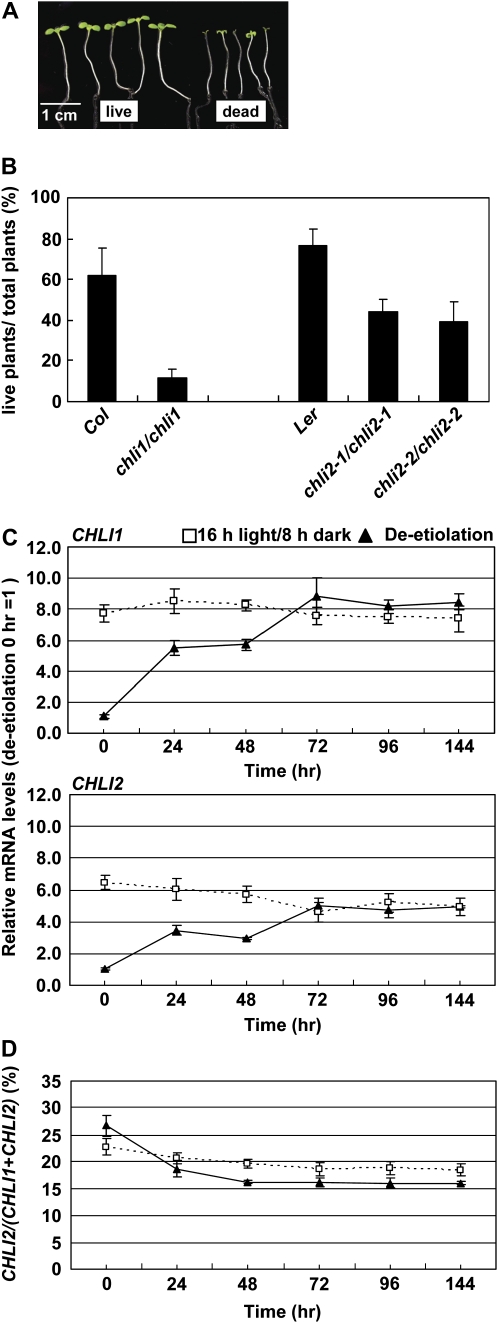
To investigate if CHLI2 contributes to plant fitness, we compared the chli mutants with the wild type in de-etiolation experiments. Seeds were imbibed at 4°C in the dark for 3 d, exposed to light at 24°C for 1 d, and then moved to the dark to germinate at 24°C for 7 d. Etiolated seedlings were then transferred to a regular 16-h-light/8-h-dark cycle, and seedling survival rates were scored 6 d later. Seedlings that failed to expand their cotyledons (i.e. the two cotyledons had a combined length less than 0.25 cm) were scored as dead (Fig. 6A). As shown in Figure 6B, the chli1/chli1 mutant had a 14% survival rate and the two alleles of chli2/chli2 mutants had a 40% survival rate. Both rates were significantly lower than that of their corresponding wild type. This result suggested that both genes contributed to increase the seedling survival rate upon de-etiolation. We then analyzed the CHLI1 and CHLI2 RNA levels during de-etiolation, comparing with the CHLI1 and CHLI2 RNA levels in seedlings that were grown under a regular 16-h-light/8-h-dark cycle after cold stratification (Fig. 6C). The results indicated that the expression of both genes was induced by light upon de-etiolation. When the RNA levels of the two genes were directly compared, CHLI1 was still severalfold higher than CHLI2, agreeing with the result that the survival rate of the chli1 mutant was more severely affected than that of the chli2 mutants. However, CHLI2 contributed to about 27% of total CHLI RNA in the dark and only about 16% of total CHLI after de-etiolation (Fig. 6D), suggesting that CHLI2 might have slightly more contribution in the dark. This might be the reason that chli2 mutants had no clear phenotype when directly grown under the light but had a lower survival rate in transitions from dark to light.
Figure 6.
The chli mutants had lower survival rates during de-etiolation. Seeds were grown on MS medium and imbibed at 4°C in the dark for 3 d, exposed to light at 22°C for 1 d, and then moved to the dark to germinate at 22°C for 7 d. Etiolated seedlings were then transferred to a 16-h-light/8-h-dark cycle for another 6 d (de-etiolation) and then photographed. A, Examples of seedlings being scored as “live” or “dead.” B, Seedling survival rates of Col, Ler, chli1/chli1, chli2-1/chli2-1, and chli2-2/chli2-2 after de-etiolation. Data shown are means ± sd of six independent experiments, each experiment containing 100 to 120 seedlings per genotype. C, Comparison of CHLI1 and CHLI2 mRNA levels in Ler during de-etiolation (black triangles and solid lines). CHLI1 and CHLI2 mRNA levels were determined by real-time quantitative RT-PCR as described in “Materials and Methods.” D, Percentage of CHLI2 in total CHLI (CHLI1+CHLI2) during de-etiolation. Amounts of CHLI1 and CHLI2 mRNA were determined by real-time quantitative RT-PCR and deduced from standard curves produced from known quantities of CHLI1 and CHLI2 plasmid DNA. For both C and D, time on the x axis indicates hours after transferring to the 16-h-light/8-h-dark cycle after etiolation. Seedlings were harvested 3 h after light came on each day. Seedlings of the same age but grown under a 16-h-light/8-h-dark cycle since stratification were analyzed as a control (white squares and dotted lines). Data shown are means ± sd of four independent samples, each sample containing 50 to 60 plants. [See online article for color version of this figure.]
The chli1/chli1 chli2/chli2 Double Mutant and the cs215/cs215 Mutant Are gun Mutants
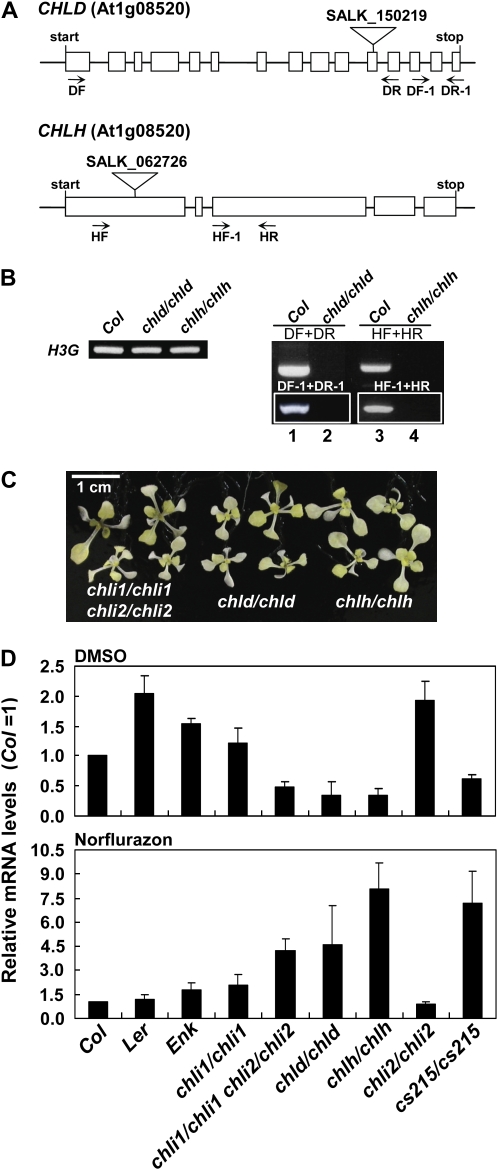
Mutants in the H and D subunits of Mg-chelatase are gun mutants (Strand et al., 2003), but two alleles of the chli1 mutants are not (Mochizuki et al., 2001). To investigate if the chli1/chli1 chli2/chli2 double mutant or the cs215/cs215 mutant would show the gun phenotype, we first obtained mutants with T-DNA insertion in the CHLD and CHLH genes for use as controls. The chld mutant allele was identical to the allele reported previously and has been shown to be a gun mutant (SALK_150219; Strand et al., 2003; Ankele et al., 2007). The chlh mutant has a T-DNA insertion in the first exon of the CHLH gene (SALK_062726; Fig. 7A). No transcripts across the T-DNA insertion site or 3′ to the T-DNA insertion site could be detected in either mutant (Fig. 7B), and both mutants had an albino phenotype similar to that of the chli1/chli1 chli2/chli2 double mutant (Fig. 7C). We then measured Lhcb1 transcript levels in these mutants after treatment with Nf. As shown in Figure 7D, the chli1/chli1 chli2/chli2 double mutant accumulated a higher level of Lhcb1 transcripts than the wild type, similar to the level found in the chld/chld mutant. Interestingly, cs215/cs215 and chlh/chlh mutants accumulated an even higher level of Lhcb1 transcripts. These results indicated that the chli1/chli1 chli2/chli2 double mutant and the cs215/cs215 mutant were gun mutants.
Figure 7.
Lhcb1 mRNA levels in Nf-treated wild type (Col, Ler, and Enk), various chli mutants, and chld/chld and chlh/chlh mutants. A, Schematic representation of the CHLD and CHLH genes. Exons are represented by white boxes, and introns are represented by lines between the boxes. The locations of T-DNA insertion sites and RT-PCR primers are indicated. B, No transcript across the T-DNA insertion site or 3′ to the insertion site could be detected in the chld/chld or chlh/chlh mutants. Total RNA extracted from wild-type and mutant plants was analyzed by RT-PCR using primers indicated in A. H3G transcripts were analyzed as a control. C, Phenotypes of the chli1/chli1 chli2/chli2, chld/chld, and chlh/chlh mutants. Plants were grown on MS medium for 12 d. D, Comparison of Lhcb1 mRNA levels in the wild type and mutants. Seedlings were grown on MS medium for 5 d. Homozygous pale-green or albino mutants (chli1/chli1, cs215/cs215, chli1/chli1 chli2/chli2, chld/chld, and chlh/chlh) were selected and, together with the chli2/chli2 and wild-type seedlings, transferred to MS medium containing either 5 μm Nf or the same volume of dimethyl sulfoxide (DMSO). Seedlings were grown for another 5.5 d under strong continuous light (100 μmol m−2 s−1) and then used for total RNA isolation. Lhcb1 mRNA levels were determined by real-time quantitative RT-PCR as described in “Materials and Methods.” Data shown are means ± sd derived from four independent samples per genotype, each sample containing 10 to 15 plants.
Absence of CHLI Causes Instability of CHLD
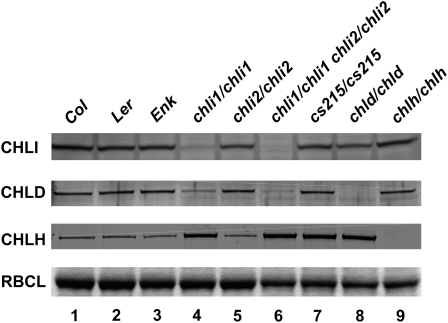
It has been shown that changes in the level of one Mg-chelatase subunit causes instability in other subunits (Hansson et al., 1999; Petersen et al., 1999). Therefore, we investigated whether the level of CHLD or CHLH proteins was affected in the various chli mutants. We used immunoblots to detect the levels of the three Mg-chelatase subunits. As shown in Figure 8, in the chli1/chli1 and chli1/chli1 chli2/chli2 mutants, CHLI was reduced or absent and the amount of CHLD protein was also greatly reduced (lanes 4 and 6). In comparison, the cs215/cs215 mutant still contained a normal amount of CHLI protein (Fig. 8, lane 7) and its amount of CHLD protein was also normal. On the other hand, the amount of CHLI was not affected by the chld mutation (Fig. 8, lane 8). These results agreed with results from barley showing that chli-null mutants also lacked CHLD but chld mutations did not affect the amount of CHLI (Petersen et al., 1999). Furthermore, the normal amount of CHLD in the cs215 mutant suggested that the cs215 mutant protein could still complex with CHLD, agreeing with previous observations in barley that semidominant mutant chli protein could still complex with CHLD (Hansson et al., 1999). In addition, the presence of CHLD and CHLH in the cs215/cs215 mutant also indicated that the gun phenotype of the cs215/cs215 mutant was not caused by the absence of CHLD or CHLH. Interestingly, loss of CHLI or CHLD activity in the chli1/chli1, chli1/chli1 chli2/chli2, cs215/cs215, and chld/chld mutants seemed to have resulted in an increase of the CHLH protein level (Fig. 8, lanes 4 and 6–8).
Figure 8.
Immunoblot analyses of the three Mg-chelatase subunits in various mutants and corresponding wild types. Total proteins extracted from leaves of 14-d-old plants were analyzed by SDS-PAGE and immunoblots with antibodies against CHLI, CHLD, and CHLH. RBCL from the same set of samples as revealed by Coomassie Brilliant Blue staining was analyzed as a control.
DISCUSSION
The expression level of CHLI2 was much lower than that of CHLI1. When driven by the CHLI1 promoter, expression of CHLI2 alone rescued the chli1/chli1 chli2/chli2 double mutant. These data indicate that CHLI2 can be functionally equivalent to CHLI1 if expressed at a sufficient level. This result also agrees with results from Kobayashi et al. (2008) showing that CHLI2 is a functional ATPase. Previous proposals of CHLI2 rapid turnover and inability in hexameric ring assembly were based on data suggesting that CHLI1 and CHLI2 had similar expression levels (Rissler et al., 2002; Apchelimov et al., 2007), which might have resulted from probe or primer cross-reactions.
The chli2/chli2 mutants had no clear phenotype when grown in the light but had a lower survival rate during de-etiolation, suggesting that the presence of a second copy of CHLI may contribute to plant fitness under certain growth conditions. Although CHLI1 was the major isoform in both light and dark, CHLI2 contributed to about 27% of total CHLI RNA in the dark and only about 16% of total CHLI after de-etiolation. These results agreed with the miniarray data showing that CHLI1 RNA was almost 3-fold of CHLI2 in 3-week-old light-grown seedling but only 2-fold of CHLI2 in 3-d-old etiolated seedlings (Matsumoto et al., 2004). These results suggest that the presence of CHLI2 may be more important in the dark or in a sudden dark-to-light transition as in the de-etiolation experiments. We searched several sequenced plant genomes and found that two copies of CHLI are present in Chlamydomonas reinhardtii, Physcomitrella patens, and Populus trichocarpa but only one copy is present in rice (Oryza sativa) and Sorghum bicolor. It is not clear why monocots seem to have lost the second copy of CHLI. It is possible that changes in the physiology of monocots rendered the second copy unnecessary.
It has been suggested that, because the N terminus of CHLD has some sequence similarity to CHLI, CHLD may interact with CHLH in the absence of CHLI and provide a low level of Mg-chelatase activity (Rissler et al., 2002). However, our data showed that both the chli1/chli1 chli2/chli2 double mutant and the cs215/cs215 mutant had no detectable chlorophylls and were indistinguishable from the chld/chld- and chlh/chlh-knockout mutants. In fact, similar to barley, the stability of CHLD seems to rely on the presence of CHLI. These results suggest that CHLD and CHLH support no significant chlorophyll biosynthesis in the absence of CHLI, agreeing with results from Rhodobacter and barley that all three subunits are required for the Mg-chelatase activity (Gibson et al., 1995; Willows et al., 1996; Kannangara et al., 1997).
We showed that the chli1/chli1 chli2/chli2 double mutant and the cs215/cs215 mutant showed the gun phenotype. Our data help remove doubts that knocking out the Mg-chelatase activity causes the gun phenotype. Indeed, mutants in all three subunits in barley have been shown to be gun mutants (Gadjieva et al., 2005). It has been shown recently that the immediate product of Mg-chelatase, Mg-protoporphyrin IX, does not accumulate under Nf treatment and therefore is unlikely to be the determinant for retrograde signaling (Mochizuki et al., 2008; Moulin et al., 2008). Therefore, it remains to be elucidated why perturbation of the tetrapyrrole biosynthesis pathway often causes the gun phenotype. It is also interesting that the cs215/cs215 and the chlh-knockout mutants showed a higher level of Lhcb1 accumulation than the chli1/chli1 chli2/chli2 double mutant and the chld/chld mutant, even though they are indistinguishable in appearance. These mutants may be useful materials to facilitate the identification of compounds whose accumulation levels correlate with the Lhcb1 transcript levels and that therefore may be candidates for the signaling molecules.
MATERIALS AND METHODS
Positional Cloning of the cs215 Locus
The cs215 mutant of Arabidopsis (Arabidopsis thaliana) is in the Enk ecotype. Heterozygous cs215 plants were crossed with wild-type Col and Ler plants. DNA from F2 seedlings with the cs215/cs215 phenotype was isolated for mapping. Simple sequence length polymorphism and cleaved-amplified polymorphic sequence markers between the Col and Ler ecotypes (http://www.Arabidopsis.org) were tested on Enk DNA to identify markers that could distinguish Enk versus Col or Enk versus Ler ecotypes.
Plant Materials, Growth Conditions, and Treatments
The cs215 (Enk ecotype) and chli1, chld, and chlh (Col ecotype) mutants were obtained from the ABRC (http://www.Arabidopsis.org/abrc/). The chli2 mutants GT13937 and GT18178 (Ler ecotype) were obtained from Cold Spring Harbor Laboratory (http://genetrap.cshl.org). The T-DNA or Ds insertion positions in the mutants were confirmed by PCR and direct sequencing of the PCR products (for primer sequences, see Supplemental Table S1). Mutants were backcrossed to their corresponding wild type, and lines with single T-DNA or Ds insertion were selected based on the kanamycin or BASTA (glufosinate ammonium) resistance segregation ratio.
Sterilized seeds were plated on Murashige and Skoog (MS) medium containing 0.3% Gelrite, 1× Murashige and Skoog salts, Gamborg's B5 vitamin, and 2% Suc. After a 3-d cold stratification, seeds were grown in growth chambers under a 16-h photoperiod with a light intensity of approximately 60 μmol m−2 s−1 at 22°C. Total chlorophyll and carotenoid contents were determined as described (Lichtenthaler, 1987). Kanamycin selection of T-DNA and Ds transposon-containing seedlings were performed on medium supplemented with kanamycin (Sigma-Aldrich) at 50 mg L−1. The pCHLI1∷CHLI2 construct was obtained by two-step PCR (primers used were KpnI-CHLI1F and KpnI-CHLI2R for the first step and P1 and P2 for the second step), cloned into the transformation vector pPZP221 (Hajdukiewicz et al., 1994), transformed into Agrobacterium tumefaciens GV3101, and introduced into the CHLI1/chli1 chli2/chli2 mutant using the floral spray method (Chung et al., 2000). Transgenic plants were screened on MS medium containing 100 mg L−1 G418.
For the chli1/chli1 de-etiolation experiment, because the chli1/chli1 mutant was sterile and could only be sown from seeds of CHLI1/chli1 heterozygous plants, seeds from CHLI1/chli1 heterozygous plants were first grown in the light to confirm the segregation ratio of the chli1/chli1 mutant, which was almost always around 1:4. The same batch of seeds was then used for the de-etiolation experiments. When calculating survival rate, the theoretical number of chli1/chli1 seedlings was deduced using the segregation ratio from light-grown seedlings. The number of survived chli1/chli1 seedlings was then counted using the yellow cotyledons of the chli1/chli1 mutant as an indication.
For Nf treatment experiments, seeds (from heterozygous plants of the albino and pale-green mutant lines and from homozygous plants of others) were sterilized and plated on MS medium. After a 3-d cold stratification, plates were moved to a growth chamber for 5 d. Albino (cs215/cs215, chli1/chli1 chli2/chli2, chld/chld, and chlh/chlh) and pale-green (chli1/chli1) mutants were selected and, together with chli2-1/chli2-1 and wild-type seedlings, were transferred to new MS medium containing 5 μm Nf or the same volume of dimethyl sulfoxide and grown for another 5.5 d under strong continuous light (100 μmol m−2 s−1).
RNA Analysis
Total RNA was isolated from Arabidopsis shoots with TRIzol reagent (Invitrogen) and treated with RQ1 RNase-Free DNase (Promega). Template cDNA was prepared using 1 μg of total RNA and the Moloney murine leukemia virus reverse transcription system (Promega). Real-time quantitative RT-PCR was performed using the LightCycler system (Roche Applied Science) and the Lightcycler-FastStart DNA Master SYBR Green I kit (Roche Diagnostics). Each PCR contained 10 to 50 ng of cDNA and 0.5 μM of each of the primer pairs. The initial denaturing step of 10 min was followed by 40 PCR cycles of 95°C for 10 s, 60°C for 5 s, and 72°C for 1 s per 25 bp of the expected product. After the PCR, the melting temperature was tested. Quantification was performed using LightCycler Relative Quantification software version 1.0. Normalization was done using the transcript level of H3G and confirmed by the transcript level of ubiquitin10.
Immunoblots
Total proteins from leaves of 14-d-old plants were extracted with SDS sample buffer (300 mm Tris-HCl, pH 8.5, 1 mm EDTA, pH 8.0, 8% SDS, and 1 mm phenylmethylsulfonyl fluoride). Forty micrograms of total proteins was separated by SDS-PAGE and transferred to Immobilon-P membrane (Millipore). Immunostaining was performed with antisera to soybean (Glycine max) CHLI and CHLH and Plectonema boryanum CHLD at a 1:1,000 dilution followed by secondary staining with an alkaline phosphatase-conjugated goat anti-rabbit serum at a 1:1,000 dilution. Colorimetric development with 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium was used to visualize protein bands.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Wei-Ning Hwang, Ming-D Yang, and Dr. Chi-Chou Chiu for initial mapping of the cs215 locus. We thank the ABRC for the cs215, chld, and chlh mutants and Cold Spring Harbor Laboratory for the chli2 mutants GT13937 and GT18178. We thank Dr. Tatsuru Masuda for providing the CHLI, CHLD, and CHLH antibodies. We thank Dr. Harry Wilson of Academia Sinica for English editing.
This work was supported by the National Science Council (grant no. NSC–97–2321–B–001–001 to H.-m.L.) and the Academia Sinica of Taiwan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hsou-min Li (mbhmli@gate.sinica.edu.tw).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ankele E, Kindgren P, Pesquet E, Strand A (2007) In vivo visualization of Mg-protoporphyrin IX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell 19 1964–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apchelimov AA, Soldatova OP, Ezhova TA, Grimm B, Shestakov SV (2007) The analysis of the ChlI 1 and ChlI 2 genes using acifluorfen-resistant mutant of Arabidopsis thaliana. Planta 225 935–943 [DOI] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd D, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18 630–634 [DOI] [PubMed] [Google Scholar]
- Chung MH, Chen MK, Pan SM (2000) Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Res 9 471–476 [DOI] [PubMed] [Google Scholar]
- Cottage AJ, Mott EK, Wang JH, Sullivan JA, MacLean D, Tran L, Choy MK, Newell C, Kavanagh TA, Aspinall S, et al (2008) GUN1 (GENOMES UNCOUPLED1) encodes a pentatricopeptide repeat (PPR) protein involved in plastid protein synthesis-responsive retrograde signaling to the nucleus. In J Allen, E Gantt, J Golbeck, B Osmond, eds, Photosynthesis: Energy from the Sun. 14th International Congress on Photosynthesis. Springer, Dordrecht, The Netherlands, pp 1201–1205
- Fitzmaurice WP, Nguyen LV, Wernsman EA, Thompson WF, Conkling MA (1999) Transposon tagging of the sulfur gene of tobacco using engineered maize Ac/Ds elements. Genetics 153 1919–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjieva R, Axelsson E, Olsson U, Hansson M (2005) Analysis of gun phenotype in barley magnesium chelatase and Mg-protoporphyrin IX monomethyl ester cyclase mutants. Plant Physiol Biochem 43 901–908 [DOI] [PubMed] [Google Scholar]
- Gibson LC, Willows RD, Kannangara CG, von Wettstein D, Hunter CN (1995) Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: reconstitution of activity by combining the products of the bchH, -I, and -D genes expressed in Escherichia coli. Proc Natl Acad Sci USA 92 1941–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25 989–994 [DOI] [PubMed] [Google Scholar]
- Hansson A, Kannangara CG, von Wettstein D, Hansson M (1999) Molecular basis for semidominance of missense mutations in the XANTHA-H (42-kDa) subunit of magnesium chelatase. Proc Natl Acad Sci USA 96 1744–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson A, Willows RD, Roberts TH, Hansson M (2002) Three semidominant barley mutants with single amino acid substitutions in the smallest magnesium chelatase subunit form defective AAA+ hexamers. Proc Natl Acad Sci USA 99 13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannangara CG, Vothknecht UC, Hansson M, von Wettstein D (1997) Magnesium chelatase: association with ribosomes and mutant complementation studies identify barley subunit Xantha-G as a functional counterpart of Rhodobacter subunit BchD. Mol Gen Genet 254 85–92 [DOI] [PubMed] [Google Scholar]
- Kjemtrup S, Sampson KS, Peele CG, Nguyen LV, Conkling MA, Thompson WF, Robertson D (1998) Gene silencing from plant DNA carried by a geminivirus. Plant J 14 91–100 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Mochizuki N, Yoshimura N, Motohashi K, Hisabori T, Masuda T (2008) Functional analysis of Arabidopsis thaliana isoforms of the Mg-chelatase CHLI subunit. Photochem Photobiol Sci 7 1188–1195 [DOI] [PubMed] [Google Scholar]
- Larkin R, Alonso J, Ecker J, Chory J (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299 902–906 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148 350–382 [Google Scholar]
- Matsumoto F, Obayashi T, Sasaki-Sekimoto Y, Ohta H, Takamiya K, Masuda T (2004) Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol 135 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A (2008) The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc Natl Acad Sci USA 105 15184–15189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M, McCormac A, Terry M, Smith A (2008) Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc Natl Acad Sci USA 105 15178–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57 739–759 [DOI] [PubMed] [Google Scholar]
- Petersen BL, Møller MG, Jensen PE, Henningsen KW (1999) Identification of the Xan-g gene and expression of the Mg-chelatase encoding genes Xan-f, s-g and -h in mutant and wild type barley (Hordeum vulgare L.). Hereditas 131 165–170 [Google Scholar]
- Rissler HM, Collakova E, DellaPenna D, Whelan J, Pogson BJ (2002) Chlorophyll biosynthesis: expression of a second chl I gene of magnesium chelatase in Arabidopsis supports only limited chlorophyll synthesis. Plant Physiol 128 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatova O, Apchelimov A, Radukina N, Ezhova T, Shestakov S, Ziemann V, Hedtke B, Grimm B (2005) An Arabidopsis mutant that is resistant to the protoporphyrinogen oxidase inhibitor acifluorfen shows regulatory changes in tetrapyrrole biosynthesis. Mol Genet Genomics 273 311–318 [DOI] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421 79–83 [DOI] [PubMed] [Google Scholar]
- Susek R, Ausubel F, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 10 787–799 [DOI] [PubMed] [Google Scholar]
- Willows RD, Gibson LC, Kanangara CG, Hunter CN, von Wettstein D (1996) Three separate proteins constitute the magnesium chelatase of Rhodobacter sphaeroides. Eur J Biochem 235 438–443 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.