Abstract
Feedback deexcitation is a photosynthetic regulatory mechanism that can protect plants from high light stress by harmlessly dissipating excess absorbed light energy as heat. To understand the genetic basis for intraspecies differences in thermal dissipation capacity, we investigated natural variation in Arabidopsis (Arabidopsis thaliana). We determined the variation in the amount of thermal dissipation by measuring nonphotochemical quenching (NPQ) of chlorophyll fluorescence in Arabidopsis accessions of diverse origins. Ll-1 and Sf-2 were selected as high NPQ Arabidopsis accessions, and Columbia-0 (Col-0) and Wassilewskija-2 were selected as relatively low NPQ accessions. In spite of significant differences in NPQ, previously identified NPQ factors were indistinguishable between the high and the low NPQ accessions. Intermediate levels of NPQ in Ll-1 × Col-0 F1 and Sf-2 × Col-0 F1 compared to NPQ levels in their parental lines and continuous distribution of NPQ in F2 indicated that the variation in NPQ is under the control of multiple nuclear factors. To identify genetic factors responsible for the NPQ variation, we developed a polymorphic molecular marker set for Sf-2 × Col-0 at approximately 10-centimorgan intervals. From quantitative trait locus (QTL) mapping with undistorted genotype data and NPQ measurements in an F2 mapping population, we identified two high NPQ QTLs, HQE1 (high qE 1, for high energy-dependent quenching 1) and HQE2, on chromosomes 1 and 2, and the phenotype of HQE2 was validated by analysis of near isogenic lines. Neither QTL maps to a gene that had been identified previously in extensive forward genetics screens using induced mutants, suggesting that quantitative genetics can be used to find new genes affecting thermal dissipation.
Plants require light energy, by definition, to drive photosynthesis. However, too much light causes photooxidative damage in plants (Barber and Andersson, 1992). Thus, plants have diverse defense mechanisms against high light stress (Niyogi, 1999). For example, chloroplasts can move to absorb less light energy (Kasahara et al., 2002), and light-harvesting antenna size in chloroplasts can be reduced (Anderson, 1986). Plants also can harmlessly dissipate excess absorbed light energy as heat (Müller et al., 2001), and they have alternative electron transport pathways to relieve overreduction of electron transport components under stress conditions (Niyogi, 2000; Ort and Baker, 2002).
Thermal dissipation is mediated by a mechanism called feedback deexcitation. Feedback deexcitation dissipates excess absorbed light energy as heat, thereby protecting plants from high-light stress (Horton et al., 1994; Niyogi, 1999). The amount of feedback deexcitation can be quantified by measuring nonphotochemical quenching (NPQ) of chlorophyll fluorescence (Müller et al., 2001; Baker, 2008). NPQ is induced by appearance of high light and is relaxed following disappearance of the high light. Based on its relaxation kinetics, NPQ can be divided into at least three components: energy-dependent quenching (qE), state-transition quenching, and photoinhibitory quenching (Maxwell and Johnson, 2000; Müller et al., 2001). Among them, qE is generally the major component in plants (Maxwell and Johnson, 2000). Biochemical and molecular genetics studies have shown that a pH gradient across the thylakoid membrane (Briantais et al., 1979; Munekage et al., 2001, 2002), the xanthophyll cycle (Demmig-Adams et al., 1990; Niyogi et al., 1998), and the PsbS protein (Li et al., 2000) of PSII are important factors involved in controlling the induction and/or extent of NPQ. Based on the semidominance of loss-of-function mutations (Li et al., 2000, 2002a) and overexpression of the psbS gene (Li et al., 2002b) in Arabidopsis (Arabidopsis thaliana), the expression level of the PsbS protein has been suggested as an important factor in determining the qE (and total NPQ) capacity of plants.
Naturally occurring variation in NPQ capacity has been observed in different plant species (Johnson et al., 1993; Demmig-Adams and Adams, 1994; Demmig-Adams, 1998). The variation of saturated NPQ values ranges from 2.5 to 4.5 in British plant species, and plants grown in open habitats tend to have larger NPQ capacity (Johnson et al., 1993). Sun-acclimated plants contain up to four times as much NPQ capacity as low-light-acclimated plants of the same species (Osmond et al., 1993; Ruban et al., 1993; Brugnoli et al., 1994; Demmig-Adams and Adams, 1994; Demmig-Adams et al., 1995; Demmig-Adams, 1998; Roberts et al., 1998). In Monstera deliciosa, for example, sun-acclimated leaves showed higher NPQ than low-light-acclimated leaves (Demmig-Adams and Adams, 1994), and this difference in NPQ is correlated with changes in PsbS protein levels (Demmig-Adams et al., 2006). Although it has been suggested that there may be a genetic basis for the variation (Horton et al., 1994), this possibility has not yet been analyzed.
This kind of natural variation in plant traits, in most cases, shows continuous variations that are under the control of polygenic factors, and quantitative genetic studies are required to understand the genetic basis of the variation (Alonso-Blanco and Koornneef, 2000). Arabidopsis has become a model system for plant quantitative trait locus (QTL) mapping (Alonso-Blanco and Koornneef, 2000; Maloof, 2003; Tonsor et al., 2005) because it has considerable trait variations among accessions, advanced molecular biological tools for efficient genotyping with molecular markers, and a fully sequenced genome. In addition, development and improvement of statistical tools for QTL mapping facilitate analyses of quantitative traits (Lander and Botstein, 1989; Zeng, 1994; Sen and Churchill, 2001). Using Arabidopsis, a number of quantitative traits have been analyzed; however, QTL mapping is still underused for photosynthesis-related traits, in spite of advantages for quantitative genetic studies, such as simple ways for quantification of photosynthetic parameters (Krause and Weis, 1991; Laisk et al., 2002; Long and Bernacchi, 2003; Baker, 2008).
In this article, we report natural variation of NPQ among Arabidopsis accessions and test the hypothesis that the variation between a high NPQ accession (Sf-2) and a low NPQ accession (Columbia-0 [Col-0]) is related to the PsbS protein. We measured induction and relaxation of NPQ in Arabidopsis accessions and divided them into high and low NPQ accessions. Biochemical and molecular biological experiments did not associate the NPQ differences with PsbS or other previously identified NPQ factors. Genetic analyses revealed that the differences are controlled by polygenic nuclear factors. To identify these factors, we performed QTL mapping using Sf-2 × Col-0 F2 progeny as a mapping population and identified two high NPQ QTLs. The significance of NPQ variation and possible roles for these QTLs in thermal dissipation are discussed.
RESULTS
NPQ in Arabidopsis Accessions
To study the natural variation of NPQ capacity, we initially measured total NPQ in 62 Arabidopsis accessions (Supplemental Table S1). The tested accessions originated from diverse growth conditions, and parental lines of recombinant inbred lines (RILs) that are available or will be available in the near future were also included (http://www.inra.fr/internet/Produits/vast/RILs.htm). To focus on the major qE component, NPQ was measured for 10 min during actinic illumination with 1500 μmol photons m−2 s−1, followed by 5 min in the dark to determine its relaxation. All NPQ measurements were done between 1 and 5 pm to exclude any possible effects of circadian control of NPQ. The results showed that there are variations in NPQ capacity among Arabidopsis accessions (Supplemental Table S1) and that most of the NPQ is qE because it relaxed very fast in the dark (data not shown). Based on these measurements, the accessions were classified into high NPQ and low NPQ Arabidopsis accessions.
Ll-1, Sf-2, Ts-1, Van-0, Mz-0, Mr-0, and Kin-0 are high NPQ Arabidopsis accessions containing total NPQ capacity higher than 3.0, whereas Col-0, Landsberg erecta (Ler), and Wassilewskija-2 (Ws-2), so-called lab accessions, are low NPQ Arabidopsis accessions having total NPQ capacity below 2.5 (Supplemental Table S1). Interestingly, the three highest accessions, Ll-1, Sf-2, and Ts-1, all originated from Spain. Except for Ll-1, the other high NPQ accessions have been used as parental lines of RILs, but the RILs were not yet available at the time these experiments were initiated. We decided to use Ll-1 and Sf-2 as high NPQ accessions and Col-0 and Ws-2 as low NPQ accessions for further measurement and characterization.
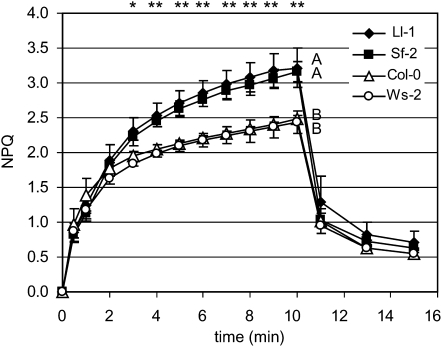
After growing these selected accessions together in identical growth conditions, we measured their NPQ in the next generation. Ll-1 and Sf-2 showed significantly higher total NPQ than Col-0 and Ws-2 after 3 min in actinic illumination (P < 0.01), and the NPQ differences became larger with time (P < 0.001) (Fig. 1).
Figure 1.
NPQ variation in high and low NPQ Arabidopsis accessions. NPQ values in high and low NPQ Arabidopsis accessions were measured on attached rosette leaves using an FMS2 fluorometer during actinic illumination with 1,500 μmol photons m−2 s−1 for 10 min, followed by relaxation in the dark for 5 min. Each data point represents the mean ± sd (n = 3). The significance of differences was tested by ANOVA (*, P < 0.01; **, P < 0.001), and the grouping (A and B) was made using multiple comparisons (Duncan).
Characterization of NPQ Factors in Selected Accessions
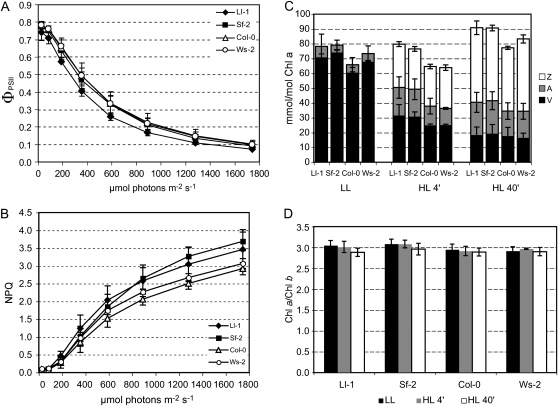
Photosynthetic efficiency and previously identified NPQ factors were compared between these high and low NPQ accessions. First of all, quantum yield of PSII (ΦPSII) measuring the proportion of energy used for photochemistry (Maxwell and Johnson, 2000) was calculated (Fig. 2A). The levels of ΦPSII in these accessions were very similar to each other, although Ll-1 had a slightly lower value than the others. In addition, the maximum quantum yield of PSII (maximum photochemical efficiency of PSII in the dark-adapted state) of the accessions was also indistinguishable (data not shown). Ll-1 and Sf-2 showed higher NPQ than Col-0 and Ws-2 in increasing light intensities; however, the differences were not statistically significant up to the highest light intensity applied (Fig. 2B), possibly due to the longer measuring time and greater contributions of other NPQ components (e.g. photoinhibitory quenching).
Figure 2.
ΦPSII, NPQ, xanthophyll cycle deepoxidation, and chlorophyll a/b ratio of high and low NPQ Arabidopsis accessions. A and B, ΦPSII and NPQ values were estimated by applying a pulse of saturating light to attached rosette leaves that had been exposed to the corresponding light intensity for 5 min. Each data point represents the mean ± sd (n = 3). C and D, Total xanthophyll cycle pigments (V, violaxanthin; A, antheraxanthin; Z, zeaxanthin) relative to chlorophyll (Chl) a, and chlorophyll a/b ratio were determined in low light (LL; 150 μmol photons m−2 s−1) and after being treated in high light (HL; 1,700 μmol photons m−2 s−1) for 4 and 40 min. Each data point represents the mean ± sd (n = 2 or 3).
To check differences in xanthophyll cycle activity, the contents of each xanthophyll cycle pigment were determined. The deepoxidated xanthophylls, antheraxanthin (A) and zeaxanthin (Z), are generated from violaxanthin (V) by V deepoxidase (VDE) in high light (Yamamoto et al., 1962). In low light (150 μmol photons m−2 s−1), Ll-1, Sf-2, Col-0, and Ws-2 contained a very small proportion of deepoxidated forms (A and Z). After being exposed to high light (1,700 μmol photons m−2 s−1) for 4 and 40 min, all accessions showed very similar levels of deepoxidation (Fig. 2C). These results indicated that the high and low NPQ accessions contain functional VDE and that there are no differences in VDE activation by high light between the two groups. Additionally, the chlorophyll a/b ratios reflecting the PSII antenna size were similar to each other (Fig. 2D).
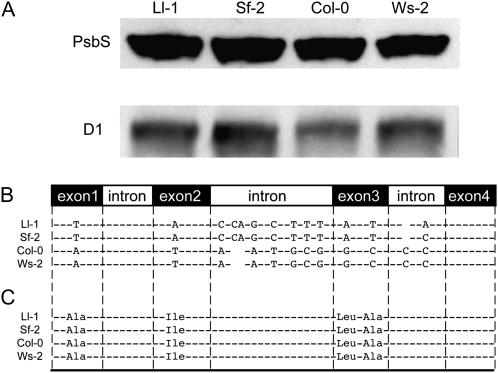
Third, the amount of the PsbS protein was compared. PsbS is required for qE, and qE (and total NPQ) is proportionally increased as the PsbS protein level increases (Li et al., 2000, 2002a, 2002b). To determine whether PsbS is responsible for the NPQ variation, the amount of PsbS in each accession was estimated by immunoblot analysis. In these accessions, the relative PsbS protein levels were very similar to each other, compared to D1 protein levels, although the D1 protein level in Col-0 was slightly lower than the others (Fig. 3A). We also determined the PsbS genomic DNA sequences and then compared their predicted amino acid sequences because changes in amino acid residues could affect PsbS function (Li et al., 2002c). Interestingly, we found DNA sequence polymorphisms between the high and the low NPQ accessions: Ll-1 and Sf-2 have nearly identical PsbS genomic DNA sequences, and the Col-0 and Ws-2 sequences are identical to each other (Fig. 3B). However, all polymorphisms in exons are silent; therefore, it was predicted that there are no amino acid differences. Taken together, these high and low NPQ accessions contained nearly identical levels of previously identified NPQ factors, including VDE activity and PsbS, suggesting that the differences in NPQ may be controlled by a novel factor or factors.
Figure 3.
PsbS protein levels and PsbS genomic DNA polymorphisms in high and low NPQ Arabidopsis accessions. A, PsbS protein levels in each accession were determined by immunoblot analysis with anti-PsbS antibody (Li et al., 2002a). D1 levels were included as a control. The image in B displays PsbS genomic DNA sequences showing DNA polymorphisms among accessions, and C shows the predicted amino acid at each polymorphic site.
NPQ of Sf-2 × Col-0 F1 and F2 Progenies
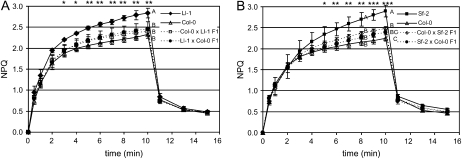
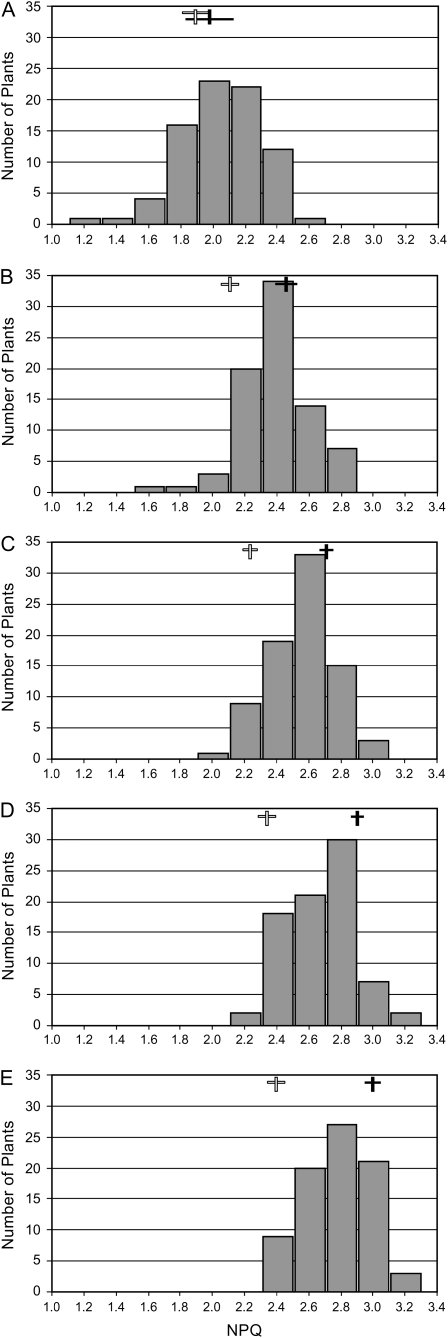
To understand the genetic basis of the NPQ variation, we crossed Ll-1 and Sf-2 to Col-0 and measured induction and relaxation of NPQ in F1 and F2 progenies. Col-0 was used as a low NPQ parental line because the genomic DNA sequences of Col-0 are completely determined, and physical map positions of molecular markers are also available (Arabidopsis Genome Initiative, 2000). NPQ levels in the F1 were significantly lower than those of Ll-1 and Sf-2 and were slightly higher than that those of Col-0, although multiple comparison tests (Duncan) indicated that F1 from both crosses were in the same group as Col-0 (Fig. 4). F1 plants from reciprocal crosses showed similar levels of NPQ and were statistically grouped together (Fig. 4). These observations indicated that the high NPQ is controlled by nuclear rather than cytoplasmic factor(s). To determine how many nuclear factors are involved, we measured NPQ in 80 Sf-2 × Col-0 F2 progeny and analyzed the frequency distribution of NPQ every 2 min during illumination with actinic light. At each time point, NPQ levels were widely distributed (Fig. 5), and a normality test (Shapiro-Wilk) showed that NPQ levels follow a normal distribution (P < 0.01), indicating that more than one factor is responsible for the NPQ variation. Between these two high NPQ accessions, Ll-1 is very similar to Ll-0 that is being used as a parental line for generation of Ll-0 × Ler RILs. Therefore, we decided to use Sf-2 and Col-0 as high NPQ and low NPQ accessions, respectively, to locate QTLs responsible for high NPQ.
Figure 4.
NPQ in F1 plants from reciprocal crosses of Ll-1 × Col-0 (A) and Sf-2 × Col-0 (B). The solid lines with closed and open symbols represent NPQ values in high and low NPQ parental lines, respectively, measured during actinic illumination with 1,500 μmol photons m−2 s−1 for 10 min, followed by relaxation in the dark for 5 min. The dotted lines represent NPQ values in F1 plants. Each data point represents the mean of two or three measurements. Significant differences and groupings (A to C) were done by ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001) followed by multiple comparisons (Duncan).
Figure 5.
Frequency distribution of NPQ in the Sf-2 × Col-0 F2 progeny. NPQ values at 2 min (A), 4 min (B), 6 min (C), 8 min (D), and 10 min (E) in actinic illumination were used to calculate the number of F2 plants in each interval. Vertical and horizontal boxes correspond to averaged NPQ values and their sd (n = 5), respectively, of Sf-2 (closed) and Col-0 (open).
Determination of Genotypes
To determine genotype in the F2 mapping population, we developed a polymorphic DNA marker set (Supplemental Table S2). Using the polymorphic markers, genotypes at each marker position were determined in 72 F2 progeny. The genotyping was almost complete, with only a few missing genotype data. From the genotype data, genotype frequencies at marker positions were calculated (Supplemental Fig. S1). The G-test (Dytham, 2003) indicated that overall, no significant segregation distortion was observed in our F2 mapping population (Supplemental Fig. S1). With the genotype data, we also confirmed the order of markers on each chromosome and estimated the distance between each two consecutive markers using MAPMAKER/EXP 3.0 (Lander et al., 1987). The reestimated marker order was the same as the marker order on the Arabidopsis physical map (Supplemental Table S2). The average distance between flanking markers was calculated as 10.7 centimorgan (cM). The longest interval was 31.9 cM between SO392 and A44575 on chromosome 1, and the shortest one was 2.2 cM between C6 and NGA126 on chromosome 3 (Supplemental Table S2).
QTL Mapping
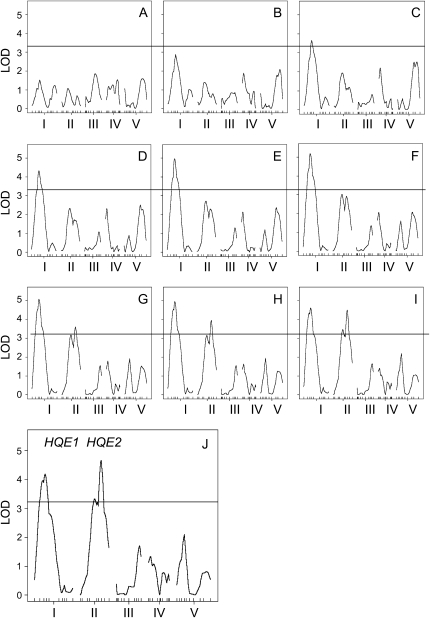
To find chromosome regions that are responsible for the high NPQ phenotype of Sf-2, we conducted QTL mapping using the R/qtl program (Broman et al., 2003). The QTL mapping with 10 min NPQ identified two high NPQ QTLs, High qE 1 (HQE1) and High qE 2 (HQE2) (Fig. 6J). The log of the odds (LOD) scores of both QTLs were higher than the threshold LOD score of 3.2 that was estimated after 1,000 permutations with 0.05 significance (Churchill and Doerge, 1994). HQE1 is 35 cM from the first marker on chromosome 1, and the nearest marker is NF19G9. HQE2 is 67.5 cM from the first marker on chromosome 2, and the nearest marker is C4H (Table I). Because all three possible genotypes are represented in the F2 mapping population, we were able to determine dominance (d) and additive (a) effects of each QTL. The degree of dominance (d/a) indicated nearly additive gene action (−0.06) of HQE1 and partly dominant gene action (0.49) of the Sf-2 allele of HQE2 (Table I; Tanksley, 1993). The estimated 1.5 LOD support intervals (Lander and Botstein, 1989) were between 12.5 and 57.5 cM on chromosome 1 for HQE1 and between 40.0 and 77.0 cM on chromosome 2 for HQE2 (Table I). Among total variance, the portion of variance explained by these two QTL was 37.8%. In addition, there was little evidence supporting interaction between HQE1 and HQE2 (data not shown).
Figure 6.
Results of high NPQ QTL mapping. QTLs for high NPQ were localized using R/qtl with NPQ values measured at 1 min (A), 2 min (B), 3 min (C), 4 min (D), 5 min (E), 6 min (F), 7 min (G), 8 min (H), 9 min (I), and 10 min (J). The horizontal line represents significance threshold (P < 0.05) estimated from 1,000 permutations.
Table I.
Results of QTL mapping for NPQ in Sf-2 × Col-0 F2 population
Ch, chromosome; a, additive effects; d, dominance effects; d/a, degree of dominance; % Var, percentage of variance explained.
| Timea | QTL | Ch | cMb | Markerc | LOD | 1.5 LOD | a | d | d/a | % Var |
|---|---|---|---|---|---|---|---|---|---|---|
| 10 min | High qE 1 (HQE1) | 1 | 35.0 | NF19G9 | 4.18 | 12.5 ∼ 57.5 | 0.146 | −0.009 | −0.06 | 12.1 |
| 10 min | High qE 2 (HQE2) | 2 | 67.5 | C4H | 4.66 | 40 ∼ 77 | 0.158 | 0.078 | 0.49 | 15.2 |
| 10 min | HQE1 + HQE2 | 37.8 | ||||||||
| 6 min | High qE 1 (HQE1) | 1 | 35.0 | NF19G9 | 5.18 | 23 ∼ 47 | 0.158 | 0.008 | 0.05 | 27.9 |
Time at which NPQ values used for QTL mapping were measured.
Position from the first marker on each chromosome.
The closest marker to the peak of corresponding QTL.
We also did QTL mapping using NPQ values measured at each minute and found interesting differences between HQE1 and HQE2 (Fig. 6, A–I). The LOD score of HQE1 became significant after 3 min in actinic illumination (Fig. 6C); thereafter, HQE1 remained as a significant high NPQ QTL during the 10-min measurement. At 6 min, HQE1 alone explained 27.9% of total NPQ variance (Table I). The support interval of HQE1 at 6 min was 23 to 47 cM, which was narrower than the interval at 10 min (Table I). In contrast, the response of HQE2 to the actinic light was relatively slower than that of HQE1. HQE2 became a significant QTL after 7 min in the light (Fig. 6G), and it remained significant until the end of the 10-min measurement. Therefore, from 3 to 6 min, only HQE1 seems to be responsible for the high NPQ, and from 7 min in the actinic light, both HQE1 and HQE2 may control the high NPQ phenotype.
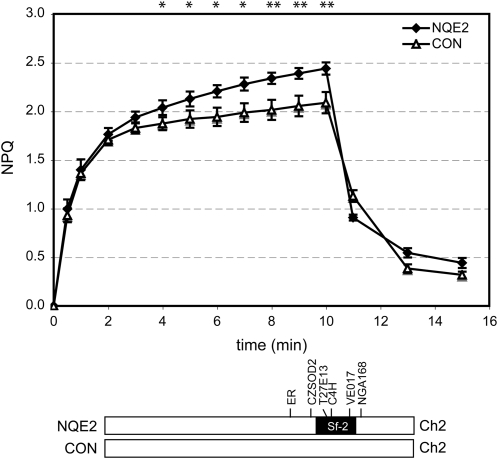
To confirm our NPQ QTL mapping, we generated near isogenic lines, NQE2, harboring the Sf-2 allele of HQE2 in the Col-0 background. F1 plants of Sf-2 and Col-0 cross were backcrossed to Col-0 four times, and lines were selected based on genotyping. NPQ measurements indicated that NQE2 showed higher NPQ than control lines containing the Col-0 allele of HQE2 (Fig. 7).
Figure 7.
NPQ levels of near isogenic lines for HQE2 (NQE2). NPQ levels were measured in near isogenic lines for HQE2 (NQE2), which contain the Sf-2 allele of HQE2 (black) in a Col-0 background (white bar). The breakpoints of the Sf-2 introgression are between the markers CZSOD2 and T27E13 on one side and between the markers VE017 and NGA168 on the other side. Segregants containing the Col-0 allele of HQE2 (CON) were used as control lines. The significance of differences between NQE2 and CON was tested using Student's t test (*, P < 0.05; **, P < 0.01). Ch2, Chromosome 2.
DISCUSSION
In our survey of natural variation of NPQ capacity in Arabidopsis, we tried to find some accessions that have much less NPQ than other accessions. In such cases, it might be relatively straightforward to determine the genetic basis for the NPQ difference, as found for other traits (Maloof et al., 2001). However, so far, we could not find Arabidopsis accessions having very low NPQ. That might be because NPQ is an important photoprotection mechanism without which plants would suffer more oxidative stress (Li et al., 2002b) and show much lower fitness estimated by lifetime seed production (Külheim et al., 2002).
Several accessions exhibited relatively high NPQ compared to the standard lab accession, Col-0. Why do the high NPQ accessions have a higher capacity for thermal dissipation than other accessions? We found a significant, but very weak correlation between latitude and NPQ among European accessions (data not shown). The latitude of geographical origin has been shown to have a significant relationship with variation of other traits in Arabidopsis (Li et al., 1998; Maloof et al., 2001; Stinchcombe et al., 2004). Instead of latitude, environmental parameters in the natural habitat may be more critical for NPQ capacity because NPQ is critical to deal with sudden changes in light intensity (Müller et al., 2001; Külheim et al., 2002). To better understand the relationship between NPQ and natural habitat, habitat information needs to be quantified (Johnson et al., 1993).
Another possible explanation for the weak relationship between latitude and NPQ is that mechanisms other than NPQ may play a major role in photoprotection in some accessions (Johnson et al., 1993). Although NPQ capacity in Cvi and Ler, for example, was very similar (Supplemental Table S1), Cvi showed higher photooxidative stress tolerance than Ler as expected from Cvi's origin, a tropical zone (Abarca et al., 2001). Interestingly, Cvi contains a different allele of chloroplastic Cu/Zn-superoxide dismutase 2 (CSD2) from that of Ler in predicted amino acid sequences and in mobility in a protein gel, and it was suggested that the Cvi-specific CSD2 is one of the factors responsible for the higher tolerance of Cvi (Abarca et al., 2001).
Strong relationships were observed between deepoxidation state and NPQ capacity among plant species as well as among plants of the same species grown in different light conditions (Johnson et al., 1993; Brugnoli et al., 1994; Demmig-Adams and Adams, 1994; Demmig-Adams et al., 1995; Demmig-Adams, 1998; Adams et al., 1999). In addition, it has been known that the PsbS protein level can determine the NPQ level and that certain amino acid residues in PsbS play an important role in NPQ (Li et al., 2002b, 2002c). However, the deepoxidation states of the xanthophyll cycle in Ll-1, Sf-2, Col-0, and Ws-2 were similar to each other (Fig. 2C), and Ll-1 and Sf-2 contained almost identical levels of PsbS compared to Col-0 and Ws-2 without any polymorphisms in predicted amino acid sequences (Fig. 3). These results suggest that the QTLs identified here, HQE1 and HQE2, do not act directly or indirectly through NPQ factors that have been found previously in extensive biochemical and genetic analyses of NPQ.
The response of HQE1 to photosynthetically active light was faster than that of HQE2 (Fig. 6, A–J). After being exposed to actinic illumination for 3 min, HQE1 became a significant high NPQ QTL (Fig. 6C), and from 7 min, both HQE1 and HQE2 were responsible for the high NPQ (Fig. 6G). These results indicated that early NPQ induction might be affected by HQE1 alone, and saturated NPQ levels might be explained by both HQE1 and HQE2 without any interaction between HQE1 and HQE2. Therefore, it could be speculated that sudden changes in light intensity would be dealt with by NPQ controlled by HQE1 and that, if the high light intensity continues, HQE2 and HQE1 together increase NPQ capacity to cope with the high light. Although HQE1 was the only identified significant QTL between 3 and 6 min, the NPQ measured during that period time was widely distributed (Fig. 5).
In conclusion, we were able to identify high and low NPQ Arabidopsis accessions and map QTLs that influence NPQ in two of these accessions. This information provides a guideline for selection of appropriate RILs and experimental design for future NPQ QTL mapping. We have also developed DNA molecular markers showing polymorphisms between Sf-2 and Col-0 that can be used for genotyping of Sf-2 × Col-0 RILs. Although we had hypothesized that the difference in NPQ between the accessions would be related to PsbS, no differences in known NPQ factors were found. Analysis of other accessions might reveal cases in which NPQ variation is related to PsbS. Identification of the high NPQ QTLs described here will eventually make it possible to clone new genes responsible for high NPQ.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were obtained from the Arabidopsis Biological Resource Center and from the seed collection of the Niyogi laboratory. Before planting on soil, the seeds were kept in water at 4°C for 1 week. Arabidopsis plants were grown in short-day conditions (10 h light/14 h dark) for 5 to 6 weeks in a light- (150 μmol photons m−2 s−1) and temperature (22°C)-controlled growth chamber before chlorophyll fluorescence measurements and leaf tissue harvests.
Chlorophyll Fluorescence Measurements
Plants were dark-adapted for 2 h before NPQ measurement. The measurement began at 1 pm, and all measurements were completed by 5 pm. Chlorophyll fluorescence parameters were measured on attached rosette leaves using an FMS2 fluorometer (Hansatech) during actinic illumination with 1,500 μmol photons m−2 s−1 for 10 min, followed by darkness for 5 min. NPQ was calculated as (Fm − Fm′)/Fm′, with Fm and Fm′ being maximum fluorescence measured after dark adaptation and in the light-adapted condition, respectively, by applying a saturating pulse of light. ΦPSII was calculated as (Fm′ − Fs)/Fm′, with Fm′ being measured by applying a saturating pulse of light at the end of each 5-min illumination at a given intensity, and Fs being steady-state fluorescence during the illumination (Maxwell and Johnson, 2000).
Pigment Analyses
Pigment analyses were done following Niyogi laboratory protocols with some modifications (Müller-Moulé et al., 2002). Leaf discs were collected with a puncher (diameter of 8 mm) from rosette leaves and floated on top of water in petri dishes. Following incubation in low light (150 μmol photons m−2 s−1), the leaf discs were exposed to high light (1,700 μmol photons m−2 s−1) for 4 and 40 min. The treated leaf discs were frozen in liquid nitrogen and ground to a fine powder. From the leaf powder, pigments were extracted twice with 200 μL of 100% (v/v) acetone, and following centrifugation supernatants were filtered through a 0.2-μm nylon filter and subjected to HPLC.
Immunoblot Analysis and DNA Sequence Determination
Total proteins were extracted from plant leaves of each accession. Leaf tissue (200 mg) was frozen in liquid nitrogen and then ground to a fine powder. To the tissue powder, 200 μL protein extraction buffer (4% [w/v] SDS, 25 mm Tris-HCl, pH 8.8, and 2.5% [v/v] glycerol) was added to the tissue powder. Following further grinding in the buffer, the protein samples were denatured in boiling water for 5 min. After centrifugation for 3 min, the supernatant was separated and then kept in a –70°C freezer. The protein concentration was determined using a Bio-Rad Protein assay kit (Bio-Rad Laboratories), and 30 μg of total proteins were loaded in each well of a precast 10% to 20% Tris-Glycine gel (Invitrogen). The separated proteins were blotted onto a nitrocellulose membrane (Schleicher & Schuell) using a semidry transfer unit (Hoefer Pharmacia Biotech). For quantification of the PsbS protein, a polyclonal antibody raised against a PsbS oligopeptide was used (Li et al., 2002a), and for the D1 protein, a polyclonal antibody raised against spinach (Spinacia oleracea) D1 (kindly provided by Prof. A. Melis at University of California, Berkeley, CA) was used. As the secondary antibody, horseradish peroxidase-labeled antibody was used, and the chemiluminescence signal was detected using ECL western blotting detection reagents (Amersham Biosciences).
Genomic DNA sequences were determined from PCR-amplified DNA. For PCR from genomic DNA, primers were designed using the web-based Primer3 program (http://www.broad.mit.edu/cgi-bin/primer/primer3_www.cgi), and PCR products were separated on agarose gels and then purified using a gel extraction kit (Qiagen). Sequencing of purified DNA was performed using the Big-Dye Terminator v3.0 ready reaction cycle sequencing kit (Applied Biosystems) and an ABI 3100 automated DNA sequencer (Applied Biosystems).
Marker Set Generation and Genotyping
PCR conditions for previously available markers were obtained from The Arabidopsis Information Resource, and restriction enzymes used for each marker are listed in Supplemental Table S2. For NQE2, CZSOD2 and T27E13 were used to determine genotypes between ER and C4H, and COP1 and VE017 were used for genotyping between C4H and NGA168. The newly generated insertion/deletion markers for Sf-2 × Col-0 were C42050 (forward: 5′-GCTGAGTATAGAGCAGGTTGGTG-3′; reverse: 5′-CGTCCTGTTTCAATTTGTCATC-3′) and D01650 (forward: 5′-CTGCTTTTAGACCGCTTTCC-3′; reverse: 5′-TCCCAATGCTAAGTTCTGCTG-3′). PCR for C42050 was annealed at 60°C and polymerized at 72°C for 30 s. Annealing for D01650 was at 59°C, and polymerization lasted for 1 min. The PCR products of C42050 and D01650 were separated on 3% and 1.2% agarose gels, respectively. The newly developed cleaved-amplified polymorphic sequence markers were SNP397C (forward: 5′-TTTGAGCTTGTTTCCTCGTG-3′; reverse: 5′-ATATCTGTGGGGTTGCTTGG-3′), A44575 (forward: 5′-CAAACCCAAAACCAAAGCTG-3′; reverse: 5′-TGCTTACATGGGGGAAAAAG-3′), SNP299C (forward: 5′-TTGAAGGCATGAGTTGTTGG-3′; reverse: 5′-CGCCGATTTAGTACGACTACG-3′), and D12320 (forward: 5′-TGGGAGAGCCTAATGTTTCTG-3′; reverse: 5′-CCAACCACCAAATCCTTCAC-3′). The PCR reactions of SNP397C and SNP299C were annealed at 58°C and polymerized for 1 min, and then the PCR products were digested with HpyCh4 IV and DdeI, respectively. Amplified A44575 DNA following PCR at 59°C annealing and 1 min polymerization was digested with HinfI. PCR for D12320 was done at 59°C annealing and 30 s polymerization, and then DdeI-digested fragments were separated on 3% agarose gels.
For genotyping, genomic DNA was extracted as follows: frozen small leaves were ground in a 1.5-mL tube and then the fine powder was thawed in 10 μL of alkali solution (0.5 n NaOH) (Klimyuk et al., 1993). Following boiling at 100°C for 30 s, 100 μL of neutralization solution (0.2 m Tris-HCl, pH 8.0, and 1 mm EDTA) were added. For a 20-μL PCR reaction, 2 μL of the leaf tissue solution was used.
Statistical Analysis and QTL Mapping
Test statistics including ANOVA, Student's t test and multiple comparison tests were calculated using Microsoft Excel and XLSTAT. Following the genotyping, a G-test was used to test whether the genotype ratio at each marker position fitted to the expected F2 segregation ratio (Supplemental Fig. S1) (Dytham, 2003). Intervals between neighboring markers were estimated using MAPMAKER/EXP 3.0 (Lander et al., 1987). The first marker on each chromosome was set at 0 cM, and each marker position was calculated from accumulated intervals between markers.
QTL mapping was performed using the R/qtl program (Broman et al., 2003). The LOD significance threshold was estimated after permutation tests that had been replicated 1,000 times (Churchill and Doerge, 1994). LOD support interval was calculated using the lodint function with 1.5 LOD unit drop to form the support interval. Additive and dominance effects of each QTL were calculated using estimations of the means of the three genotype groups that were in the output of scanone function with the Expectation Maximization method in the R/qtl program (Lander and Botstein, 1989; Tanksley, 1993). Estimation of percentage of variance explained by each QTL was obtained from a drop-one-term analysis of results in an additive model. The additive model was calculated using the fitqtl function with imputation method in the R/qtl program (Sen and Churchill, 2001).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genotype segregation of Sf-2 × Col-0 F2 progeny at each marker position.
Supplemental Table S1. List of NPQ values in Arabidopsis accessions.
Supplemental Table S2. List of markers, positions, types, and PCR conditions.
Supplementary Material
Acknowledgments
We thank Jae Pasari and Carsten Külheim for technical support and Prof. Anastasios Melis for providing the D1 antibody. This manuscript was improved by the critical comments of Ben Gutman, Heidi Ledford, and Xiao-Ping Li. Our special thanks go to Dr. Dan Kliebenstein, who gave us initiative for this study.
This work was supported by the Office of Basic Energy Sciences, Chemical Sciences Division, U.S. Department of Energy (contract DE–AC03–76SF000098).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Krishna K. Niyogi (niyogi@nature.berkeley.edu).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Abarca D, Roldán M, Martín M, Sabater B (2001) Arabidopsis thaliana ecotype Cvi shows an increased tolerance to photo-oxidative stress and contains a new chloroplastic copper/zinc superoxide dismutase isoenzyme. J Exp Bot 52 1417–1425 [DOI] [PubMed] [Google Scholar]
- Adams WW III, Demmig-Adams B, Logan BA, Barker DH, Osmond CB (1999) Rapid changes in xanthophyll cycle-dependent energy dissipation and photosystem II efficiency in two vines, Stephania japonica and Smilax australis, growing in the understory of an open Eucalyptus forest. Plant Cell Environ 22 125–136 [Google Scholar]
- Alonso-Blanco C, Koornneef M (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci 5 22–29 [DOI] [PubMed] [Google Scholar]
- Anderson JM (1986) Photoregulation of the composition, function, and structure of thylakoid membranes. Annu Rev Plant Physiol 37 93–136 [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Baker NR (2008) Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol 59 89–113 [DOI] [PubMed] [Google Scholar]
- Barber J, Andersson B (1992) Too much of a good thing: light can be bad for photosynthesis. Trends Biochem Sci 17 61–66 [DOI] [PubMed] [Google Scholar]
- Briantais JM, Vernotte C, Picaud M, Krause GH (1979) A quantitative study of the slow decline of chlorophyll a fluorescence in isolated chloroplasts. Biochim Biophys Acta 548 128–138 [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19 889–890 [DOI] [PubMed] [Google Scholar]
- Brugnoli E, Cona A, Lauteri M (1994) Xanthophyll cycle components and capacity for non-radiative energy dissipation in sun and shade leaves of Ligustrum ovalifolium exposed to conditions limiting photosynthesis. Photosynth Res 41 451–463 [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative triat mapping. Genetics 138 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B (1998) Survey of thermal energy dissipation and pigment composition in sun and shade leaves. Plant Cell Physiol 39 474–482 [Google Scholar]
- Demmig-Adams B, Adams WW (1994) Capacity for energy dissipation in the pigment bed in leaves with different xanthophyll cycle pools. Aust J Plant Physiol 21 575–588 [Google Scholar]
- Demmig-Adams B, Adams WW, Heber U, Neimanis S, Winter K, Krüger A, Czygan FC, Bilger W, Björkman O (1990) Inhibition of zeaxanthin formation and of rapid changes in radiationless energy dissipation by dithiothreitol in spinach leaves and chloroplasts. Plant Physiol 92 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW, Logan BA, Verhoeven AS (1995) Xanthophyll cycle-dependent energy dissipation and flexible photosystem II efficiency in plants acclimated to light stress. Aust J Plant Physiol 22 249–260 [Google Scholar]
- Demmig-Adams B, Ebbert V, Mellman DL, Mueh KE, Schaffer L, Funk C, Zarter CR, Adamska I, Jansson S, Adams WW III (2006) Modulation of PsbS and flexible vs sustained energy dissipation by light environment in different species. Physiol Plant 127 670–680 [Google Scholar]
- Dytham C (2003) Choosing and Using Statistics: A Biologist's Guide, Ed 2. Blackwell Publishing, Malden, MA
- Horton P, Ruban AV, Walters RG (1994) Regulation of light harvesting in green plants. Indication by nonphotochemical quenching of chlorophyll fluorescence. Plant Physiol 106 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GN, Young AJ, Scholes JD, Horton P (1993) The dissipation of excess excitation energy in British plant species. Plant Cell Environ 16 673–679 [Google Scholar]
- Kasahara M, Kagawa T, Oikawa K, Suetsugu N, Miyao M, Wada M (2002) Chloroplast avoidance movement reduces photodamage in plants. Nature 420 829–832 [DOI] [PubMed] [Google Scholar]
- Klimyuk V, Carroll B, Thomas C, Jones J (1993) Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J 3 493–494 [DOI] [PubMed] [Google Scholar]
- Krause GH, Weis E (1991) Chlorophyll fluorescence and photosynthesis: the basics. Annu Rev Plant Physiol Plant Mol Biol 42 313–349 [Google Scholar]
- Külheim C, Ågren J, Jansson S (2002) Rapid regulation of light harvesting and plant fitness in the field. Science 297 91–93 [DOI] [PubMed] [Google Scholar]
- Laisk A, Oja V, Rasulov B, Ramma H, Eichelmann H, Kasparova I, Pettai H, Padu E, Vapaavuori E (2002) A computer-operated routine of gas exchange and optical measurements to diagnose photosynthetic apparatus in leaves. Plant Cell Environ 25 923–943 [Google Scholar]
- Lander E, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1 174–181 [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Suzuki JI, Hara T (1998) Latitudinal variation in plant size and relative growth rate in Arabidopsis thaliana. Oecologia 115 293–301 [DOI] [PubMed] [Google Scholar]
- Li XP, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403 391–395 [DOI] [PubMed] [Google Scholar]
- Li XP, Gilmore AM, Niyogi KK (2002. a) Molecular and global time-resolved analysis of a psbS gene dosage effect on pH- and xanthophyll cycle-dependent nonphotochemical quenching in photosystem II. J Biol Chem 277 33590–33597 [DOI] [PubMed] [Google Scholar]
- Li XP, Müller-Moulé P, Gilmore AM, Niyogi KK (2002. b) PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc Natl Acad Sci USA 99 15222–15227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Phippard A, Pasari J, Niyogi KK (2002. c) Structure–function analysis of photosystem II subunit S (PsbS) in vivo. Funct Plant Biol 29 1131–1139 [DOI] [PubMed] [Google Scholar]
- Long SP, Bernacchi CJ (2003) Gas exchange measurements, what can they tell us about the underlying limitations to photosynthesis? Procedures and sources of error. J Exp Bot 54 2393–2401 [DOI] [PubMed] [Google Scholar]
- Maloof JN (2003) Genomic approaches to analyzing natural variation in Arabidopsis thaliana. Curr Opin Genet Dev 13 576–582 [DOI] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC, et al (2001) Natural variation in light sensitivity of Arabidopsis. Nat Genet 29 441–446 [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence—a practical guide. J Exp Bot 51 659–668 [DOI] [PubMed] [Google Scholar]
- Müller P, Li XP, Niyogi KK (2001) Non-photochemical quenching. A response to excess light energy. Plant Physiol 125 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Moulé P, Conklin PL, Niyogi KK (2002) Ascorbate deficiency can limit violaxanthin de-epoxidase activity in vivo. Plant Physiol 128 970–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T (2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110 361–371 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Takeda S, Endo T, Jahns P, Hashimoto T, Shikanai T (2001) Cytochrome b6f mutation specifically affects thermal dissipation of absorbed light energy in Arabidopsis. Plant J 28 351–359 [DOI] [PubMed] [Google Scholar]
- Niyogi KK (1999) Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50 333–359 [DOI] [PubMed] [Google Scholar]
- Niyogi KK (2000) Safety valves for photosynthesis. Curr Opin Plant Biol 3 455–460 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Björkman O (1998) Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort DR, Baker NR (2002) A photoprotective role for O2 as an alternative electron sink in photosynthesis? Curr Opin Plant Biol 5 193–198 [DOI] [PubMed] [Google Scholar]
- Osmond CB, Ramus J, Levavasseur G, Franklin LA, Henley WJ (1993) Fluorescence quenching during photosynthesis and photoinhibition of Ulva rotundata Blid. Planta 190 97–106 [Google Scholar]
- Roberts A, Borland A, Maxwell K, Griffiths H (1998) Ecophysiology of the C3-CAM intermediate Clusia minor L. in Trinidad: seasonal and short-term photosynthetic characteristics of sun and shade leaves. J Exp Bot 49 1563–1573 [Google Scholar]
- Ruban AV, Young AJ, Horton P (1993) Induction of nonphotochemical energy dissipation and absorbance changes in leaves. Evidence for changes in the state of the light-harvesting system of photosystem II in vivo. Plant Physiol 102 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Churchill GA (2001) A statistical framework for quantitative trait mapping. Genetics 159 371–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, Purugganan MD, Schmitt J (2004) A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc Natl Acad Sci USA 101 4712–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD (1993) Mapping polygenes. Annu Rev Genet 27 205–233 [DOI] [PubMed] [Google Scholar]
- Tonsor SJ, Alonso-Blanco C, Koornneef M (2005) Gene function beyond the single trait: natural variation, gene effects, and evolutionary ecology in Arabidopsis thaliana. Plant Cell Environ 28 2–20 [Google Scholar]
- Yamamoto HY, Nakayama TOM, Chichester CO (1962) Studies on the light and dark interconversions of leaf xanthophylls. Arch Biochem Biophys 97 168–173 [DOI] [PubMed] [Google Scholar]
- Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136 1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.