Abstract
Neotyphodium uncinatum and Neotyphodium siegelii are fungal symbionts (endophytes) of meadow fescue (MF; Lolium pratense), which they protect from insects by producing loline alkaloids. High levels of lolines are produced following insect damage or mock herbivory (clipping). Although loline alkaloid levels were greatly elevated in regrowth after clipping, loline-alkaloid biosynthesis (LOL) gene expression in regrowth and basal tissues was similar to unclipped controls. The dramatic increase of lolines in regrowth reflected the much higher concentrations in young (center) versus older (outer) leaf blades, so LOL gene expression was compared in these tissues. In MF-N. siegelii, LOL gene expression was similar in younger and older leaf blades, whereas expression of N. uncinatum LOL genes and some associated biosynthesis genes was higher in younger than older leaf blades. Because lolines are derived from amino acids that are mobilized to new growth, we tested the amino acid levels in center and outer leaf blades. Younger leaf blades of aposymbiotic plants (no endophyte present) had significantly higher levels of asparagine and sometimes glutamine compared to older leaf blades. The amino acid levels were much lower in MF-N. siegelii and MF-N. uncinatum compared to aposymbiotic plants and MF with Epichloë festucae (a closely related symbiont), which lacked lolines. We conclude that loline alkaloid production in young tissue depleted these amino acid pools and was apparently regulated by availability of the amino acid substrates. As a result, lolines maximally protect young host tissues in a fashion similar to endogenous plant metabolites that conform to optimal defense theory.
Loline alkaloids (LAs; Hofmeister, 1892; Siegel et al., 1990; TePaske et al., 1993; Blankenship et al., 2001) are protective secondary metabolites produced by some Epichloë and Neotyphodium spp. (epichloae), fungi that live as systemic symbionts in many cool season grasses (Poaceae subfamily Pooideae). The lolines are active against a broad spectrum of insects (Schardl et al., 2007) and are derived from l-Pro (Pro) and l-homoserine (Hse; Blankenship et al., 2005). Mock herbivory (clipping plants) is reported to induce higher levels of lolines in several grass-epichloë symbiota (Craven et al., 2001; Bultman et al., 2004; Gonthier et al., 2008), suggesting that the epichloae have evolved to regulate their metabolism in a manner appropriate for defense of their hosts. However, little is known of the regulation of LA synthesis in symbio and whether these symbionts follow prevailing models for how plants deploy chemical defenses against herbivores (McKey, 1979; Rhoades, 1979; Barto and Cipollini, 2005).
The loline-alkaloid biosynthesis (LOL) gene cluster contains nine genes likely to direct LA production (Spiering et al., 2005). Neotyphodium uncinatum contains two highly similar LOL clusters (LOL1 and LOL2), and a single LOL cluster has been found in each of the LA-producing species, Neotyphodium coenophialum, Neotyphodium siegelii, and some strains of Epichloë festucae, among others (Spiering et al., 2005; Kutil et al., 2007). Fermentation cultures of N. uncinatum produce lolines, and studies involving application of labeled precursors and intermediates have almost completely elucidated the LA biosynthetic pathway (Blankenship et al., 2005; Spiering et al., 2005; Faulkner et al., 2006; Schardl et al., 2007). Putative roles of the LOL gene products—based on sequence relationships to known enzyme classes—fit well with the pathway. Furthermore, an RNA interference knockdown of lolC reduces LA levels, and a lolP knockout prevents conversion of N-methylloline to N-formylloline (Spiering et al., 2005, 2008). Expression kinetics of the LOL genes are tightly correlated with each other and with the LA production phase in N. uncinatum cultures (Zhang et al., 2009). This finding raises the question whether and how LOL gene expression in symbio relates to changes in LA levels in response to development and stresses in host plants.
LA production in symbio may be influenced by physiological differences among plant tissues and developmental stages, as well as differences in nutritional status and environmental stresses (Kennedy and Bush, 1983; Belesky et al., 1987; Justus et al., 1997; Tong et al., 2006). Given the anti-insect activity of lolines, effects of plant damage on LA levels are of particular interest. Mock herbivory (clipping of leaves) leads to apparent increases in LA concentrations in regrowth tissues of tall fescue (TF; Lolium arundinaceum) symbiotic with N. coenophialum (Bultman et al., 2004; Sullivan et al., 2007) and of meadow fescue (MF; Lolium pratense) symbiotic with N. uncinatum or N. siegelii (Craven et al., 2001). Despite the higher LA levels, however, clipping or damage of TF-N. coenophialum by the herbivore Spodoptera frugiperda (fall armyworm) was reported to elicit only minor, marginally significant (P = 0.052) effects on expression of lolC (Sullivan et al., 2007). A study of the Glyceria striata-Epichloë glyceriae symbiotum demonstrated significantly higher expression of lolC and higher LA production when the grass was artificially damaged, whereas the effect of damage by S. frugiperda on LA concentrations and lolC expression was not significant (Gonthier et al., 2008).
Prevailing concepts about how plants deploy chemical defenses include the optimal defense theory (ODT; McKey, 1979; Rhoades, 1979) and the growth differentiation balance hypothesis (GDBH; Barto and Cipollini, 2005). The ODT addresses the distribution of chemical defenses in the plant, predicting that such defenses will be concentrated in tissues that have relatively little means to physically inhibit herbivory (e.g. in young tissues) and are important in the fitness of the plant. The GBDH addresses the location of biosynthesis and predicts that mature tissues are more likely to produce secondary metabolites than are actively growing tissues, which instead need to use resources for biomass production. It is intriguing to consider whether the epichloae obey the predictions of ODT and GDBH, considering that many epichloae protect their hosts by synthesizing insecticidal alkaloids, but they are also evolutionarily derived from plant-pathogenic fungi (Moon et al., 2004) and do not always enhance host fitness (Faeth et al., 2004). In order to address these questions, it is necessary to understand how secondary metabolism of the epichloae is regulated in symbio. The production of lolines in MF-N. uncinatum and MF-N. siegelii is an ideal test case because the lolines accumulate to very high levels—up to 1.9% dry weight—in regrowth of clipped plants (Craven et al., 2001). Here, we test the hypotheses that LOL gene expression and substrate availability correlate with LA levels in younger versus older leaf tissues and in response to clipping in MF-N. uncinatum and MF-N. siegelii symbiota.
RESULTS
LAs in Regrowth and Center and Outer Leaf Blades
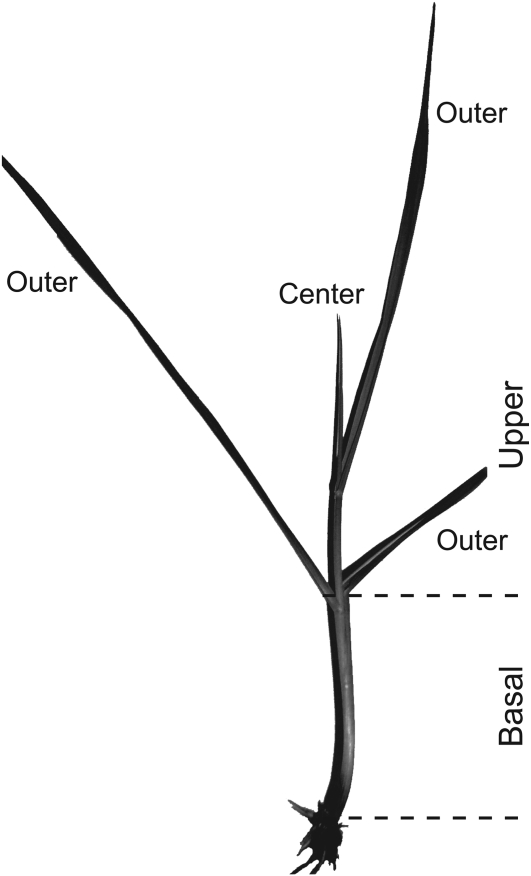
To assess the distribution of lolines in MF-N. siegelii and MF-N. uncinatum, plants were clipped approximately at the junction between the pseudostems and leaf blades, and this “initial harvest” was separated into outer (older) and center (younger) leaf blades (Fig. 1). To assess the effect of clipping on LA accumulation, newly emerging leaf blades (regrowth) as well as the basal portions (between soil level and the tops of leaf sheathes) were sampled every 3 d until day 15 postclipping (PC).
Figure 1.
Photograph of a MF tiller, with sampled tissues indicated.
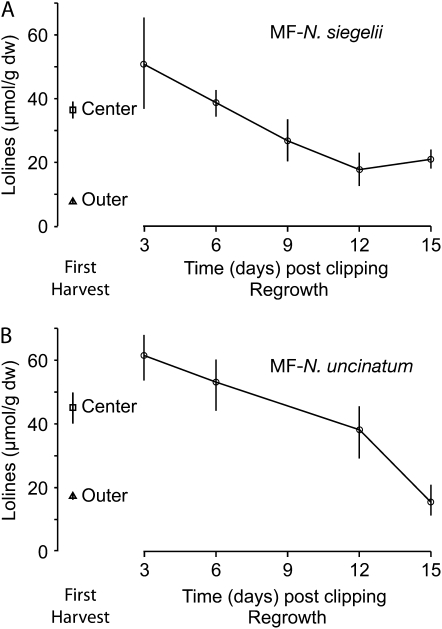
In MF-N. siegelii, LA concentrations in the basal tissues throughout the 15-d time course ranged between 20 and 34 μmol/g dry weight and showed no significant differences from basal tissues of unclipped control plants (P > 0.05; t test, two tailed, equal variance; data not shown). Regrowth tissues at day 3 had slightly higher LA levels compared to the center leaves from the initial harvest and much higher levels compared to the outer leaves, which constituted the bulk of tissue from the initial harvest (Fig. 2A). Similarly, LA levels in regrowth at day 3 were 3.6- to 6.1-fold higher than the combined upper tissue of unclipped control plants harvested throughout the time course (data not shown). At day 6 PC, LA levels in regrowth were similar to levels in the center leaf blades of unclipped plants (Fig. 2A). By days 12 and 15 PC, levels had declined and were approaching the LA levels in outer leaf blades of the initial harvest. Thus, the high LA levels in young regrowth tissue reflected the high LA content of young leaves of unclipped plants.
Figure 2.
LA concentrations in MF with N. siegelii (A) or MF with N. uncinatum (B), including outer and center leaf blades of initial harvest (n = 10 for MF-N. siegelii; n = 9 for MF-N. uncinatum), and in a time course of LA levels in regrowth leaf blades (n = 2 for each time point). Data are not shown for day 9 of MF-N. uncinatum because only one plant was analyzed. Bars are ses of the mean. dw, Dry weight.
Similar results were obtained with the MF-N. uncinatum symbiota. The basal pseudostem tissue at all time points PC had an LA content of 22 to 38 μmol/g dry weight, which was not significantly different from that in basal tissues of unclipped control plants (data not shown). In the initial harvest, LA levels in the center leaf blades were approximately 3-fold higher than in the outer leaf blades. In the regrowth tissues at days 3 and 6 PC, levels of LA were 44 to 68 μmol/g dry weight, slightly higher than the LA level in center leaf blades (Fig. 2B). From day 9 to 15 PC, as the regrowth tissue matured, the concentration of LA decreased gradually, whereas unclipped control plants showed no significant changes in LA concentrations. Thus, younger leaf blades contained higher LA than older leaf blades; clipping did not change LA levels in basal tissues, and the high LA levels in young regrowth reflected the high LA content of younger leaf blades of unclipped plants.
Gene Expression in MF-N. siegelii
Expression levels of five LOL genes were compared in basal tissues between unclipped control and clipped plants over the time course. Gene expression of lolC, lolO, lolA, lolE, and lolT in basal tissues suggested no trends in response to clipping (Supplemental Fig. S1A). Expression of the LOL genes was also assayed in regrowth leaf blades over the 15-d time course and compared to expression in upper tissue of unclipped control plants (Supplemental Fig. S1B). Also assayed were fungal genes for biosynthesis of the proximal LA precursors, Pro (proC, encoding pyrroline-5-carboxylate reductase) and O-acetylhomoserine (metE, encoding homoserine O-acetyl transferase), and the housekeeping gene, tubB (encoding β-tubulin; Supplemental Fig. S1C). Expression profiles of the LOL genes and other genes lacked definite trends, and overall changes in gene expression were minor, within a range of 0.5- to 2.0-fold difference from their respective median mRNA levels. Thus, in the MF-N. siegelii symbiotum, high LA in early regrowth tissues was not reflected in LOL gene expression levels.
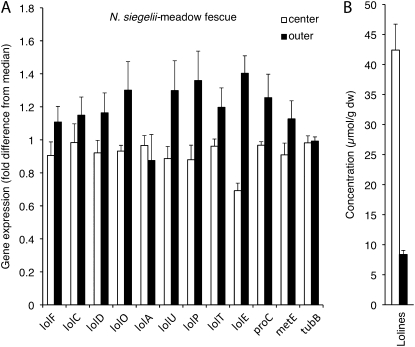
Comparisons of center and outer leaf blades from the same plants provided strictly controlled comparisons of younger and older tissues. Expression of eight LOL genes, metE, proC, and tubB were assayed for the outer and center tissues of MF-N. siegelii symbiota. A trend of slightly higher LOL gene expression in the outer leaf blades than in center leaf blades was observed (Fig. 3A); however, this difference was <0.5-fold for most of the genes and was not significant (P > 0.05, two tailed, pairwise t tests). The slightly higher expression of the LOL genes in the outer leaf blade was in contrast to the much higher (approximately 5-fold) LA concentrations in the center leaf blades (Fig. 3B).
Figure 3.
Relative expression of N. siegelii genes (A) and LA concentrations (B) in center leaf blades (white bars) and outer leaf blades (black bars) of MF. Gene expression is indicated as fold-difference from the median for each gene in the experiment. Error bars show ses of the mean (n = 4). dw, Dry weight.
Gene Expression in MF-N. uncinatum
Similar studies of gene expression were conducted for N. uncinatum in MF, for which assays included LOL genes, proC, metE, and a gene designated C2H2, predicted to encode a transcription factor that preliminary tests suggested may affect LOL gene expression in N. uncinatum cultures (D.-X. Zhang and C.L. Schardl, unpublished data). Expression of LOL genes, proC, metE, and C2H2 in regrowth tissues of the MF-N. uncinatum symbiota did not exhibit significant differences compared to their expression in upper parts of unclipped controls (t tests, homoscedastic, P > 0.05; Supplemental Fig. S2). Thus, as in MF-N. siegelii, the higher LA concentrations in early regrowth tissues were not reflected in gene expression levels in MF-N. uncinatum.
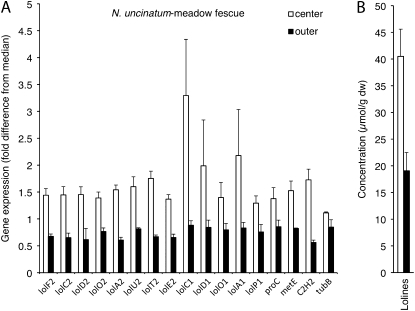
In contrast to N. siegelii, N. uncinatum exhibited significantly higher (t tests, pairwise, P < 0.05) expression of LOL genes as well as amino acid biosynthesis genes that corresponded to the higher LA concentrations in center than outer leaf blades (Fig. 4).
Figure 4.
Relative expression of N. uncinatum genes (A) and LA concentrations (B) in center leaf blades (white bars) and outer leaf blades (black bars) of MF. Gene expression is indicated as fold-difference from the median for each gene in the experiment. Error bars show ses of the mean, where n = 2 for lolC1, lolA1, lolO1, and lolD1 center leaves, and n = 4 for all others. dw, Dry weight.
Fungal Biomass Estimation Based on Housekeeping Gene Expression
The relative metabolically active fungal biomass in different tissues was estimated by measuring fungal housekeeping gene mRNA levels, with the plant translation elongation factor 1-α (EF1-α) mRNA as reference. In both MF-N. siegelii and MF-N. uncinatum symbiota, expression levels of the fungal genes tefA (encoding translation elongation factor 1-α) and tubB were similar between the regrowth tissues from clipped plants and upper tissues from unclipped plants and also similar between center and outer leaf blades (data not shown).
Water Content in Outer and Center Leaf Blades
Since the LAs are water-soluble compounds and the LA concentrations were calculated based on tissue dry weight, the possible effect of water content differences on LA concentrations was considered. In MF-N. siegelii, the dry-to-fresh weight ratio was 0.129 in center leaf blades and 0.174 in outer leaf blades; in MF-N. uncinatum, the average ratio of dry-to-fresh weight in center leaf blades was 0.184 and in the outer leaf blades was 0.208. Therefore, the water content ratio between center and outer leaf blades was 1.05 for MF-N. siegelii symbiotum and 1.03 for MF-N. uncinatum. These ratios were very low compared to LA concentration differences between these two tissue types, which were approximately 5-fold for MF-N. siegelii (Fig. 3) and 2-fold for MF-N. uncinatum (Fig. 4).
Amino Acids in Leaf Tissues
The two LA precursor amino acids, Pro and Hse, are metabolically linked to two groups of amino acids. One group, hereafter called the Gln group, includes various interconvertible amino acids, Pro, Glu, Gln, Arg, Orn, and citrulline. Of these, levels of Orn and citrulline were consistently low or undetectable, and the other amino acids were quantified. The other group, hereafter called the Asn group, includes interconvertible amino acids Asn and Asp, the Asp derivatives Lys and Hse, and the Hse derivatives Thr, Met, and Ile. Of these, Hse, Met, Ile, and Lys were consistently low or undetectable, so Asn, Asp, and Thr were quantified.
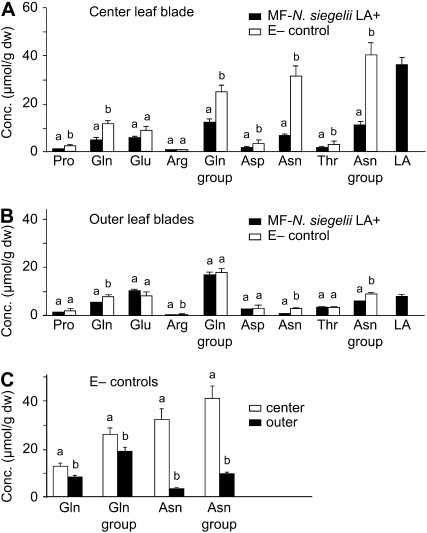
In center leaf blades of the MF-N. siegelii symbiotum, levels of the Gln-group and Asn-group amino acids were significantly lower than in aposymbiotic plants (E–, no endophyte present; Fig. 5A). In contrast, in outer leaf blades (Fig. 5B), there was much less difference in amino acid levels between MF-N. siegelii and E– control leaf blades, and although some comparisons were significant (Gln, Asn, and the Asn group), the differences were much less than in the center leaf blades. In the E– plants, center leaf blades had significantly higher levels of Gln, Gln-group, Asn, and Asn-group amino acids than in outer leaf blades (Fig. 5C). The differences for Asn and the Asn-group amino acids were most pronounced with levels in center leaves approximately 8- and 4-fold higher, respectively, than in outer leaves.
Figure 5.
Amino acid and LA concentrations in MF with N. siegelii (n = 7) and endophyte-free (E–) control plants (n = 8). A to C, The Gln group of amino acids includes Pro, Gln, Glu, and Arg, and the Asn group includes Asp, Asn, and Thr. Error bars indicate ses of the mean. Same letters above bars indicate nonsignificance for that amino acid among the plants. A, Amino acid and LA profiles in center (younger) leaf blades. B, Amino acid and LA profiles in outer (older) leaf blades. C, Comparison of amino acid levels in center and outer leaf blades of E– MF control plants. Conc., Concentration; dw, dry weight.
Interestingly, in center leaf blades of MF-N. siegelii, the concentration of LA was much higher than the concentrations of precursor amino acids, yet was comparable to the concentrations of Gln-group and Asn-group amino acids in the E– controls (Fig. 5A).
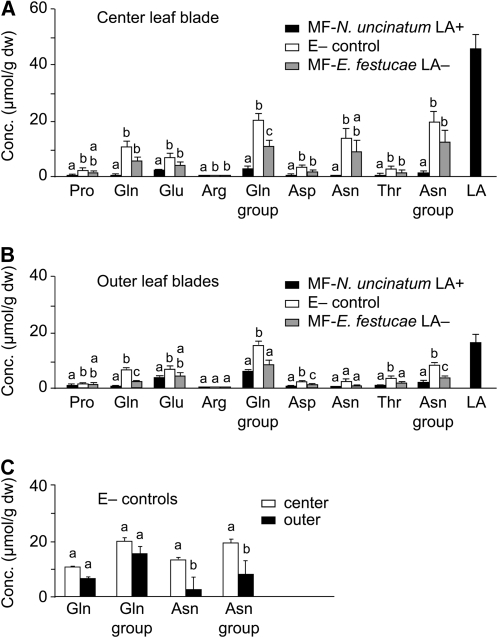
Similarly to MF-N. siegelii, MF-N. uncinatum symbiota had significantly lower Gln-group and Asn-group amino acids in center leaf blades than did E– MF controls (Fig. 6A). These were also compared to MF-E. festucae symbiota, which had endophyte strains that lack LOL genes and produce no lolines (LA–). Although MF-E. festucae LA– tended to have lower amino acid levels than E– plants, the difference was not significant except for the combined Gln-group levels. Compared to MF-N. uncinatum, center leaf blades of MF-E. festucae LA– had significantly higher Gln-group and Asn-group amino acid levels. Comparisons of amino acid levels in outer leaves indicated a similar trend in that MF-E. festucae LA– averaged higher amino acid levels than MF-N. uncinatum (Fig. 6B). This difference was significant for the Asn group but not for the Gln group amino acids. In the E– control plants, center leaf blades averaged higher levels of Gln, Gln-group, Asn, and Asn-group amino acids than in outer leaf blades, but the differences were significant only for Asn and Asn-group amino acids (Fig. 6C).
Figure 6.
Amino acid and LA concentrations in MF with N. uncinatum, LA nonproducing strains of E. festucae (LA–), and endophyte-free (E–) control plants. A to C, The Gln group of amino acids includes Pro, Gln, Glu, and Arg, and the Asn group includes Asp, Asn, and Thr. Error bars indicate ses of the mean (n = 8). Same letters above bars indicate nonsignificance for that amino acid among the plants. A, Amino acid and LA profiles in center (younger) leaf blades. B, Amino acid and LA profiles in outer (older) leaf blades. C, Comparison of amino acid levels in center and outer leaf blades of E– MF control plants. Conc., Concentration; dw, dry weight.
The average molar concentration of lolines in center leaves of MF-N. uncinatum was 16- and 32-fold higher than those of the Gln-group and Asn-group amino acids, respectively (Fig. 6A). This LA concentration even exceeded the amino acid concentrations in E– controls by approximately 2-fold and those in MF-E. festucae LA– by approximately 4-fold.
DISCUSSION
Elevated LA levels have been characterized as an induced antiherbivore response to wounding and insect damage in grass-endophyte symbiota (Craven et al., 2001; Bultman et al., 2004; Gonthier et al., 2008). The highest levels of LA known are those in the MF symbiota with N. siegelii and N. uncinatum, following clipping (Craven et al., 2001). Here, we report that the effect of clipping on LA levels is attributable to a consistent difference between younger and older leaf blades. This pattern indicates that, in these defensive mutualisms, the symbionts obey a prediction of the ODT (McKey, 1979; Rhoades, 1979) whereby younger tissues relatively lacking in physical defenses are provided greater chemical defenses than older tissues. Among the possible causes we investigated for this pattern of LA accumulation, there was no evidence that relative water content or differences in the amounts of metabolically active fungus were involved. Differences in LOL gene expression were another possibility, but there was no indication that clipping induced higher LOL gene expression in regrowth or in the subtending basal tissues. Comparison of older and younger leaf blades in the MF-N. siegelii symbiotum also showed no significant differences in LOL gene expression between the younger and older tissues. Only in comparisons of center and outer leaf blades of the MF-N. uncinatum symbiota did relative LOL gene expression reflect the difference in LA concentrations. These findings indicated that LA production was mainly controlled by other means, possibly by substrate availability. Indeed, in E– MF, the differences between younger and older leaf tissues in levels of LA precursors were consistent with this possibility. Furthermore, comparisons of amino acid levels in symbiota and E– MF, particularly in the young leaf blades, suggested that LA production was a major sink for these amino acids. Therefore, substrate availability appears to be an important factor in determining LA levels, and the fact that the LA precursors are readily derived from amino acids that are mobilized to young leaves helps to ensure that the endophytes provide greatest chemical protection to those tissues.
The lolines are well suited as protective metabolites in grass-epichloë symbiota because their expression in host tissues appears to fit ODT predictions. The lolines provide protection to younger tissues that clearly lack the level of physical defense of the older leaf blades, so greater abundance of lolines in the younger leaves is expected. Such was the case in our study, as well as in a study of TF-N. coenophialum (Eichenseer et al., 1991). Also, Justus et al. (1997) observed that, in imbibed seeds of MF-N. uncinatum, LA concentrations were approximately twice as high in the embryo than in the rest of the seed. Embryos would of course constitute the tissues most relevant to the fitness of the plant. Although the pattern of LA expression appears to fit the ODT, the GDBH predicts a cost in growth potential in younger tissues if they devote a large proportion of their resources to biosynthesis of secondary metabolites (Barto and Cipollini, 2005). In keeping with this prediction, MF with epichloë endophyte (species undetermined) and TF-N. coenophialum symbiota are reported to produce lower biomass than E– MF or TF when nitrogen is limiting (Cheplick et al., 1989; Wäli et al., 2008). Production of abundant LA by these epichloae may at least partly account for this effect.
ODT and GDBH can be reconciled if the older tissues produce the defensive compounds, which are then translocated to younger tissues (Cronin and Hay, 1996). Lolines can be translocated (Schardl et al., 2007), so if ODT and GDBH are both operative, the endophytes might elevate LA production in older tissues provided there is a mechanism to concentrate them in younger tissues. The amino acid and gene expression data reported here indicated that this is not the case. In MF-N. uncinatum, both LOL gene expression and LA levels were approximately twice as high in younger compared to older leaf blades. Surprisingly, in MF-N. siegelii, expression of LOL genes was similar or slightly less in the center compared to outer leaf blades. Nevertheless, in both MF-N. siegelii and MF-N. uncinatum, the precursor amino acid pools were depleted in molar amounts comparable to LA levels in the respective tissues, with the effect most apparent in center leaf blades. This result indicated that both N. uncinatum and N. siegelii actively produce LA in the younger host tissues and deplete free amino acid pools in the process, contrary to predictions of GDBH.
It is tempting to attribute most of the reduction in amino acids in center leaf blades to diversion into the LA pathway, but other metabolic processes and growth of the endophytes probably also contributed to the effect. Indeed, the presence of LA– E. festucae was associated with lower amino acid levels compared to E– plants. Nevertheless, MF-N. uncinatum had significantly and dramatically lower amino acid levels in the center leaf blades than did MF-E. festucae LA–. This result strongly supports the possibility that LA production is a major sink for free amino acids, even in the actively growing young leaf blades.
Dynamic correlations among amino acids and alkaloids have been reported in plants. High levels of quinolizidine alkaloids in bitter lupin (Lupinus spp.) seeds are accompanied with low basic amino acid levels, whereas sweet seeds have a low level of quinolizidine alkaloids and high amino acid levels (Aniszewski et al., 2001). Park et al. (2002) have recently demonstrated antisense suppression of the berberine bridge enzyme in California poppy (Eschscholzia californica) cells, reducing the amount of benzophenanthridine alkaloids but increasing the levels of several amino acids. Observations described herein are in keeping with such a dynamic flow in which a high level of alkaloids may result in a low level of certain precursor amino acids.
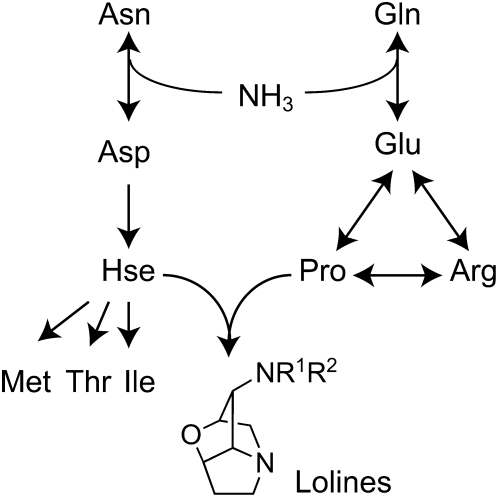
Asn and Gln are considered to be especially important in mobilization of fixed N from older to younger plant tissues, where their amide N is incorporated into other amino acids and nitrogenous metabolites (Nakano et al., 2000). Derivatives of Asn and Gln include the LA precursors Pro and Hse (Fig. 7). However, the epichloae can grow on minimal medium with sugar and inorganic nitrogen, indicating that they can synthesize all of their required amino acids (Chung and Schardl, 1997; Blankenship et al., 2001). Furthermore, elevated LOL gene expression in MF-N. uncinatum center leaves corresponded with higher expression of the two amino acid biosynthesis genes analyzed: proC for synthesis of Pro and metE for synthesis of O-acetylhomoserine, the intermediate in pathways to Met, Ile, Thr, and lolines. Nevertheless, isotopically labeled precursor amino acids, Glu, Pro, Orn, Asp, and Hse, are readily taken up by N. uncinatum in minimal medium culture and incorporated into lolines (Blankenship et al., 2005; Faulkner et al., 2006). Thus, the LA biosynthetic pathway makes use of free amino acids that are specifically mobilized to younger plant tissues. An interesting area of future research would be the flux of amino acids and other nitrogen sources in the compartments of the young leaf tissue during LA biosynthesis. For example, it is conceivable that the endophyte induces an increase of amino acid efflux from the grass symplast into the apoplast or a decrease of amino acid influx from apoplast into the grass symplast. Alternatively, the relative flux of amino acids into the fungal versus plant symplast may be simply determined by differences in affinities and activities of the plant and fungal transporters.
Figure 7.
Simplified pathway diagram for synthesis of Asn-group and Gln-group amino acids and LAs. Translocation of Asn and Gln transports fixed nitrogen from older to younger tissues (Nakano et al., 2000). The lolines are derived from the Gln derivative, Pro, and the Asn-derivative, l-homoserine (Hse), probably via conjugation of its derivative, O-acetyl-Hse, to the ring nitrogen atom of Pro (Blankenship et al., 2005; Faulkner et al., 2006). In fungi, O-acetyl-Hse is the precursor of Thr, Ile, and Met, whereas in plants, O-phospho-Hse plays the same role.
Although N. siegelii exhibits a dramatically higher level of LA production in younger than older leaf blades, there is no indication that gene regulation plays a role. It appears more likely that substrate levels exert primary control on LA production. In E– plants, Asn-group and, to a lesser extent, Gln-group amino acids are significantly higher in the center than outer leaf blades. In symbiota, the dramatically higher flux of Asn-group amino acids may, by mass action, promote higher LA production rates in those tissues. Considering that the result is a greater protection of young tissues, in keeping with ODT predictions, we speculate that in evolution of metabolic capabilities of epichloae, the lolines were selectively favored because of the availability of precursors in the young plant tissues.
We further speculate that regulation of LA production was originally based on substrate availability alone, but in some endophytes, such as N. uncinatum, transcriptional regulation also evolved. For appropriate transcriptional regulation, the fungus needs somehow to respond to the physiological state of the plant. The nature of plant-to-endophyte communication is unknown and well worth exploring. One possibility is that relative amino acid levels available to the endophyte are crucial signals affecting transcriptional responses. In support of this possibility, differences in LA production and LOL gene expression by N. uncinatum have been demonstrated in different culture conditions, including minimal media with different N sources (Blankenship et al., 2001; Spiering et al., 2002). However, failure to obtain lolines in cultures of N. siegelii, N. coenophialum, and other endophytes suggests that more specific plant signals may also be involved in regulating LOL gene expression.
CONCLUSION
MF (L. pratense) and many related grass species typically host symbiotic epichloae (Epichloë and Neotyphodium species) that produce alkaloids with anti-insect properties. Indications that N. uncinatum and N. siegelii are mutualistic endophytes include their dependence on vertical transmission in host seeds, the absence of any apparent symptoms in host plants infected with these endophytes, and the high levels of the potently insecticidal LAs in these symbiota (Craven et al., 2001). However, these endophytes may exact a cost in depleting amino acids by synthesizing the lolines. We observed that the loline levels are particularly high in younger compared to older leaf blades. Furthermore, whereas young leaf blades of aposymbiotic plants have high levels of Asn and Gln, these and metabolically related amino acids are depleted in young leaf blades of plants with the loline-producing endophytes. Much less depletion is apparent in MF symbiotic with genotypes of the related endophyte, E. festucae, that do not produce lolines. Elevated LA levels in young leaf blades were not generally attributable to increased LOL gene expression, since no significant increase in LOL gene expression was observed in regrowth or basal tissues after clipping. Although young leaves of MF-N. uncinatum had higher LOL gene expression than older leaves, no such difference was observed in MF-N. siegelii. Taken together, these observations indicate that availability of amino acids that are precursors of lolines is likely to control the different levels of lolines in plant tissues. As a consequence, younger leaf blades have higher levels of these protectants, as would be expected from ODT.
MATERIALS AND METHODS
Biological Materials and Sampling
The MF-Neotyphodium uncinatum symbiota were derived from cv Predix seedlings inoculated with N. uncinatum CBS 102646 by the method of Latch and Christensen (1985). Uninoculated plants grown from the same seed lot and propagated together with the MF-N. uncinatum symbiota served as E– controls. Also, MF-Epichloë festucae symbiota (Wilkinson et al., 2000) were established by inoculation of seeds from the same seed lot. The MF symbiotum with Neotyphodium siegelii ATCC 74483 (plant number 955 in our collection) and its uninoculated control (E– plant number 953) were similarly derived from a common seed lot of cv Predix. The presence or absence of endophyte in each plant was tested by tissue print immunoblot (An et al., 1993). Endophytes were cultured from the symbiotic plants and confirmed as N. uncinatum, N. siegelii, or E. festucae by morphological evaluation.
The MF-N. siegelii symbiotum, plant 955, and the E– control plant number 953 were derived from a different seed lot and exhibited different morphologies from the MF-N. uncinatum and MF-E. festucae symbiota. Therefore, the MF-N. siegelii symbiotum was compared only to E– plant 953. The MF-N. uncinatum and MF-E. festucae symbiota, plus a set of E– MF plants, were derived from a common seed lot and exhibited similar morphologies, so comparisons between them were considered valid.
All plants were grown in the greenhouse with heating and supplemental lighting to provide stable environmental conditions. Conditions were 14 h light at 24°C and 10 h dark at 21°C, with watering as needed. Fertilization was biweekly with 20-10-20 Peat-Lite fertilizer (J.R. Peters) at 150 ppm.
To test effects of clipping on LA levels, 16 MF-N. siegelii plants were randomly allocated to treatments, and 10 plants were clipped on day 0, whereas six plants were unclipped controls. Similarly, nine MF-N. uncinatum plants were clipped on day 0, and four plants served as unclipped controls. Plants were clipped at the base of the leaf blades directly above the pseudostems, giving the initial harvest of “upper” tissue (Fig. 1). The remaining pseudostems above the soil were the “basal” tissue, and “regrowth” tissue refers to emerging leaf blades above the initial cutting site from the clipped plants. For the unclipped control plants, upper and basal tissues were harvested simultaneously from each of two plants on days 0 and 6 (for MF-N. siegelii) or days 0, 6, and 12 (for MF-N. uncinatum), corresponding to the days PC of the clipped plants. The regrowth and basal tissues of clipped plants were sampled from two plants per day on days 3, 6, 9, 12, and 15 PC (except that MF-N. uncinatum only had one clipped-plant sample on day 9). The upper tissue collected from these plants when clipped (initial harvest) was separated into outer and center leaf blades (Fig. 1). All samples were flash-frozen in liquid nitrogen immediately after collection and stored at –80°C until further analysis.
LA Extraction and Quantitation
LA were extracted from 40 to 100 mg freeze-dried plant tissues and quantified by gas chromatography (GC) as described by Blankenship et al. (2001), with slight modifications. One-tenth volume of 1 n NaOH (instead of saturated sodium bicarbonate) was added to the freeze-dried finely ground tissue. After CHCl3 was added, tubes were shaken by hand until milky and then set at room temperature for 1 h. The CHCl3 layer was pipetted into a glass vial for GC analysis. The LA amount reported is the sum of all detected LAs, namely, loline, N-acetylnorloline, N-formylloline, and N-acetylloline.
RNA Extraction and Quantitation
Approximately 100 mg fresh weight of flash-frozen plant material was ground in liquid nitrogen to a fine powder with mortar and pestle, and total RNA was extracted with the RNeasy Plant Minikit (Qiagen). Removal of contaminating DNA by DNase treatment was performed as described by Spiering et al. (2002). Integrity of the total RNA was routinely checked by gel electrophoresis and visualizing of the RNA after ethidium bromide staining. Quantitation of RNA was carried out in 96-well 300-μL black polystyrene microplates (Whatman) by following the instructions of Quant-iT RNA Assay Kit (5–100 ng range; Invitrogen). Each sample was set up in triplicate, and each plate was read three times on a Spectra Max Gemini XS microplate spectrofluorometer (Molecular Devices) with fluorescence excitation/emission at 644/673 nm. The average fluorescence value for each sample was used to calculate the RNA concentration based on an RNA standard curve. All RNA samples were diluted to 10 ng/μL prior to use.
TaqMan Primer and Probe Design
Primers and probes for lolC1, lolC2, and tubB were the same as used by Spiering et al. (2005). All other TaqMan (TQM) primers and probes were designed in this study and supplied by Integrated DNA Technologies. All TQM probes were 5′-labeled with a 6-carboxyfluorescein reporter and 3′-labeled with black hole quencher 1, except a TQM tefA probe, for which the 5′-label was tetrachlorofluorescin. Primer and probe sequences designed in this study are listed in Table I. The cDNA sequence of each gene in the LOL2 cluster (GenBank accession no. AY723750) was aligned with that of the LOL1 cluster (AY723749) using the BLAST 2 sequences tool (National Center for Biotechnology Information [NCBI]) to identify single nucleotide polymorphisms (SNPs) for primer and probe design to specifically quantify expression of LOL2 or LOL1 cluster genes. Selected cDNA fragments (<300 bp) containing SNPs were input to PrimerQuest under IDT SciTools on the IDT Web site (http://www.idtdna.com). Primer/probe search parameters were set to real-time PCR, primer and probe quest, and optimum settings of primer size to 24 nucleotides, primer temperature 60°C, primer GC % of 50%, product range 80 to 200 bp, probe size 25 nucleotides, probe temperature 65°C, and probe GC % of 50%. Primer/probe sets were chosen based on these criteria: product size <150 bp if possible; forward and reverse primer temperature difference within 1°C; probe temperature at least 1°C higher than primer temperature; containing SNPs and no matches to other genomic regions when the primer or probe sequences were BLAST searched in the NCBI nonredundant database (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) with the organism set to Neotyphodium. Whenever possible, the two primers were designed so that the product crossed an intron boundary to identify potential genomic DNA contamination in RNA samples or the probe or one primer was designed to cross over an intron region to eliminate potential for any genomic DNA to yield a PCR product. Primers were diluted in water to 5 μm working solution and probes to 25 μm. Each primer pair was tested with reverse transcription (RT)-PCR; cDNA was synthesized from total RNA with the Monster Script first-strand cDNA synthesis kit (Epicentre Biotechnologies) following the manufacturer's protocol. The cDNA (0.5 μL) was then used in a 25-μL reaction containing 0.2 μm of each primer, 2.5 units AmpliTaq Gold, AmpliTaq Gold PCR buffer with MgCl2 (1.5 mm final concentration) supplied by the manufacturer (Applied Biosystems), 200 μm each of dATP, dCTP, dGTP, and dTTP, and a temperature regime of 95°C for 9 min, followed by 40 cycles of 95°C for 25 s, 62°C for 30 s, and 72°C for 30 s.
Table I.
Primer and probe sequences used in this study
| Name of Primer or Probe | Sequence |
|---|---|
| TQM tefA fwd | TCTACCACCACCGGTCACTTGATT |
| TQM tefA rev | TGAGCTTGTCAAGAACCCACGCAT |
| TQM tefA Probe | TET-TGCGGTGGAATTGACAAGCGTACCAT--BHQ_1 |
| TQM lolF2 fwd | CTACAGACTGCGAAAGCTCGGATT |
| TQM lolF2 rev | GGAGAAGCTCTGCATCATAGAACTGG |
| TQM lolF2 Probe | 6-FAM--CTTATCCGGGAGCAGCAGTCGACAG--BHQ_1 |
| TQM lolD2 fwd | TCTTTGTTGCCGACTTGAACGACG |
| TQM lolD2 rev | ATGCCCAAGGACAGGATCAACTCA |
| TQM lolD2 Probe | 6-FAM--AAAGCAGCTATGATCGACGGCTGATCCA--BHQ_1 |
| TQM lolO2 Fwd | TTCTTGCACCAGACGAATGCTTCC |
| TQM lolO2 rev | AATACTTGCGACAGCTTGACGAGG |
| TQM lolO2 Probe | 6-FAM-ATAACATAGACGGCTCCGTGATGGCT-BHQ_1 |
| TQM lolA2 fwd | TCGCCACCATGGATGCCAATGATA |
| TQM lolA2 rev | TTTAGCAGTGTGCTGCTCCGAGAT |
| TQM lolA2 Probe | 6-FAM-TTCTCACGGTCATGATTTCGCACGAC-BHQ_1 |
| TQM lolU2 fwd | ATGACAACGACGTTCAAGCCTCCT |
| TQM lolU2 rev | ACTTTCTGGCTTCCGTCATGGAGA |
| TQM lolU2 Probe | 6-FAM-AACTCCTGGAGAAGACTTTCGCGCA-BHQ_1 |
| TQM lolP1 fwd | ACCTGTCGACTTCTCTCGTCTGAT |
| TQM lolP1 rev | AGGTAGGTCAGCATCTTGTCAACG |
| TQM lolP1 Probe | 6-FAM-AAGACGGAGACGTGTTCGGCTACGT-BHQ_1 |
| TQM lolT2 fwd | TAGCCACTTGTGGCAATCAGAGAC |
| TQM lolT2 rev | GCGTATGCCAGAAGGAATGCATCA |
| TQM lolT2 Probe | 6-FAM-TGCAGCTCCTGGAGATTGACCTCAAA-BHQ_1 |
| TQM lolE2 fwd | TGGAGCCTAACAAGACGGACCAAA |
| TQM lolE2 rev | TGAGCCGGTGGGCGTAGAATTTAT |
| TQM lolE2 Probe | 6-FAM-AGCCGTCTTTGGCACCTACCACTTT-BHQ_1 |
| TQM C2H2 TF fwd | AGCCTGGCCATACGTTTCGATGTA |
| TQM C2H2 TF rev | AGCCCAGGAAATCACAAGGGTAGA |
| TQM C2H2 TF Probe | 6-FAM-ACAAGAGCTATTCTCGCGCAGGACAT-BHQ_1 |
| ProC Efes fwd | GCATGGCGATTTCACCATGACG |
| ProC Efes rev | CATCCAGTGGTGTTGAATCTGG |
| TQM ProC fwd | CGTCCATGTTGTTCAATGGCAGGT |
| TQM ProC rev | AACACTGCTGCTCTCATACGGGAA |
| TQM ProC Probe | 6-FAM-TATCCAGGTAACCAGGGCCTTGGATT-BHQ_1 |
| MetE EFes fwd | AGCCCAGTAACTGCGAAAGATGGA |
| MetE Efes rev | ATCATGGCCCTCAGGACTGTCAAT |
| TQM MetE fwd | ACTTGGAGGCATGTTCGTTCTGGA |
| TQM MetE rev | ATGCTTTGTCGTTGTGCTTCACCC |
| TQM MetE Probe | 6-FAM-ATACGATGCATCGTTCCCATTGCCACGT-BHQ_1 |
| TQM lolF1 fwd | CAATCTTGGAGAAGCTCCGCATCA |
| TQM lolF1 rev | CTCGCTGTCTACAATGACTACAGA |
| TQM lolF1 Probe | 6-FAM-AGAAGGGAAACAGGCTGTCGACTGCT-BHQ_1 |
| TQM lolE1 fwd | TGGATCCTAACAAGACGGACCAAA |
| TQM lolE1 rev | AGCCGGTGGGCGTAGAATTTG |
| TQM lolE1 probe | 6-FAM-AGCTGTCTTTGGCACCTACCACTTTGA-BHQ_1 |
| TQM lolA1 fwd | CCACCATGGATGCCAATGATATTCC |
| TQM lolA1 rev | TTTAGCAGTGTACTGCTCCGAGAT |
| TQM lolA1 probe | 6-FAM-TTCCCACGGTCATGATTTCGCACGAC-BHQ_1 |
| TQM lolU1 fwd | GTGACAACAACGTTCAAGATTCCTTCG |
| TQM lolU1 rev | ACTTTCTGGCTCCCGTCATGGA |
| TQM lolU1 probe | 6-FAM-AGCTCCTGGAGAAGACTGTCGCGCA-BHQ_1 |
| TQM lolO1 fwd | TTCTTGCACCAGGCGAATGCTTC |
| TQM lolO1 rev | AATACTTGCGACAGCTCGACGAGG |
| TQM lolO1 probe | 6-FAM-ATAACGTAGATGGCTCCGTGATGGCTC-BHG_1 |
| TQM lolD1 fwd | CCGACTTGAACGACATTGTTCGTAAGT |
| TQM lolD1 rev | ATGCCCAAGGAGAGGATCAACTCAA |
| TQM lolD1 probe | 6-FAM-AAAGCAGCTACGATCGACGGCTGATCCA-BHQ_1 |
| TQM lolT1 fwd | TAACCACTTGTGGCAATCAGAGAC |
| TQM lolT1 rev | GTATGCCAGAAGGAAGGCATCA |
| TQM lolT1 probe | 6-FAM-TGCAGCTCCTGGAGATTGACCTCGAA-BHQ_1 |
| Lolium EF1-α fwd | TTGAGAGGTCCACCAACCTTGACT |
| Lolium EF1-α rev | GCACAGTTCCAATGCCACCAATCT |
| Lolium EF1-α probe | 6-FAM-TTGAGGCTCTTGACCAGATCAATGAGC -IABLFQ |
For quantification of mRNA of N. uncinatum tefA, encoding translation elongation factor 1-α, and C2H2 (NCBI GenBank accessions AF308131 and AY789054, respectively), a similar approach as described above was applied to design TQM primers and probes.
Since the sequences of N. uncinatum proC (encoding pyrroline-5-carboxylate reductase) and metE (encoding homoserine O-acetyl transferase) were unknown, the exons of gene orthologs identified in the E. festucae E2368 genome (http://www.endophyte.uky.edu/) were used to design primers (Table I) for amplification and sequencing of proC (ProC Efes forward and ProC Efes reverse) and metE (MetE Efes forward and MetE Efes reverse) cDNA fragments from N. uncinatum RNA. PCR (95°C for 9 min, followed by 40 cycles of 95°C for 25 s, 62°C for 30 s, and 72°C for 1 min) was performed in reaction conditions as described above. PCR products were sequenced with the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems), and reaction products were analyzed on an Applied Biosystems model 3730xl. Primer and probe sets for quantitative RT-PCR of proC and metE were then designed based on these cDNA sequences.
A primer and probe set for the plant housekeeping gene EF1-α (encoding plant translation elongation factor 1-α) was designed as follows: EST sequencing of the normalized cDNA libraries from stromata and inflorescences of MF-E. festucae made available 77,000 plant sequence reads (U. Hesse and C.L. Schardl, unpublished data), and the cDNA sequences of plant EF1-α were submitted for BLASTn in NCBI with restriction to the genus Lolium. Appropriate primer-probe sequences were designed from the 100% match regions.
Specificities of the primer and probe sets for N. uncinatum LOL2 genes were tested on RNA from the MF-N. siegelii symbiotum. The LOL gene sequences from N. siegelii are expected to be most similar to LOL1 genes of N. uncinatum, based on comparisons of lolC1 sequences (Spiering et al., 2002, 2005). Therefore, LOL2 should be poorly amplified from N. siegelii with LOL1-specific primers. Conversely, specificity of the primer and probe sets for N. uncinatum LOL1 genes was checked with RNA from the TF-Neotyphodium coenophialum symbiotum because the N. coenophialum LOL gene sequences are nearly identical to the N. uncinatum LOL2 genes (Kutil et al., 2007).
LOL1 and LOL2 genes have shown similar expression patterns in N. uncinatum culture (Zhang et al., 2009). Because sample RNA was limiting, not all LOL1 and LOL2 gene expression profiles could be analyzed, so we investigated all LOL2 genes (except the pseudogene lolP2Δ; Spiering et al., 2008), plus five LOL1 genes, along with housekeeping and other biosynthesis genes for N. uncinatum. Primer and probe sets designed for LOL1 genes were used to investigate LOL gene expression in N. siegelii.
Real-Time Quantitative RT-PCR
Real-time quantitative RT-PCR was carried out using the TaqMan One-Step RT-PCR Master Mix reagents kit (Applied Biosystems) in 96-well PCR plates on a PRISM 7900HT instrument (Applied Biosystems) with cycling conditions as previously described (Spiering et al., 2005) with slight modifications: each 25-μL reaction contained 50 ng RNA, 400 nm of each primer, and 200 nm probe, and the program was run for 45 cycles. Amplification efficiency of each primer and probe set was determined from the slope of the standard curve. Each sample was run in triplicate on a plate for standard curves and as duplicates for relative quantification of the gene transcript.
Levels of target gene and the endogenous control gene (tefA) transcripts were analyzed on the same plate to control for plate-to-plate variation. Ct (cycle threshold) values were automatically calculated by SDS 2.3 software of the PRISM 7900HT, and the default baseline setting (cycles 3–15) was used.
Expression of all tested genes was calculated with the relative comparative Ct method (ΔΔCt = normalized Ct as ΔCt – calibrator, where ΔCt = Ct of target gene – Ct of tefA, and calibrator = median of ΔCt; Anonymous, 2007), using tefA as the reference gene for normalization. The relative level of gene expression was then converted into fold-difference relative to the calibrator as 2−ΔΔCt.
The method was validated for the LOL and other genes based on the values of slope and R2 from the standard curves of quantitative RT-PCR. Most gene standard curves had R2 > 0.98 and within 0.1 slope difference from that of the reference gene, tefA. Standard curve slopes for several genes, namely, lolC2, lolA2, and lolE2 from MF-N. uncinatum, and lolA and lolU from MF-N. siegelii, differed by 0.2 to 0.3 from that of the reference gene. This range is still acceptable for a relative comparative method according to error estimation (Zhang et al., 2009). However, the exception was lolF in MF-N. siegelii, for which the slope of −4.33 (R2 = 0.962) was significantly different from that of tefA (slope = –3.41, R2 = 0.983). Also, lolF2 in MF-N. uncinatum amplification plots showed a Ct >40, even though its slope was within the appropriate range, which suggested a very low level of lolF transcripts. Limited amounts of symbiotum RNA precluded development of another primer and probe set for N. siegelii, so data from using this lolF primer and probe set are reported but are interpreted with caution. Also, due to limiting symbiotum RNA, standard curves were not run for tubB, C2H2, and plant gene EF1-α. However, for all of these genes, the amplification plots showed good geometric phases compared to the other genes, which indicated efficient amplification.
Specificity tests of N. uncinatum LOL1 and LOL2 primer and probe sets were conducted by using them in quantitative RT-PCR for RNA from MF-N. siegelii and TF-N. coenophialum because the N. siegelii LOL cluster is similar to N. uncinatum LOL1, and the N. coenophialum LOL cluster is similar to LOL2 (Kutil et al., 2007; M.J. Spiering and C.L. Schardl, unpublished data). Results of these tests indicated that lolA2, lolC2, lolE2, lolU2, lolC1, and lolO1 were highly specific; lolD2, lolT2, lolD1, lolT1, and lolA1 were generally specific; and lolO2, lolU1, and lolE1 were not specific. Therefore, in studies of N. uncinatum, data from primer-probe sets for lolA2, lolC2, lolE2, lolU2, lolC1, lolO1, and lolP1 would represent expression from the individual homologs, whereas data from primer-probe sets for lolD2, lolD1, lolT2, lolT1, and lolA1 could be influenced by expression of the other homolog, and data from lolO2, lolU1, and lolE1 primer and probe sets may represent the total expression of homologs. Specificity for lolF primer and probe sets could not be determined. The primer sequences designed for N. uncinatum lolF1 and lolF2 showed no similarity to N. coenophialum lolF gene sequence; therefore, TF-N. coenophialum was not applicable for this estimation.
Water Content Measurement in Outer and Center Tissues
Water content was measured for the freshly clipped outer and center leaf blades. A portion of each frozen ground sample was weighed as fresh weight, then freeze-dried, and the dry weight was recorded.
Amino Acid Analysis in Plant Tissues by Liquid Chromatography-Mass Spectrometry
Plant tissues were freeze-dried and processed for amino acid analysis. Regrowth tissues, uncut control upper tissues, center leaf blades, and outer leaf blades were sampled as described above from MF-N. siegelii and MF-N. uncinatum. Center and outer leaf blades were also collected from aposymbiotic (E–) control plants for each symbiotum and from MF with E. festucae strains previously determined to lack LOL genes and to be non-LA producers (LA–; Wilkinson et al., 2000). Freeze-dried tissue was finely ground, and 20 mg was extracted under optimized conditions with 0.5 mL of 86% ethanol. Then, 100 μL of the supernatant was combined with 100 μL internal standard (containing the mixture of 0.2 m each of homoarginine, Met-d3, and homophenylalanine, provided in the kit), and the mixture was cleaned up and derivatized with the EZ:faast kit (Phenomenex). Amino acid analysis was performed by liquid chromatography-mass spectrometry with a dual-pump ProStar 210 HPLC with 1200 L quadrupole MS-MS (Varian).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FJ480953, FJ480952, FJ480951, FJ464778, FJ464780, and FJ664515.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Time course of N. siegelii gene expression in MF basal and regrowth tissues after clipping, compared to unclipped controls.
Supplemental Figure S2. Time course of N. uncinatum gene expression in MF basal and regrowth tissues after clipping, compared to unclipped controls.
Supplementary Material
This work was supported by U.S. Department of Agriculture Grants 200506271031 and 200710021743. The Epichloë festucae genome sequence was determined with support of National Science Foundation Grant EF–0523661, and meadow fescue cDNA sequences were obtained with support of U.S. Department of Agriculture National Research Initiative Grant 20053531916141.
Kentucky Agricultural Experiment Station publication number 09–12–048 published with the approval of the director.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Christopher L. Schardl (schardl@uky.edu).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- An ZQ, Siegel MR, Hollin W, Tsai HF, Schmidt D, Schardl CL (1993) Relationships among non-Acremonium sp. fungal endophytes in five grass species. Appl Environ Microbiol 59 1540–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniszewski T, Ciesiolka D, Gulewicz K (2001) Equilibrium between basic nitrogen compounds in lupin seeds with differentiated alkaloid content. Phytochemistry 57 43–50 [DOI] [PubMed] [Google Scholar]
- Anonymous (2007) PRISM ABI 7900 Sequence Detection System User Guide. Applied Biosystems, Foster City, CA
- Barto E, Cipollini D (2005) Testing the optimal defense theory and the growth-differentiation balance hypothesis in Arabidopsis thaliana. Oecologia 146 169–178 [DOI] [PubMed] [Google Scholar]
- Belesky DP, Robbins JD, Stuedemann JA, Wilkinson SR, Devine OJ (1987) Fungal endophyte infection-loline derivative alkaloid concentration of grazed tall fescue. Agron J 79 217–220 [Google Scholar]
- Blankenship JD, Houseknecht JB, Pal S, Bush LP, Grossman RB, Schardl CL (2005) Biosynthetic precursors of fungal pyrrolizidines, the loline alkaloids. ChemBioChem 6 1016–1022 [DOI] [PubMed] [Google Scholar]
- Blankenship JD, Spiering MJ, Wilkinson HH, Fannin FF, Bush LP, Schardl CL (2001) Production of loline alkaloids by the grass endophyte, Neotyphodium uncinatum, in defined media. Phytochemistry 58 395–401 [DOI] [PubMed] [Google Scholar]
- Bultman TL, Bell G, Martin WD (2004) A fungal endophyte mediates reversal of wound-induced resistance and constrains tolerance in a grass. Ecology 85 679–685 [Google Scholar]
- Cheplick GP, Clay K, Marks S (1989) Interactions between infection by endophytic fungi and nutrient limitation in the grass Lolium perenne and Festuca arundinacea. New Phytol 111 89–97 [Google Scholar]
- Chung KR, Schardl CL (1997) Vegetative compatibility between and within Epichloë species. Mycologia 89 558–565, 976 [Google Scholar]
- Craven KD, Blankenship JD, Leuchtmann A, Hignight K, Schardl CL (2001) Hybrid fungal endophytes symbiotic with the grass Lolium pratense. Sydowia 53 44–73 [Google Scholar]
- Cronin G, Hay ME (1996) Within-plant variation in seaweed palatability and chemical defenses: optimal defense theory versus the growth-differentiation balance hypothesis. Oecologia 105 361–368 [DOI] [PubMed] [Google Scholar]
- Eichenseer H, Dahlman DL, Bush LP (1991) Influence of endophyte infection, plant age and harvest interval on Rhopalosiphum padi survival and its relation to quantity of N-formyl and N-acetyl loline in tall fescue. Entomol Exp Appl 60 29–38 [Google Scholar]
- Faeth SH, Helander ML, Saikkonen KT (2004) Asexual Neotyphodium endophytes in a native grass reduce competitive abilities. Ecol Lett 7 304–313 [Google Scholar]
- Faulkner JR, Hussaini SR, Blankenship JD, Pal S, Branan BM, Grossman RB, Schardl CL (2006) On the sequence of bond formation in loline alkaloid biosynthesis. ChemBioChem 7 1078–1088 [DOI] [PubMed] [Google Scholar]
- Gonthier DJ, Sullivan TJ, Brown KL, Wurtzel B, Lawal R, VandenOever K, Buchan Z, Bultman TL (2008) Stroma-forming endophyte Epichloe glyceriae provides wound-inducible herbivore resistance to its grass host. Oikos 117 629–633 [Google Scholar]
- Hofmeister F (1892) The active constituents of Lolium temulentum. Arch Exp Pathol Pharmakol 30 203–230 [Google Scholar]
- Justus M, Witte L, Hartmann T (1997) Levels and tissue distribution of loline alkaloids in endophyte-infected Festuca pratensis. Phytochemistry 44 51–57 [Google Scholar]
- Kennedy CW, Bush LP (1983) Effect of environmental and management factors on the accumulation of N-acetyl and N-formyl loline alkaloids in tall fescue. Crop Sci 23 547–552 [Google Scholar]
- Kutil BL, Greenwald C, Liu G, Spiering MJ, Schardl CL, Wilkinson HH (2007) Comparison of loline alkaloid gene clusters across fungal endophytes: Predicting the co-regulatory sequence motifs and the evolutionary history. Fungal Genet Biol 44 1002–1010 [DOI] [PubMed] [Google Scholar]
- Latch GCM, Christensen MJ (1985) Artificial infections of grasses with endophytes. Ann Appl Biol 107 17–24 [Google Scholar]
- McKey D (1979) The distribution of secondary compounds within plants. In GA Rosenthal, DH Janzen, eds, Herbivores: Their Interactions with Secondary Plant Metabolites. Academic Press, Orlando, FL, pp 56–134
- Moon CD, Craven KD, Leuchtmann A, Clement SL, Schardl CL (2004) Prevalence of interspecific hybrids amongst asexual fungal endophytes of grasses. Mol Ecol 13 1455–1467 [DOI] [PubMed] [Google Scholar]
- Nakano K, Suzuki T, Hayakawa T, Yamaya T (2000) Organ and cellular localization of asparagine synthetase in rice plants. Plant Cell Physiol 41 874–880 [DOI] [PubMed] [Google Scholar]
- Park SU, Yu M, Facchini PJ (2002) Antisense RNA-mediated suppression of benzophenanthridine alkaloid biosynthesis in transgenic cell cultures of California poppy. Plant Physiol 128 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades DF (1979) Evolution of plant chemical defense against herbivores. In GA Rosenthal, DH Janzen, eds, Herbivores: Their Interactions with Secondary Plant Metabolites. Academic Press, Orlando, FL, pp 4–55
- Schardl CL, Grossman RB, Nagabhyru P, Faulkner JR, Mallik UP (2007) Loline alkaloids: currencies of mutualism. Phytochemistry 68 980–996 [DOI] [PubMed] [Google Scholar]
- Siegel MR, Latch GCM, Bush LP, Fannin FF, Rowan DD, Tapper BA, Bacon CW, Johnson MC (1990) Fungal endophyte-infected grasses: alkaloid accumulation and aphid response. J Chem Ecol 16 3301–3315 [DOI] [PubMed] [Google Scholar]
- Spiering MJ, Faulkner JR, Zhang DX, Machado C, Grossman RB, Schardl CL (2008) Role of the LolP cytochrome P450 monooxygenase in loline alkaloid biosynthesis. Fungal Genet Biol 45 1307–1314 [DOI] [PubMed] [Google Scholar]
- Spiering MJ, Moon CD, Wilkinson HH, Schardl CL (2005) Gene clusters for insecticidal loline alkaloids in the grass-endophytic fungus Neotyphodium uncinatum. Genetics 169 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiering MJ, Wilkinson HH, Blankenship JD, Schardl CL (2002) Expressed sequence tags and genes associated with loline alkaloid expression by the fungal endophyte Neotyphodium uncinatum. Fungal Genet Biol 36 242–254 [DOI] [PubMed] [Google Scholar]
- Sullivan TJ, Rodstrom J, Vandop J, Librizzi J, Graham C, Schardl CL, Bultman TL (2007) Symbiont-mediated changes in Lolium arundinaceum inducible defenses: evidence from changes in gene expression and leaf composition. New Phytol 176 673–679 [DOI] [PubMed] [Google Scholar]
- TePaske MR, Powell RG, Clement SL (1993) Analyses of selected endophyte-infected grasses for the presence of loline-type and ergot-type alkaloids. J Agric Food Chem 41 2299–2303 [Google Scholar]
- Tong DW, Wang JY, Brain P, Gooneratne R (2006) Seasonal change of loline alkaloids in endophyte-infected meadow fescue. Agric Sci China 5 793–797 [Google Scholar]
- Wäli PR, Helander M, Nissinen O, Lehtonen P, Saikkonen K (2008) Endophyte infection, nutrient status of the soil and duration of snow cover influence the performance of meadow fescue in sub-artic conditions. Grass Forage Sci 63 324–330 [Google Scholar]
- Wilkinson HH, Siegel MR, Blankenship JD, Mallory AC, Bush LP, Schardl CL (2000) Contribution of fungal loline alkaloids to protection from aphids in a grass-endophyte mutualism. Mol Plant Microbe Interact 13 1027–1033 [DOI] [PubMed] [Google Scholar]
- Zhang DX, Stromberg AJ, Spiering MJ, Schardl CL (2009) Coregulated expression of loline alkaloid-biosynthesis genes in Neotyphodium uncinatum cultures. Fungal Genet Biol (in press) [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.