Abstract
Ectopic expression of defined transcription factors can reprogram somatic cells to induced pluripotent stem (iPS) cells, but the utility of iPS cells is hampered by the use of viral delivery systems. Small molecules offer an alternative to replace virally transduced transcription factors with chemical signaling cues responsible for reprogramming. In this report we describe a small-molecule screening platform applied to identify compounds that functionally replace the reprogramming factor Klf4. A series of small-molecule scaffolds were identified that activate Nanog expression in mouse fibroblasts transduced with a subset of reprogramming factors lacking Klf4. Application of one such molecule, kenpaullone, in lieu of Klf4 gave rise to iPS cells that are indistinguishable from murine embryonic stem cells. This experimental platform can be used to screen large chemical libraries in search of novel compounds to replace the reprogramming factors that induce pluripotency. Ultimately, such compounds may provide mechanistic insight into the reprogramming process.
Keywords: chemical biology, high-throughput screening, Nanog
Recently it has been demonstrated that forced expression of combinations of 4 transcription factors—Oct4, Klf4, Sox2, and c-Myc or Oct4, Sox2, Nanog, and Lin28—can reprogram somatic cells into induced pluripotent stem (iPS) cells that closely resemble embryonic stem (ES) cells (1–4). The induction of pluripotency in somatic cells by ectopic expression of these 4 transcription factors has created new opportunities in the stem cell field. For example, patient-derived pluripotent cells ultimately may provide a personalized source of tissue to replace cells lost to degenerative and age-associated diseases (5). However, the clinical use of iPS cells is hindered by the use of virally transduced transcription factors and proto-oncogenes.
Optimization of the original reprogramming method has achieved reprogramming of both mouse and human somatic cells with the 3-factor combination of Oct4, Sox2, and Klf4 in the absence of c-Myc (6–8). Other methods to reduce the number of reprogramming factors have taken advantage of endogenously expressed reprogramming factors, thereby precluding the need for ectopic expression of these factors, such as Sox2 (9–11). More recently, iPS cells have been generated using excisable vectors (12–14), nonintegrating vectors (15, 16), and transient transfection approaches (17). Although these methods have solved some of the limitations posed by the presence of multiple proviruses in the genome of iPS cells, they either remain inefficient or fail to further our understanding of the mechanistic details of epigenomic reprogramming.
Small molecules that induce epigenetic changes also have been used to increase the efficiency of reprogramming and/or replace reprogramming factors (18–22). Recently, the histone deacetylase inhibitor valproic acid was found to allow for 2-factor reprogramming (Oct4 and Sox2) of human fibroblasts in the absence of Klf4 and c-Myc (18, 19). This method relies on the use of known molecules that target known mechanisms. The identification of novel small molecules that induce pluripotency could provide useful chemical tools to unravel and study the molecular mechanisms governing this process. Furthermore, application of such molecules ultimately may lead to useful new therapeutics for treating disease. Toward this end, we have developed a high-throughput screening strategy to identify small molecules that can functionally replace reprogramming transcription factors. In this paper we describe the identification of a compound class that can substitute for the reprogramming factor Klf4 in the formation of iPS cells. Our results provide proof of principle that high-throughput screening is a robust vehicle for identifying small molecules that can replace the transcription factors used to reprogram somatic cells.
Results
Nanog-Luciferase Reporter Mouse Strain.
The induction of pluripotency in fibroblasts by the ectopic expression of Oct4, Sox2, Klf4, and c-Myc requires a minimum of 8–12 days and occurs at low frequency, with <0.1% of the somatic cells giving rise to iPS cells (4, 23). The slow kinetics of reprogramming impedes the development of robust assays to screen large chemical libraries with a reliable readout that can be captured on a well-to-well basis in a miniaturized format. To overcome these limitations, we relied on the high sensitivity and quantitative readout capabilities of luciferase to develop a screening platform. To establish a sensitive readout for iPS cell formation, the firefly luciferase gene was inserted into the Nanog locus by homologous recombination. We chose Nanog because it plays important roles in maintaining the undifferentiated state (24, 25), is completely inactivated in somatic lineages, and iPS cells selected for Nanog reactivation demonstrate complete developmental potential (1, 2, 4).
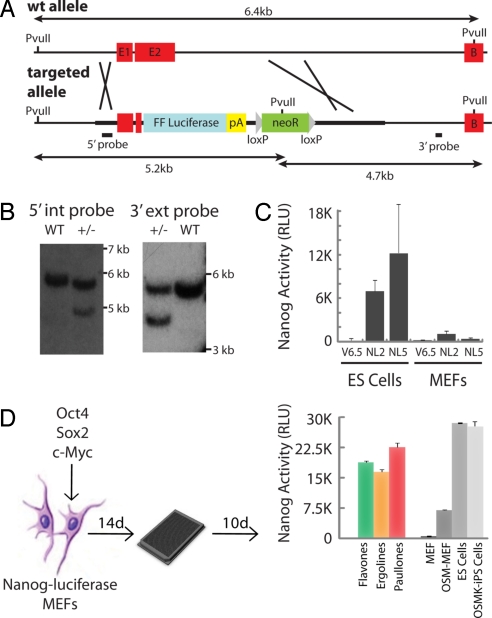
A targeting vector was constructed using 1.2 kb of the Nanog promoter as a 5′ arm and 1.4 kb of Nanog intron 2 as a 3′ arm. The firefly luciferase gene from pGL3-basic and a floxed pgk-neo cassette were inserted between the targeting arms (Fig. 1A). Nanog-luciferase (NL) ES cell clones that carried the targeting construct in the Nanog locus, as confirmed by Southern blot analysis (Fig. 1B), gave rise to a signal 10–30 times greater than that of wild-type V6.5 ES cells (Fig. 1C). The NL ES cells were introduced into blastocysts to generate chimeric mice, which were bred to generate both transgenic mice and mouse embryonic fibroblasts (MEFs). NL-MEFs produced negligible amounts of luciferase (Fig. 1C) and were used for subsequent experiments.
Fig. 1.
NL screening platform. (A) Scheme for targeting the firefly (FF) luciferase gene to the Nanog locus. (B) Southern blot hybridization analysis of the NL targeted allele. The targeted allele–specific 5.2-kb and 4.7-kb PvuII fragments were detected with the 5′ internal and 3′ external probes, respectively, shown in (A). (C) NL activity of correctly targeted ES cell clones (NL2 and NL5) versus NL-MEFs. Differentiated NL tissue shows a significant reduction in luciferase expression. V6.5 ES cells and MEFs are provided as a luciferase-negative control. (D) Small-molecule screening strategy. NL-MEFs were transduced with a reduced reprogramming cocktail (Oct4, Sox2, and c-Myc [OSM]), expanded for 2 weeks, plated into 1,536-well plates, treated with compound, and assayed for luciferase expression 10 days after plating: flavones (7-hydroxyflavone, 20 μM), ergolines (lysergic acid ethylamide, 2 μM), and paullones (kenpaullone, 5 μM). Nanog activity is reported in relative light units (RLUs) and was read in a 96-well (B) or 384-well (D) format. Error bars indicate SD (n = 3).
High-Throughput Chemical Screening Strategy: Klf4 Replacement.
In this work, we customized our screening strategy to identify compounds that could replace Klf4. It has been suggested that Klf4 plays a major role in chromatin remodeling, and recent evidence demonstrates that Klf4 is an important mediator of the undifferentiated ES cell state (26, 27). In addition, overexpression of Klf4 can promote breast cancer (28) and squamous cell carcinomas (29), ascribing a potent oncogenic role to this transcription factor. Thus, replacement of Klf4 is a critical step in the eventual application of iPS cells as a therapeutically viable strategy.
To identify a chemical replacement for Klf4, NL-MEFs were transduced with a subset of reprogramming factors comprising Oct4, Sox2, and c-Myc (OSM). To obtain the ≈109 cells needed to screen large chemical libraries required expansion of OSM-transduced NL-MEFs for 3 passages before screening. At passage 4, OSM-transduced NL-MEFs were seeded into 1,536-well plates at 500 cells/well in mouse ES cell growth media supplemented with leukemia inhibitory factor (LIF; 20 ng/mL) and screened against a large (500,000 compounds), chemically diverse small-molecule library (30). Compounds were added at a final concentration of 2.2 μM (20 nL of 1 mM stock solutions in 9 μL of media) immediately after plating, and Nanog-driven luciferase expression was assayed 10 days later (Fig. 1D). Analysis of a focused library in pilot experiments demonstrated a high degree of homogeneity of luciferase response from well to well—104 molecules in triplicate [supporting information (SI) Fig. S1]. Furthermore, normalization of these data using the “jackknife” z-score method—which accounts for plate-to-plate variation and does not rely on positive control readings—and subsequent analysis using the 1-sample Kolmogorov-Smirnov test (P = .54) indicated little difference between the empirical and theoretical inverse gamma distributions. These statistical analyses confirmed both that the screening platform was robust and that the output data were highly reliable (31).
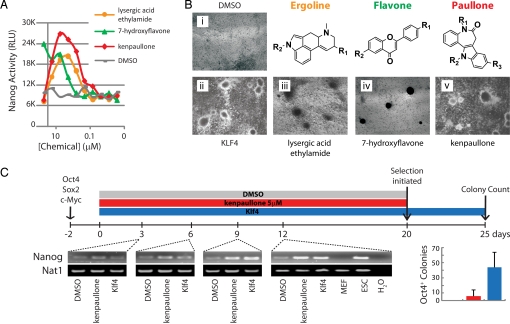
Nanog activity from OSM transduced NL-MEFs was 1 order of magnitude greater than that in nontransduced cells and 4-fold less than the activity in pluripotent NL reporter cells (Fig. 1D). From the primary screen, compounds that gave rise to a signal >2.2-fold (≈5,000 compounds) over the plate average—assuming that most compounds are inactive and can serve as controls (31)—were selected for reanalysis in triplicate. Measurements with these putative hit compounds were collected and averaged, and those that activated the luciferase signal 2.5-fold over DMSO-treated controls (≈1,000 compounds) were counterscreened in a cell-based SV40-driven firefly luciferase assay to rule out false-positives that directly and nonspecifically activate luciferase reporters (32). Compounds that reproducibly hit in the Nanog assay but did not hit in the SV40 assay were further analyzed in a dose–response format to identify the optimal concentration for activity (Fig. 2A). Collectively, this strategy further narrowed our hit compounds to 135 small molecules (≈0.03% of the initial set), which grouped in 3 lead structural classes: flavones, lysergamides, and paullones (Fig. 2B).
Fig. 2.
Activity of lead chemical scaffolds. (A) Dose-dependent Nanog activation of a lead compound from each chemical class: lysergic acid ethylamide (ergolines), 7-hydroxyflavone (flavones), and kenpaullone (paullones). Above 10 μM, kenpaullone and lysergic acid ethylamide are toxic. (B) OSM-MEFs took on colony morphology upon application of lead compounds (i, DMSO; ii, Klf4; iii, 7-hydroxyflavone 20 μM; iv, lysergic acid ethylamide 2 μM; v, kenpaullone 5 μM). (C) O4N-MEFs were transduced with OSM on day -2 and treated with DMSO, retrovirally delivered Klf4, or kenpaullone 2 days later (day 0). RT-PCR of samples collected at days 3, 6, 9, and 12 confirmed the reactivation of Nanog observed using NL-MEFs. At day 20, kenpaullone and DMSO were removed, and neomycin was added to the culture media to initiate selection of iPS cells. Neomycin-resistant colonies (indicating reactivation of endogenous Oct4) were counted at day 25. Kenpaullone is shown in red; Klf4, in blue. Error bars indicate SD (n = 4 wells of a 6-well plate). Dose–response curves are representative of at least 6 independent experiments and were read in a 384-well format. Images were captured at × 40 magnification.
Activity of Lead Compounds.
To distinguish compounds that replace Klf4 from those that simply activate Nanog expression, colony formation was used as a secondary assay. Representative compounds from each lead structural class were added to OSM-transduced MEFs and analyzed for the presence of colonies 10 days later. Members of each structural class induced the OSM-MEFs to form colonies (Fig. 2B); however, colonies that stained positive for alkaline phosphatase (AP) were observed only in the presence of kenpaullone and lysergic acid ethylamide (data not shown). Kenpaullone proved to be the most robust reprogramming molecule based on this series of assays, and thus was studied in more detail.
Although AP-positive colony formation is consistent with the conversion of somatic cells to iPS cells, it does not prove establishment of a pluripotent state. To determine whether the iPS cells derived with kenpaullone were fully reprogrammed, we transduced MEFs carrying the gene encoding neomycin resistance in the Oct4 locus, O4N-MEF (4), with OSM vectors and treated these cells with retrovirally delivered Klf4, vehicle control, or kenpaullone 2 days later (Fig. 2C). Nanog expression was analyzed by semiquantitative RT-PCR at 3-day intervals to confirm the reactivation observed using the NL reporter line. Consistent with observations using NL-MEFs, Nanog expression was activated in kenpaullone-treated OSM-transduced MEFs in a time-dependent manner (Fig. 2C and Fig. S2).
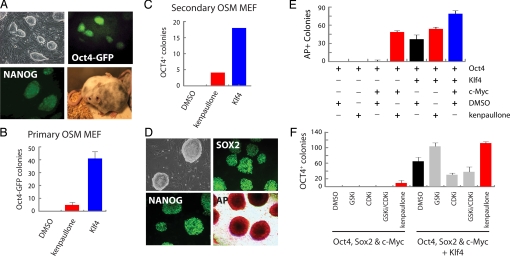
After 20 days of concurrent kenpaullone treatment, colonies expressing Oct4 from their endogenous locus were selected upon supplementation of the culture media with neomycin (Fig. 2C). Kenpaullone was able to replace Klf4, as demonstrated by the appearance of Oct4-neo colonies, although the reprogramming was less efficient than after transduction with a Klf4 vector. Likewise, OSM transduction and kenpaullone treatment of MEFs carrying a GFP reporter in the Oct4 locus, O4G-MEF (33), gave rise to GFP-positive colonies (Fig. 3A) with similar efficiency and kinetics as observed in the O4N-MEFs (Fig. 3B). The DMSO-treated OSM-transduced O4G-MEFs did not display GFP-positive cells or colony formation even after >40 days in culture (data not shown).
Fig. 3.
Characterization of iPS cells derived with kenpaullone. (A) iPS cells generated with OSM and 5 μM kenpaullone express the pluripotency-associated markers Oct4 and Nanog from their endogenous loci and are capable of contributing to chimeric mice. (B) Chemical complementation of Klf4 with kenpaullone is less efficient than retroviral delivery of Klf4, based on the number of colonies (per 10,000 MEFs) that express GFP from the endogenous Oct4 locus. Error bars indicate SD (n = 3). (C) Secondary MEFs harboring proviral copies of OSM were treated with kenpaullone (5 μM) or retrovirally delivered Klf4. Colonies were scored based on expression of OCT4. (D) Kenpaullone-treated Oct4/c-Myc-NPCs gave rise to iPS cells that expressed pluripotency-associated proteins (Nanog, Sox2, and AP) from endogenous loci. (E) NPCs were transduced with Oct4 +/− Klf4 and c-Myc and treated with kenpaullone (5 μM) or vehicle control. Data represent colonies that expressed AP after 8 days (per 100,000 NPCs). Error bars indicate SD (n = 3). (F) O4N-MEFs were transduced with OSM +/− Klf4 and treated with a GSK-3β–specific inhibitor (GSKi; CHIR99021, 3 μM), a general CDK inhibitor (CDKi; purvalanol A, 3 μM), both the GSKi and the CDKi, or kenpaullone. Colonies were scored for neomycin resistance (per 50,000 MEFs) at day 25. Error bars indicate SD (n = 4).
Characterization of iPS Cells.
iPS cells generated by OSM and kenpaullone treatment were indistinguishable from ES cells by morphological criteria and expressed the pluripotency-associated markers Oct4 and Nanog from the endogenous loci (Fig. 3A). However, the reprogramming efficiency of kenpaullone-treated OSM-transduced O4G-MEFs was 10-fold lower than that of Klf4-infected controls (Fig. 3B). To confirm the activity of kenpaullone in an independent culture system, we used “secondary” MEFs that carry doxycycline-inducible proviruses encoding Oct4, Sox2, and c-Myc (34). The results, shown in Fig. 3C, indicate that kenpaullone induced reprogramming of secondary OSM-MEFs with a similar efficiency as primary infected OSM-MEFs. The reprogramming kinetics of kenpaullone-treated iPS cell formation was delayed, however; doxycycline-independent iPS cells appeared only after 25–30 days, compared with the 15 days observed for the 4-factor control iPS cells. This slight kinetic delay in reprogramming suggests that kenpaullone is less efficient in epigenetic reprogramming than Klf4.
Once established, Klf4-free iPS cells grew in the absence of doxycycline and kenpaullone while maintaining ES cell morphology and Nanog and Oct4 expression. To test for pluripotency, the cells were injected into blastocysts and were found to generate germline-competent chimeras (Fig. 3A and data not shown).
It has been shown that mouse neural progenitor cells (NPCs) that express endogenous Sox2 can be reprogrammed to a pluripotent state with the addition of Oct4 and Klf4 +/− c-Myc (9, 11) or simply Oct4 (10), albeit with delayed kinetics and low efficiency. To determine the functionality of kenpaullone in the reprogramming of neural cells, NPCs were transduced with all combinations of Oct4, Klf4, and c-Myc and treated with 5 μM kenpaullone or vehicle control. Eight days later, discernible AP-positive colonies had formed in Oct4/c-Myc/kenpaullone-treated wells that, when passaged, appeared morphologically indistinguishable from ES cells and stained positive for the pluripotency-associated markers Nanog, Sox2, and AP (Fig. 3D). Furthermore, these cells no longer expressed GFP from the virally delivered c-Myc vector (pCMV-Myc-IRES-GFP; Fig. S3), consistent with retroviral silencing observed in reprogrammed cells (4). The efficiency of iPS cell formation from NPCs was lower with kenpaullone than with Klf4 gene transduction, although not as low as in MEFs (Fig. 3 B and E). It is also noteworthy that neither MEFs nor NPCs reprogrammed in the absence of c-Myc when kenpaullone was substituted for Klf4. Collectively, these results suggest that kenpaullone requires transduction of c-Myc for reprogramming and emphasizes that kenpaullone may not completely recapitulate the role of Klf4.
Biological Activity of Kenpaullone.
Previous work with kenpaullone has demonstrated a wide range of biological utility, extending from maintenance of pancreatic β cell survival and proliferation (35) to the induction of apoptosis in cancer cells (36). Kenpaullone's diverse range of activity is a direct result of its kinase inhibition promiscuity (SI Text, Tables S1 and S2). It is a potent inhibitor of GSK-3β (23 nM), CDK1/cyclin B (400 nM), CDK2/cyclin A (680 nM), and CDK5/p35 (850 nM) and exhibits inhibitory activity toward various other kinases at higher concentrations (36, 37).
To determine whether kenpaullone's reprogramming activity results from its well-established role as a GSK-3β or CDK inhibitor, a highly selective small-molecule inhibitor of GSK-3β (CHIR99021), a promiscuous inhibitor of CDKs (purvalanol A), or both were applied to OSM-transduced O4N-MEFs and compared against kenpaullone. As shown in Fig. 3F, known inhibitors of these kinases were unable to replace Klf4 in the reprogramming of murine fibroblasts and had negligible activity in the NL reporter MEF line (Fig. S4). It is interesting that kenpaullone, like CHIR99021, increased the efficiency of 4-factor reprogramming (Fig. 3F). This observation is consistent with reports indicating that GSK-3β inhibitors can help facilitate the self-renewal of mouse ES cells (38) and the reprogramming of mouse NPCs back to the pluripotent state (11). But because other, more specific GSK-3β inhibitors were not able to activate Nanog expression (Fig. S4 and Table S3) or to replace Klf4 (Fig. 3F), this activity likely is independent of the reprogramming activity of kenpaullone.
In addition to small-molecule–mediated inhibition, short hairpin RNA (shRNA)-targeted ablation of each of the canonical kenpaullone kinase targets individually, combinatorially or in a pooled format did not result in colony formation or an increase in Nanog-driven luciferase expression in OSM-transduced MEFs (Fig. S5). Collectively, these data strongly suggest that kenpaullone's activity does not result from its well-documented role as a GSK-3β or cell cycle inhibitor. Finally, kenpaullone does not directly activate Klf4 expression at the mRNA or protein level (Fig. S6), suggesting that kenpaullone functions through an entirely novel mechanism.
Discussion
In this paper we have described the development and application of an unbiased, high-throughput (>500,000 compounds) cell-based screening technology that can be applied to identify small molecules to replace virally transduced reprogramming transcription factors. In a proof of concept application, we have identified 3 classes of molecules that activate Nanog expression in murine fibroblasts during direct reprogramming in the absence of Klf4, and have characterized one such molecule, kenpaullone, in detail. We have shown that kenpaullone is able to replace Klf4 in the reprogramming of primary and secondary fibroblasts and NPCs. OSM iPS cells derived in the presence of kenpaullone display the characteristics of pluripotent ES cells, including the ability to contribute to chimeric mice.
Previous studies with small molecules that induce epigenetic changes have suggested that chromatin remodeling is a rate-limiting step in the conversion from a somatic to a pluripotent epigenetic state (18–22). For example, treatment of MEFs with a small-molecule inhibitor of DNA methyltransferase (5-azacytidine) facilitates reprogramming during a brief temporal window by removing demethylation marks, thereby lowering the kinetic barrier to the transition to pluripotency (20, 22). Histone deacetylase inhibitors and histone methyl transferase inhibitors also have been shown to increase the efficiency of reprogramming (18, 19, 21). In particular, the histone deacetylase inhibitor valproic acid was shown to dramatically increase 3-factor reprogramming efficiency in the absence of c-Myc in both mouse and human cells and to allow 2-factor reprogramming (Oct4 and Sox2) of human fibroblasts in the absence of Klf4 and c-Myc (18, 19).
Although modulation of known epigenetic control elements has provided significant progress toward reprogramming that does not require genetic manipulations, such techniques rely on specific compounds to target known mechanisms. With the study of induced pluripotency still in its infancy, the mechanisms governing reprogramming remain largely unknown. It is likely that the activation of different alternative pathways by small molecules will enhance the efficiency of vector-free reprogramming. Furthermore, identifying chemicals with novel mechanisms of action to modulate this process is of great interest. This work demonstrates that unbiased, cell-based screens can identify small molecules from large chemical libraries to replace transcription factors for reprogramming. Moreover, the identification of these and other molecules provides novel tools to study the molecular mechanisms at play during epigenome overhaul and ultimately may help bring iPS cell technology one step closer to clinical application.
Materials and Methods
Chemicals.
Kenpaullone, purvalanol A, and 7-hydroxyflavone were purchased from Sigma, and CHIR99021 was purchased from Axon MedChem.
Gene Targeting and Blastocyst Injection.
Generation of the NL targeting vector is described in detail in SI Materials and Methods. For targeting, the NL vector was linearized and electroporated into ES cells. G418 selection (350 μg/mL) was started 24 h later and maintained for 10–12 days. Resistant clones were selected and expanded for screening by Southern blot analysis. Injection of NL ES or iPS cells into either BDF2 or BALB/C host blastocysts was carried out as reported previously (39).
Cell Culture and Viral Infections.
Transgenic MEFs were isolated from NL, secondary OSM (34), O4N (4), or O4G (33) mice, selected from E13.5 embryos and cultivated in DMEM containing 15% FBS, L-glutamine, beta-mercaptoethanol, and nonessential amino acids. ES and iPS cells were cultivated on irradiated MEFs in the aforementioned media plus 20 ng/mL LIF (Chemicon).
NPCs were derived from ES cells and expanded 4–5 passages, as described previously (40). MEFs and NPCs were infected with pooled viral supernatant generated by transfection of HEK293T cells (Fugene; Roche) with a VSV-G vector and doxycycline-inducible lentiviral (23) or Maloney retroviral vectors (pMXs; Addgene, generously deposited by S. Yamanaka) containing the cDNAs of Oct4, Sox2, and c-Myc with or without Klf4. Detailed experimental conditions concerning the generation of iPS cells from NPCs, O4N, O4G, and secondary MEFs are described in SI Materials and Methods. Media components were purchased from Invitrogen unless specified otherwise.
Chemical Screening and Nanog Assay.
The screening protocol is described in detail in SI Materials and Methods. In brief, NL-MEFs (≈108 cells) were transduced with concentrated VSV-G–psuedotyped lentiviral particles containing Oct4, Sox2, and c-Myc in 15-cm dishes. OSM NL-MEFs were expanded 4 passages, plated into 1,536-well plates at 500 cells/well, treated (2.2 μM), and then analyzed for luciferase activity 10 days later via the addition of Bright-Glo reagent (Promega)
Counterscreening and dose–response experiments were run in 384-well plates (Greiner). Cells were plated at 1,000 cells/well in ES cell growth media and assayed 10 days later. All experiments were run in duplicate and repeated at least 3 times.
RT-PCR and Immunostaining.
Reverse-transcription and immunostaining were performed as described previously (41), using gene-specific primers (Table S4) and the following antibodies (diluted 1:500): Sox2 (polyclonal mouse; Santa Cruz Biotechnology), Oct4 (monoclonal mouse; Santa Cruz Biotechnology), and Nanog (polyclonal rabbit; Santa Cruz Biotechnology).
Supplementary Material
Acknowledgments.
This work was supported by a National Science Foundation Predoctoral Fellowship (to C.A.L.), a Human Frontier Science Program Long-Term Fellowship (to J.S.), a Canadian Institute for Health Research Post-Doctoral Fellowship (to L.L.L.), and grants from the Skaggs Institute of Chemical Biology of The Scripps Research Institute (to P.G.S.), the Novartis Research Foundation (P.G.S.), and the National Institute of Health (RO1-HD045022 and R37-CA084198, to R.J.). We thank the Novartis chemical screening facility for technical assistance and Dr. Anthony Orth and Chang Liu for helpful discussions.
Footnotes
Conflict of interest: R.J. is an advisor to Stemgent and Fate.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903860106/DCSupplemental.
References
- 1.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 2.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES cell–like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 5.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2007;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 7.Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensible for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:1–3. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Hockemeyer D, et al. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eminli S, Utikal J, Arnold K, Jaenisch R, Hochedlinger K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells. 2008;26:2467–2474. doi: 10.1634/stemcells.2008-0317. [DOI] [PubMed] [Google Scholar]
- 10.Kim JB, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Silva J, et al. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaji K, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woltjen K, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soldner F, et al. Parkinson's disease patient–derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009 Mar 26; doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 18.Huangfu D, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huangfu D, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 20.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y, et al. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brambrink T, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 25.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang J, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 28.Foster KW, et al. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–6495. [PubMed] [Google Scholar]
- 29.Foster KW, et al. Induction of KLF4 in basal keratinocytes blocks the proliferation–differentiation switch and initiates squamous epithelial dysplasia. Oncogene. 2005;24:1491–1500. doi: 10.1038/sj.onc.1208307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plouffe D, et al. In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc Natl Acad Sci USA. 2008;105:9059–9064. doi: 10.1073/pnas.0802982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 32.Auld DS, Thorne N, Maguire WF, Inglese J. Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression. Proc Natl Acad Sci USA. 2009;106:3585–3590. doi: 10.1073/pnas.0813345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lengner CJ, et al. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markoulaki S, et al. Transgenic mice with defined combinations of drug-inducible reprogramming factors. Nat Biotechnol. 2009;27:169–171. doi: 10.1038/nbt.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stukenbrock H, et al. 9-Cyano-1-azapaullone (cazpaullone), a glycogen synthase kinase-3 (GSK-3) inhibitor activating pancreatic beta cell protection and replication. J Med Chem. 2008;51:2196–2207. doi: 10.1021/jm701582f. [DOI] [PubMed] [Google Scholar]
- 36.Zaharevitz DW, et al. Discovery and initial characterization of the paullones, a novel class of small-molecule inhibitors of cyclin-dependent kinases. Cancer Res. 1999;59:2566–2569. [PubMed] [Google Scholar]
- 37.Knockaert M, et al. Intracellular targets of paullones: Identification following affinity purification on immobilized inhibitor. J Biol Chem. 2002;277:25493–25501. doi: 10.1074/jbc.M202651200. [DOI] [PubMed] [Google Scholar]
- 38.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 40.Bibel M, Richter J, Lacroix E, Barde YA. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat Protoc. 2007;2:1034–1043. doi: 10.1038/nprot.2007.147. [DOI] [PubMed] [Google Scholar]
- 41.Lyssiotis CA, et al. Inhibition of histone deacetylase activity induces developmental plasticity in oligodendrocyte precursor cells. Proc Natl Acad Sci USA. 2007;104:14982–14987. doi: 10.1073/pnas.0707044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.