Abstract
Environmental stochasticity is known to play an important role in life-history evolution, but most general theory assumes a constant environment. In this paper, we examine life-history evolution in a variable environment, by decomposing average individual fitness (measured by the long-run stochastic growth rate) into contributions from average vital rates and their temporal variation. We examine how generation time, demographic dispersion (measured by the dispersion of reproductive events across the lifespan), demographic resilience (measured by damping time), within-year variances in vital rates, within-year correlations between vital rates and between-year correlations in vital rates combine to determine average individual fitness of stylized life histories. In a fluctuating environment, we show that there is often a range of cohort generation times at which the fitness is at a maximum. Thus, we expect ‘optimal’ phenotypes in fluctuating environments to differ from optimal phenotypes in constant environments. We show that stochastic growth rates are strongly affected by demographic dispersion, even when deterministic growth rates are not, and that demographic dispersion also determines the response of life-history-specific average fitness to within- and between-year correlations. Serial correlations can have a strong effect on fitness, and, depending on the structure of the life history, may act to increase or decrease fitness. The approach we outline takes a useful first step in developing general life-history theory for non-constant environments.
Keywords: generation time, demographic dispersion, stochastic growth rate, reproduction, survival, life history
1. Introduction
Identifying rules to categorize life histories and to predict their evolution continues to be a major challenge for evolutionary biology. Fisher (1930) and Cole (1954) first argued that the evolution of life histories should be understood in terms of their effect on average individual fitness measured as population growth rate. Stearns (1976, 1977) used this view to discuss inter-species differences in the age distribution of reproduction and survival as adaptations constrained by trade-offs due to the costs of reproduction (Williams 1966; Gadgil & Bossert 1970). Another theme in life-history analysis uses allometric scaling to organize life histories (Calder 1996) and to identify invariants, parameters that do not vary among life histories (Charnov 1993). Most of this work assumes constant environmental conditions and uses the tools of classical demography (Keyfitz 1977; Caswell 2001).
A distinct selective force that shapes and constrains possible life histories is stochastic environmental variation (Levins 1968; Schaffer 1974). Empirical studies show that environmental fluctuations rather than trade-offs can generate negative correlations between components of a life history (Knops et al. 2007), and determine whether costs of reproduction are detectable (Tavecchia et al. 2005). Changes in the pattern of environmental fluctuations, due to, for example, climate, generate selective pressure on life histories both in the short run (Boyce et al. 2006) and over geological time (Ruzzante et al. 2008). To understand how environmental fluctuations shape the evolution of life histories, we have to use stochastic demography (Tuljapurkar 1990; Caswell 2001). The evolutionary consequences of fluctuating environments have been examined for some life-history patterns including life cycle delays (Tuljapurkar & Wiener 2000; Koons et al. 2008), semelparity and iteroparity (Orzack & Tuljapurkar 1989; Orzack 1993), and longevity (Morris et al. 2008). They have also been used to study and compare empirical patterns of environmental variability in populations of many species, including mammals (Gaillard et al. 2000) and plants (Tuljapurkar et al. 2003; Morris et al. 2006).
Our goal in this paper is to examine the ‘fitness landscape’ of age-structured life histories in fluctuating environments, i.e. the mapping between life-history phenotypes and fitness. We begin by discussing the evolution of life histories and explaining the significance of this landscape, as well as the difference between life histories in constant and variable environments. We next use general theory to identify key parameters of a life history, which determine fitness in fluctuating environments. These include the usual suspects, generation time (a measure of the speed of the life history over an absolute time scale, Gaillard et al. 2005) and net reproductive rate (a measure of reproductive output that corresponds to the average number of female offspring a female will produce during her lifetime). But other parameters matter in the presence of temporal variation. The first is the temporal variance of vital rates in relation to their elasticity (elasticity measures the potential impact that a given change in a vital rate will make on population growth). Elasticity and variability may be negatively correlated due to buffering as suggested by Pfister (1998) or environmental canalization as suggested by Gaillard & Yoccoz (2003). The second is the dispersion of reproduction over age, which indicates the level of iteroparity in a given life history. The third thing that matters is the correlations (positive or negative) of vital rates both within a year and between years. The last parameter is the resilience of the demography of the life history to past environmental perturbations (measured by damping time of the average life history) in relation to serial correlation. To understand the quantitative effect of such key parameters, we explore life histories numerically (modelled after mammals and long-lived birds) for a range of environmental variabilities. We close with a summary and discussion of our main findings.
2. Life-history evolution
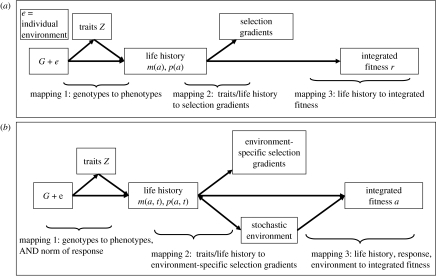
Consider a population living in constant environmental conditions, say in a laboratory. Here the life history is a population-averaged set of vital rates, such as age-specific fertility and mortality. Life-history evolution may be viewed as involving three biological processes, which we call ‘mappings’, shown in figure 1a. Mapping 1 describes how genotypes G and individual developmental environments e produce life-history traits Z (such as, say, birth weight). The nature of this mapping is revealed by the methods of gene mapping, the analysis of quantitative trait loci and perhaps most commonly in natural populations, by the methods of quantitative genetics (Kruuk et al. 2008). Mapping 2 describes the selective consequences of variation in life-history traits and is usually explored by studying selection gradients (following Lande & Arnold 1983). Mapping 3 integrates these selection components into a fitness measure, which in this setting is the Lotka growth rate r (equivalently the Malthusian parameter). Mapping 3 has been explored by using classical demography to examine how changes in the life history produce change in r (Caswell 2001). We say that mapping 3 describes the fitness landscape over which evolution can drive life histories when environments are constant over time. To actually follow the evolution of life histories, we need to understand all three mappings. Given such insight, we may relate genetic variation (via mapping 1) to phenotypic variation, relate the latter to selection on components of the life history (via mapping 2), relate variation in life-history components to variation in fitness r (via mapping 3) and then use dynamic equations to describe genotypic and phenotypic evolution. The last step is based on theory showing that allele frequency dynamics are driven by variation among genotypes in their realized fitness r (Lande 1982; Charlesworth 1994).
Figure 1.
The mappings from genotypes to fitness for (a) constant environments and (b) temporally varying environments.
Now consider the realistic case of a natural population living in temporally varying environmental conditions. From one year to the next, there is variation in environmental conditions that affect all individuals in every year t. These environmental conditions may include many factors, e.g. temperature, rain or the abundance of other species (plant food for a herbivore or prey for a predator). Now mortality and fertility may vary not just by age but also in response to environmental conditions in each year. These responses may be correlated within a year (e.g. environments that lead to poor survival may also lead to poor reproduction). There may also be lagged responses that result in correlations between years, e.g. if environments in a particular year lead to high fertility, there may be a reduction in energy storage so that in the following year individuals cannot exploit good environments should they occur.
Thus in a temporally variable environment, a life history includes (i) the time averages of age-specific mortality and fertility, (ii) the response of annual mortality and fertility to the environment, including correlations between these responses within a year, and (iii) lagged (also called serial) correlations, in which the effects of environments in a given year carry over to future years. These many dimensions are reflected in the appropriate fitness measure for stochastic environments, which is not r but the stochastic growth rate a. The stochastic growth rate is the long-run growth rate of a population in a temporally varying environment (Tuljapurkar 1990). Returning to our three mappings, the lower half of figure 2 shows that mapping 1 now includes the genetic determination of both trait values and the norm of reaction of trait values to the environment. Mapping 2 now includes environment-specific selection gradients, because selection on traits can be different in different environmental conditions. In mapping 3, fitness a is determined not just by the life-history components, but also by environmental change over time. The relationship between a life history (including its environmental response) and a variable environment, on the one hand, and fitness a on the other hand describes the fitness landscape for life histories in fluctuating environments. This identification rests on theory showing that genotypic variation in a drives the dynamics of rare alleles (Tuljapurkar 1982; Charlesworth 1994) in stochastic structured populations, and also drives the dynamics of alleles in stochastic unstructured populations (Lande 2007; Lande et al. 2009).
Figure 2.
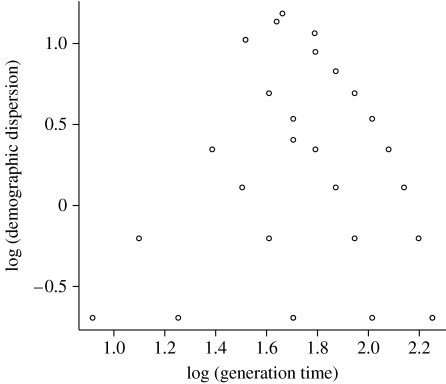
The lack of association between the natural log of generation time and the natural log of demographic dispersion is one way of demonstrating the substantial differences between the stylized life histories we analyse.
Other papers in this special issue of this journal describe evolutionary and ecological studies that explore particular aspects of the three mappings in figure 1. Population genetic studies typically focus on mapping 1, ecological studies of selection on mapping 2 and theoretical studies on mapping 3. Ezard et al. (2009) explore the effects of mappings 2 and 3 in the case of large mammal dynamics, whereas Zheng et al. (2009) combine mapping 1 for allelic variants at a single gene with mappings 2 and 3 to describe the effects on population and evolutionary dynamics. Our decomposition of the evolutionary processes into mappings was inspired by earlier theoretical work by Coulson et al. (2006). We use theory for populations in variable environments to explore mapping 3, the fitness landscape that shows how life history and environment determine fitness.
3. Describing life histories
We now define the main quantities that we use to describe life histories; most mathematical details are in the electronic supplementary material. We use discrete (annual) time-steps denoted t and discrete age classes labelled by an index i.
(a) Life histories in a constant environment
In an unchanging environment, a life history is described by the values of age-specific survival and reproduction. The survival rate from age class i to age class i+1 is denoted by p(i) and the average fertility of age class i is denoted by m(i). Survivorship (the probability of living to age at least x) is given by
| (3.1) |
The life-history consequences of these vital rates are effectively described by six parameters that we now describe.
First, the net reproductive rate is the expected lifetime reproduction of a newborn. Second, the cohort generation time is the average age of reproduction in a cohort,
| (3.2) |
In species reproducing at several ages, the extent of iteroparity is described by the dispersion of reproduction with respect to age. Our third parameter is the demographic dispersion σd, which measures the dispersion of reproduction events across age and is defined as
| (3.3) |
To understand the last two quantities, consider a semelparous life history in which all individuals reproduce at one fixed age A; then Tc=A and σd=0. With iteroparous reproduction spread evenly from age A to age B>A (i.e. l(i)m(i) is equal at these ages and zero at other ages), we have
and the dispersion is
Here we can see clearly how σd measures the age dispersion of reproduction. Thus, the value of σd measures the extent of iteroparity, and also whether individuals often skip breeding years.
To follow population dynamics, we use the vital rates to construct a population projection matrix (also called a Leslie matrix) A (see Caswell (2001) for details). This matrix has a dominant eigenvalue λ and determines our fourth parameter, the average individual fitness r=log λ in a constant environment. A population has growth rate exactly r only when its age composition equals the stable age distribution in which the proportion of individuals at age i is given by
| (3.4) |
measured relative to newborns for whom u(1)=1. In a stable population, the age distribution of reproduction is determined by the proportion of individuals u(i) at every age i and their fertility m(i), and our fifth parameter, the generation time in a stable population T, is defined as
| (3.5) |
T measures the weighted mean age of mothers (sensu Leslie 1966) and differs from cohort generation time Tc in equation (3.2), but the two are close in value when R0 is close to 1, i.e. when r is close to 0.
In practice, even in a constant environment, there may be perturbations to the age composition, e.g. due to in- or outmigration, or disease. Suppose that at a particular time t=0, a population's age composition is not stable and is, say, y(i,0)≠u(i) at each age i. As time passes, the age composition changes, becoming y(i,1) at t=1, y(i,2) at t=2, and so on. As t increases, the difference between y(i,t) and u(i) goes to zero: this is demographic convergence (also called demographic ergodicity). This demographic convergence is exponential: the difference between y(i,t) and u(i) decreases as e−t/τ where τ is a damping time. With the passage of every τ years, the ‘distance’ to the stable age distribution decreases by a factor of . A population with a large damping time τ takes a long time to reach stable proportions, and vice versa. The damping time τ is our sixth parameter. It is known that τ decreases when σd increases (Keyfitz & Caswell 2005).
(b) Life histories in a variable environment
When environments vary over time, and vital rates change in response, the first thing we can do is record time averages of age-specific survival and fertility. If we simply call these p(i) and m(i), respectively, at age i, we can characterize the average life history by the corresponding values of the six summary parameters R0, Tc, T, r, σd and τ as defined above. Next, we have to describe variability in vital rates over time. We could do this in steps, by first describing variation in the environment, then the response of vital rates to environment and then computing the variation in the rates (Lacey et al. 1983). We will short-cut this by working directly with fluctuations in the vital rates.
Suppose that we observe fertility m(i,t) at age i for several years t. The time average of these is m(i) and the variance of the set is an estimate of the annual variance, call it σm(i). The coefficient of variation of this fertility is c=(σm(i)/m(i)). Within-year variances σm(i), σp(i) are described in this way for every survival and fertility rate. We are also interested in within-year correlations, which tell us whether two different rates change in a correlated way. For example, if a cost of reproduction is paid in current-year survival, we expect a negative within-year correlation, call it ρ, between m(i,t) and p(i,t). Finally, we must consider between-year correlations. For example, a cost of reproduction may be paid not in the year when reproduction occurs but the following year, in which case we expect a negative between-year correlation, call it ξ, between m(i,t) and s(i,t+1). To fully describe a temporally varying life history, we specify all within-year correlations such as ρ and all between-year correlations such as ξ.
4. From life history to fitness
We focus on populations that are growing slowly over the long run, meaning that the average fertility and mortality yield a growth rate r close to zero and the stochastic growth rate a is also close to zero. Hence we take values of net reproductive rate R0 to be close to 1. We can use the small-noise approximation for the fitness a (Tuljapurkar 1982). As is well known (Morris & Doak 2002; Lande et al. 2003), this approximation provides robust qualitative results for fairly large fluctuations.
(a) A formula for fitness
Under our assumptions, the fitness of a life history is given to a good approximation (Tuljapurkar 1982) by
| (4.1) |
The four terms on the right-hand side describe the effects of the four main dimensions of the life-history phenotype:
r is the growth rate determined by the average fertility and mortality,
Vs is the contribution of within-year variances of all vital rates,
Vc is the contribution of within-year covariances of all vital rates, and
S is the contribution of between-year serial correlations of all vital rates.
The quantity r is well approximated (Keyfitz & Caswell 2005) by
| (4.2) |
The next two terms in (4.1) require the sensitivities of λ=er to the average survival and fertility rates as defined by
| (4.3) |
The elasticity to, say, survival rate at age i is ep(i)=(p(i)/λ)sp(i).
The full expressions for Vs, Vc and S are written out in the electronic supplementary material, appendix. Here we need to note only that Vs is a sum, over all survival and fertility rates, of their variances weighted by the squares of their sensitivities (or equivalently of the squares of their coefficients of variation weighted by the squares of their elasticities). Vc is a similar sum but over distinct pairs of rates with each term also weighted by the correlation between each pair. S describes transient dynamics and correlations between vital rates at all time lags.
(b) The determinants of fitness
We now use our theory to examine how fitness a changes among life histories. We suppose that genetic change (via mapping 1; figure 1) produces life-history phenotypes that vary with respect to one or more of the parameters that we have discussed in the previous section. Our task is to describe the effect of such changes on fitness a (via mapping 3; figure 1). The general features of the fitness landscape are the following:
An increase in Tc (and equivalently in T) corresponds by definition to shifting the mean age of reproduction to later ages. This decreases r in increasing populations (see equation (4.2)) and hence also decreases a.
The sensitivities sm, sp are inversely proportional to T (Caswell 2001). Hence Vs and Vc are inversely proportional to T2 while S is inversely proportional to T. An increase in T reduces Vs and Vc and thus increases a. There is a smaller increase in the magnitude of the term S, although the sign of S may be positive or negative, so the effect on a is not clear.
Thus an increase in T has two opposing effects: decreasing a via the effect on r and increasing a via the effect on Vs and Vc. So we predict that a will be maximized at intermediate values of generation time.
Increasing within-year variance in vital rates will increase Vs and Vc and thus decrease a. Variability matters most in those rates that have the highest sensitivity. As shown by Pfister (1998) and Gaillard & Yoccoz (2003), there is evidence for a negative correlation between variability and elasticity.
When there are no within-year correlations among vital rates, Vs=0. When within-year correlations are all negative, the value of Vs will be negative, and conversely. Hence negative within-year correlations increase a, by damping the overall effect of variance on fitness. Positive within-year correlations amplify variance, and thus decrease a.
Increasing the demographic dispersion σd increases r (see equation (4.2)) and thus a. A distinct effect of increasing σd is to reduce damping time τ. Faster damping means that the population quickly ‘forgets’ fluctuations produced by the environment, so we expect that increasing σd will reduce the magnitude of S. The net effect on a is unclear since S may be positive or negative.
5. A phenotype space of life histories
To flesh out these general relationships between life history and a, we explored numerically a biologically motivated set of model life histories. Our approach was inspired by that in Orzack & Tuljapurkar (1989), but we consider a far larger set of phenotypes to obtain a broad and rich picture of the fitness landscape. We describe below our construction of life histories and the analytical expression for fitness a that follows from applying equation (4.1), and in the next section present and discuss numerical results.
(a) A family of model life histories
We consider life histories with a first age of reproduction of 2 years or more, a last age of reproduction of 10 years or less, and equal fertility at all fertile ages, but many distinct age patterns of reproduction. We assume that the populations we study are large enough that we may ignore demographic stochasticity. Demographic stochasticity does affect extinction probabilities and matters to the fate of individuals in individual-based models, but does not affect the long-run growth rate (Athreya & Karlin 1971). We include year-skipping, for example a life history with equal fertility at ages of 2–10, and a life history with reproduction at ages of only 3 and 10. We excluded true semelparity that occurs when there is only one breeding attempt during lifetime. The value of R0 reflects long-run growth and should be very close to 1 so that a is close to zero; note that R0 is not a maximal reproductive rate. We set R0=1.01 for every life history; values as high as R0=1.1 have little qualitative effect on our results. Without such a constraint, the fitness landscape would be merely a single peak at the highest possible survival and fertility. We set survival rate to be p(1)=0.7 for the youngest age class and p(i)=1 for all ages i≥2. Figure 2 displays the wide range of values of cohort generation time Tc and demographic dispersion σd spanned by the average phenotypes we consider.
We suppose that stochastic fluctuations occur in only fertility with the same coefficient of variation c at every fertile age. The within-year correlation between every pair of fertilities has the same value ρ. If there is a total of J fertile ages in a life history, then
because the variance–covariance matrix of fertilities must be positive semi-definite.
Between-year correlations are assumed to follow an autoregressive process with a serial correlation coefficient ξ. We compute fitness for many values of c, ρ, ξ, but focus on results with c=0.5. We report mainly on three values of ρ (0, large negative and large positive), referring to these simply as the cases of negative, zero and positive correlations, respectively. We report mainly on results for ξ=−0.5, 0, +0.5, and refer to these simply as the cases of negative, zero and positive serial correlations, respectively.
For a particular choice of average fertility and mortality, we first construct a Leslie matrix of average rates and compute Tc, σd, and r. Next, we compute Vs in equation (4.1). We then set the within-year correlation ρ to each of six values (0.9 and 0.5 times the most negative possible value, zero, +0.25, +0.5 and +0.75), and compute corresponding values of Vc in equation (4.1). Finally, for each of these within-year correlations, we set the between-year correlation ξ to each of seven equally spaced values between −0.75 and +0.75, and compute corresponding values of S in equation (4.1). Note that although we did not explicitly consider density-dependent responses of average fitness, our scenarios encompass situations in which the action of density dependence may be captured in terms of changes in within- and between-year variances. For each average age distribution of fertility and each c, we have nine contrasting stochastic life histories and corresponding fitness values. We report on results from 1218 such combinations.
(b) The components of fitness
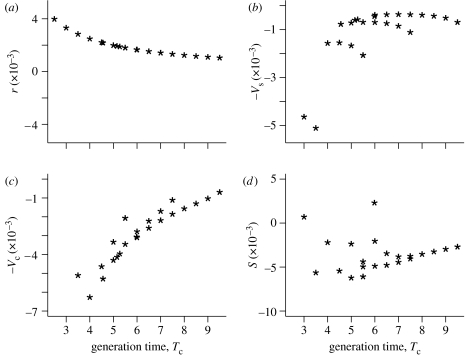
To illustrate the fitness decomposition a=r−Vs−Vc+S, we present in figure 3 values of the four components for coefficient of variation c=0.5, positive within-year correlation ρ=+0.25 and positive between-year correlation ξ=+0.25. Figure 3a shows r declining with Tc across life histories. Figure 3b shows the contributions of within-year variances. These are all negative and their magnitude declines with increasing Tc as we expect. At a given generation time Tc, there is much more scatter between the contributions of within-year variance than between values of r, showing that differences in σd matter here. Figure 3c shows the contributions of positive within-year correlation between fertilities. The magnitude of these negative contributions also decreases as Tc increases. Finally, figure 3d shows the contributions produced by positive serial correlation added to the within-year positive covariance; note that some of these are positive and others negative, showing that between-year correlations can act to reduce or increase fitness.
Figure 3.
The association between cohort generation time Tc and (a) r, (b) variation in demographic rates, Vs, (c) within-year covariation between demographic rates Vc and (d) between-year covariation in demographic rates, S. For each life history, c=0.5 and within- and between-year covariances between vital rates have been set to +0.25.
6. Exploring the fitness landscape
For convenience, we discuss separately the effects of average rates, within-year variation and between-year variation, although these effects are not independent; for example, the average life history determines sensitivities and hence the response to fluctuations. The terms Vs, Vc, S, respectively, are zero at different ‘boundaries’ of phenotype space. With zero variance c=0, so there is no variability and Vs=Vc=S=0. With variability c>0 but no within-year correlation, we have ρ=0 and hence Vc=S=0. With variability c>0 and within-year correlation ρ>0 but no between-year correlation, we have ξ=0 and so S=0.
(a) Effects of average life history
Calculations on our model life histories confirm equation (4.2), showing that r∝(1/Tc). More generally, we expect r to scale with body weight with an allometric exponent of −0.25 (e.g. Hennemann 1983) and Tc to scale with body weight with an allometric exponent of +0.25 (e.g. Millar & Zammuto 1983), so that the product rTc should be independent of body weight. Hence we expect that the product rTc is a dimensionless number (Charnov 1993) that will differ between populations and species mainly on account of differences in average environmental conditions, biotic and abiotic.
In our set of model life histories, there is no correlation between Tc and σd (r=−0.028, p=0.886; figure 2). The variation in Tc accounted for less than 0.1 per cent of the variation observed in σd. The only pattern we found was that σd was the lowest at both the shortest and the longest Tc, with peak dispersion at intermediate values (i.e. 5–6 years) of Tc. Contrary to the intuitive expectation that the level of iteroparity (demographic dispersion) should increase with generation length, in our model life histories a low variance of reproductive ages can occur at any generation time, so we do not build in any coupling between the level of iteroparity and generation time.
As σd is a biological time (sensu Lindstedt 1981), and r has dimension (1/time), we also investigated the relationship between σd and r on a log scale. We did not find any linear relationship between σd and r, and variation in σd accounted for less than 0.1 per cent of the observed variation in r. We confirmed that higher dispersion produces a higher r, as expected from equation (4.2), but the numerical contribution of dispersion to r is negligible compared with the effect of Tc, because log R0≪1. A multiple regression using standardized measures of Tc and σd showed that a given change in Tc had at least 650 times more influence on log r than the same change in σd.
From a life-history perspective, these results suggest that the large among-species variation that occurs in average population growth rate has been shaped mainly by variation in the speed of the life history, although demographic dispersion does affect the response to fluctuations as shown below. This suggests that pioneering work on life-history evolution (Charnov & Schaffer 1973; Charlesworth & Leon 1976; Williams 1996) which focused on iteroparity–semelparity continuum might fruitfully be generalized by focusing on the fast–slow continuum.
(b) Within-year and between-year correlations
Negative within-year correlation means that fertilities at different ages respond differently to the same environmental conditions. Such negative correlations can arise if young and old individuals pay a higher cost of reproduction than prime-age individuals, or if there is age-dependent variation in the efficiency and/or ability of foraging. Positive within-year correlation means that fertility at all ages responds in the same way to environmental conditions, and may result from age-independent costs or foraging abilities, or age-independent physiological responses to environments.
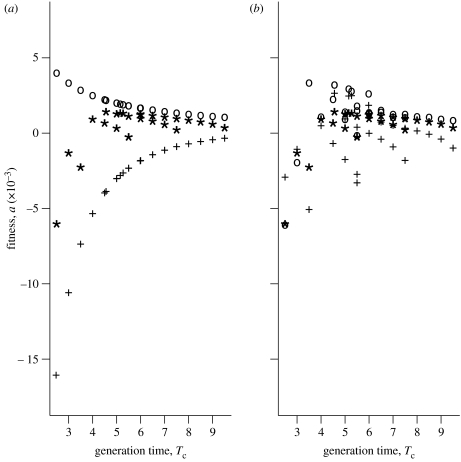
Figure 4a shows the strong effect of within-year correlation on the relationship between fitness a and Tc. We display values for negative (i.e. the most negative possible), zero and positive within-year correlations ρ, and zero serial correlation (ξ=0).
With no within-year correlation (points indicated by asterisks), fitness a rises with Tc and reaches a maximum at intermediate cohort generation times of 5 years. The peak is modest and fitness declines slowly at higher values of Tc. This behaviour is what we predicted in §4b. Here ‘fast’ life histories (with low Tc) and high r suffer a high cost because they experience substantial fluctuations. Somewhat ‘slower’ life histories are buffered against fluctuations. Very slow life histories are even more buffered but pay too high a cost in reduced r.
With positive within-year correlation (points indicated by pluses), fitness a rises steadily with increasing Tc and there is no peak. Here the advantage in r for ‘fast’ life histories (with low Tc) is more than offset by the high cost of fluctuations relative to ‘slow’ life histories.
When within-year correlation is most negative (points indicated by circles), these correlations alone significantly dampen the effect of fluctuations. As a result, there is little fitness gained through buffering at long generation times. Fitness a declines steadily as Tc increases, and fast life histories have the edge.
Figure 4.
The sum plotted against generation time. The effect of (a) positive within-year covariance with zero serial covariance (circles: ρ<0, ξ=0; asterisks: ρ=0, ξ=0; pluses: ρ>0, ξ=0) and (b) positive within-year covariance with positive between-year covariance (circles: ξ<0, ρ=0; asterisks: ξ=0, ρ=0; pluses: ξ>0, ρ=0).
Next, consider between-year correlations in fertility. Negative between-year correlation means that high fertilities in the current year will probably generate low fertilities in following years and may result from, e.g., depletion of energy stores in years when fertility is higher than average. Organisms with capital breeder tactics (sensu Jonsson 1997) might exhibit such negative serial correlations. By contrast, positive serial correlation may result if years with above-average fertility are also years in which energy is acquired and stored at above-average rates, as could be observed in resource-pulse systems (Ostfeld & Keesing 2000). Figure 4b displays changes in fitness a with increasing Tc for negative (−0.75), zero and positive (+0.75) between-year correlations, with no within-year correlations. The starred points in this figure are exactly the same as the starred points in figure 4a.
For all values of between-year correlation, fitness increases with Tc with a visible peak of approximately 5–6 years and then falls slowly as Tc increases further.
When generation times are under 5–6 years, positive between-year correlation can increase or decrease fitness of life histories with similar generation times (we explain this below). But at longer generation times, negative between-year correlation increases a, whereas positive between-year correlation decreases a. The magnitude of the effect of ξ on a is much larger at short generation times.
(c) Effects of reproductive dispersion
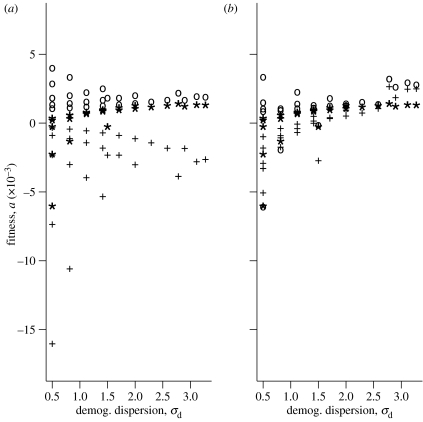
There are two reasons why we expect demographic dispersion σd to affect a. Variance in fertilities affects a via the term Vs in equation (4.1), which is a weighted sum of variances in all fertilities. Denote the weight on the variance at age i by z2(i). It turns out (see the electronic supplementary material, appendix for details) that z(i) is just the fraction of reproduction, on average that occurs at age i. When σd is large, only a few ages reproduce, so each z(i) is large. By contrast, when σd is small, each z(i) is small, the weights are even smaller and Vs is small. Hence we expect that Vs will decrease as σd increases. Next, the damping time of a life history is known to decrease as σd increases (Keyfitz & Caswell 2005). If the average life history has very rapid damping, the effect of correlated fluctuations will not persist for long and will not accumulate over time. Hence as σd increases, we expect that the magnitude of S will decrease.
Figure 5a, for ξ=0 and different ρ, and figure 5b, for ρ=0 and different ξ, show how σd affects a. Recall from figure 1 that a given σd is shared by several life histories with different Tc. Consider the points (indicated by asterisks) in figure 5a, for which within-year correlation ρ=0 and between-year correlation ξ=0. For small values of σd there is considerable scatter in the corresponding fitness values. As σd increases, we would predict (based on the arguments of the preceding paragraph) that all life histories will be buffered against fluctuations, and thus the scatter among fitness values should decrease. This is indeed what happens, for every value of within-year correlation ρ. We expect a similar pattern in figure 5b. When σd is small and within-year correlation ρ=0, we expect that the effects of between-year correlation ξ (via S in equation (4.1)) will be large, and thus expect high scatter among fitness values for any σd. But at larger values of σd, the effects of between-year correlation are smaller for all life histories, and we expect much less scatter in fitness values. The figure supports this argument.
Figure 5.
The effect of changing the sign of the within-year covariances on the association between cohort generation length and average fitness with no serial correlation. Pluses represent positive within covariances, asterisks represent no correlation and circles represent negative correlations. (a) Circles: ρ<0, ξ=0; asterisks: ρ=0, ξ=0; pluses: ρ>0, ξ=0. (b) Circles: ξ<0, ρ=0; asterisks: ξ=0, ρ=0; pluses: ξ>0, ρ=0.
7. Conclusions
(a) The fitness landscape
Our numerical and analytical results show that temporal variability in life histories strongly influences the fitness landscape. To summarize our findings, we fitted a series of statistical models to the difference (a−r) between stochastic growth rate and the growth rate without fluctuations. We examined the effects of the parameters Tc (the speed of the life history), ρ (the within-year correlation), ξ (the between-year correlation) and σd (the demographic dispersion of the life history), as well as their interactions.
Our theoretical analysis shows that generation time has effects proportional to (1/Tc) and . Thus, we considered models with each of these factors separately and both together. The best model among these included only Tc and yielded an R2=0.51 and highly significant (here and below all significant coefficients in the formulae have p<3×10−4) coefficients with
This model captures the major effect of the speed of life histories on a, especially the way in which an intermediate generation time can lead to the highest difference (a−r) between stochastic and deterministic growth rates.
Since our numerical analysis clearly shows a strong interaction of within-year correlation ρ with Tc, and between-year correlation ξ with Tc, we next fit a series of models of these interactions. The best fitting of these increased R2 to 0.90, with a change in the Akaike's information criterion (ΔAIC) of −2130 compared with the model above, and
This model shows clearly the importance of interactions between the correlation patterns and generation time. In our family of life histories, the direct effects of positive ρ and positive ξ are negative. But note that the interaction coefficient between ρ and has a sign opposite to that between ξ and
The next factor that we include is the demographic dispersion σd, which (without interactions) yields a model with R2=0.94 and a change in the AIC from the preceding model by −599, yielding the formula
The main difference between this model and the preceding model without demographic dispersion is that the coefficient on (1/Tc) decreased by approximately two-thirds and that on decreased by approximately one-third. Instead, we now see a positive dependence on the demographic dispersion of the life history. We have pointed out earlier that σd has little effect on r, so the last term in this model expresses a property of stochastic growth rate.
The last step was to include interactions between σd and the correlations ρ and ξ based on our results in figure 5. This model yielded a slightly better R2=0.95 and a further change in the AIC of −61. Even though we have more parameters than in the preceding model, we obtain a better overall fit as supported by the change in AIC. We found a significant coefficient for an interaction between within-year correlations ρ and σd but not for an interaction between serial correlations ξ and σd. Compared with the preceding model, there is little change in most of the coefficients, with the important exception that in this final model the coefficient for a direct effect of ρ is no longer different from 0 (p<0.159). It is particularly striking that the direct effect of σd does not change, so the new interaction terms represent an additional dimension of variation in the data. Our final model, including the coefficient on ρ, is
This last equation above summarizes the main features of the fitness landscape. Remember from our discussion that r is rather accurately estimated by (log R0/Tc). Fluctuations drive a to values above or below r, depending on the life history and the characteristics of the fluctuations. The speed of the life history, as measured by Tc, has a strong direct effect, in that slower life histories tend to be buffered against the effects of fluctuations. But Tc has equally important indirect effects, interacting with within-year correlation ρ, so that negative ρ increases fitness relative to zero or positive ρ. There is also a positive interaction between serial correlations ξ and Tc. The direct effect of serial correlation ξ is negative so that negative serial correlation will increase fitness and positive serial correlation will tend to decrease it. Demographic dispersion plays a significant role: increasing σd has a direct positive effect on fitness. Dispersion also interacts negatively with within-year correlation and may interact weakly but positively with serial correlation.
To conclude our description of the fitness landscape, we note several striking features that are demonstrated by our graphical analysis. First, in a fluctuating environment, there is often a range of cohort generation times at which the average fitness a of a life history is at a maximum. Thus, we should expect to find optimal phenotypes in fluctuating environments that are quite distinct from optimal phenotypes in constant environments. Second, the average fitness of a life history in stochastic environments is directly and strongly affected by demographic dispersion, even when the average fitness in deterministic environment is not. Demographic dispersion also determines the response of the average fitness of a life history to within-year fluctuations. Third, negative correlations between vital rates act to boost fitness, whereas positive serial correlations act to reduce fitness. Finally, serial correlations can have a strong effect on fitness, and, depending on the structure of the life history, may act to increase or decrease its average fitness.
(b) Moving forward
An important conclusion is that an appropriate treatment of environmental stochasticity is required to understand how the diversity of life histories observed in Nature has arisen. Our results also show that conclusions based on assumptions of a constant environment are not a useful or reliable guide to the stochastic case. This message might not come as a surprise to many life-history researchers. However, the novelty of our results is in characterizing the profound effect of the sign and strength of within-year correlations between vital rates, and temporal autocorrelation in the environment on the association between generation length, demographic dispersion and average individual fitness. To achieve this, we have developed a powerful framework to decompose fitness. Here we have used stylized life histories that we have carefully selected to cover the range of those observed. In reality, observed life histories contain further sources of variation we do not consider—especially, change in survival rates with age. Further work will apply the methods developed here to real-world cases. In particular, the increased availability of time-series of vital rates reliably estimated from longitudinal studies of vertebrate populations should soon offer the possibility of performing such comparative life-history analyses in stochastic environments. Such analyses might lead to a change in our current understanding of life-history evolution based on works that invariably assumed constant environments (e.g. Brown & Sibly (2006) for a recent example).
Acknowledgements
We thank Fanie Pelletier, Dany Garant and Andrew Hendry for their unceasing patience in awaiting submission of this manuscript to their special symposium, and their subsequent rapid handling of the manuscript. We also thank Marco Festa-Bianchet, an anonymous referee, and Fanie Pelletier for their many and useful criticisms. S.T. was funded by the NIA and NSF, J-.M.G. by CNRS and T.C. by NERC.
Footnotes
One contribution of 14 to a Theme Issue ‘Eco-evolutionary dynamics’.
Supplementary Material
Mathematical formulas
References
- Athreya K.B., Karlin S. Branching processes with random environments. II. Limit theorems. Ann. Math. Stat. 1971;42:1843–1858. doi:10.1214/aoms/1177693051 [Google Scholar]
- Boyce M.S., Haridas C.V., Lee C.T. Demography in an increasingly variable world. Trends Ecol. Evol. 2006;21:141–148. doi: 10.1016/j.tree.2005.11.018. doi:10.1016/j.tree.2005.11.018 [DOI] [PubMed] [Google Scholar]
- Brown J.H., Sibly R.M. Inaugural article: life-history evolution under a production constraint. Proc. Natl Acad. Sci. USA. 2006;103:17 595–17 599. doi: 10.1073/pnas.0608522103. doi:10.1073/pnas.0608522103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder W.A. Courier Dover; Mineola, NY: 1996. Size, function, and life history. [Google Scholar]
- Caswell H. 2nd edn. Sinauer; Sunderland, MA: 2001. Matrix population models: construction, analysis and interpretation. [Google Scholar]
- Charlesworth B. Cambridge University Press; Cambridge, UK: 1994. Evolution in age-structured populations. [Google Scholar]
- Charlesworth B., Leon J.A. The relation of reproductive effort to age. Am. Nat. 1976;110:449–459. doi:10.1086/283079 [Google Scholar]
- Charnov E.L. Oxford University Press; New York, NY: 1993. Life history invariants: some explorations of symmetry in evolutionary ecology. [Google Scholar]
- Charnov E.L., Schaffer W.M. Life-history consequences of natural selection: Cole's result revisited. Am. Nat. 1973;107:791–793. doi:10.1086/282877 [Google Scholar]
- Cole L.C. The population consequences of life history phenomena. Q. Rev. Biol. 1954;29:103–137. doi: 10.1086/400074. doi:10.1086/400074 [DOI] [PubMed] [Google Scholar]
- Coulson T., Benton T.G., Lundberg P., Dall S.R.X., Kendall B.E. Putting evolutionary biology back in the ecological theatre: a demographic framework mapping genes to communities. Evol. Ecol. Res. 2006;8:1155–1171. [Google Scholar]
- Ezard T.H.G., Côté S.D., Pelletier F. Eco-evolutionary dynamics: disentangling phenotypic, environmental and population fluctuations. Phil. Trans. R. Soc. B. 2009;364:1491–1498. doi: 10.1098/rstb.2009.0006. doi:10.1098/rstb.2009.0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.A. Clarendon Press; Oxford, UK: 1930. The genetical theory of natural selection. [Google Scholar]
- Gadgil M., Bossert W.H. Life historical consequences of natural selection. Am. Nat. 1970;104:1–24. doi:10.1086/282637 [Google Scholar]
- Gaillard J.M., Yoccoz N.G. Temporal variation in survival of mammals: a case of environmental canalization? Ecology (Durham) 2003;84:3294–3306. doi:10.1890/02-0409 [Google Scholar]
- Gaillard J.M., Festa-Bianchet M., Yoccoz N.G., Loison A., Toigo C. Temporal variation in fitness components and population dynamics of large herbivores. Annu. Rev. Ecol. Syst. 2000;31:367–393. doi:10.1146/annurev.ecolsys.31.1.367 [Google Scholar]
- Gaillard J.M., Yoccoz N.G., Lebreton J.D., Bonenfant C., Devillard S., Loison A., Pontier D., Allaine D. Generation time: a reliable metric to measure life-history variation among mammalian populations. Am. Nat. 2005;166:119–123. doi: 10.1086/430330. doi:10.1086/430330 [DOI] [PubMed] [Google Scholar]
- Hennemann W.W. Relationship among body mass, metabolic rate and the intrinsic rate of natural increase in mammals. Oecologia. 1983;56:104–108. doi: 10.1007/BF00378224. doi:10.1007/BF00378224 [DOI] [PubMed] [Google Scholar]
- Jonsson K.I. Capital and income breeding as alternative tactics of resource use in reproduction. Oikos. 1997;78:57–66. doi:10.2307/3545800 [Google Scholar]
- Keyfitz N. Addison-Wesley; Reading, MA: 1977. Introduction to the mathematics of population. [Google Scholar]
- Keyfitz N., Caswell H. Springer; New York, NY: 2005. Applied mathematical demography. [Google Scholar]
- Knops J.M.H., Koenig W.D., Carmen W.J. Negative correlation does not imply a tradeoff between growth and reproduction in California oaks. Proc. Natl Acad. Sci. USA. 2007;104:16 982–16 985. doi: 10.1073/pnas.0704251104. doi:10.1073/pnas.0704251104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koons D.N., Metcalf C.J.E., Tuljapurkar S. Evolution of delayed reproduction in uncertain environments: a life-history perspective. Am. Nat. 2008;172:797–805. doi: 10.1086/592867. doi:10.1086/592867 [DOI] [PubMed] [Google Scholar]
- Kruuk L.E.B., Slate J., Wilson A.J. New answers for old questions: the evolutionary quantitative genetics of wild animal populations. Annu. Rev. Ecol. Evol. Syst. 2008;39:525–548. doi:10.1146/annurev.ecolsys.39.110707.173542 [Google Scholar]
- Lacey E.P., Real L., Antonovics J., Heckel D.G. Variance models in the study of life histories. Am. Nat. 1983;122:114–131. doi:10.1086/284122 [Google Scholar]
- Lande R. A quantitative theory of life history evolution. Ecology. 1982;63:607–615. doi:10.2307/1936778 [Google Scholar]
- Lande R. Expected relative fitness and the adaptive topography of fluctuating selection. Evolution. 2007;61:1835–1846. doi: 10.1111/j.1558-5646.2007.00170.x. doi:10.1111/j.1558-5646.2007.00170.x [DOI] [PubMed] [Google Scholar]
- Lande R., Arnold S.J. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. doi:10.2307/2408842 [DOI] [PubMed] [Google Scholar]
- Lande R., Engen S., Sæther B.-E. Oxford University Press; Oxford, UK: 2003. Stochastic population dynamics in ecology and conservation. [Google Scholar]
- Lande R., Engen S., Sæther B.-E. An evolutionary maximum principle for density-dependent population dynamics in a fluctuating environment. Phil. Trans. R. Soc. B. 2009;364:1511–1518. doi: 10.1098/rstb.2009.0017. doi:10.1098/rstb.2009.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie P.H. The intrinsic rate of increase and the overlap of successive generations in a population of guillemots (Uria aalge Pont.) J. Anim. Ecol. 1966;35:291–301. doi:10.2307/2396 [Google Scholar]
- Levins R. Princeton University Press; Princeton, NJ: 1968. Evolution in changing environments. [Google Scholar]
- Lindstedt S.L. Body size, physiological time, and longevity of homeothermic animals. Q. Rev. Biol. 1981;56:1–15. doi:10.1086/412080 [Google Scholar]
- Millar J.S., Zammuto R.M. Life histories of mammals: an analysis of life tables. Ecology. 1983;64:631–635. doi:10.2307/1937181 [Google Scholar]
- Morris W.F., Doak D.F. Sinauer; Sunderland, MA: 2002. Quantitative conservation biology. [Google Scholar]
- Morris W.F., Tuljapurkar S., Haridas C.V., Menges E.S., Horvitz C.C., Pfister C.A. Sensitivity of the population growth rate to demographic variability within and between phases of the disturbance cycle. Ecol. Lett. 2006;9:1331–1341. doi: 10.1111/j.1461-0248.2006.00988.x. doi:10.1111/j.1461-0248.2006.00988.x [DOI] [PubMed] [Google Scholar]
- Morris W.F. Longevity can buffer plant and animal populations against changing climatic variability. Ecology. 2008;89:19–25. doi: 10.1890/07-0774.1. doi:10.1890/07-0774.1 [DOI] [PubMed] [Google Scholar]
- Orzack S.H. Life history evolution and population dynamics in variable environments: some insights from stochastic demography. Lect. Notes Biomath. 1993;98:63–104. [Google Scholar]
- Orzack S.H., Tuljapurkar S. Population dynamics in variable environments. VII. The demography and evolution of iteroparity. Am. Nat. 1989;133:901–923. doi:10.1086/284959 [Google Scholar]
- Ostfeld R.S., Keesing F. Pulsed resources and community dynamics of consumers in terrestrial ecosystems. Trends Ecol. Evol. 2000;15:232–237. doi: 10.1016/s0169-5347(00)01862-0. doi:10.1016/S0169-5347(00)01862-0 [DOI] [PubMed] [Google Scholar]
- Pfister C.A. Patterns of variance in stage-structured populations: evolutionary predictions and ecological implications. Proc. Natl Acad. Sci. USA. 1998;95:213–218. doi: 10.1073/pnas.95.1.213. doi:10.1073/pnas.95.1.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzante D.E., Walde S.J., Gosse J.C., Cussac V.E., Habit E., Zemlak T.S., Adams E.D.M. Climate control on ancestral population dynamics: insight from Patagonian fish phylogeography. Mol. Ecol. 2008;17:2234–2244. doi: 10.1111/j.1365-294X.2008.03738.x. doi:10.1111/j.1365-294X.2008.03738.x [DOI] [PubMed] [Google Scholar]
- Schaffer W.M. Optimal reproductive effort in fluctuating environments. Am. Nat. 1974;108:783–790. doi:10.1086/282954 [Google Scholar]
- Stearns S.C. Life-history tactics: a review of the ideas. Q. Rev. Biol. 1976;51:3–47. doi: 10.1086/409052. doi:10.1086/409052 [DOI] [PubMed] [Google Scholar]
- Stearns S.C. The evolution of life history traits: a critique of the theory and a review of the data. Annu. Rev. Ecol. Syst. 1977;8:145–171. doi:10.1146/annurev.es.08.110177.001045 [Google Scholar]
- Tavecchia G., Coulson T., Morgan B.J.T., Pemberton J.M., Pilkington J.C., Gulland F.M.D., Clutton-Brock T.H. Predictors of reproductive cost in female Soay sheep. J. Anim. Ecol. 2005;74:201–213. doi:10.1111/j.1365-2656.2005.00916.x [Google Scholar]
- Tuljapurkar S.D. Population dynamics in variable environments. III. Evolutionary dynamics of r-selection. Theor. Popul. Biol. 1982;21:141–165. doi:10.1016/0040-5809(82)90010-7 [Google Scholar]
- Tuljapurkar S.D. Lecture Notes in Biomathematics. vol. 85. Springer; New York, NY: 1990. Population dynamics in variable environments. [Google Scholar]
- Tuljapurkar S., Wiener P. Escape in time: stay young or age gracefully? Ecol. Model. 2000;133:143–159. doi:10.1016/S0304-3800(00)00288-X [Google Scholar]
- Tuljapurkar S., Horvitz C.C., Pascarella J.B. The many growth rates and elasticities of populations in random environments. Am. Nat. 2003;162:489–502. doi: 10.1086/378648. doi:10.1086/378648 [DOI] [PubMed] [Google Scholar]
- Williams G.C. Princeton University Press; Princeton, NJ: 1966. Adaptation and natural selection: a critique of some current evolutionary thought. [Google Scholar]
- Williams G.C. Princeton University Press; Princeton, NJ: 1996. Adaptation and natural selection: a critique of some current evolutionary thought. [Google Scholar]
- Zheng C., Ovaskainen O., Hanski I. Modelling single nucleotide effects in phosphoglucose isomerase on dispersal in the Glanville fritillary butterfly: coupling of ecological and evolutionary dynamics. Phil. Trans. R. Soc. B. 2009;364:1519–1532. doi: 10.1098/rstb.2009.0005. doi:10.1098/rstb.2009.0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mathematical formulas