Abstract
Several regulators of G protein signaling (RGS) proteins contain a G protein γ-subunit-like (GGL) domain, which, as we have shown, binds to Gβ5 subunits. Here, we extend our original findings by describing another GGL-domain-containing RGS, human RGS6. When RGS6 is coexpressed with different Gβ subunits, only RGS6 and Gβ5 interact. The expression of mRNA for RGS6 and Gβ5 in human tissues overlaps. Predictions of α-helical and coiled-coil character within GGL domains, coupled with measurements of Gβ binding by GGL domain mutants, support the contention that Gγ-like regions within RGS proteins interact with Gβ5 subunits in a fashion comparable to conventional Gβ/Gγ pairings. Mutation of the highly conserved Phe-61 residue of Gγ2 to tryptophan, the residue present in all GGL domains, increases the stability of the Gβ5/Gγ2 heterodimer, highlighting the importance of this residue to GGL/Gβ5 association.
The “regulators of G protein signaling” (RGS) protein family consists of at least 20 mammalian gene products that act as GTPase activating proteins on the α-subunits of heterotrimeric, signal-transducing G proteins (1–3). By accelerating the inactivation of GTP-bound Gα subunits, RGS proteins serve as negative regulators of G protein-mediated signaling pathways. Additionally, two RGS proteins, p115–RhoGEF and PDZ–RhoGEF, can also act as effectors, coupling GTP-bound Gα12 and/or Gα13 to Rho activation (4, 5).
Regions within certain RGS proteins that lie outside the RGS domain interact with other components of G protein signal transduction machinery. For example, the N-terminal PDZ domain of RGS12 associates in vitro with the C termini of G protein-coupled receptors (6), and the N-terminal domain of RGS4 mediates receptor-selective inhibition of G protein-mediated Ca2+ signaling in pancreatic acinar cells (7). We recently identified a G protein γ subunit-like (GGL) domain within four mammalian RGS family members and identified its role in mediating specific interaction of RGS7 and RGS11 with G protein β5-subunits (8). In a complementary study, a native RGS7/Gβ5 complex has been isolated from bovine retina (9). Our discovery of RGS7/Gβ5 and RGS11/Gβ5 associations tripled the number of known interacting partners for this outlier Gβ subunit, which previously had been known to interact only with Gγ2 (10–13). Here, we describe the cloning, expression, and Gβ binding selectivity of RGS6, another RGS protein possessing the GGL domain. Residues critical for Gβ subunit binding specificity have been identified by mutagenesis of Gγ2 and the GGL domains of RGS6, RGS7, and RGS11.
MATERIALS AND METHODS
Expression Constructs.
cDNAs for RGS7, RGS11, and G protein subunits have been described (8). RGS6 was isolated by reverse transcription–PCR by using sense (5′-GCG GCC GCA TGG CTC AAG GAT CCG GGG ATC AAA G-3′) and antisense (5′-TCT AGA CTG GGA TCA GGG CCT CTT AGC GAG-3′) primers as described (14) and subcloned with an N-terminal hemagglutinin (HA)-epitope tag into pcDNA3.1 (Invitrogen). G protein β-subunit cDNAs were subcloned with an N-terminal myc-epitope tag into pcDNA3.1 (Invitrogen). Mutagenesis was performed as described (6). The Gγ2 (F61W)/RGS fusion was created by adding a 17-aa linker (PRAAASVMDICRIRPWYP; derived from pcDNA3.1 polylinker) C-terminal to amino acid 70 of the Gγ2 (F61W) point mutant, followed by the RGS domain of rat RGS12 (amino acids 716–838; GenBank acc. no. U92280).
In Vitro Transcription/Translation.
Reactions were performed, and translation products were immunoprecipitated and analyzed by SDS/PAGE as described (8), except for variations in detergent conditions. “Low-detergent” immunoprecipitations were performed and washed in buffer D [50 mM NaCl/10 mM MgCl2/50 mM Tris, pH 8.0/1 mM EDTA/10 mM 2-mercaptoethanol/20% (vol/vol) glycerol/Complete protease inhibitors; Roche Diagnostics] containing 0.05% C12E10, whereas “high-detergent” immunoprecipitations were performed in buffer D containing 0.1% Triton X-100 and washed in RIPA-500 buffer containing 500 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, and 0.1% SDS.
Transient Transfection and Immunoprecipitation.
COS-7 cell lysates were prepared and immunoprecipitated by using RIPA-150 buffer as described (8); proteins were separated by SDS/PAGE, electroblotted onto nitrocellulose, and detected with primary antibodies conjugated to horseradish peroxidase (HRP) and chemiluminescence (Amersham Pharmacia). The HRP-conjugated anti-HA mAb 3F10 was obtained from Roche Diagnostics; HRP-conjugated anti-myc mAb was purchased from Invitrogen.
Model Generation and Analysis.
Published alignments (15) allowed for replacement of residues at the interface of Gβ1 and Gγ1 with the corresponding residues of Gβ5 and the GGL domain of hRGS11 by using the program o (16). Where necessary, alternate allowed rotamers were chosen to limit steric overlap. Conjugate gradient energy minimization of the initial model was done with x-plor (17) by using standard energy functions and a coordinate restraint term applied to Cα-positions to compensate for the lack of experimental restraints. Cavity determination was performed by using the program voidoo (18). Structures were presented by using insight (Molecular Simulations, Waltham, MA). Secondary structure and coiled-coil predictions were performed with predictprotein (www.embl-heidelberg.de; ref. 19).
RESULTS AND DISCUSSION
RGS6 Has a DEP/GGL/RGS Domain Structure and Binds Gβ5.
To extend our findings on the Gβ binding specificity of RGS7 and RGS11 to other uncharacterized RGS proteins, we cloned human RGS6. A partial clone of RGS6 was described originally as S194, a brain-derived mRNA identified during mapping of the human AD3 locus at chromosome 14q24.3 (20). Database searches with S194 identified additional human RGS6 sequence records (e.g., GenBank acc. nos. H09621, AA351873, and AF073920). Oligonucleotides flanking the predicted ORF of full-length RGS6 were used to amplify RGS6 mRNA by reverse transcription–PCR from human brain total RNA. Two forms of RGS6 cDNA were isolated (GenBank acc. nos. AF107619 and AF107620), encoding ORFs of 490 or 472 aa, respectively; the difference between forms is the presence or absence of 18 contiguous amino acids (457-PESEQGRRTSLEKFTRSV-474) C-terminal to the RGS domain, likely a result of alternative splicing.
RGS6 is most similar to RGS7 (21–23). Both RGS6 isoforms encode an N-terminal DEP (Disheveled, EGL-10, Pleckstrin) domain (amino acids 39–121) 81% identical to the mouse RGS7 DEP domain, a C-terminal RGS domain (amino acids 333–447) 80% identical to the RGS box of human RGS7, and a GGL domain (amino acids 254–317) 57% identical to the mouse RGS7 GGL domain. In comparison to Gγ subunits, the central portion of the RGS6 GGL domain (amino acids 262–309) is most similar to bovine Gγ2 (41% identity).
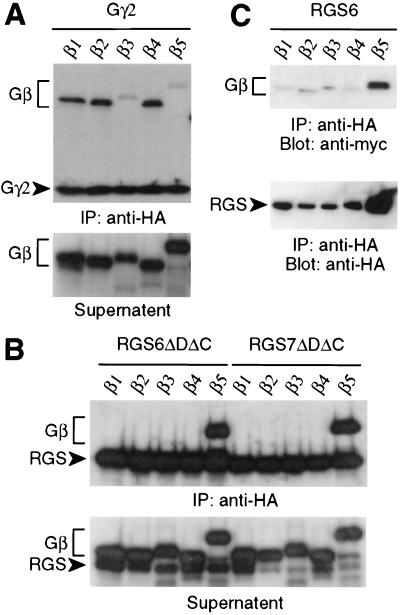
As the RGS7 and RGS11 GGL domains bind only Gβ5 subunits (8), we tested the RGS6 GGL domain for Gβ binding specificity. Gβ subunits (Gβ1–Gβ5) were produced by in vitro transcription/translation along with HA-epitope-tagged RGS proteins or Gγ subunits to detect possible interacting pairs. An anti-HA mAb was used to immunoprecipitate 35S-labeled Gγ and RGS proteins; associated 35S-labeled Gβ subunits were detected by SDS/PAGE and autoradiography. Gγ2 bound to Gβ1, Gβ2, and Gβ4 but not Gβ5 (Fig. 1A), as previously described (8, 24). The Gβ5/Gγ2 complex is reported to be abnormally sensitive to low levels of detergent compared with other Gβ/Gγ pairings (25); we therefore believe that the minimal detergent (0.05% C12E10) necessary during immunoprecipitation results in disruption of the Gβ5/Gγ2 complex.
Figure 1.
Gβ binding specificity of RGS6 and RGS7. Gβ subunits were cotranslated in reticulocyte lysates with (A) HA-tagged Gγ2 or (B) HA-tagged, truncated RGS6 (ΔDΔC, where ΔD indicates a DEP domain deletion and ΔC indicates a C-terminal deletion; amino acids 255–456) or truncated RGS7 (ΔDΔC; amino acids 202–395, SwissProt P49802) protein. Lysates were immunoprecipitated (IP) in low detergent with anti-HA mAb, and immunoprecipitated proteins and clarified supernatants were visualized separately by SDS/PAGE and autoradiography. (C) Expression vectors for full-length, HA-tagged RGS6 and individual, myc-tagged Gβ subunits were transiently cotransfected into COS-7 cells. Cell lysates were immunoprecipitated with anti-HA mAb, and coimmunoprecipitated Gβ subunits were detected by immunoblotting (Blot) with anti-myc-HRP or anti-HA-HRP conjugates.
In contrast to the Gβ binding specificity of Gγ2, a truncated RGS6 protein containing both GGL and RGS domains (RGS6ΔDΔC) did not interact with Gβ1–Gβ4 but bound Gβ5, the same specificity observed with the analogously truncated RGS7 (Fig. 1B) and RGS11 (see Fig. 3D and ref. 8). To determine the specific nature of RGS6/Gβ5 association in a cellular context, COS-7 cells were cotransfected with HA-tagged, full-length RGS6 and one of five myc-tagged Gβ subunits. Lysates from transfected cells were immunoprecipitated with anti-HA mAb, and associated Gβ subunits were detected with anti-myc antibody. Of the five Gβ subunits tested, only Gβ5 coimmunoprecipitated with full-length RGS6 (Fig. 1C). We routinely observe greater levels of RGS6 and Gβ5 proteins in cell lysates when they are coexpressed, perhaps reflective of the instability of unpaired Gβ and GGL subunits similar to the instability seen in classical Gβ and Gγ subunits (26–28), including Gβ5 (11).
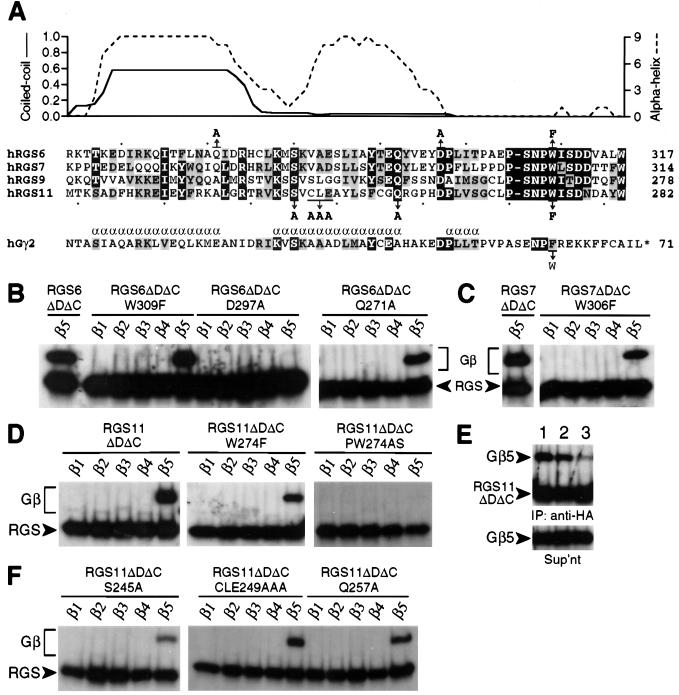
Figure 3.
In vitro Gβ binding specificity of GGL domain mutants. (A) Secondary structure predictions for the RGS6 GGL domain and sequence alignment between Gγ2, RGS6, RGS7, RGS9, and RGS11. Identical residues are in black boxes; conserved residues are shown in shaded boxes. For RGS6, probabilities of α-helical character (indexed to a maximum of 9; ref. 19) and coiled-coil interaction (indexed to a maximum of 1.0; ref. 33) are plotted above the primary sequence of the GGL domain (x axis). α-Helices within Gγ2 (ref. 32) are indicated by an α above the sequence. The position and nature of point mutations are denoted above or below the sequence line with arrows. Individual Gβ subunits were cotranslated in reticulocyte lysates with wild-type or mutant RGS6 (B), RGS7 (C), and RGS11 proteins (D–F). HA-tagged RGS or Gγ proteins were immunoprecipitated in low detergent (except as noted in E) with anti-HA mAb, and associated Gβ proteins were visualized by SDS/PAGE and autoradiography. (E) Immunoprecipitations (IP) of cotranslated Gβ5 and wild-type (lane 1) or W274F mutant (lanes 2 and 3) RGS11ΔDΔC proteins were performed in high-detergent (lanes 1 and 3) or low-detergent (lane 2) conditions and visualized separately from clarified supernatants (Sup’nt) as above.
We conclude that, within the DEP/GGL/RGS subfamily of RGS proteins, RGS6 is most closely related to RGS7 in primary sequence and shares Gβ5 binding selectivity with both RGS7 and RGS11.
RGS6 and Gβ5 mRNAs Have Overlapping Distribution.
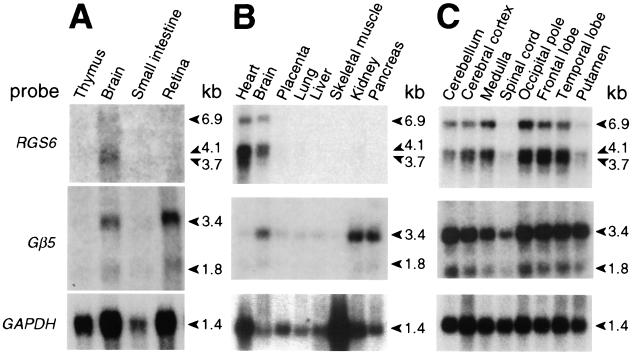
Unlike the widespread expression of other Gβ subunits, Gβ5 mRNA is expressed in a tissue-restricted manner: mouse Gβ5 is expressed predominantly in the brain and retina (10, 29), whereas human Gβ5 mRNA is found in brain, retina, kidney, and pancreas (8, 30). To test whether RGS6 and Gβ5 are coexpressed in human tissues, we compared expression patterns of both genes in Northern blot analyses. RGS6 mRNA was detected in whole brain (Fig. 2 A and B) and in brain anatomical regions with an expression pattern overlapping that of Gβ5; however, RGS6 expression is barely detectable in the caudate putamen and spinal cord (Fig. 2C). This pattern of expression is in contrast with that reported by Gold et al. (31) for RGS6 in the rat brain; such a cross-species difference in brain expression patterns also was observed for RGS11 (8, 31). Unlike the expression pattern of human RGS11, human RGS6 mRNA is not seen in the retina or pancreas but is observed in the heart (Fig. 2 A and B). The observed coexpression of RGS6 and Gβ5 mRNA in the human brain suggests that an RGS6/Gβ5 complex may play a role in G protein-mediated neuronal signaling.
Figure 2.
Comparison of RGS6 and Gβ5 expression patterns. Northern blots of 20 μg total RNA (A) or 2 μg poly(A+) RNA from various human tissues (B and C) were serially hybridized with a Gβ5 cDNA probe (ref. 8), with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probe as a control for RNA loading and quality, and with an RGS6 cDNA probe. kb, kilobase.
GGL Domain Fidelity: Testing the “Trp-274 Hypothesis.”
An assumption inherent in our original report (8) is that sequence homology between GGL domains and Gγ subunits reflects a similarity at the secondary and quaternary structure levels—i.e., a predominantly α-helical extended chain forming extensive contacts, including an N-terminal parallel coiled coil (15), with Gβ5 subunits. Indeed, secondary structure predictions for RGS6 (Fig. 3A), RGS7, and RGS11 (data not shown) suggest an α-helical character in regions analogous to the first two α-helices of Gγ1 and Gγ2 (15, 32); for the RGS9 GGL domain, only the N-terminal region (amino acids 216–233) is predicted to be α-helical (data not shown). Furthermore, for both RGS6 and RGS7, the putative N-terminal α-helix within the GGL domain is predicted to participate in a coiled-coil structure (Fig. 3A and data not shown); its putative counterpart, the N terminus of Gβ5, is also predicted to form a coiled coil by the same program (ref. 33; data not shown).
In our initial report (8), we predicted an important role for Trp-274 of the RGS11 GGL domain in Gβ5 selectivity (the Trp-274 hypothesis), given that (i) all Gγ subunits have a smaller aromatic residue (Phe) at the analogous position and (ii) Gβ1–Gβ4 have a larger residue (Asn-340) than that of Gβ5 (Ala-353) at the position predicted to interact with Trp-274 of RGS11 (8). To test this prediction, we created GGL-domain mutants by replacing tryptophan with phenylalanine in RGS6 (W309F), RGS7 (W306F), and RGS11 (W274F) and assessed Gβ binding by coexpression with Gβ1–Gβ5 in vitro. None of the Trp-to-Phe mutants acquired affinity toward Gβ1–Gβ4 (Fig. 3 B–D), and the mutations had only a modest impact on Gβ5 affinities in low-detergent conditions (0.05% C12E10). However, the Trp-to-Phe mutations destabilized the GGL/Gβ5 interactions in high-detergent conditions; for example, when washed with 1% Triton X-100/0.1% SDS/0.5% sodium cholate, very little Gβ5 remained bound to the W274F mutant of RGS11 (Fig. 3E, lane 3) in comparison to robust Gβ5 binding observed for the wild-type protein (Fig. 3E, lane 1). Similarly destabilized Gβ5 interactions were observed with RGS6 and RGS7 Trp-to-Phe mutants in high-detergent conditions (data not shown). This region is clearly critical for Gβ binding, as mutation of tryptophan and the preceding proline to serine and alanine, respectively, eliminates Gβ5 binding by RGS11 (PW274AS; Fig. 3D). This result is entirely consistent with the known importance of this dipeptide motif in the Gβ1/Gγ1 and Gβ1/Gγ2 crystal structures, in which the proline “kinks” the Cα-chain inward toward Gβ1, positioning the phenylalanine residue into a hydrophobic pocket (15, 32).
GGL Domain Fidelity: Role of Other Residues.
Other amino acids within RGS6 and RGS11 were also modified, singly or in tandem, to test their roles in both Gβ5 binding affinity and Gβ subunit binding selectivity. Ser-245 of RGS11 and Asp-297 of RGS6 represent two positions conserved among all GGL domains and Gγ subunits (8). The analogous positions within Gγ1 (Ser-34 and Asp-51) and Gγ2 (Ser-31 and Asp-48) form hydrogen bonds with Asp-27 and Ser-279/Ser-281 of Gβ1, respectively (15, 32). Conversion of Ser-245 (RGS11) and Asp-297 (RGS6) to alanine confirmed their importance to GGL domain function; considerably reduced Gβ5 binding was seen with the RGS11 S245A mutant (Fig. 3F), whereas the RGS6 D297A mutant did not bind Gβ5 at all, either in vitro (Fig. 3B) or in cell cotransfection studies (data not shown). In contrast, Gln-271 of RGS6 is conserved only in the highly related RGS7; as expected, mutation of Gln-271 to alanine (the residue present within RGS9 and RGS11) had no discernable effect on Gβ5 binding by RGS6 (Fig. 3B).
Gln-257 of RGS11 is conserved among all GGL domains, including that of EGL-10 (8), whereas the analogous position within Gγ1 (Glu-46) and Gγ2 (Ala-43) is neither well conserved nor involved in interactions with Gβ1 (15, 32). Hence, as predicted by the lack of importance to Gγ/Gβ interactions, replacement of Gln-257 with alanine yielded no change in the Gβ binding behavior of RGS11 (Q257A; Fig. 3F). To this point, therefore, GGL domain mutations affect GGL/Gβ5 assembly when made in positions analogous to Gγ/Gβ contact points. Conversely, GGL domain mutations do not affect GGL/Gβ5 assembly when made in positions analogous to those not involved in Gγ/Gβ contacts. These findings support our assumption that the GGL domain interacts with Gβ5 in a manner similar to conventional Gγ/Gβ pairings.
GGL Domain Fidelity: Comparison to Gγ1’s Selectivity for Gβ1.
Studies of Gγ1 and Gγ2 binding specificities have identified a five-residue region of Gγ1 (amino acids 36–40) critical for selective dimerization with Gβ1 and not Gβ2 (34, 35); replacement of three residues of Gγ1, Cys-36–Cys-37–Glu-38, with the corresponding residues of Gγ2, Ala-33–Ala-34–Ala-35, is sufficient to allow binding of Gγ1 to Gβ2 (35). Therefore, we replaced the corresponding residues of RGS11 (Cys-247–Leu-248–Glu-249) with three alanines and tested Gβ subunit association. The triple-alanine mutation, either alone (CLE249AAA) or in combination with mutation to Trp-274 (W274F + CLE249AAA; not shown), neither engendered additional Gβ binding specificity nor abrogated association with Gβ5 (Fig. 3F) and Gβ5L (data not shown), suggesting that, unlike Gγ1 and Gγ2, this region of the GGL domain does not control Gβ binding selectivity.
Fidelity of Gγ1 and Gγ2: Revisiting the Trp-274 Hypothesis.
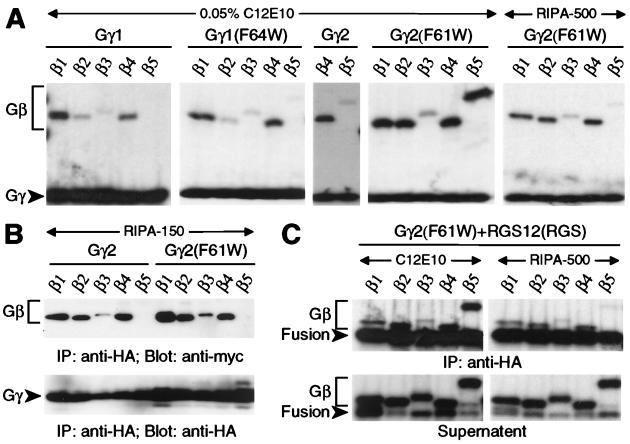
We tested a corollary of the Trp-274 hypothesis espoused above; namely, that conversion of the analogous position within Gγ1 and Gγ2 from phenylalanine to tryptophan would prevent or weaken binding to Gβ1–Gβ4 and/or allow Gβ5 association. The Gβ binding characteristics of Gγ1 were unaffected by this mutation (F64W), as compared with wild-type Gγ1 (Fig. 4A). In contrast, the same mutation to Gγ2 (F61W), although it did not affect affinity toward Gβ1–Gβ4, engendered binding to Gβ5 when tested in vitro in low-detergent conditions (Fig. 4A). Under high-detergent conditions, Gβ5 binding was not detected, whereas binding to other Gβ subunits was unaffected [Gγ2 (F61W) RIPA-500 vs. C12E10; Fig. 4A].
Figure 4.
Gβ binding specificity of Gγ1 and Gγ2 mutants. HA-tagged Gγ proteins (wild-type or mutated as indicated) were either cotranslated in vitro (A and C) or cotransfected into COS-7 cells (B) with individual Gβ subunits, immunoprecipitated (IP) with anti-HA mAb in either low detergent (0.05% C12E10) or high detergent (RIPA), and visualized by SDS/PAGE and autoradiography (A and C) or immunoblotting (B) with indicated antisera (Blot). “Fusion” denotes chimeric protein composed of HA-tagged Gγ2 (F61W) subunit fused to the rat RGS12 RGS domain.
Of all mutants tested for Gβ5 interaction, only the Gγ2 (F61W) mutant and the cognate GGL domain Trp-to-Phe mutants were affected by detergent conditions; all other GGL domain mutants (Fig. 3A) had the same affinity (or lack thereof) for Gβ5 isoforms in cotranslation studies with low- or high-detergent conditions (data not shown). We believe this detergent sensitivity explains our inability to detect Gβ5 binding by Gγ2 (F61W) after exposure to RIPA-150 buffer in cell cotransfection experiments (Fig. 4B). This conclusion is consistent with the extreme detergent sensitivity of the wild-type Gβ5/Gγ2 complex (25). Additionally, in vitro cotranslation/immunoprecipitation studies with high-detergent RIPA-500 buffer disrupt RGS7/Gβ5 association in contrast to robust interaction observed in low-detergent conditions (Fig. 1B and data not shown). The in vitro association of RGS6 and RGS11 with Gβ5 isoforms has far more tolerance to high-detergent conditions, allowing for detection of such complexes upon COS-7 cotransfection and RIPA-buffer lysis (Fig. 1C and ref. 8).
Lack of Steric Hindrance from the RGS Domain.
To test the possibility that GGL-domain-containing RGS proteins are prevented from binding Gβ1–Gβ4 because of steric hindrance from a C-terminal RGS domain absent in conventional Gγ subunits, we created a chimeric Gγ2 (F61W)/RGS protein that mimics the spatial orientation of GGL and RGS domains. As observed for the Gγ2 (F61W) protein, the larger Gγ2 (F61W)/RGS12 fusion protein associated with Gβ1, Gβ2, Gβ4, and Gβ5 subunits in vitro in low-detergent conditions, with a loss only of Gβ5 association apparent in high-detergent conditions (Fig. 3C). This result suggests that the lack of interaction between Gβ1–Gβ4 subunits and GGL-domain-containing RGS proteins is not the result of steric hindrance from the RGS domain but rather is intrinsic to the GGL domain.
Molecular Modeling of the Interaction of RGS11 Trp-274 with Gβ5.
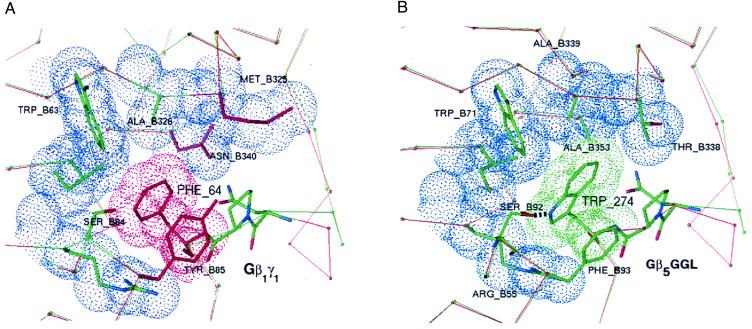
To understand the role of the Trp-274 position in differential Gβ binding affinities of GGL domains and Gγ subunits, we returned to our RGS11/Gβ5 interface model (8), which is based on side-chain replacement of the Gβ1/Gγ1 crystal coordinates (15). The highlighted Gβ5/GGL interface (Fig. 5B) has greater complementarity than the equivalent region in Gβ1/Gγ1 (Fig. 5A). In the Gβ1/Gγ1 crystal structure (15), packing defects around Phe-64 produce a ≈3.0-Å3 cavity, whereas no equivalent cavity surrounds Trp-274 in the modeled Gβ5/GGL complex. In addition, Thr-338 and Ala-353 in Gβ5 are smaller than the corresponding residues of Gβ1 (Met-325 and Asn-340) and serve to accommodate the larger bulk of Trp-274 relative to Phe-64. Further enhancing the specificity of the Gβ5/GGL complex, the hydroxyl group of Ser-92 in Gβ5 potentially forms a hydrogen bond with the indole nitrogen of Trp-274; no similar hydrogen bond is possible in the Gβ1/Gγ1 structure.
Figure 5.
Specificity-determining residues at the interface of Gβ1 and Gγ1 compared with equivalent regions of modeled Gβ5 and the GGL domain of RGS11. Highlighted in blue are van der Waals surfaces of Gβ1 contacting Phe-64 of Gγ1 (A) or similar contacts between Gβ5 and Trp-274 of the RGS11 GGL domain (B). Residues colored red in the Gβ1/Gγ1 structure differ from equivalent residues in the Gβ5/GGL model. Except for the conserved tripeptide motif (NPF or NPW), thin red and green lines trace the Cα-backbone of Gβ1/Gγ1 and Gβ5/GGL, respectively. Just before the conserved tripeptide motif, the Cα-traces diverge because of the insertion of a single residue in Gγ1 relative to the GGL domain.
The presence of a small cavity in Gβ1/Gγ1 around Phe-64 suggests that this pocket could accommodate a larger residue, thereby allowing Gγ1 (F64W) and Gγ2 (F61W) mutants to maintain binding to their normal Gβ partners (Fig. 4A). If this pocket within Gβ5 specifies a Trp residue in GGL domains by virtue of increased cavity size and potential hydrogen bonding with Ser-92, then this specification could partially explain why Gγ2 (F61W) has increased affinity for Gβ5 as a binding partner. The reverse mutation to Phe in the GGL domain would not fit as snugly into the pocket specifying Trp, thereby giving a relative loss of binding energy, which was observed as lowered in vitro affinity for Gβ5 by RGS6(W309F), RGS7(W306F), and RGS11(W274F) under high-detergent conditions. We await experimentally derived structural data on the GGL/Gβ5 complex to confirm the relative importance of this region to Gβ binding specificity.
We extend our discovery of a GGL domain within RGS7 and RGS11 to include RGS6. Structural predictions and mutagenesis data strengthen our belief that the GGL domain binds to Gβ5 subunits in a manner similar to that of conventional Gβ/Gγ associations. The molecular basis for the absolute selectively of the GGL domain for Gβ5 subunits remains to be determined. However, we have been able to increase the relative strength of the Gβ5/Gγ2 association by mutating a single residue within Gγ2, Phe-61. This mutation is presumed to mimic atomic interactions unique to wild-type Gβ5/GGL domain associations. Continued study of the nature of Gβ5 association by GGL domains relative to that of Gγ subunits should assist our understanding of the role(s) of the DEP/GGL/RGS proteins in modulating G protein-coupled signaling.
ABBREVIATIONS
- ΔD
DEP domain deletion
- ΔC
C-terminus deletion
- GGL
G protein γ-subunit-like
- HA
hemagglutinin
- HRP
horseradish peroxidase
Note
While this manuscript was under review, two additional reports of RGS/Gβ5 association were published (36, 37). One reported the discovery of a tight association between RGS9 and Gβ5L in rod photoreceptors (36). This discovery validates our original prediction based on the presence of a GGL domain within the primary sequence of RGS9.
Footnotes
References
- 1.Siderovski D P, Hessel A, Chung S, Mak T W, Tyers M. Curr Biol. 1996;6:211–212. doi: 10.1016/s0960-9822(02)00454-2. [DOI] [PubMed] [Google Scholar]
- 2.Dohlman H G, Thorner J. J Biol Chem. 1997;272:3871–3874. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- 3.Berman D M, Gilman A G. J Biol Chem. 1998;273:1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- 4.Hart M J, Jiang X, Kozasa T, Roscoe W, Singer W D, Gilman A G, Sternweis P C, Bollag G. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- 5.Fukuhara S, Murga C, Zohar M, Igishi T, Gutkind J S. J Biol Chem. 1999;274:5868–5879. doi: 10.1074/jbc.274.9.5868. [DOI] [PubMed] [Google Scholar]
- 6.Snow B E, Hall R A, Krumins A M, Brothers G M, Bouchard D, Brothers C A, Chung S, Mangion J, Gilman A G, Lefkowitz R J, et al. J Biol Chem. 1998;273:17749–17755. doi: 10.1074/jbc.273.28.17749. [DOI] [PubMed] [Google Scholar]
- 7.Zeng W, Xu X, Popov S, Mukhopadhyay S, Chidiac P, Swistok J, Danho W, Yagaloff K A, Fisher S L, Ross E M, et al. J Biol Chem. 1998;273:34687–34691. doi: 10.1074/jbc.273.52.34687. [DOI] [PubMed] [Google Scholar]
- 8.Snow B E, Krumins A M, Brothers G M, Lee S-F, Wall M A, Chung S, Mangion J, Arya S, Gilman A G, Siderovski D P. Proc Natl Acad Sci USA. 1998;95:13307–13312. doi: 10.1073/pnas.95.22.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabrera J L, de Frietas F, Satpaev D K, Slepak V Z. Biochem Biophys Res Commun. 1998;28:898–902. doi: 10.1006/bbrc.1998.9218. [DOI] [PubMed] [Google Scholar]
- 10.Watson A J, Aragay A M, Slepak V Z, Simon M I. J Biol Chem. 1996;271:28154–28160. doi: 10.1074/jbc.271.45.28154. [DOI] [PubMed] [Google Scholar]
- 11.Zhang S, Coso O A, Lee C, Gutkind J S, Simonds W F. J Biol Chem. 1996;271:33575–33579. doi: 10.1074/jbc.271.52.33575. [DOI] [PubMed] [Google Scholar]
- 12.Bayewitch M L, Avidor-Reiss T, Levy R, Pfeuffer T, Nevo I, Simonds W F, Vogel Z. J Biol Chem. 1998;273:636–644. doi: 10.1074/jbc.273.4.2273. [DOI] [PubMed] [Google Scholar]
- 13.Lindorfer M A, Myung C-S, Savino Y, Yasuda H, Khazan R, Garrison J C. J Biol Chem. 1998;273:34429–34436. doi: 10.1074/jbc.273.51.34429. [DOI] [PubMed] [Google Scholar]
- 14.Snow B E, Antonio L, Suggs S, Siderovski D P. Gene. 1998;206:247–253. doi: 10.1016/s0378-1119(97)00593-3. [DOI] [PubMed] [Google Scholar]
- 15.Sondek J, Bohm A, Lambright D G, Hamm H E, Sigler P B. Nature (London) 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 16.Jones T A, Kjeldgard M. o. Uppsala Univ., Uppsala: Dept. Cell Mol. Biol.; 1996. , Version 6.1. [Google Scholar]
- 17.Brunger A T. x-plor. Yale Univ., New Haven, CT: Dept. Mol. Biophys. Biochem.; 1993. , Version 3.1. [Google Scholar]
- 18.Kleywegt G J, Jones T A. Acta Crystallogr D. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 19.Rost B. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 20.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Nature (London) 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 21.Koelle M R, Horvitz H R. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- 22.He W, Cowan C W, Wensel T G. Neuron. 1998;20:95–102. doi: 10.1016/s0896-6273(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 23.Elmore T, Rodriguez A, Smith D P. DNA Cell Biol. 1998;17:983–989. doi: 10.1089/dna.1998.17.983. [DOI] [PubMed] [Google Scholar]
- 24.Yan K, Kalyanaraman V, Gautam N. J Biol Chem. 1996;271:7141–7146. doi: 10.1074/jbc.271.12.7141. [DOI] [PubMed] [Google Scholar]
- 25.Fletcher J E, Lindorfer M A, Defilippo J M, Yasuda H, Guilmard M, Garrison J C. J Biol Chem. 1998;273:636–644. doi: 10.1074/jbc.273.1.636. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt C J, Neer E J. J Biol Chem. 1991;266:4538–4544. [PubMed] [Google Scholar]
- 27.Pronin A N, Gautam N. FEBS Lett. 1993;328:89–93. doi: 10.1016/0014-5793(93)80971-v. [DOI] [PubMed] [Google Scholar]
- 28.Higgins J B, Casey P J. J Biol Chem. 1994;269:9067–9073. [PubMed] [Google Scholar]
- 29.Watson A J, Katz A, Simon M I. J Biol Chem. 1994;269:22150–22156. [PubMed] [Google Scholar]
- 30.Jones P G, Lombardi S J, Cockett M I. Biochim Biophys Acta. 1998;1402:288–291. doi: 10.1016/s0167-4889(98)00017-2. [DOI] [PubMed] [Google Scholar]
- 31.Gold S J, Ni Y G, Dohlman H G, Nestler E J. J Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wall M A, Posner B A, Sprang S R. Structure (London) 1998;6:1169–1183. doi: 10.1016/s0969-2126(98)00117-8. [DOI] [PubMed] [Google Scholar]
- 33.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 34.Lee C, Murakami T, Simonds W F. J Biol Chem. 1995;270:8779–8784. doi: 10.1074/jbc.270.15.8779. [DOI] [PubMed] [Google Scholar]
- 35.Meister M, Dietrich A, Gierschik P. Eur J Biochem. 1995;234:171–177. doi: 10.1111/j.1432-1033.1995.171_c.x. [DOI] [PubMed] [Google Scholar]
- 36.Makino E R, Handy J W, Li T, Arshavsky V Y. Proc Natl Acad Sci USA. 1999;96:1947–1952. doi: 10.1073/pnas.96.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levay K, Cabrera J L, Satpaev D K, Slepak V Z. Proc Natl Acad Sci USA. 1999;96:2503–2507. doi: 10.1073/pnas.96.5.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]