Abstract
Understanding of the burden of Chlamydia trachomatis infection and its clinical sequelae is hampered by the absence of accurate, well-characterized tests using serological methods to determine past exposure to infection. An “in-house” immunoglobulin G (IgG) enzyme-linked immunosorbent assay (ELISA) based on the C. trachomatis-specific antigen Pgp3 was produced and evaluated against three commercial ELISAs derived from the major outer membrane protein: the Medac pELISA plus, the Savyon SeroCT-IgG ELISA, and the Ani Labsystems IgG enzyme immunoassay. Sensitivities and specificities were determined using sera from both male and female patients (n = 356) for whom C. trachomatis had been detected in the lower genital tract at least 1 month prior to the testing of the sample and from 722 Chlamydia-negative children aged 2 to 13 years. The Pgp3 ELISA was significantly more sensitive (57.9% [95% confidence interval {95% CI}, 52.7 to 62.9%]) than the Ani Labsystems (49.2% [95% CI, 44.0 to 54.3%]; P = 0.003), SeroCT (47.2% [95% CI, 42.1 to 52.4%]; P < 0.0005), and Medac (44.4% [95% CI, 39.3 to 49.6%]; P < 0.0005) ELISAs. The Pgp3, Ani Labsystems, and SeroCT assays, but not the Medac assay, had significantly higher sensitivity for female specimens than for male specimens (73.8 versus 44.2%, 59.8 versus 40.5%, 55.5 versus 40%, and 45.7 versus 43.7%, respectively). For female patients, the Pgp3 assay was 14.0% (95% CI, 5.5 to 22.5%) more sensitive than the next most sensitive ELISA, the Ani Labsystems assay (P = 0.001). There was no significant difference in specificity between the Pgp3 (97.6% [95% CI, 96.2 to 98.6%]), Ani Labsystems (99% [95% CI, 97.7 to 99.6%]), SeroCT (97.2% [95% CI, 95.7 to 98.2%]), and Medac (96% [95% CI, 94.3 to 97.2%]) ELISAs. None of the ELISAs showed evidence of cross-reactivity with antibodies to Chlamydia pneumoniae.
Chlamydia trachomatis is the commonest sexually transmitted bacterial infection in developed countries, with national surveillance programs consistently showing rising rates of diagnosed infections over the past decade. In the United Kingdom, figures based on cases diagnosed in departments of genitourinary medicine (GUM) suggest a population rate of 190 per 100,000 men and 187 per 100,000 women (52). Reported rates are highly dependent on the level of testing at different clinics, with the probability that many Chlamydia cases are not diagnosed. The population prevalence of uncomplicated genital Chlamydia in 16- to 24-year-olds in the United Kingdom is thought to be between 2% and 6% in both men and women (17, 33), while the opportunistic National Chlamydia Screening Programme (2008) indicates a higher prevalence of around 10%, likely due to selective testing of higher-risk individuals (15). Nucleic acid amplification tests, commonly used in GUM clinics, identify infection only when the organism is present. Once infection has been resolved, these tests provide no information on past exposure. While detection rates are rising, due in part to increased screening and testing, the overall prevalence of past C. trachomatis exposure is not known.
The prevalence of past exposure to genital C. trachomatis and changes over time in age-specific prevalence can be explored serologically. For instance, in Finland, Lyytikäinen et al. (32) studied pregnant women under the age of 29 using a commercial enzyme-linked immunosorbent assay (ELISA) based on C. trachomatis-specific peptides derived from the major outer membrane protein (MOMP). However, for wider application, confidence in the sensitivity and specificity of available antibody tests is critical (24). None of the current ELISAs have ever been rigorously evaluated against large numbers of well-defined serologically positive and negative control sera; hence, their sensitivity and specificity remain open to question.
Chlamydia trachomatis is from the same family, Chlamydiaceae, as Chlamydia pneumoniae, a common respiratory pathogen with which it shares genetic homology (26). Sera from patients exposed to C. trachomatis show diverse serological profiles against immunodominant C. trachomatis antigens (2, 18, 42, 47, 49), many of which are cross-reactive with sera from patients exposed to other chlamydial species, in particular C. pneumoniae (2, 5, 19, 44). In addition, C. trachomatis antigens, such as the 60-kDa heat shock protein (hsp60) and lipopolysaccharide (LPS), cross-react with other bacterial species (35, 50, 57). Microimmunofluorescence (MIF) (54), which detects antibodies to chlamydial elementary bodies (EB), has long been considered the “gold standard” for the serodiagnosis of chlamydial infections (55). However, the procedure lacks standardization and is subjective; moreover, its specificity is considered suspect because of cross-reactivity with other chlamydial species (5, 27, 40, 44). A number of ELISAs are also commercially available, including several based on peptides of MOMP, which makes up 60% of the total outer membrane protein and is highly immunogenic (8).
In addition to MOMP, the Pgp3 protein, expressed by open reading frame 5 of the chlamydial plasmid and secreted into the host cell cytosol, is a promising C. trachomatis-specific immunogen (11, 31), since the plasmid is rarely found in C. pneumoniae isolates (51), and its sequence is highly conserved (<1% divergence) between strains (7, 10, 22). Sensitivities of 50 to 60% and specificities of 80 to 90% have been reported for Pgp3 ELISAs when sera from acutely C. trachomatis infected patients and from healthy blood donors, respectively, have been assayed (1-3).
The aim of this study was to produce a sensitive and specific C. trachomatis Pgp3 ELISA for fast throughput of large numbers of sera, to be used particularly in epidemiological studies and potentially as a method for assessing the population impact of Chlamydia screening programs (24). Its performance was evaluated against those of three commercially available ELISAs using well-characterized sera from people who have or have never been exposed to C. trachomatis.
MATERIALS AND METHODS
Patient specimens.
In order to assess the sensitivity and specificity of the assay, sera from well-defined exposed and unexposed populations were assayed. The study was approved by the North Somerset Research Ethics Committee.
Chlamydia-positive control serum samples.
Three hundred fifty-six patients (including 190 men, 164 women, and 2 individuals of unknown sex) attending two GUM departments (Milne Centre, Bristol, United Kingdom, and the Jefferiss Wing, London, United Kingdom) were recruited. Written informed consent was obtained from all individuals. For inclusion in the study, patients had to have been diagnosed as positive for a Chlamydia organism at least 1 month previously. The majority of patients were diagnosed as Chlamydia positive at the department from which they were recruited. The Milne Centre has used the Gen-Probe APTIMA Combo 2 assay since 2005; prior to that, it used the polymer conjugate-enhanced enzyme immunoassay (EIA) (Dako, Ely, Cambridgeshire, United Kingdom), with PCR confirmation using the Cobas PCR assay (Roche Diagnostics Inc., Branchburg, NJ). The Jefferiss Wing has used the Becton Dickinson ProbeTec ET strand displacement assay since 2001; prior to that, it used the Abbott LCx assay. A standardized questionnaire in which the following information was detailed was completed: the patient's recent sexual behavior, current clinical presentation, the results of Chlamydia testing, and clinical presentation at the time of the previous Chlamydia-positive test(s). Patients provided a 3.5-ml blood sample. Patients under the age of 16 years were excluded, as were those unable to give informed consent. Serum samples for this study were collected at both centers between May 2006 and January 2008. Sera were stored at −70°C within 6 h of collection and were transported to the laboratory at St Mary's on dry ice. All assays were performed on samples that had undergone at least one, and no more than two, freeze-thaw cycles.
Chlamydia-negative control serum samples.
We accessed sera from 747 children (ages, 2 to 13 years) held at two serum archives, one at the Department of Diagnostic Virology, St Mary's Hospital (D. A. Muir), and the other at the Health Protection Agency (HPA), Colindale, London, United Kingdom (D. W. Brown). These children were assumed not to have been exposed to C. trachomatis. Sera held at the St Mary's Hospital archive were collected between January 2005 and December 2007. Sera held at the HPA were collected between 1996 and 2006. Samples from both archives were stored at −70°C and transported on dry ice. These samples had undergone two (St Mary's) or three (HPA) freeze-thaw cycles at the point of assay.
DNA constructs for Pgp3 protein expression.
A construct for the production of Pgp3 antigen was produced with an N-terminal glutathione S-transferase (GST) tag and a thrombin cleavage site for GST removal. A DNA fragment encoding Pgp3 was PCR amplified from the LGV-1 (strain 440) plasmid-encoding construct pCTL1 2A (22) using Pfu DNA polymerase (Stratagene, La Jolla, CA) and primers 5′-CGTAGGATCCATGGGAAATTCTGGTTT-3′ and 5′-CGTACTCGAGTTAAGCGTTTGTTTGAGGT-3′. The PCR amplicon was digested with BamHI and XhoI and was ligated into the multiple cloning site of pGEX-4T-1 (Stratagene).
Recombinant protein purification and analysis.
Endonuclease A-deficient Escherichia coli strain PC2 [BL21(DE3) endA::Tetr T1r pLysS] (9) was used to produce the Pgp3-GST fusion protein. Following transformation and induction with 0.25 mM isopropyl-thio-β-d-galactopyranoside (Sigma-Aldrich, Dorset, United Kingdom), the fusion protein was fractionated by sonication in buffer A (200 mM NaCl, 50 mM Tris-HCl [pH 7.4], 0.5 mM EDTA) containing 0.5 mM phenylmethylsulfonyl fluoride and 5 mM dithiothreitol (DTT). Crude extracts precleared by centrifugation were incubated with glutathione Sepharose (GE Healthcare Life Sciences, Little Chalfont, Buckinghamshire, United Kingdom), and the resin was extensively washed in buffer A. The GST tag was cleaved from the protein by thrombin (3 U per mg of protein) for 4 h at 25°C, and the protein was eluted in buffer A. Protein-containing fractions diluted with 3 volumes of 50 mM Tris-HCl (pH 7.4) were injected into a 5-ml HiTrap Q FF column (GE Healthcare) on an Akta purifier (GE Healthcare). Bound protein was eluted with a linear gradient of 0.15 to 0.3 M NaCl in 50 mM Tris-HCl (pH 7.4), and then 5 mM DTT was added to each 1-ml fraction. Aliquots (10 μl) of each fraction were denatured in 2× sodium dodecyl sulfate (SDS) buffer (28) for 5 min at 95°C and analyzed by polyacrylamide gel electrophoresis (PAGE) on a 10% gel. Pooled protein-rich fractions were further concentrated two- to fivefold by ultrafiltration in a Centriprep Ultracel YM-3 (3,000-molecular-weight cutoff) column (Millipore, Watford, United Kingdom). Protein concentrations were determined by the Bradford assay with a bovine serum albumin standard (Bio-Rad, Hertfordshire, United Kingdom).
Pgp3 Western blotting.
Purified Pgp3 (75 ng/well), denatured in 2× SDS buffer as described above, was analyzed by PAGE on a 12.5% gel and transferred to a polyvinylidene difluoride (PVDF) Immobilon membrane (Millipore) in transfer buffer (25 mM Tris-HCl [pH 8.3]-192 mM glycine-20% methanol) using the Bio-Rad Trans-Blot Semi-Dry cell at 125 mA for 1 h. After transfer, the membrane was washed in PBST-A (phosphate-buffered saline [PBS], pH 7.2, containing 0.1% Tween 20; Sigma), blocked in PBSM (PBST-A with 5% nonfat milk powder; Marvel, Premier Foods, St Albans, United Kingdom) at 4°C overnight, and incubated with serum (diluted 1:100 in PBSM) for 2 h at room temperature with gentle agitation. Following three washes in PBST-A, the membrane was incubated with a horseradish peroxidise (HRP)-labeled secondary antibody (goat anti-human immunoglobulin G [IgG] [Fc fragment]-HRP; Sigma) (diluted 1:10,000 in PBSM) for 1 h, as described above, and was further washed three times with PBST-A. Protein bands were visualized by the chemiluminescence system (ECL; GE Healthcare).
Western blotting of EB.
C. trachomatis EB (strain LGV2/434/Bu) were prepared in Buffalo green monkey kidney cells and Urografin gradient (Bayer HealthCare, Uxbridge, United Kingdom) and were purified by modifying the method of Pickett et al. (45). Purified EB were denatured in 2× SDS buffer, separated by SDS-PAGE on a 10% or 12.5% gel, and then transferred and immunoblotted as described above. For pediatric sera, the HRP-labeled secondary antibody (goat anti-human IgG [Fc fragment]-HRP; Sigma) was diluted 1:1,000, and peroxidase activity was visualized by incubation with a 3,3′,5,5′-tetramethylbenzidine (TMB) substrate solution (Promega, Southampton, United Kingdom).
Indirect ELISA.
C. trachomatis ELISA conditions were initially established with purified untagged Pgp3 protein and sera collected from a cohort of patients with nonspecific urethritis and with known titers of primary antibodies against C. trachomatis and C. pneumoniae, previously determined by MIF (23). The initial concentration of the coating antigen (Pgp3), the blocking buffer, the test serum dilutions, and the concentration of the goat anti-human IgG (Fc fragment)-HRP secondary antibody, together with the incubation times, were all optimized in a series of preliminary experiments. The AMPAK ELISA amplification system (Dako) was investigated as a means of increasing sensitivity, but the signal-to-noise ratio was not improved (data not shown). The final procedure, before refinement with checkerboard titrations (detailed in Results), is described.
Immunosorb 96-well microtitration plates (Nunc, Roskilde, Denmark) were coated with Pgp3 protein for 1 h at 37°C (20 ng per well in 100 mM sodium carbonate buffer, pH 9.6). The bound protein was washed with PBST-B (PBS, pH 7.2, containing 0.05% Tween 20), blocked with 200 μl of 1% Hammersten casein (GE Healthcare) in PBST-B for 2 h at 37°C, and washed three times. All sera were then assayed in duplicate at a 1:100 dilution in the blocking buffer. After 1 h at 37°C, the bound protein and antibody were washed three times, and 100 μl of an HRP-labeled goat anti-human antibody (Fc fragment) (Sigma) diluted 1:8,000 was added. After 1 h at 37°C, unbound antibody was removed by six washes with PBST-B; then 100 μl TMB solution (Bio-Rad) was added, and the mixture was incubated for 10 min at 25°C. The reaction was stopped with 50 μl 2 M H2SO4, and absorbance was read at 450 nm. Readings were corrected for background by subtracting the average absorbance of two (blank) wells with no serum.
Commercial C. trachomatis assays.
Sera were assayed by the Pgp3 ELISA and four commercial assays. Three were MOMP peptide-based ELISAs: the C. trachomatis-IgG-pELISA plus Medac assay (Medac, Wedel, Germany), the SeroCT-IgG ELISA (Savyon Diagnostics, Ashdod, Israel), and the C. trachomatis IgG EIA (Ani Labsystems, Vantaa, Finland). The fourth was the C. pneumoniae IgG/IgM MIF test kit (Ani Labsystems, Finland). There were insufficient pediatric sera in some cases (HPA cohort) to undertake the Ani Labsystems EIA. In each case, sera were assayed precisely according to the manufacturer's instructions. For MIF, sera were considered positive for C. pneumoniae if IgG antibodies were detectable at a dilution of 1:32, as recommended by the manufacturer. To assay for anti-C. trachomatis antibody by MIF, sera were diluted 1:8, 1:16, and 1:32. Those sera with anti-C. trachomatis IgG antibody detectable at a dilution of 1:32 were judged to be positive.
Statistical analysis.
Statistical analysis, including receiver operating characteristic (ROC) analysis and multiple logistic regression, was conducted using Stata, version 10.1 (Stata Corporation, College Station, TX). Specificity and sensitivity were calculated for each assay, with 95% Agresti-Coull confidence intervals (CI). Comparisons of sensitivity and specificity between assays used methods for paired proportions (two-sided McNemar's test), while comparisons between males and females used methods for independent proportions (two-sided Fisher's exact test); differences in sensitivity/specificity were described with 95% CI. A one-sided Fisher's exact test was used for comparison of C. trachomatis test positivity between sera testing positive and negative for C. pneumoniae, assuming that anti-C. pneumoniae antibodies could only increase and not decrease the chance of a positive C. trachomatis test result.
RESULTS
Characterization of recombinant Pgp3 protein.
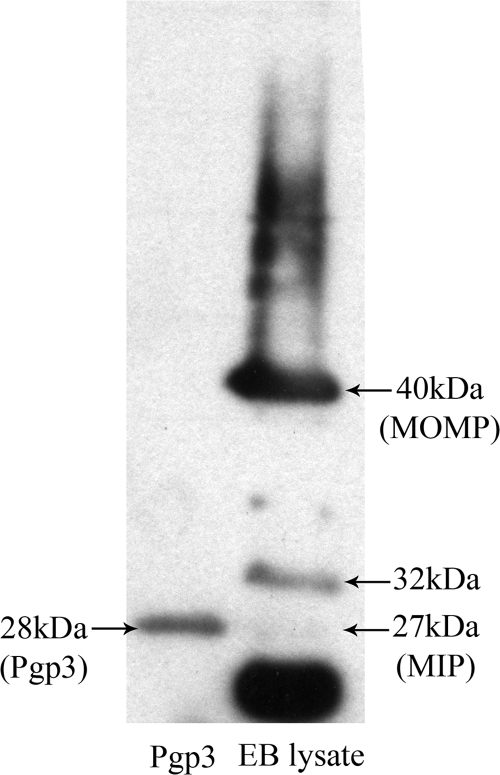
The purity and antigenicity of the untagged Pgp3 protein were assessed by Western blotting with sera that were known to be positive for anti-C. trachomatis IgG by all commercial assays and that reacted with more than eight known C. trachomatis EB proteins in Western blots (2). All sera tested bound to the 28-kDa Pgp3 protein band (an example is shown in Fig. 1).
FIG. 1.
Purification and characterization of Pgp3. Samples of the purified Pgp3 and lysed C. trachomatis EB proteins (diluted 1:10 in SDS buffer) were separated on a 12.5% polyacrylamide gel and transferred to a PVDF membrane before incubation with a 1:100 dilution of a serum sample from the positive cohort. Bound IgG antibody was detected by an HRP-conjugated secondary antibody and ECL chemiluminescence. Known C. trachomatis EB proteins, including MOMP and the macrophage infectivity potentiator (MIP) protein, are shown.
Detection of recombinant Pgp3 protein by ELISA.
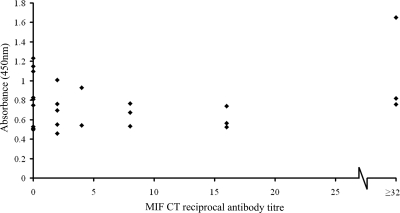
A first-generation ELISA was performed as described above. Sera from individuals known (by MIF) to be seronegative or seropositive showed a positive correlation between the MIF antibody titers and the absorbance values, and the negative samples gave low absorbance readings, implying that the Pgp3 coating antigen showed little evidence of cross-reactivity with nonspecific antibodies (Fig. 2).
FIG. 2.
First-generation Pgp3 ELISA. Pgp3 was used as the coating antigen (20 ng/well in sodium carbonate buffer [pH 9.6]) in an indirect ELISA. A panel of sera for which anti-C. trachomatis antibody titers had previously been assessed by MIF were tested at a dilution of 1:100. The scatter plot shows the absorbance at 450 nm (y axis) for each sample against the anti-C. trachomatis antibody titer determined by MIF (x axis). Samples with anti-C. trachomatis antibodies detectable by MIF at a reciprocal dilution of ≥32 are grouped together.
The Pgp3 antigen and MIF-positive and -negative sera were diluted 25-fold and then serially diluted 2-fold. The HRP-conjugated anti-human secondary antibody was diluted 1,000-fold initially and thereafter in 2-fold dilutions. To optimize the ELISA, the dilutions of the antigen, sera, and HRP-conjugated antibody were assessed in checkerboard titrations. The best signal-to-noise ratio of the positive (signal) and negative-control (noise) sera was obtained by applying the purified antigen at a concentration of 100 ng/well and using a serum dilution of 1:100 and a secondary-antibody dilution of 1:8,000. All other conditions were as described for the first-generation ELISA, except for the coating of the plates with the antigen, for which the Pgp3 was diluted in 100 mM sodium carbonate buffer (pH 8.4). All ELISA plates included positive and negative reference sera, as well as wells with no sera.
Chlamydia-negative control serum samples and specificity.
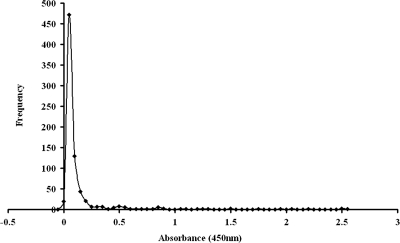
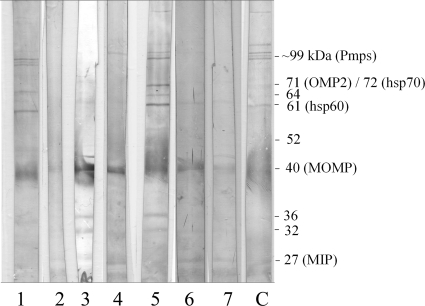
All 747 serum samples from children were assayed by the Pgp3 ELISA (Fig. 3). Of these, 25 gave positive results for anti-C. trachomatis antibodies at a 1:32 dilution by the MIF assay. Twenty of these potentially antibody positive samples were investigated for immune activity against C. trachomatis EB proteins by Western blotting (the remaining five sera had insufficient volume for the procedure). Seven of these samples bound to more than eight EB proteins (Fig. 4) and were considered definitively positive (2); to support this result, these sera tested positive by most of the commercial ELISAs (Table 1). These 7 sera were excluded from further analysis, leaving 740 pediatric serum samples.
FIG. 3.
Analysis of pediatric samples by the Pgp3 ELISA. The frequency distribution curve of the Pgp3 ELISA with all 747 pediatric samples is shown.
FIG. 4.
Western blot of C. trachomatis-positive pediatric sera. Sera with Pgp3 assay absorbance values above 0.2 were assayed by Western blotting. Lysed C. trachomatis EB proteins were separated on a 12.5% polyacrylamide gel and then transferred to a PVDF membrane. Pediatric sera (n = 75) diluted 1:100 were assayed, and those considered positive (>8 bands reactive) are shown. A positive-control serum sample (C) is included. Common C. trachomatis immunodominant antigens are shown, including the several ∼99-kDa polymorphic membrane proteins (Pmps), the 72-kDa hsp70, the 71-kDa outer membrane protein 2 (OMP2), the 61-kDa hsp60, MOMP, and the macrophage infectivity potentiator (MIP) protein.
TABLE 1.
Reactivities of Western blot-positive pediatric sera with the Pgp3 ELISA and commercial serological assays
| Sample no.a | Result by the:
|
|||
|---|---|---|---|---|
| Pgp3 assay (absorbance at 450 nm) | Medac pELISAb (AU/ml) | SeroCT ELISAc (COI) | Ani Labsystems EIAd (S/CO) | |
| 1 | 2.49 | 10.6 | 3.89 | ND |
| 2 | 1.92 | 54.7 | 2.06 | 2.40 |
| 3 | 2.01 | 313.9 | 4.08 | 4.36 |
| 4 | 1.70 | 90.5 | 3.30 | 3.70 |
| 5 | 2.59 | 145.6 | 5.02 | 4.52 |
| 6 | 2.46 | 67.4 | 2.88 | 2.31 |
| 7 | 2.18 | 48.7 | 1.57 | 2.00 |
All seven samples were positive for C. trachomatis and C. pneumoniae by MIF at a dilution of 1:32.
Results were considered negative at ≤28 AU/ml and positive at >28 AU/ml.
COI, cutoff index. Results were considered negative at ≤1.1 and positive at >1.1.
S/CO, signal-to-cutoff ratio. Results were considered negative at ≤1.4 and positive at >1.4. ND, not determined (insufficient serum available).
Further analysis of the association between MIF for C. trachomatis, MIF for C. pneumoniae, and the Pgp3 assay for these 740 sera (discussed below) led to the conclusion that the remaining 18 sera positive for C. trachomatis by MIF were likely to be true positives for anti-C. trachomatis antibodies. For the calculation and comparison of the specificities of the Pgp3 assay and three commercially available C. trachomatis ELISAs, these 18 sera were both included (to be more conservative) and excluded (Table 2). The only significant difference in specificity in either case was between the Pgp3 and Ani Labsystems ELISAs when the 18 pediatric sera positive for C. trachomatis by MIF were included (P = 0.001).
TABLE 2.
Comparative specificities of the Pgp3 ELISA and commercial ELISAs
| Assay | No. of samples | Specificity (%) (95% CI) | Difference in specificity between the Pgp3 assay and the commercial assaya (%) (95% CI) | McNemar's P value |
|---|---|---|---|---|
| Before exclusion of 18 sera positive for C. trachomatis by MIF | ||||
| Pgp3 | 740 | 96.1 (94.4 to 97.3) | ||
| Ani Labsystems | 595b | 98.7 (97.3 to 99.4) | −3.2 (−5.2 to −1.2) | 0.001 |
| SeroCT | 740 | 96.9 (95.4 to 97.9) | −0.8 (−2.7 to 1.1) | 0.45 |
| Medac | 740 | 95.8 (94.1 to 97.1) | 0.3 (−1.8 to 2.3) | 0.89 |
| After exclusion of 18 sera positive for C. trachomatis by MIF | ||||
| Pgp3 | 722 | 97.6 (96.2 to 98.6) | ||
| Ani Labsystems | 580b | 99.0 (97.7 to 99.6) | −1.6 (−3.3 to 0.2) | 0.08 |
| SeroCT | 722 | 97.2 (95.7 to 98.2) | 0.4 (−1.3 to 2.1) | 0.73 |
| Medac | 722 | 96.0 (94.3 to 97.2) | 1.7 (−0.2 to 3.6) | 0.09 |
The specificity of the commercial assay was subtracted from the specificity of the Pgp3 assay.
HPA pediatric serum samples had insufficient volume available for the Ani Labsystems assay.
Determination of Pgp3 ELISA cutoff.
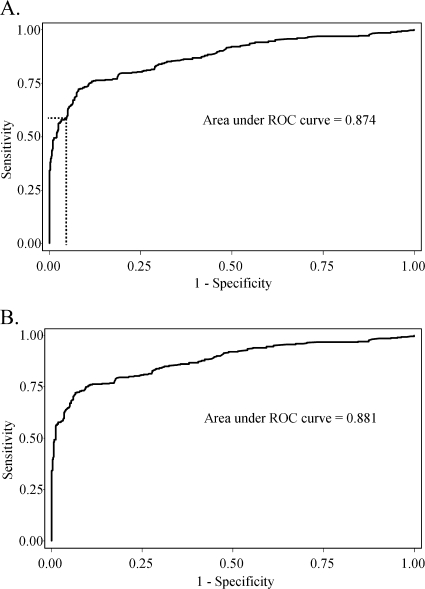
A cutoff value for absorbance at 450 nm (≥0.473) was selected for the Pgp3 ELISA to give a specificity of ≥96% for the 740 pediatric sera left after the exclusion of the 7 samples that tested positive for C. trachomatis EB by Western blotting. Figure 5A shows the corresponding ROC curve obtained by using these 740 negative sera and the sera from the 356 C. trachomatis-positive patients.
FIG. 5.
ROC curves for 356 C. trachomatis-positive sera and either 740 pediatric sera (after exclusion of sera that were positive for EB by Western blotting) (A) or 722 pediatric sera (after the additional exclusion of sera that were positive for C. trachomatis by MIF) (B). The broken line identifies the point on the ROC curve in panel A corresponding to the absorbance at 450 nm, chosen as the final cutoff for the assay.
The specificities of the various tests were compared using the 722 C. trachomatis-negative pediatric sera left after the exclusion of a further 18 samples positive for C. trachomatis by MIF; the specificities of the Pgp3 ELISA and other ELISAs were, naturally, slightly higher in this group (Table 2). Figure 5B shows the ROC curve using these 722 sera negative for C. trachomatis by MIF along with the 356 C. trachomatis-positive sera.
Sensitivity of the Pgp3 assay compared with those of commercial C. trachomatis ELISAs.
The detection of anti-C. trachomatis antibody by the Pgp3 ELISA was compared to that by three commercially available immunoassays. The Pgp3 ELISA was significantly more sensitive (57.9%) than the Ani Labsystems (49.2%; P = 0.003), SeroCT (47.2%; P < 0.0005), and Medac (44.4%; P < 0.0005) assays (Table 3). The Pgp3, Ani Labsystems, and SeroCT assays, but not the Medac assay, had significantly higher sensitivity on female specimens than on male specimens (73.8 versus 44.2%, 59.8 versus 40.5%, 55.5 versus 40%, and 45.7 versus 43.7%, respectively) (Table 3). For female patients, the Pgp3 assay was 14.0 percentage points more sensitive than the next most sensitive ELISA, the Ani Labsystems ELISA (P = 0.001).
TABLE 3.
Comparative sensitivities of the Pgp3 assay and commercial ELISAs
| Patient group and assay | No. of samples from C. trachomatis- positive patients | Sensitivity (%) (95% CI) | Difference in sensitivity between the Pgp3 assay and the commercial assaya (%) (95% CI) | McNemar's P value |
|---|---|---|---|---|
| All patients | ||||
| Pgp3 | 356 | 57.9 (52.7-62.9) | ||
| Ani Labsystems | 356 | 49.2 (44.0-54.3) | 8.7 (2.9-14.5) | 0.003 |
| SeroCT | 356 | 47.2 (42.1-52.4) | 10.7 (4.9-16.5) | <0.0005 |
| Medac | 356 | 44.4 (39.3-49.6) | 13.5 (7.0-20.0) | <0.0005 |
| Femalesb | ||||
| Pgp3 | 164 | 73.8 (66.5-79.9) | ||
| Ani Labsystems | 164 | 59.8 (52.1-67.0) | 14.0 (5.5-22.5) | 0.001 |
| SeroCT | 164 | 55.5 (47.8-62.9) | 18.3 (10.1-26.5) | <0.0005 |
| Medac | 164 | 45.7 (38.3-53.4) | 28.0 (18.9-37.2) | <0.0005 |
| Malesb | ||||
| Pgp3 | 190 | 44.2 (37.3-51.3) | ||
| Ani Labsystems | 190 | 40.5 (33.8-47.6) | 3.7 (−4.5-11.8) | 0.42 |
| SeroCT | 190 | 40.0 (33.3-47.1) | 4.2 (−4.1-12.6) | 0.36 |
| Medac | 190 | 43.7 (36.8-50.8) | 0.5 (−8.6-9.6) | 1.00 |
The sensitivity of the commercial assay was subtracted from that of the Pgp3 assay.
P values for differences in sensitivity on male versus female sera are as follows: for the Pgp3 test, <0.0005; for the Ani Labsystems assay, <0.0005; for the SeroCT assay, 0.004; and for the Medac assay, 0.75.
Impact of C. pneumoniae exposure on ELISA performance.
After 7 samples positive for EB by Western blotting were excluded, 18 of the 740 remaining children's sera were positive for C. trachomatis by MIF. We investigated the possibility that these might be false-positives resulting from cross-reaction with antibodies against C. pneumoniae. Twelve of these sera were positive for C. pneumoniae by MIF, while six showed no evidence of anti-C. pneumoniae antibodies. We also looked for evidence of cross-reaction between tests for C. trachomatis and C. pneumoniae in the Chlamydia-positive patients' sera.
Of the 722 children's sera that were negative for C. trachomatis by MIF, 191 (26.5%) were positive for anti-C. pneumoniae antibody, as were 246 (69.1%) of the 356 C. trachomatis-positive patient sera. We compared the positivity of C. trachomatis tests between serum samples that were positive and negative for C. pneumoniae by MIF separately for these exposed and unexposed groups (Table 4). There was no significant difference in ELISA positivity between sera that were positive or negative for C. pneumoniae by MIF in either of the patient groups (one-sided Fisher's exact test), but there was a positive association between the MIF tests for C. trachomatis and C. pneumoniae. Thus, there was no evidence of cross-reactivity with C. pneumoniae for any of the C. trachomatis ELISAs. However, these results confirmed the expected cross-reactivity between the MIF assays for C. trachomatis and C. pneumoniae. Multiple logistic regression in the C. trachomatis-positive group likewise confirmed that the MIF assay for C. trachomatis was a significant independent predictor of C. pneumoniae positivity by MIF (odds ratio [OR], 2.4; P = 0.001) but the Pgp3 assay was not (OR, 0.7; P = 0.22).
TABLE 4.
C. trachomatis antibody status in samples positive and negative for C. pneumoniae
| Group and assay | No. of samples tested by each C. trachomatis assay with the following C. pneumoniae result by MIF:
|
% of patient group positive for C. trachomatis by each assay and with the following C. pneumoniae result by MIF:
|
P value by Fisher's exact test | ||
|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | ||
| Children not exposed to C. trachomatisb | |||||
| Pgp3 | 531 | 191 | 1.9 | 3.7 | 0.14 |
| Ani Labsystems | 436a | 144 | 1.1 | 0.7 | 0.54 |
| SeroCT | 531 | 191 | 2.8 | 2.6 | 0.56 |
| Medac | 531 | 191 | 3.8 | 4.7 | 0.35 |
| C. trachomatis-exposed patients | |||||
| Pgp3 | 110 | 246 | 55.5 | 58.9 | 0.31 |
| Ani Labsystems | 110 | 246 | 46.4 | 50.4 | 0.28 |
| SeroCT | 110 | 246 | 44.5 | 48.4 | 0.29 |
| Medac | 110 | 246 | 38.2 | 47.2 | 0.07 |
| MIF for C. trachomatis | 110 | 246 | 42.7 | 60.2 | 0.0017 |
HPA pediatric serum samples had insufficient volume available for the Ani Labsystems assay.
Results exclude the 18 sera positive for C. trachomatis by MIF.
Among 740 children's sera, including those that were C. trachomatis positive by MIF, however, univariable logistic regression showed a moderate association of both C. trachomatis positivity by MIF and Pgp3 assay positivity with C. pneumoniae positivity by MIF (ORs, 5.6 and 3.4, respectively; P, 0.001 in both cases). These associations were weakened in a multivariable model (ORs, 3.5 and 2.1; P, 0.031 and 0.09, respectively), as is typical of highly correlated predictors. Conversely, a multivariable model of Pgp3 positivity showed that a positive result for C. trachomatis by MIF was extremely strongly associated with a positive result by the Pgp3 assay (OR, 66; P < 0.0005), while a positive result for C. pneumoniae by MIF was not independently associated with a positive result for C. trachomatis by the Pgp3 assay (OR, 2.1; P = 0.09). These results are most consistently explained by the presence of true anti-C. trachomatis antibodies in some of the sera: both MIF for C. trachomatis and the Pgp3 assay react strongly with these antibodies.
Reproducibility of the Pgp3 ELISA and qualitative interpretation of the data.
Interassay reproducibility was determined by assaying five samples in 20 independent Pgp3 assays, giving coefficients of variation between 3.1% and 13.2%. Intra-assay variation was tested by analyzing five serum samples 16 times. Coefficients of variation were found to be between 1.9% and 5.8% for this ELISA (Table 5).
TABLE 5.
Inter- and intra-assay variation in the Pgp3 ELISA
| Sample | Interassay variation
|
Intra-assay variation
|
||||
|---|---|---|---|---|---|---|
| Mean absorbance at 450 nm | SD | Coefficient of variation (%) | Mean absorbance at 450 nm | SD | Coefficient of variation (%) | |
| 1 | 2.647 | 0.0807 | 3.1 | 2.523 | 0.0645 | 2.6 |
| 2 | 1.414 | 0.0555 | 3.9 | 1.493 | 0.0280 | 1.9 |
| 3 | 0.455 | 0.0303 | 6.7 | 0.447 | 0.0125 | 2.8 |
| 4 | 0.097 | 0.0128 | 13.2 | 0.112 | 0.0051 | 4.6 |
| 5 | 0.081 | 0.0075 | 9.3 | 0.077 | 0.0045 | 5.8 |
We ran the Pgp3 ELISA twice on 166 of the C. trachomatis organism-positive patient sera and 595 of the pediatric sera. One sample in each of these two groups testing positive in the first assay was negative in the second, and one sample from each group testing negative in the first assay tested positive in the second. All four of these samples had Pgp3 ELISA absorbance values at 450 nm close to the cutoff of 0.473.
DISCUSSION
The sensitivities of the three commercial ELISAs were similar, ranging from 44 to 49%, while the Pgp3 ELISA was significantly (P < 0.003) more sensitive, at 57.9% (95% CI, 52.7 to 62.9%), than all three commercial assays. The sensitivity of the Pgp3 ELISA was greater for females (73.8%) than for males (44.2%). The specificity of the Pgp3 ELISA, calculated after exclusion of the 18 sera positive for C. trachomatis by MIF, was 97.6% and not significantly different from those of the commercial assays (96.0 to 99.0%). Even when a more conservative estimate was used, the specificity of the Pgp3 ELISA (96.1%) was similar to those of the other ELISAs (95.8 to 98.7%).
This is the largest published study evaluating C. trachomatis serological ELISAs using well-characterized sera and taking into consideration the anti-C. pneumoniae antibody status. The importance of using reliable control sera with which to establish a serological assay cannot be overstated. Our positive-control sera came from patients confirmed as Chlamydia organism positive at least 1 month previously. Children were chosen as the source of Chlamydia-negative control sera because adults can resolve infection spontaneously in the absence of treatment (37, 39), and thus, adults who are currently negative for C. trachomatis organisms may have experienced past infection.
Although children are unlikely to have been exposed to C. trachomatis, infection can result from vertical transmission (48), sexual abuse (14), consensual sexual intercourse, or exposure to C. trachomatis eye disease (13). The patient profile of St Mary's Hospital includes a large proportion of children from Africa, the Middle East, and northern India, where trachoma is hyperendemic (46) and occurs as early as the age of 2 to 3 years (56). Hence, it was expected that a small percentage of the children's sera would have antibodies to C. trachomatis.
Indeed, of the 747 pediatric sera, 25 (3.3%) were positive for C. trachomatis by MIF. Of these, seven were also found to be positive by Western blotting using EB, and these were excluded. The Ani Labsystems MIF kit, which tests for antibodies to C. pneumoniae, C. trachomatis, and Chlamydia psittaci, was used in this study. In this assay, the C. pneumoniae and C. trachomatis EB dots are depleted of LPS in order to reduce cross-reaction. We chose a serum dilution of 1:32, because the higher the dilution, the more likely it is that true anti-C. trachomatis antibodies are being detected. However, even the use of LPS-depleted EB at a serum dilution of 1:32 does not exclude the possibility of cross-reactive anti-C. pneumoniae antibodies. The Pgp3 protein has no equivalent orthologue in C. pneumoniae and therefore is not expressed in this chlamydial species. We would not anticipate an association between Pgp3 antibody status and the MIF for C. pneumoniae, but this was observed if sera positive for C. trachomatis by MIF were included. Logistic regression analysis provided strong evidence that this is most probably due to the presence of genuine antibody to C. trachomatis in sera positive for C. trachomatis by MIF cross-reacting with the test for C. pneumoniae. For purposes of comparison, we have provided the specificities of the ELISAs with children's sera positive for C. trachomatis by MIF included (Table 2).
The criteria for defining positive and negative patient cohorts have differed in previous studies comparing commercial assays. With no “gold standard”' against which to standardize Chlamydia serology, calculations of sensitivities and specificities are subject to misclassification errors. The value of the present study lies in the fact that each assay has been subjected to scrutiny against the same carefully defined control samples.
The specificities determined for the commercial ELISAs in this study are similar to those reported by the manufacturers, who also used sera from children (41). Other published studies have used samples from adults who were presumed to be at low risk, e.g., blood donors. Our specificities are comparable to those of some (25, 38), but not all (2, 4, 53), of these studies.
The sensitivities reported by the ELISA manufacturers and published in the literature are generally higher than those that we find. The manufacturers have all used sera from Chlamydia culture-positive individuals to determine their assays' sensitivities. Elsewhere, the assays have been characterized using sera from patients who have tested seropositive by MIF (19, 38), from women only (4, 29, 38), or from individuals positive for Chlamydia at the time of serum collection (53), who will often have symptomatic disease.
The use of Pgp3 in an ELISA has been reported previously, although only on small cohorts. Using MIF-positive samples, Comanducci et al. (12) found IgG antibodies to Pgp3 in 80% of samples. A sensitivity of 92% was reported for 50 patients with symptomatic cervical chlamydial disease or nongonococcal urethritis (21). Our results for sensitivity (58%) are in agreement with those of two studies by Bas et al., which found sensitivities of 53% and 57%, respectively (2, 3). However, as mentioned previously, blood donors were used as negative controls in these studies, contributing to their lower results for specificity (80% [2] and 89% [3] compared to 97.6%). A further study by Bas et al. looking at Chlamydia exposure in relation to reactive arthritis found that their Pgp3 ELISA had a specificity of 90% (on 20 patients with arthropathies independent of C. trachomatis) and a sensitivity of 59% (on 17 patients with C. trachomatis reactive arthritis) (1). It should be noted that, in other studies with Pgp3, efforts were made to retain Pgp3 in its native form in order to preserve conformational epitopes (1-3, 12, 21). In this study, purified Pgp3 was stored in the presence of the reducing agent DTT but was diluted 1:1,000 for the assay. Batches of Pgp3 stored without DTT did not react differently in the ELISA (data not shown).
There was clear evidence of a sex difference in the sensitivities of all the assays, except for the Medac pELISA (Table 3). This was most marked with the Pgp3 ELISA, which demonstrated a 30% increase in sensitivity among women over men. Sex differences in the humoral immune response have been reported. Närvänen et al. (41), using the Ani Labsystems ELISA, found that among culture-positive patients, antibody could be detected in 84.2% of women and 61.3% of men. They found that when suspected C. trachomatis infection could not be confirmed by culture, frequently an antibody response was detectable by the ELISA: 45.3% of the Finnish women compared to 38.3% of the Finnish men had anti-C. trachomatis antibodies. Other studies observed that antibody prevalence and titers were higher for women than for men, even within couples (6, 30, 43). This may reflect a higher antigenic load in women than in men (16, 20, 36) or a more marked immune response where an infection higher in the genital tract is involved. The availability of a more sensitive, well-characterized ELISA allows further investigation of gender differences in response to C. trachomatis infection.
We have confirmed that our new Pgp3 ELISA and the three commercial C. trachomatis MOMP peptide-based ELISAs are >95% specific and are not cross-reactive with C. pneumoniae. Broadly, the commercial ELISAs were less than 50% sensitive, while the Pgp3 ELISA had a sensitivity close to 60%, which was higher still (74%) when female sera were assayed. This is in spite of evidence for plasmid-free isolates of C. trachomatis, which appear to be rare, at least in the United Kingdom (34). There is interest in using serology to monitor changes in the age-specific anti-C. trachomatis antibody prevalence and to improve understanding of the epidemiology of chlamydial infection. This is a potential method for evaluating the population impact of C. trachomatis control programs (24, 32), since it would allow estimates of the changing prevalence of past exposure to C. trachomatis rather than only current infection. However, a prerequisite for this is better-characterized and better-performing assays. The Pgp3 ELISA is both sensitive and specific and performs significantly better than the Chlamydia-specific MOMP peptide-based assays. The way is now open to explore the epidemiology of C. trachomatis genital tract infection by serological means.
Acknowledgments
We thank Shaw Taylor, Angela Taylor, and the staff of the Milne Centre for Sexual Health, Bristol, United Kingdom. We also thank the staff of the Jefferiss Wing of St Mary's Hospital, London, United Kingdom. Invaluable technical advice was provided by Ian Clarke, Alan Herring, Anna Helander, and Peter Cherepanov.
This work was funded by the Medical Research Council, United Kingdom. We are also grateful for support from the NIHR Biomedical Research Centre funding scheme.
Footnotes
Published ahead of print on 8 April 2009.
REFERENCES
- 1.Bas, S., S. Genevay, M. C. Schenkel, and T. L. Vischer. 2002. Importance of species-specific antigens in the serodiagnosis of Chlamydia trachomatis reactive arthritis. Rheumatology 411017-1020. [DOI] [PubMed] [Google Scholar]
- 2.Bas, S., P. Muzzin, B. Ninet, J. E. Bornand, C. Scieux, and T. L. Vischer. 2001. Chlamydial serology: comparative diagnostic value of immunoblotting, microimmunofluorescence test, and immunoassays using different recombinant proteins as antigens. J. Clin. Microbiol. 391368-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bas, S., P. Muzzin, and T. L. Vischer. 2001. Chlamydia trachomatis serology: diagnostic value of outer membrane protein 2 compared with that of other antigens. J. Clin. Microbiol. 394082-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bax, C. J., J. A. E. M. Mutsaers, C. L. Jansen, J. B. Trimbos, P. J. Dörr, and P. M. Oostvogel. 2003. Comparison of serological assays for detection of Chlamydia trachomatis antibodies in different groups of obstetrical and gynecological patients. Clin. Diagn. Lab. Immunol. 10174-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergström, K., M. Domeika, D. Vaitkiene, K. Persson, and P. A. Mårdh. 1996. Prevalence of Chlamydia trachomatis, Chlamydia psittaci and Chlamydia pneumoniae antibodies in blood donors and attendees of STD clinics. Clin. Microbiol. Infect. 1253-260. [DOI] [PubMed] [Google Scholar]
- 6.Bessho, H., and A. Matsumoto. 1990. Comparison of antibody titers to Chlamydia trachomatis in men and women. Kansenshogaku Zasshi 641024-1029. [DOI] [PubMed] [Google Scholar]
- 7.Black, C. M., R. C. Barnes, K. A. Birkness, B. P. Holloway, and L. W. Mayer. 1989. Nucleotide sequence of the common plasmid of Chlamydia trachomatis serovar L2: use of compatible deletions to generate overlapping fragments. Curr. Microbiol. 1967-74. [Google Scholar]
- 8.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 311161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherepanov, P. 2007. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 35113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comanducci, M., S. Ricci, R. Cevenini, and G. Ratti. 1990. Diversity of the Chlamydia trachomatis common plasmid in biovars with different pathogenicity. Plasmid 23149-154. [DOI] [PubMed] [Google Scholar]
- 11.Comanducci, M., R. Cevenini, A. Moroni, M. M. Giuliani, S. Ricci, V. Scarlato, and G. Ratti. 1993. Expression of a plasmid gene of Chlamydia trachomatis encoding a novel 28 kDa antigen. J. Gen. Microbiol. 1391083-1092. [DOI] [PubMed] [Google Scholar]
- 12.Comanducci, M., R. Manetti, L. Bini, A. Santucci, V. Pallini, R. Cevenini, J. M. Sueur, J. Orfila, and G. Ratti. 1994. Humoral immune response to plasmid protein Pgp3 in patients with Chlamydia trachomatis infection. Infect. Immun. 625491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darougar, S. 1981. Chlamydial ocular infection. J. Antimicrob. Chemother. 8350-354. [DOI] [PubMed] [Google Scholar]
- 14.Dattel, B. J., D. V. Landers, K. Coulter, J. Hinton, R. L. Sweet, and J. Schachter. 1988. Isolation of Chlamydia trachomatis from sexually abused female adolescents. Obstet. Gynecol. 72240-242. [PubMed] [Google Scholar]
- 15.Denholm, R., and J. Riha. 31 July 2008, posting date. Chlamydia testing activity in laboratories across England. Report on pilot survey using data from 1st April-31st December 2007. National Chlamydia Screening Programme, HIV/STI Department, Health Protection Agency, Centre for Infections, London, United Kingdom. http://www.chlamydiascreening.nhs.uk/ps/assets/pdfs/lab/Chlamydia%20_testing_activity_final_310708.pdf.
- 16.Eckert, L. O., R. J. Suchland, S. E. Hawes, and W. E. Stamm. 2000. Quantitative Chlamydia trachomatis cultures: correlation of chlamydial inclusion-forming units with serovar, age, sex, and race. J. Infect. Dis. 182540-544. [DOI] [PubMed] [Google Scholar]
- 17.Fenton, K. A., C. Korovessis, A. M. Johnson, A. McCadden, S. McManus, K. Wellings, C. H. Mercer, C. Carder, A. J. Copas, K. Nanchahal, W. Macdowall, G. Ridgway, J. Field, and B. Erens. 2001. Sexual behaviour in Britain: reported sexually transmitted infections and prevalent genital Chlamydia trachomatis infection. Lancet 3581851-1854. [DOI] [PubMed] [Google Scholar]
- 18.Follmann, F., A. W. Olsen, K. T. Jensen, P. R. Hansen, P. Andersen, and M. Theisen. 2008. Antigenic profiling of a Chlamydia trachomatis gene-expression library. J. Infect. Dis. 197897-905. [DOI] [PubMed] [Google Scholar]
- 19.Frikha-Gargouri, O., R. Gdoura, A. Znazen, N. Ben Arab, J. Gargouri, M. Ben Jemaa, and A. Hammami. 2008. Evaluation and optimization of a commercial enzyme linked immunosorbent assay for detection of Chlamydophila pneumoniae IgA antibodies. BMC Infect. Dis. 898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisler, W. M., R. J. Suchland, W. L. Whittington, and W. E. Stamm. 2001. Quantitative culture of Chlamydia trachomatis: relationship of inclusion-forming units produced in culture to clinical manifestations and acute inflammation in urogenital disease. J. Infect. Dis. 1841350-1354. [DOI] [PubMed] [Google Scholar]
- 21.Ghaem-Maghami, S., G. Ratti, M. Ghaem-Maghami, M. Comanducci, P. E. Hay, R. L. Bailey, D. C. Mabey, H. C. Whittle, M. E. Ward, and D. J. Lewis. 2003. Mucosal and systemic immune responses to plasmid protein Pgp3 in patients with genital and ocular Chlamydia trachomatis infection. Clin. Exp. Immunol. 132436-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatt, C., M. E. Ward, and I. N. Clarke. 1988. Analysis of the entire nucleotide sequence of the cryptic plasmid of Chlamydia trachomatis serovar L1. Evidence for involvement in DNA replication. Nucleic Acids Res. 164053-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horner, P. J., D. Cain, M. McClure, B. J. Thomas, C. Gilroy, M. Ali, J. N. Weber, and D. Taylor-Robinson. 1997. Association of antibodies to Chlamydia trachomatis heat-shock protein 60 kD with chronic nongonococcal urethritis. Clin. Infect. Dis. 24653-660. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, A. M., and P. Horner. 2008. A new role for Chlamydia trachomatis serology? Sex. Transm. Infect. 8479-80. [DOI] [PubMed] [Google Scholar]
- 25.Jones, C. S., P. A. Maple, N. J. Andrews, I. D. Paul, and E. O. Caul. 2003. Measurement of IgG antibodies to Chlamydia trachomatis by commercial enzyme immunoassays and immunofluorescence in sera from pregnant women and patients with infertility, pelvic inflammatory disease, ectopic pregnancy, and laboratory diagnosed Chlamydia psittaci/Chlamydia pneumoniae infection. J. Clin. Pathol. 56225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21385-389. [DOI] [PubMed] [Google Scholar]
- 27.Kutlin, A., N. Tsumura, U. Emre, P. M. Roblin, and M. R. Hammerschlag. 1997. Evaluation of Chlamydia immunoglobulin M (IgM), IgG, and IgA rELISAs Medac for diagnosis of Chlamydia pneumoniae infection. Clin. Diagn. Lab. Immunol. 4213-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 29.Land, J. A., A. P. Gijsen, A. G. Kessels, M. E. Slobbe, and C. A. Bruggeman. 2003. Performance of five serological Chlamydia antibody tests in subfertile women. Hum. Reprod. 182621-2627. [DOI] [PubMed] [Google Scholar]
- 30.Levett, P. N. 1994. Seroepidemiology of chlamydial infection among a sexually-active population in Barbados. West Indian Med. J. 4380-83. [PubMed] [Google Scholar]
- 31.Li, Z., D. Chen, Y. Zhong, S. Wang, and G. Zhong. 2008. The chlamydial plasmid-encoded protein Pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect. Immun. 763415-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyytikäinen, E., M. Kaasila, P. Koskela, M. Lehtinen, T. Patama, E. Pukkala, K. Tasanen, H. M. Surcel, and J. Paavonen. 2008. Chlamydia trachomatis seroprevalence atlas of Finland 1983-2003. Sex. Transm. Infect. 8419-22. [DOI] [PubMed] [Google Scholar]
- 33.Macleod, J., C. Salisbury, N. Low, A. McCarthy, J. A. Sterne, A. Holloway, R. Patel, E. Sanford, A. Morcom, P. Horner, G. Davey-Smith, S. Skidmore, A. Herring, O. Caul, F. D. Hobbs, and M. Egger. 2005. Coverage and uptake of systematic postal screening for genital Chlamydia trachomatis and prevalence of infection in the United Kingdom general population: cross sectional study. BMJ 330940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magbanua, J. P., B. T. Goh, C. E. Michel, A. Aguirre-Andreasen, S. Alexander, I. Ushiro-Lumb, C. Ison, and H. Lee. 2007. Chlamydia trachomatis variant not detected by plasmid based nucleic acid amplification tests: molecular characterisation and failure of single dose azithromycin. Sex. Transm. Infect. 83339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maurin, M., F. Eb, J. Etienne, and D. Raoult. 1997. Serological cross-reactions between Bartonella and Chlamydia species: implications for diagnosis. J. Clin. Microbiol. 352283-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michel, C. E., C. Sonnex, C. A. Carne, J. A. White, J. P. Magbanua, E. C. Nadala, Jr., and H. H. Lee. 2007. Chlamydia trachomatis load at matched anatomic sites: implications for screening strategies. J. Clin. Microbiol. 451395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molano, M., C. J. Meijer, E. Weiderpass, A. Arslan, H. Posso, S. Franceschi, M. Ronderos, N. Munoz, and A. J. van den Brule. 2005. The natural course of Chlamydia trachomatis infection in asymptomatic Colombian women: a 5-year follow-up study. J. Infect. Dis. 191907-916. [DOI] [PubMed] [Google Scholar]
- 38.Morré, S. A., C. Munk, K. Persson, S. Krüger-Kjaer, R. van Dijk, C. J. Meijer, and A. J. van den Brule. 2002. Comparison of three commercially available peptide-based immunoglobulin G (IgG) and IgA assays to microimmunofluorescence assay for detection of Chlamydia trachomatis antibodies. J. Clin. Microbiol. 40584-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morré, S. A., A. J. van den Brule, L. Rozendaal, A. J. Boeke, F. J. Voorhorst, S. de Blok, and C. J. Meijer. 2002. The natural course of asymptomatic Chlamydia trachomatis infections: 45% clearance and no development of clinical PID after one-year follow-up. Int. J. STD AIDS 13(Suppl. 2)12-18. [DOI] [PubMed] [Google Scholar]
- 40.Moss, T. R., S. Darougar, R. M. Woodland, M. Nathan, R. J. Dines, and V. Cathrine. 1993. Antibodies to Chlamydia species in patients attending a genitourinary clinic and the impact of antibodies to C. pneumoniae and C. psittaci on the sensitivity and the specificity of C. trachomatis serology tests. Sex. Transm. Dis. 2061-65. [DOI] [PubMed] [Google Scholar]
- 41.Närvänen, A., M. Puolakkainen, W. Hao, K. Kino, and J. Suni. 1997. Detection of antibodies to Chlamydia trachomatis with a peptide-based species-specific enzyme immunoassay. Infect. Dis. Obstet. Gynecol. 5349-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newhall, W. J., B. Batteiger, and R. B. Jones. 1982. Analysis of the human serological response to proteins of Chlamydia trachomatis. Infect. Immun. 381181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ni, A. P., G. Y. Lin, L. Yang, H. Y. He, C. W. Huang, Z. J. Liu, R. S. Wang, J. S. Zhang, J. Y. Yu, N. Li, J. B. Wang, and H. Y. Yang. 1996. A seroepidemiologic study of Chlamydia pneumoniae, Chlamydia trachomatis and Chlamydia psittaci in different populations on the mainland of China. Scand. J. Infect. Dis. 28553-557. [DOI] [PubMed] [Google Scholar]
- 44.Patel, H. C., B. T. Goh, N. D. Viswalingam, and J. D. Treharne. 1995. Interpretation of Chlamydia trachomatis antibody response in chlamydial oculogenital infection. Genitourin. Med. 7194-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pickett, M. A., J. S. Everson, P. J. Pead, and I. N. Clarke. 2005. The plasmids of Chlamydia trachomatis and Chlamydophila pneumoniae (N16): accurate determination of copy number and the paradoxical effect of plasmid-curing agents. Microbiology 151893-903. [DOI] [PubMed] [Google Scholar]
- 46.Polack, S., S. Brooker, H. Kuper, S. Mariotti, D. Mabey, and A. Foster. 2005. Mapping the global distribution of trachoma. Bull. W. H. O. 83913-919. [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez-Campillo, M., L. Bini, M. Comanducci, R. Raggiaschi, B. Marzocchi, V. Pallini, and G. Ratti. 1999. Identification of immunoreactive proteins of Chlamydia trachomatis by Western blot analysis of a two-dimensional electrophoresis map with patient sera. Electrophoresis 112269-2279. [DOI] [PubMed] [Google Scholar]
- 48.Schachter, J., M. Grossman, R. L. Sweet, J. Holt, C. Jordan, and E. Bishop. 1986. Prospective study of perinatal transmission of Chlamydia trachomatis. JAMA 2553374-3377. [PubMed] [Google Scholar]
- 49.Sharma, J., Y. Zhong, F. Dong, J. M. Piper, G. Wang, and G. Zhong. 2006. Profiling of human antibody responses to Chlamydia trachomatis urogenital tract infection using microplates arrayed with 156 chlamydial fusion proteins. Infect. Immun. 741490-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sziller, I., S. S. Witkin, M. Ziegert, Z. Csapo, A. Ujhazy, and Z. Papp. 1998. Serological responses of patients with ectopic pregnancy to epitopes of the Chlamydia trachomatis 60 kDa heat shock protein. Hum. Reprod. 131088-1093. [DOI] [PubMed] [Google Scholar]
- 51.Thomas, N. S., M. Lusher, C. C. Storey, and I. N. Clarke. 1997. Plasmid diversity in Chlamydia. Microbiology 1431847-1854. [DOI] [PubMed] [Google Scholar]
- 52.UK Collaborative Group for HIV and STI Surveillance. November 2007, posting date. Testing times. HIV and other sexually transmitted infections in the United Kingdom: 2007. Health Protection Agency, Centre for Infections, London, United Kingdom. http://www.hpa.org.uk/web/HPAwebFile/HPAweb_C/1203496897276.
- 53.Verkooyen, R. P., M. F. Peeters, J. H. van Rijsoort-Vos, W. I. van der Meijden, and J. W. Mouton. 2002. Sensitivity and specificity of three new commercially available Chlamydia trachomatis tests. Int. J. STD AIDS 13(Suppl. 2)23-25. [DOI] [PubMed] [Google Scholar]
- 54.Wang, S. P., and J. T. Grayston. 1974. Human serology in Chlamydia trachomatis infection with microimmunofluorescence. J. Infect. Dis. 130388-397. [DOI] [PubMed] [Google Scholar]
- 55.Wang, S. P., and J. T. Grayston. 1984. Micro-immunofluorescence serology of Chlamydia trachomatis, p. 87-118. In L. M. de la Maza and E. M. Peterson (ed.), Medical virology III: proceedings of the 1983 International Symposium on Medical Virology. Elsevier Biomedical Press, Amsterdam, The Netherlands.
- 56.West, S., B. Muñoz, V. Turner, B. Mmbaga, and H. Taylor. 1991. The epidemiology of trachoma in central Tanzania. Int. J. Epidemiol. 201088-1092. [DOI] [PubMed] [Google Scholar]
- 57.Yuan, Y., K. Lyng, Y.-X. Zhang, D. D. Rockey, and R. P. Morrison. 1992. Monoclonal antibodies define genus-specific, species-specific, and cross-reactive epitopes of the chlamydial 60-kilodalton heat shock protein (hsp60): specific immunodetection and purification of chlamydial hsp60. Infect. Immun. 602288-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]