Abstract
We describe a high-throughput assay using PCR coupled to electrospray ionization-mass spectrometry (PCR/ESI-MS) to determine the genotypes of Staphylococcus aureus isolates. The primer sets used in the PCR/ESI-MS assay were designed to amplify the same genes analyzed in multilocus sequence typing (MLST). The method was used to identify the clonal complex and USA type of each isolate and is suitable for use in a clinical or public-health setting. The method was validated using a panel of diverse isolates from the Centers for Disease Control and Prevention that were previously characterized by MLST and pulsed-field gel electrophoresis (PFGE). Clinical isolates from two geographically distinct hospitals were characterized, and the clustering results were in agreement with those for repetitive-element PCR and PFGE. The PCR/ESI-MS method enables genotyping of over 180 samples of S. aureus per day in an automated fashion.
Microbial genotyping methods are used to study global evolution of pathogens and to reveal genetic clonality, which is used to determine point sources during epidemiological investigations (1, 2). There are a variety of molecular genotyping methods available, each with certain strengths and weaknesses (4). The methods that are most valuable for understanding pathogen evolution over the long term rely on techniques such as multilocus sequence typing (MLST). This genotyping method is suitable for tracking global pathogen evolution over decades because it focuses on stable housekeeping genes and provides information that places isolates into genetic categories that provide perspective on a global scale (10, 12). For real-time outbreak investigations in health care settings, the molecular genotyping method used must focus on identification of recent genetic changes. Generally, techniques such as pulsed-field gel electrophoresis (PFGE) (27), repetitive-element PCR (rep-PCR) (32), or Staphylococcus protein A typing (19, 20) are used to support these investigations. These methods provide genetic signatures indicative of a microbe's recent history. However, electrophoretic methods are based on gel mobility and therefore provide analog rather than digital signatures. This means that it may be difficult to compare data between laboratories, and the techniques do not reliably reveal where an isolate fits with respect to the global evolutionary history of a pathogen.
Yet, a third category of molecular genotyping is direct PCR analysis of an isolate. Direct PCR is aimed at determining the presence and specific types of certain acquired genetic elements in the microbial genome. The regions targeted for amplification are those portions of the genome known to change over very short periods of time. Examples of such elements include virulence factors, genes that mediate antimicrobial resistance, and housekeeping genes that encode proteins that are the target of antibiotics. This category of molecular genotyping is pathogen specific, as acquired genetic elements tend to be unique to each pathogen (28).
Here, we describe the application of PCR coupled with electrospray ionization-mass spectrometry (PCR/ESI-MS) (7, 16, 24) to Staphylococcus aureus genotyping. The primer sets used in the PCR/ESI-MS assay were designed to amplify the same genes analyzed in MLST. This assay allows us to determine the molecular genotype, as is done with MLST, and to directly characterize specific methicillin (meticillin)-resistant S. aureus (MRSA) genetic elements.
MATERIALS AND METHODS
Bacterial isolates.
Thirty isolates of Staphylococcus aureus were selected to represent the diversity of isolates from a Centers for Disease Control and Prevention (CDC) national database, including a variety of health care-associated and community-acquired strain types (21, 28). The isolates obtained from Johns Hopkins Hospital were a convenience sample of 240 clinical isolates of S. aureus from the following sources: blood samples (n = 31), wounds/skin cultures (n = 43), nasal surveillance sites (n = 57), respiratory nonsurveillance sites (n = 78), urine samples (n = 10), sterile body sites (n = 10), and medical devices, such as catheter tips (n = 11). The 47 clinical isolates from the University of Arizona Medical Center were obtained from various sources, including wounds, abscesses, drainages, sputum samples, cardiovascular catheter tips, and tissues.
PCR/ESI-MS.
General methods for genotypic characterization of isolates by multilocus PCR/ESI-MS using the commercially available Ibis T5000 instrument (Ibis Biosciences) have previously been described (9, 13). For strain differentiation and genotyping of S. aureus, a set of unique sequences from the seven genes commonly used for MLST of S. aureus was assembled (10, 12). These reference alignments were then used for the design of 22 primer pairs for PCR/ESI-MS analysis. The resolution provided by PCR/ESI-MS analysis was calculated, starting with the primer pair providing the best sequence resolution, using theoretical amplicon base compositions determined for each one of 710 S. aureus sequence types (http://saureus.mlst.net). Comparison of these base composition signatures defined the number of sequence types (STs) that were incompatible with any particular type at this particular locus. The average proportion of sequence types excluded by PCR/ESI-MS, with the corresponding standard deviations, was compared against the number of loci used in the analysis. This process was repeated, using base composition signatures extended by one additional locus at a time. A final panel of eight primer pairs was chosen to maximize average resolution with a minimum number of primer pairs. The final choice of primer pairs (Table 1) did not include all original gene targets used in sequence-based MLST but was instead chosen on the basis of strain resolution ability, amplification specificity, and amplification sensitivity with laboratory strains and sample throughput considerations of the PCR/ESI-MS platform. DNA from isolated colonies was distributed into each of 8 wells of a 96-well plate that contained pairs of PCR primers targeted to eight regions of six different MLST alleles. Twelve colonies were analyzed on each microtiter plate. The resulting amplicons were desalted and injected into a mass spectrometer by using a robotic autosampler as described previously (6). Spectra were generated and analyzed using automated software to calculate the base composition (A, G, C, and T counts) in each strand of the amplicon. The data from each of the eight spectra were concatenated to produce a 32-position PCR/ESI-MS type. This experimentally derived base composition was then compared to the calculated base compositions derived from the sequences of the alleles in the MLST database to identify the STs and associated clonal complexes.
TABLE 1.
Primer pair sets for PCR/ESI-MS analysis of S. aureus
| Primer pair code | Gene target | Coordinates | Primer sequence |
|---|---|---|---|
| BCT3025 | arcC | 2724791 to 2724920 | 5′-TGAATAGTGATAGAACTGTAGGCACAATCGT-3′ |
| 5′-TGCGCTAATTCTTCAACTTCTTCTTTCGT-3′ | |||
| BCT2149 | aroE | 1674546 to 1674697 | 5′-TGGGGCTTTAAATATTCCAATTGAAGATTTTCA-3′ |
| 5′-TACCTGCATTAATCGCTTGTTCATCAA-3′ | |||
| BCT2150 | aroE | 1674392 to 1674523 | 5′-TGATGGCAAGTGGATAGGGTATAATACAG-3′ |
| 5′-TAAGCAATACCTTTACTTGCACCACCTG-3′ | |||
| BCT2156 | gmk | 1191206 to 1191337 | 5′-TCACCTCCAAGTTTAGATCACTTGAGAGA-3′ |
| 5′-TGGGACGTAATCGTATAAATTCATCATTTC-3′ | |||
| BCT2157 | pta | 629121 to 629229 | 5′-TCTTGTTTATGCTGGTAAAGCAGATGG-3′ |
| 5′-TGGTACACCTGGTTTCGTTTTGATGATTTGTA-3′ | |||
| BCT2161 | tpi | 830671 to 830799 | 5′-TCCCACGAAACAGATGAAGAAATTAACAAAAAAG-3′ |
| 5′-TGGTACAACATCGTTAGCTTTACCACTTTCACG-3′ | |||
| BCT2163 | yqi | 379057 to 379199 | 5′-TGAATTGCTGCTATGAAAGGTGGCTT-3′ |
| 5′-TCGCCAGCTAGCACGATGTCATTTTC-3′ | |||
| BCT2166 | yqi | 379190 to 379311 | 5′-TAGCTGGCGGTATGGAGAATATGTCT-3′ |
| 5′-TCCATCTGTTAAACCATCATATACCATGCTATC-3′ |
Correlation of base composition signatures with clonal complexes.
The set of 853 distinct STs available from the S. aureus MLST isolate database (http://saureus.mlst.net/portable/portable.xls) as of 7 July 2008 was analyzed with the BURST algorithm (http://eburst.mlst.net/) to assign clonal complexes by using the default stringency method, where every member of a complex shares six or seven alleles with at least one other member. A total of 682 MLST types were assigned to clonal complexes, and 171 were not. With the exception of four distinct cases of base composition profiles that correlate with MLST types belonging to two different clonal complexes, theoretical base composition signatures were correctly grouped. The four pairs of sequence types that belong to two different clonal complexes that generated the same base composition signature are as follows: (i) sequence types 673 (clonal complex 8) and 717 (clonal complex 12), (ii) sequence types 514 and 550 (clonal complex 1) and 221 and 402 (clonal complex 5), (iii) sequence types 437 (clonal complex 25) and 117 (clonal complex 97), and (iv) sequence types 247 (clonal complex 5) and 615 (clonal complex 8). There were 32 base composition signatures (out of a total of 368) that correlated with at least one sequence type not assigned to any clonal complex.
rep-PCR.
The DiversiLab System (bioMérieux, Durham, NC) combines standardized DNA extraction methods, primers, and reagents with postamplification microfluidics-based electrophoresis to perform bacterial strain typing of MRSA. MRSA strains were cultured in trypticase soy broth incubated overnight at 35°C. DNA from each isolate was extracted using an UltraClean microbial DNA isolation kit (Mo Bio Laboratories, Solana Beach, CA). The extracted DNA (25 to 50 ng/ml) was amplified using a DiversiLab Staphylococcus fingerprinting kit, Taq polymerase, and 10× PCR buffer per the manufacturer's instructions. During PCR, the rep-PCR primers bind repetitive DNA sequences and generate amplicons of different sizes. Amplicons are size fractionated using disposable microfluidics chips that contain a copolymer matrix and an intercalating dye, which allows charge separation of the DNA fragments in a microseparation channel in the chip placed when electrophoresis occurs in an Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA). Analysis of amplicon fragments and subsequent data interpretation were accomplished via Web-based software which classifies unique fragment patterns into rep-PCR strain types (14, 15, 30). Sample similarities were calculated by two methods: the Pearson correlation coefficient and the Kullbeck-Leibler coefficient. Both methods compare all possible pairs of samples in a data set with respect to band presence or absence and position as well as band intensity. DiversiLab dendrograms are created from a distance matrix by using the unweighted-pair group method with arithmetic means. Known pulsed-field types (PFT) USA100 to USA1200 were inferred from the rep-PCR strain types by comparison with DiversiLab's PFT reference library.
PFGE methods.
Isolates from Johns Hopkins were genotyped using PFGE by comparison of the pulsed-field patterns with those of reference strains obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA, Reston, VA), supported under NIAID, NIH, contract no. N01-AI-95359. The NARSA reference numbers corresponding to clones USA100 to USA800 are NRS 382, NRS 383, NRS 384, NRS 123, NRS 385, NRS 22, NRS 386, and NRS 387, respectively. PFGE was performed using a protocol developed at the Johns Hopkins Hospital Division of Medical Microbiology. Overnight broth cultures of clinical strains of S. aureus as well as two well-characterized nosocomial MRSA control strains were pelleted, and bacterial DNA was extracted from agarose plugs by using a solution containing lysostaphin and lysozyme. Restriction enzyme digestion was performed using SmaI. Restriction endonuclease fragments were analyzed by PFGE using a contour-clamped homogeneous electric field DR-II apparatus (Bio-Rad Laboratories, Inc., Hercules, CA) set at 14°C, with the following settings: initial switch, 5 s; final switch, 50 s; and time, 23.5 h. After electrophoresis, gels were stained with ethidium bromide. Macrorestriction DNA banding patterns were digitized and analyzed using Molecular Analyst DNA fingerprinting software (Bio-Rad). Patterns were compared to the USA control strains, and an isolate with three or fewer band differences from the USA clone was presumed to be that strain (27).
RESULTS
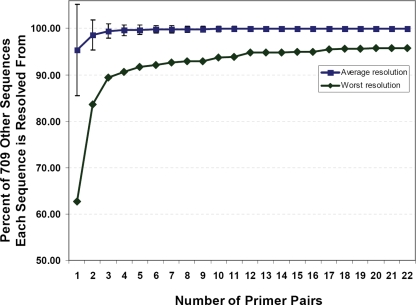
Multilocus PCR/ESI-MS follows the logic of MLST. PCR amplicons were generated from the same genes used in MLST analysis. In the case of PCR/ESI-MS, amplicons were analyzed by mass spectrometry to determine amplicon base composition as described previously (7). The final set of eight primer pairs used for the assay was selected to maximize MLST discrimination with a minimum number of primer pairs. The primer pair set provided an average discrimination of 99.83% ± 0.69% when assessed using a set of 710 distinct STs (Fig. 1). The eight primer pairs that make up the Staphylococcus aureus genotyping panel are shown in Table 1. The process of associating mass spectrometry-derived base composition signatures to STs and clonal complexes is illustrated in Fig. 2. The mass spectrometry measurements required approximately 45 s per PCR, so analysis of each S. aureus colony required about 6 min of mass spectrometry time. Thus, PCR/ESI-MS analysis can be carried out as a highly automated, high-throughput process. One instrument is capable of analysis of the nucleic acid extracts from 180 bacterial colonies (15 microtiter plates) per day.
FIG. 1.
Cumulative resolution of 710 concatenated Staphylococcus aureus MLST sequences from each other, using PCR product base compositions from 1 to 22 pairs of primers. The blue line shows the average resolution (± standard deviation) of each sequence from the 709 others as increasing numbers of primer pairs are added. The green line shows the worst resolution for each number of primer pairs. For example, with the best combined set of eight primer pairs, the 710 sequences are broken into 320 groups. Sequences on average are each different from 707.78 other sequences, or 99.83% of the other 709 sequences (± 0.69). With the same primer pair set, the largest unresolved group of sequences has 51 members. The worst resolution of any sequence from the other 709 sequences for primer pairs is therefore 92.95%.
FIG. 2.
Flow scheme for PCR/ESI-MS genotyping of S. aureus. Isolated DNA is distributed into eight wells of a microtiter plate, allowing 12 samples to be analyzed per plate. Each reaction mixture contains a pair of primers targeted to a gene used in MLST. Following PCR and desalting, amplicons are analyzed by ESI-MS. Amplicon masses are used to calculate base compositions—the A, G, C, and T counts—of the PCR products. Comparison to a database of calculated base compositions derived from the sequences in the MLST database allows assignment of clonal complexes and USA types.
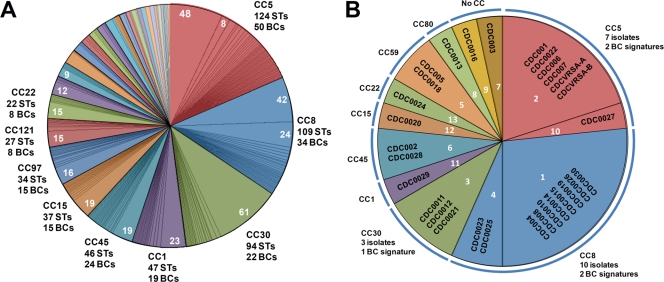
Because base composition analysis is not as information rich as sequencing, a theoretical analysis of the S. aureus MLST database was conducted to determine the resolving power of base composition analysis compared with that of sequencing. Each of the STs in the database were analyzed by calculating the expected base compositions of each of the primer pair target sites used in the PCR/ESI-MS assay and concatenating the base composition signatures from the alleles to create a theoretical PCR/ESI-MS type for each MLST sequence type. Figure 3A shows the comparison of sequence types and PCR/ESI-MS types of the S. aureus MLST database organized by clonal complexes.
FIG. 3.
Distribution of PCR/ESI-MS base composition signatures compared to that of clonal complexes determined with MLST STs. (A) The distribution of STs in clonal complexes (CCs) is shown as colored pie slices. Subdivisions within each clonal complex represent the distribution of theoretical base composition signatures for the PCR/ESI-MS S. aureus genotyping panel. Labels outside the chart indicate the number of MLST types assigned to that complex and the number of distinct base composition signatures (BCs). Numbers within the pie slices indicate the number of distinct STs yielding the same theoretical base composition signature. Minor unlabeled complexes are, in clockwise order from the first (orange) unlabeled pie slice, as follows: 59, 89, 25, 80, 398, 12, 7, 20, 50, 101, 96, 130, 395, 705, 49, 126, 182, 295, and 305. These groups contain from three to 13 STs and from one to five base composition signatures each. (B) The distribution of characterized isolates is shown as differently colored pie slices. The distribution of CDC isolates into different PCR/ESI-MS base composition signatures is shown by segmentation of the pie slices, and the PCR/ESI-MS number groupings are indicated in white. Each set of CDC isolates sharing a specific base composition signature is displayed within a segment. In all cases, PCR/ESI-MS base composition signatures correlated with sequence types belonging to only one clonal complex. There were isolated cases where a base composition signature was also consistent with STs not assigned to a clonal complex: signature 1 is consistent with STs 380 and 619; signature 4 is consistent with ST 157; signature 2 is consistent with ST 529; signature 3 is consistent with STs 42, 353, and 847; signature 5 is consistent with ST 559; signature 11 is consistent with STs 339 and 774; and signature 13 is consistent with ST 498.
The PCR/ESI-MS base composition analysis cannot distinguish the order of nucleotides in the sequence, and the entire region sequenced in MLST is not encompassed within the PCR/ESI-MS amplicon. Therefore, there was typically more than one sequence type consistent with each PCR/ESI-MS type. For example, clonal complex 5, the most abundantly populated clonal complex in the MLST database, comprises 124 sequence types that reduce to 50 PCR/ESI-MS types. Clonal complex 8, the next-most-abundant clonal complex, comprises 109 sequence types that produce 34 unique PCR/ESI-MS types. Although PCR/ESI-MS analysis does not unambiguously assign an isolate to a unique sequence type, 98.8% of 682 STs that were assigned to clonal complexes by eBurst analysis had base composition signatures that correlated with only the correct clonal complex. There were only four pairs of STs (of 682 assigned to clonal complexes) where the theoretical PCR/ESI-MS types were consistent with more than one clonal complex (Fig. 3A). Thus, while PCR/ESI-MS provides less information than sequence type, assignments of STs to clonal complexes upon the basis of theoretical base composition signatures were in agreement with MLST assignments for 99% of analyzed sequence types. The PCR/ESI-MS technique also provides a substantial degree of resolution within each complex (as shown by the pie slices in Fig. 3A).
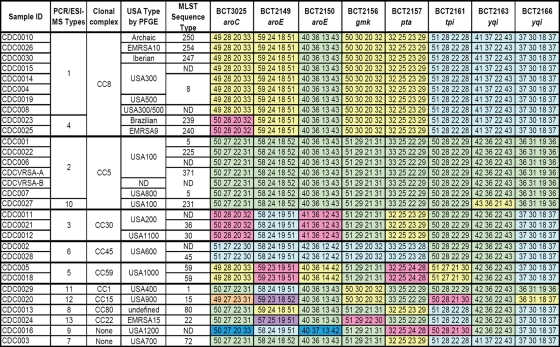
PCR/ESI-MS was then used to analyze 30 blinded isolates of S. aureus isolates from the CDC that had previously been characterized by PFGE and MLST (21). Twenty-eight of the 30 isolates had previously been associated with clonal complexes. One of the 30 isolates, a methicillin-susceptible S. aureus isolate had not been characterized by MLST. The distribution of isolates by clonal complexes is shown in Fig. 3B, and the detailed PCR/ESI-MS signatures are shown in Fig. 4. As anticipated from the theoretical analysis of the MLST database, when PCR/ESI-MS was used, all isolates were correctly assigned to clonal complexes and were sometimes parsed further within the complex. For example, clonal complex 8 had two PCR/ESI-MS types and eight PFGE types. PFGE is known to parse S. aureus isolates more finely than MLST. For example, USA300 and USA500 are both MLST type 8; these PFGE types were not resolved by PCR/ESI-MS and are also sometimes difficult to distinguish based only on PFGE (B. Limbago, unpublished results). Thus, PCR/ESI-MS provided resolution at the level of clonal complexes for the diverse CDC S. aureus panel, with somewhat finer subdividing capability within a clonal complex.
FIG. 4.
PCR/ESI-MS signatures of the CDC panel of S. aureus isolates. The eight columns on the right show base compositions (given as the numbers of A, G, C, and T counts) of amplicons from the MLST target genes (Table 1). The results shown in the clonal complex, PFGE type, and MLST sequence type columns were determined previously. Base compositions that are the same within a column are colored the same. ND, not determined.
The PCR/ESI-MS approach using a diverse set of isolates from the CDC was successfully validated as described above. Subsequently, analysis was performed using 47 unique isolates cultured from clinical specimens that included wounds, abscesses, drainages, sputum samples, cardiovascular catheter tips, and tissues collected at the University of Arizona Medical Center. The purpose of this sample set was to evaluate the correlation of PCR/ESI-MS with an independent typing method (rep-PCR in this case) in an outbreak setting. Another 240 S. aureus isolates obtained from Johns Hopkins Hospital in Maryland were also analyzed by PCR/ESI-MS. These isolates represented clinical strains recovered from patients from various sources, including blood cultures, upper- and lower-respiratory-tract specimens, wounds, sterile body fluids, tissues, and intravascular catheter tips, and were chosen to evaluate the PCR/ESI-MS method in a diverse set of samples acquired in the context of a local setting with high MRSA endemicity.
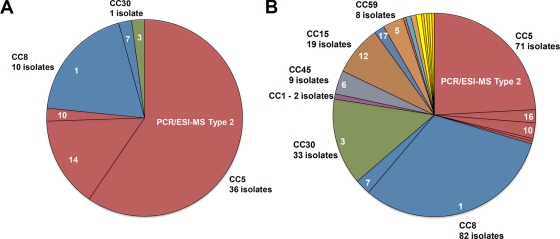
The distribution of clonal complexes and PCR/ESI-MS genotypes for the Arizona isolates are shown in Fig. 5A. The Arizona isolates were predominantly clonal complex 5 (36 isolates), which were subdivided into PCR/ESI-MS types 2, 14, and 10 (Fig. 5A), and clonal complex 8 (10 isolates), comprising PCR/ESI-MS types 1 and 7. The 240 Johns Hopkins Hospital isolates (Fig. 5B) were also predominantly of clonal complex 8 (82 isolates), subdivided into PCR/ESI-MS types 1 and 7, and clonal complex 5 (71 isolates), subdivided into PCR/ESI-MS types 2, 16, 10, 19, and 23. There were also a substantial number of representatives from clonal complexes 30, 15, 45, and 59 and smaller numbers of isolates from other clonal complexes (for minor genotypes, see the legend to Fig. 5B). Thus, most of the hospital isolates were from major clonal complex 5, which corresponds to USA100 genotypes, and from clonal complex 8, corresponding to the USA300/500 genotypes.
FIG. 5.
Genotypes and assignment into clonal complexes by PCR/ESI-MS of Arizona and Johns Hopkins Hospital isolates. (A) Distribution of clonal complexes (colors) and PCR/ESI-MS types for 47 isolates from the University of Arizona. (B) Distribution of clonal complexes (CCs) and PCR/ESI-MS types for 240 isolates from Johns Hopkins Hospital. The unlabeled minor isolates were CC97/ESI-MS type 17 (four isolates); ESI-MS type 9 (two isolates); two isolates each of CC12/ESI-MS type 18, CC1/ESI-MS type 21, and CC7/ESI-MS type 24; and one isolate each of ESI-MS types 11, 20, 22, 25, 27, and 28 (not assigned to clonal complexes).
To compare PCR/ESI-MS with gel mobility-based methods, a subset of 37 Arizona isolates was analyzed by rep-PCR, and 87 isolates from Johns Hopkins Hospital were analyzed by PFGE. Of the 37 Arizona isolates, 26 were characterized as PCR/ESI-MS type 2 or 14, which correspond to the common hospital-acquired USA100 strain type, and 9 isolates were characterized as PCR/ESI-MS type 1, which corresponds to USA300/500 (which cannot be distinguished by PCR/ESI-MS or MLST, as described earlier). The remaining two isolates were classified by PCR/ESI-MS as types 3 and 7, which correspond to USA200/1100 and USA700, respectively (Fig. 6A, inner ring). By rep-PCR, 23 of the 26 isolates determined to be USA100 by PCR/ESI-MS were a match to USA100 (Fig. 6A, outer ring), 9 isolates were characterized by PCR/ESI-MS as USA300/500, and 6 isolates were characterized as USA300 or USA300-like by rep-PCR. The remaining seven isolates did not match a sufficient number of bands in the DiversiLab database to be identified; therefore, the rep-PCR data from these isolates could not be compared with PCR/ESI-MS data. The isolate identified by PCR/ESI-MS as type 3(USA200) was discrepant with rep-PCR. Thus, when they could be compared, the PCR/ESI-MS and gel mobility-based methods were in agreement, with a single exception.
FIG. 6.
Comparison of PCR/ESI-MS with rep-PCR and PFGE. (A) Comparison of PCR/ESI-MS and rep-PCR (Arizona isolates). The inner ring represents PCR/ESI-MS results, and the outer ring represents the corresponding isolates identified by rep-PCR. The unlabeled isolates in the inner ring (clockwise) were one each of PCR/ESI-MS 3 (USA700, orange) and PCR/ESI-MS type 7 (USA200/1100, green). The gray isolates in the outer ring (labeled NM) had no match in the DiversiLab database. The unlabeled blue isolate in the outer ring was USA200-like by rep-PCR and corresponded to USA200/1100 by PCR/ESI-MS. (B) Comparison of PCR/ESI-MS and PFGE (Johns Hopkins Hospital isolates). Red slices represent USA100, medium blue (inner ring) represents USA300/500, and light blue and dark blue (outer ring) represent USA300 and USA500, respectively. The isolates unlabeled by PCR/ESI-MS (inner ring) were USA200 USA1100 (green, three isolates), USA700 (yellow, two isolates), USA1000 (purple, one isolate), and undetermined isolates (gray, three isolates). The corresponding data for isolates unlabeled by PFGE indicated, in clockwise order, USA200 (green, three isolates), USA700 (yellow, two isolates), one unique PFGE pattern (gray) corresponding to the isolate correlated with USA1000 by PCR/ESI-MS, USA800 (orange, one isolate), and two unique PFGE types (gray) corresponding to types undetermined by PCR/ESI-MS.
The 87 isolates from Johns Hopkins were genotyped using PFGE by comparison of the pulsed-field patterns with those of reference strains (see above). Agreement between PCR/ESI-MS and PFGE was 100% for the USA100 (35 isolates) and USA300/500 (43 isolates) (Fig. 6B). Of the four remaining isolates, three showed unique patterns in PFGE and the fourth was one that was not assigned to a clonal complex by PCR/ESI-MS.
DISCUSSION
S. aureus is a devastating health care-associated infectious agent that leads to widespread mortality and morbidity (3, 18, 25). New tools are needed to aid in its identification and management. Molecular genotyping tools are useful for managing infections in health care settings, as they provide a method for tracking of outbreaks. These techniques may also be useful for prediction of the phenotypic properties of an infectious agent from the genotype. For example, the USA300 clone has been extensively studied with respect to pathogenicity and antimicrobial susceptibility (17, 22, 26); it is often associated with aggressive infections (18). The knowledge that an infection is caused by a particular genotype can be helpful when making treatment decisions, provided that accurate information can be obtained rapidly.
MRSA has been extensively studied using a variety of molecular genotyping tools. Robinson and Enright conducted MLST analysis of a large collection of isolates from around the world and established the evolutionary history of MRSA (23). They determined that there are 11 major MRSA clones that fall into five major groups of related genotypes, designated clonal complexes (11). MLST is particularly useful for molecular evolution studies because it provides unambiguous digital readouts that are portable over the Internet and amenable to database population. However, MLST requires multiple PCRs, sequencing, and multiple sequence analysis; these techniques are difficult to perform and validate in a clinical laboratory setting.
L. K. McDougal and colleagues from the CDC conducted an extensive PFGE analysis of S. aureus clones from around the United States and determined that there were eight major lineages, designated PFT USA100 through USA800, categorizations that have now been extended to the USA1200 designation (29). By codetermination of the MLST and PFGE types of these isolates, correlation with the previously established MLST nomenclature was established (21). More recently, CDC scientists directly characterized a set of specific genetic elements found in MRSA strains that cause most community-acquired MRSA infections. These strains are designated USA300. The relevant genetic elements are found in MLST type 8 isolates and contain SCCmec type IV and the Panton-Valentine leukocidin toxin gene (28). Thus, the CDC approach to characterization of strains combines general molecular genotyping methods (i.e., PFGE) with direct molecular detection and typing of genetic elements that are characteristic of MRSA strains. Coupling of general genotyping methods with direct analysis of specific genetic elements is necessary for an accurate view of the strain genealogy because gene transfer between different strains of microbes and acquisition of mobile genetic elements occur over relatively short time intervals.
The clinical utility of a molecular genotyping or characterization method depends upon the value of the information that it provides as well as practical issues, such as technical difficulty, labor requirements, speed, throughput, and portability of data. Knowing the detailed molecular characteristics of an isolate can guide intervention decisions during an outbreak and enable prudent use of infection control resources. Despite some debate, pending health care legislation aimed at health care-associated infections may necessitate the ability to prove or disprove hospital-based acquisition in order to obtain insurance reimbursement (31).
Although base composition analysis as described here does not achieve the strain resolution of PFGE analysis, the PCR/ESI-MS approach described here is simple, straightforward, and automated and can correctly assign isolates to clonal complexes with a high degree of agreement (∼99%) with MLST analysis. It could be argued that both PCR/ESI-MS and MLST analyses lack discriminatory ability relative to PFGE. However, this does not make a strong statement, given that, for both MLST and PCR/ESI-MS, multiple loci, each displaying multiple alleles, are combined for inference of types. The probability of two unrelated genotypes having the same sequence type is low (11). It may even happen that the rapid accumulation of genetic variation that can be indexed by PFGE could lead to differences in PFGE patterns among the descendants of ancestral genotypes, resulting in some S. aureus clones being unnecessarily subdivided.
Here, we describe a new, high-throughput method for genotyping of S. aureus that is similar to traditional MLST strain typing but provides analysis of PCR amplicons via ESI-MS rather than standard DNA sequencing. The base compositions of the PCR amplicons, determined from PCR product masses, provide a high-resolution digital signature that is directly related to sequences of known genotypes. Digital genomic signatures, such as a nucleotide sequence or base composition of a selected region of a microbe's genome, are precise and comparable among laboratories (5, 8, 12). Another advantage of the PCR/ESI-MS method is automation and instrument throughput: a specimen is analyzed in 6 min of mass spectrometry time, and 180 samples can be analyzed per day. The instrument used in the PCR/ESI-MS studies is a prototype and not the version expected to be placed in clinical microbiology laboratories. This paper reports only on the utility of the genotyping component of the assay. The assay will also have the capability of identifying staphylococci directly from clinical samples while simultaneously allowing for strain characterization, using a multiplexed assay. The ability of the assay to identify staphylococci to the species level while also determining resistance genes such a mecA and mupA and virulence factors such as Panton-Valentine leukocidin will be reported in a separate paper (D. Wolk, L. Blyn, T. Hall, R. Sampath, R. Renken, C. Ivy, R. Melton, H. Matthews, N. White, F. Li, V. Harpin, D. Ecker, B. Limbago, L. M. McDougal, V. Wysock, M. Cai, and K. Carroll, submitted for publication).
In this report, we first analyzed a diverse panel of MRSA isolates obtained from a national surveillance study containing representative clonal complexes and PFGE groups (21, 28). The PCR/ESI-MS method correctly grouped all the isolates into clonal complexes previously assigned by MLST and provided additional resolution within the clonal complexes. When used to analyze two sets of MRSA isolates from hospitals in different geographic locations, PCR/ESI-MS grouped the isolates in excellent agreement with complexes determined by PFGE (26) or rep-PCR. Each of these methods measures different genetic variations in S. aureus. However, since all three methods measure signals grounded in the DNA sequences of the microbes, related isolates should generally group together, as was observed. Here, we focused on genotyping, but the PCR/ESI-MS method is also amenable to more-targeted genomic analysis of S. aureus, such as determination of the presence or absence of virulence factors or acquired genes or mutations that mediate antibiotic resistance (Wolk et al., submitted).
In summary, our PCR/ESI-MS approach to genotyping of S. aureus follows the logic of MLST but uses mass spectrometry to analyze the PCR products instead of sequencing. The method correctly assigned 100% of the isolates from a CDC validation panel and two collections of hospital isolates into the clonal complexes. The PCR/ESI-MS approach provides a rapid, high-throughput, and automated method of genotyping S. aureus isolates suitable for use in a clinical or public-health setting.
Acknowledgments
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the CDC.
The work at the University of Arizona was partially supported by grant AI065359.
Footnotes
Published ahead of print on 18 March 2009.
REFERENCES
- 1.Blanc, D. S. 2004. The use of molecular typing for epidemiological surveillance and investigation of endemic nosocomial infections. Infect. Genet. Evol. 4193-197. [DOI] [PubMed] [Google Scholar]
- 2.Blanc, D. S., C. Petignat, A. Wenger, G. Kuhn, Y. Vallet, D. Fracheboud, S. Trachsel, M. Reymond, N. Troillet, H. H. Siegrist, S. Oeuvray, M. Bes, J. Etienne, J. Bille, P. Francioli, and G. Zanetti. 2007. Changing molecular epidemiology of methicillin-resistant Staphylococcus aureus in a small geographic area over an eight-year period. J. Clin. Microbiol. 453729-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucher, H. W., and G. R. Corey. 2008. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 46(Suppl. 5)S344-S349. [DOI] [PubMed] [Google Scholar]
- 4.Caugant, D. A. (ed.) Molecular epidemiology of microorganisms, in press. Methods in molecular biology, vol. 551. Humana Press, Totowa, NJ.
- 5.Ecker, D. J., and K. C. Carroll. 2005. Investments in high-payoff technologies could reduce toll of infections. ASM News 71576-581. [Google Scholar]
- 6.Ecker, D. J., J. Drader, J. Gutierrez, A. Gutierrez, J. Hannis, A. Schink, R. Sampath, J. A. Ecker, L. B. Blyn, M. W. Eshoo, T. A. Hall, M. Tobarmosquera, Y. Jiang, K. Sannes-Lowery, L. Cummins, B. Libby, D. J. Walcott, C. Massire, R. Ranken, S. M. Manalili, C. Ivy, R. Melton, H. Levene, V. Harpin, F. Li, N. White, M. Pear, V. Samant, D. Knize, D. Robbins, K. Rudnick, F. Hajjar, and S. A. Hofstadler. 2006. The Ibis T5000 universal biosensor—an automated platform for pathogen identification and strain typing. JALA 11341-351. [Google Scholar]
- 7.Ecker, D. J., R. Sampath, L. B. Blyn, M. W. Eshoo, C. Ivy, J. A. Ecker, B. Libby, V. Samant, K. Sannes-Lowery, R. E. Melton, K. Russell, N. Freed, C. Barrozo, J. Wu, K. Rudnick, A. Desai, E. Moradi, D. J. Knize, D. W. Robbins, J. C. Hannis, P. M. Harrell, C. Massire, T. A. Hall, Y. Jiang, R. Ranken, J. J. Drader, N. White, J. A. McNeil, S. T. Crooke, and S. A. Hofstadler. 2005. Rapid identification and strain-typing of respiratory pathogens for epidemic surveillance. Proc. Natl. Acad. Sci. USA 1028012-8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ecker, D. J., R. Sampath, C. Massire, L. B. Blyn, T. A. Hall, M. W. Eshoo, and S. A. Hofstadler. 2008. Ibis T5000: a universal biosensor approach for microbiology. Nat. Rev. Microbiol. 6553-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ecker, J. A., C. Massire, T. A. Hall, R. Ranken, T. T. Pennella, C. Agasino Ivy, L. B. Blyn, S. A. Hofstadler, T. P. Endy, P. T. Scott, L. Lindler, T. Hamilton, C. Gaddy, K. Snow, M. Pe, J. Fishbain, D. Craft, G. Deye, S. Riddell, E. Milstrey, B. Petruccelli, S. Brisse, V. Harpin, A. Schink, D. J. Ecker, R. Sampath, and M. W. Eshoo. 2006. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J. Clin. Microbiol. 442921-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 381008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 997687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7482-487. [DOI] [PubMed] [Google Scholar]
- 13.Hannis, J. C., S. M. Manalili, T. A. Hall, R. Ranken, N. White, R. Sampath, L. B. Blyn, D. J. Ecker, R. E. Mandrell, C. K. Fagerquist, A. H. Bates, W. G. Miller, and S. A. Hofstadler. 2008. High-resolution genotyping of campylobacter species by use of PCR and high-throughput mass spectrometry. J. Clin. Microbiol. 461220-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Healy, M., J. Huong, T. Bittner, M. Lising, S. Frye, S. Raza, R. Schrock, J. Manry, A. Renwick, R. Nieto, C. Woods, J. Versalovic, and J. R. Lupski. 2005. Microbial DNA typing by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 43199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Healy, M., K. Reece, D. Walton, J. Huong, S. Frye, I. I. Raad, and D. P. Kontoyiannis. 2005. Use of the DiversiLab System for species and strain differentiation of Fusarium species isolates. J. Clin. Microbiol. 435278-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofstadler, S. A., R. Sampath, L. B. Blyn, M. W. Eshoo, T. A. Hall, Y. Jiang, J. J. Drader, J. C. Hannis, K. A. Sannes-Lowery, L. L. Cummins, B. Libby, D. J. Walcott, A. Schink, C. Massire, R. Ranken, N. White, V. Samant, J. A. McNeil, D. Knize, D. Robbins, K. Rudnik, A. Desai, E. Moradi, and D. J. Ecker. 2005. TIGER: the universal biosensor. Int. J. Mass Spectrom. 24223-41. [Google Scholar]
- 17.King, M. D., B. J. Humphrey, Y. F. Wang, E. V. Kourbatova, S. M. Ray, and H. M. Blumberg. 2006. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann. Intern. Med. 144309-317. [DOI] [PubMed] [Google Scholar]
- 18.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, and S. K. Fridkin. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2981763-1771. [DOI] [PubMed] [Google Scholar]
- 19.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koreen, L., S. V. Ramaswamy, S. Naidich, I. V. Koreen, G. R. Graff, E. A. Graviss, and B. N. Kreiswirth. 2005. Comparative sequencing of the serine-aspartate repeat-encoding region of the clumping factor B gene (clfB) for resolution within clonal groups of Staphylococcus aureus. J. Clin. Microbiol. 433985-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 415113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moran, G. J., A. Krishnadasan, R. J. Gorwitz, G. E. Fosheim, L. K. McDougal, R. B. Carey, and D. A. Talan. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355666-674. [DOI] [PubMed] [Google Scholar]
- 23.Robinson, D. A., and M. C. Enright. 2004. Multilocus sequence typing and the evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 1092-97. [DOI] [PubMed] [Google Scholar]
- 24.Sampath, R., S. A. Hofstadler, L. Blyn, M. Eshoo, T. Hall, C. Massire, H. Levene, J. Hannis, P. M. Harrell, B. Neuman, M. J. Buchmeier, Y. Jiang, R. Ranken, J. Drader, V. Samant, R. H. Griffey, J. A. McNeil, S. T. Crooke, and D. J. Ecker. 2005. Rapid identification of emerging pathogens: coronavirus. Emerg. Infect. Dis. 11373-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spellberg, B., R. Guidos, D. Gilbert, J. Bradley, H. W. Boucher, W. M. Scheld, J. G. Bartlett, and J. Edwards, Jr. 2008. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 46155-164. [DOI] [PubMed] [Google Scholar]
- 26.Stevens, D. L., A. L. Bisno, H. F. Chambers, E. D. Everett, P. Dellinger, E. J. Goldstein, S. L. Gorbach, J. V. Hirschmann, E. L. Kaplan, J. G. Montoya, and J. C. Wade. 2005. Practice guidelines for the diagnosis and management of skin and soft-tissue infections. Clin. Infect. Dis. 411373-1406. [DOI] [PubMed] [Google Scholar]
- 27.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover, F. C., L. K. McDougal, R. V. Goering, G. Killgore, S. J. Projan, J. B. Patel, and P. M. Dunman. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenover, F. C., R. R. Vaughn, L. K. McDougal, G. E. Fosheim, and J. E. McGowan, Jr. 2007. Multiple-locus variable-number tandem-repeat assay analysis of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 452215-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 196823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber, S. G., S. S. Huang, S. Oriola, W. C. Huskins, G. A. Noskin, K. Harriman, R. N. Olmsted, M. Bonten, T. Lundstrom, M. W. Climo, M. C. Roghmann, C. L. Murphy, and T. B. Karchmer. 2007. Legislative mandates for use of active surveillance cultures to screen for methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci: position statement from the Joint SHEA and APIC Task Force. Infect. Control Hosp. Epidemiol. 28249-260. [DOI] [PubMed] [Google Scholar]
- 32.Woods, C. R., Jr., J. Versalovic, T. Koeuth, and J. R. Lupski. 1992. Analysis of relationships among isolates of Citrobacter diversus by using DNA fingerprints generated by repetitive sequence-based primers in the polymerase chain reaction. J. Clin. Microbiol. 302921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]