Abstract
The new GenoType Mycobacterium tuberculosis drug resistance second line (MTBDRsl) assay (Hain Lifescience, Nehren, Germany) was tested on 106 clinical isolates and directly on 64 sputum specimens for the ability to detect resistance to fluoroquinolones, injectable drugs (amikacin or capreomycin), and ethambutol in Mycobacterium tuberculosis strains. A total of 63 strains harboring fluoroquinolone, amikacin/capreomycin, or ethambutol resistance and 43 fully susceptible strains were comparatively analyzed with the new MTBDRsl assay, by DNA sequencing, and by conventional drug susceptibility testing in liquid and solid media. No discrepancies were obtained in comparison with the DNA sequencing results. Fluoroquinolone resistance was detected in 29 (90.6%) of 32, amikacin/capreomycin resistance was detected in 39/39 (84.8%/86.7%) of 46/45, and ethambutol resistance was detected in 36 (69.2%) of 52 resistant strains. A total of 64 sputum specimens (42 smear positive, 12 scanty, and 10 smear negative) were tested with the new MTBDRsl assay, and the results were compared with those of conventional drug susceptibility testing. Fluoroquinolone resistance was detected in 8 (88.9%) of 9, amikacin/capreomycin resistance was detected in 6/7 (75.0%/87.5%) of 8, and ethambutol resistance was detected in 10 (38.5%) of 26 resistant strains. No mutation was detected in susceptible strains. The new GenoType MTBDRsl assay represents a reliable tool for the detection of fluoroquinolone and amikacin/capreomycin resistance and to a lesser extent also ethambutol resistance. In combination with a molecular test for detection of rifampin and isoniazid resistance, the potential for the detection of extensively resistant tuberculosis within 1 to 2 days can be postulated.
The worldwide emergence of extensively drug-resistant tuberculosis (XDR TB, resistant at least to rifampin and isoniazid, a fluoroquinolone [FLQ], and one of the three injectable second-line drugs amikacin [AM], kanamycin [KM], and capreomycin [CM]) is a serious global health problem (20, 25). In the World Health Organization fourth global report on drug resistance (25), it was documented that more than 45 countries have reported XDR cases. The actual incidence could be underestimated, because second-line drug susceptibility testing (DST) is not available in many countries. To avoid a progressive development similar to that observed in multidrug-resistant TB (resistant at least to rifampin and isoniazid) worldwide, now having the highest rate ever at 5.3%, timely identification of resistant Mycobacterium tuberculosis complex (MTBC) strains is mandatory.
Conventional DST for XDR strains is performed sequentially in a two-step procedure beginning with a culture and first-line drug testing, proceeding to further drug testing in the case of multidrug resistance. The time needed for testing, even with the most rapid liquid methods, is still around 1 week per test, constrained by the relatively slow growth of M. tuberculosis (15, 18). The required time can be shortened by fast molecular methods to 1 day per test (3, 8, 21). Since recently broad-based knowledge about mutations that cause resistance to ethambutol (EMB) and some second-line drugs is available. Resistance to FLQs, AM-CM, and EMB in M. tuberculosis is most frequently attributed to mutations in the gyrA, rrs, and embB genes, respectively. First investigations have shown that by targeting mutations in codons 90, 91, and 94 in the gyrA gene, approximately 70 to 90% of all FLQ-resistant strains can be correctly detected (2, 13, 24). Previous reports have linked mutations A1401G, C1402T, and G1484T in the rrs gene to AM, CM, and KAN resistance (1, 11, 12), each of them being responsible for a specific resistance pattern. Mutations G1484T and A1401G were found to cause high-level resistance to all drugs, whereas C1402T causes resistance to only CM and KAN.
Furthermore, mutations at embB codon 306 are found in 30 to 68% of EMB-resistant clinical strains (16, 17, 26).
PCR-based techniques provide new possibilities for the rapid diagnosis of first- and second-line drug resistance; however, not all mycobacterial laboratories have access to DNA-sequencing facilities. As an alternative, DNA strip assays for the detection of rifampin (INNO-LiPA Rif. TB; Innogenetics, Ghent, Belgium) or rifampin and isoniazid resistance of M. tuberculosis in a single assay (GenoType MTBDR; Hain Lifescience, Nehren, Germany) are now commercially available. These assays have been evaluated for M. tuberculosis cultures and specimens (3, 7, 8, 10, 21). The DNA strip assays are based on PCR or multiplex PCR in combination with reverse hybridization. The existence of a resistant strain is signaled either by the omission of a wild-type band or the appearance of bands representing specific mutations.
In order to increase the capacity to detect further drug resistance in M. tuberculosis, the GenoType Mycobacterium tuberculosis drug resistance second line (MTBDRsl) assay was developed with a specific focus on the most prevalent gyrA, rrs, and embB gene mutations.
The aim of the present study was to determine the sensitivity and specificity of the new MTBDRsl assay for the detection of FLQ, AM, CM, and EMB resistance-associated mutations in culture specimens and directly in smear-positive and -negative clinical specimens.
MATERIALS AND METHODS
Strains.
A set of 63 M. tuberculosis clinical isolates selected for FLQ, AM, CM, and/or EMB resistance was analyzed. The DNA preparation method used is described elsewhere (7). As a control, 43 randomly chosen, previously characterized, fully susceptible MTBC strains were used.
Specimens.
Sixty-four sputum specimens routinely sent to the National Reference Laboratory in Borstel, Germany, were chosen by being either acid-fast bacterium positive or acid-fast bacterium negative but culture positive. Specimens were processed by the conventional N-acetyl-l-cysteine-NaOH method (1% final NaOH concentration) (5). After decontamination, the concentrated sediments were suspended in 1.0 to 1.5 ml sterile phosphate buffer (pH 7.0) and smears were prepared by the Ziehl-Neelsen staining method (9). After inoculation onto solid medium and into liquid medium for growth detection, the leftover sediments of the decontaminated sputum specimens were stored at −20°C. After growth of cultures and, in case of MTBC, identification, species differentiation and DST were performed. The leftover sediments of the chosen specimens were thawed and used for MTBDRsl testing. Sample volumes of 500 μl were centrifuged for 15 min at 10,000 × g, the supernatant was discarded, and the pellet was resuspended in 100 μl distilled water. Subsequently, the suspension was boiled for 20 min and incubated in a sonic water bath at room temperature for 15 min.
Identification of MTBC from clinical specimens.
For the identification and differentiation of MTBC strains from the grown cultures, the GenoType MTBC assay was performed according to the instructions of the manufacturer.
DST.
DST for ofloxacin (OFL), AM, CM, and EMB was performed by the BACTEC MGIT 960 method (MGIT 960; Becton Dickinson Diagnostic Systems, Sparks, MD) and the proportion method on Löwenstein-Jensen medium (LJ) (4). Tests were performed with the standard critical concentrations for OFL (2 μg/ml for MGIT and LJ), AM (1 μg/ml for MGIT and 40 μg/ml for LJ), CM (2.5 μg/ml for MGIT and 40 μg/ml for LJ), and EMB (5 μg/ml for MGIT and 2 μg/ml for LJ). In the case of any results discrepant with respect to those of the molecular assay, DST was repeated by the MGIT method and with determination of the MICs on LJ (4).
Genotypic characterization.
All 106 strains derived from cultures were analyzed by DNA sequencing of the key regions involved in the development of resistance (gyrA, rrs, and embB). Primers gyrA-F (5′-GATGACAGACACGACGTTGC-3′) and gyrA-R (5′-GGGCTTCGGTGTACCTCAT-3′) target a 398-bp fragment of the M. tuberculosis H37Rv gyrA gene (19, 22). Primers rrs F 5′-AAACCTCTTTCACCATCGAC-3′ and rrs R 5′-GTATCCATTGATGCTCGCAA-3′ were used to amplify a 1,329-bp fragment of the rrs (16S rRNA) gene, and primer PR31 5′-GTTCGGATCGGGGTCTGCAA-3′, located at positions 1291 to 1310, was used for DNA sequencing of the relevant region (19). PCR primers OG240 (5′-CGTTCCGGCCTGCAT-3′) and OG243 (5′-CACCTCACGCGACAGCA-3′) were used to analyze an amplified 344-bp fragment of the embB gene (nucleotide positions 764 to 1107) (16).
The sequences of the target genes in M. tuberculosis H37Rv are listed in the GenBank database with accession no. NC_000962, L27512 (gyrA), Z83862 (rrs), and U68480 (embB).
Direct sequencing of PCR products was carried out with an ABI Prism 3100 capillary sequencer (Applied Biosystems, Foster City, CA) and the ABI Prism BigDye Terminator kit v.1.1 according to the manufacturer's instructions. In the case of the 64 clinical sputum specimens, sequencing of specific DNA fragments was performed only if any discrepancy was observed between the conventional DST and the MTBDR assays.
GenoType MTBC and MTBDRsl assays.
For all samples derived from culture suspensions, the strip assays were performed as recommended by the manufacturer. Briefly, for amplification, 35 μl of a primer-nucleotide mixture (provided with the kit), amplification buffer containing 2.5 mM MgCl2, 1.25 U Hot Start Taq DNA polymerase (Qiagen, Hilden, Germany), and 5 μl of the preparation of mycobacterial DNA in a final volume of 50 μl were used. The amplification protocol consisted of 15 min of denaturing at 95°C; 10 cycles comprising 30 s at 95°C and 120 s at 58°C; an additional 20 cycles comprising 25 s at 95°C, 40 s at 53°C, and 40 s at 70°C; and a final extension at 70°C for 8 min. Hybridization and detection were performed in an automated washing and shaking device (Profiblot; Tekan, Maennedorf, Switzerland). Steps taken to avoid amplicon contamination were manual pipetting of the amplicon, use of separate wells and tubes for each stripe, and extensive rinsing after each use. The program was started after mixing 20 μl of the amplification products with 20 μl of denaturing reagent (provided with the kit) for 5 min in separate troughs of a plastic well. Automatically, 1 ml of prewarmed hybridization buffer was added, followed by a stop to put the membrane strips into each trough. The hybridization procedure was performed at 45°C for 0.5 h, followed by two washing steps. For colorimetric detection of hybridized amplicons, streptavidin conjugated with alkaline phosphatase and substrate buffer was added. After a final washing, strips were air dried and fixed on paper. To control cross contamination, a no-template control was included in each run.
For the sputum specimens, an altered amplification protocol was applied which consisted of 15 min of denaturing at 95°C; 10 cycles comprising 30 s at 95°C and 120 s at 58°C; an additional 30 cycles comprising 25 s at 95°C, 40 s at 53°C, and 40 s at 70°C; and a final extension at 70°C for 8 min. Hybridization and detection were done as described above.
The MTBDRsl strip contains 22 probes, including two amplification and hybridization controls to verify the test procedures. Besides an M. tuberculosis-specific probe, PCR control bands for all targeted regions (gyrA, rrs, and embB) are present. For the detection of FLQ resistance, three gyrA wild-type probes (WT1, WT2, and WT3) encompass the region of the gene encoding amino acids 85 to 97. Six probes (gyrA MUT1 A90V, gyrA MUT2 S91P, gyrA MUT3A D94A, gyrA MUT3B D94N/Y, gyrA MUT3C D94G, gyrA MUT3D D94H) specifically target the most common mutations. For the detection of AM-CM resistance, two probes cover the wild-type region of rrs, while two others (rrs MUT1 and MUT2) are designed to assess nucleotide exchanges A1401G and G1484T. For the detection of EMB resistance, embB wild-type probe WT1 targets embB codon 306, while the embB MUT1A and MUT1B probes were designed to bind to nucleotide exchanges ATG/ATA (M306I) and ATG/GTG (M306V). The omission of a wild-type probe and/or the staining of a mutant probe are indications of a resistant strain.
RESULTS
Results of the GenoType MTBDRsl assay with DNA of OFL-, AM-, CM-, and/or EMB-resistant and pansusceptible M. tuberculosis strains.
A total of 63 multidrug-resistant (MDR) isolates (32 with additional resistance to OFL, 46 with additional resistance to AM, 45 with additional resistance to CM, and 52 with additional resistance to EMB) and 43 pansusceptible isolates previously characterized by conventional DST (Table 1) were included in this study. All of the strains were tested with the new MTBDRsl assay (Fig. 1), and DNA sequencing of the respective regions in the target genes was considered the reference standard (Table 2). The MTBDRsl assay produced interpretable results for all of the isolates, and the MTBC-specific control band appeared accurately.
TABLE 1.
Resistance patterns of 106 M. tuberculosis strains tested with the GenoType MTBDRsla
| No. of strains | RMP | INH | OFL | AM | CM | EMB |
|---|---|---|---|---|---|---|
| 16 | R | R | R | R | R | R |
| 2 | R | R | R | R | R | S |
| 12 | R | R | R | S | S | R |
| 2 | R | R | R | S | S | S |
| 20 | R | R | S | R | R | R |
| 7 | R | R | S | R | R | S |
| 1 | R | R | S | R | S | R |
| 3 | R | R | S | S | S | R |
| 43 | S | S | S | S | S | S |
R, resistant; S, susceptible; RMP, rifampin; INH, isoniazid. The strains in the first and second lines (bold) are classified, per definitionem, as XDR.
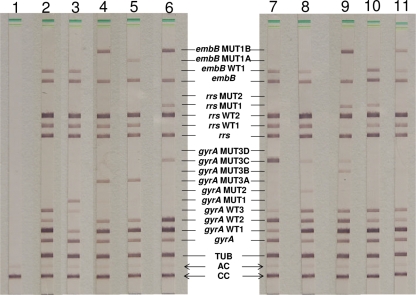
FIG. 1.
Representative patterns of a pansusceptible strain (lane 2), resistant strains (lanes 3 to 6), and mixtures of strains (lanes 7 to 11), as well as a negative control (lane 1). The positions of the oligonucleotides are given. The targeted genes and specificity are shown from the bottom to the top as follows. The MTBDRsl assay: CC, conjugate control; AC, amplification control; TUB, MTBC-specific control; gyrA amplification control; gyrA wild-type probes WT1 to -3 located at codons 85 to 97; gyrA mutant probes MUT1, -2, and -3A to -D with mutations in gyrA codons A90V (GCG-GTG), S91P (TCG-CCG), D94A (GAC-GCC), D94N/Y (GAC-AAC/TAC), D94G (GAC-GGC), and D94H (GAC-CAC); rrs amplification control; rrs wild-type probes WT1 and WT2; rrs mutation probes MUT1 with a A1401G exchange and MUT2 with a G1484T exchange; embB amplification control; embB gene wild-type probe WT1 spanning the region around codon 306; embB mutant probes MUT1A with mutation M306I (ATG-ATA) and MUT1B with mutation M306V (ATG-GTG). The pansusceptible isolate (lane 2) was positive with all WT gyrA, rrs, and embB probes of the MTBDRsl assay. The isolates in lane 3 to 6 showed FLQ resistance by different positive gyrA MUT probes. Two isolates (lanes 4 and 5) show further EMB resistance, and the isolate in lane 6 shows resistance to EMB, AM, and CM. The latter is an XDR strain, since rifampin and isoniazid resistance was also found (data not shown). In lanes 7 to 11, mixtures of strains are shown as follows: lane 7, gyrA WT and gyrA MUT3C; lane 8, gyrA WT, gyrA MUT2, and gyrA MUT3C; lane 9, gyrA WT, gyrA MUT3B, and gyrA MUT3C; lane 10, rrs WT1 and rrs MUT1; lane 11, rrs WT1 and rrs MUT1B.
TABLE 2.
GenoType MTBDRsl test results for the detection of FLQ, AM-CM, and EMB resistance in 106 M. tuberculosis strainsa
| Resistance and MTBDRsl pattern (gyrA, rrs, embB)b | MTBDRsl result | Sequencing data, amino acid change (nucleotide changes)c | DST resultc | No. (%) of strains |
|---|---|---|---|---|
| FLQ | ||||
| gyrA ΔWT3, MUT3C | FlQr | D94G (GAC-GGC) | OFLr | 13 (12.3) |
| gyrA ΔWT3, MUT3A | FlQr | D94A (GAC-GCC) | OFLr | 4 (3.8) |
| gyrA ΔWT2, MUT1 | FlQr | A90V (GCG-GTG) | OFLr | 3 (2.8) |
| gyrA ΔWT3, MUT3B | FlQr | D94N (GAC-AAC) | OFLr | 1 (0.9) |
| gyrA ΔWT2, MUT2 | FlQr | S91P (TCG-CCG) | OFLr | 1 (0.9) |
| gyrA (WT2), MUT1 | FlQr | A90V (GCG-GTG) + WT | OFLr | 1 (0.9) |
| gyrA (WT3), ([MUT3A]) | FlQr | WT + ([D94A]) (GAC-GCC) | OFLr | 1 (0.9) |
| gyrA (WT3), MUT3B + MUT3C | FlQr | D94N/G (GAC-AAC/GGC) + WT? | OFLr | 2 (1.9) |
| gyrA (WT3), MUT1 + MUT3C | FlQr | D94G (GAC-GGC), A90V (GCG-GTG) + WT | OFLr | 1 (0.9) |
| gyrA (WT3), MUT2 + MUT3C | FlQr | D94G (GAC-GGC), S91P (TCG-CCG) + WT | OFLr | 1 (0.9) |
| gyrA ΔWT3, MUT3A + (MUT3B) | FlQr | D94A/Y (GAC-GCC/TAC) + WT? | OFLr | 1 (0.9) |
| gyrA WT | FlQs | WT | OFLr | 3 (2.8) |
| gyrA WT | FlQs | WT | OFLs | 74 (69.8) |
| AM-CM | ||||
| rrs ΔWT1, MUT1 | AMr CMr | A1401G | AMr CMr | 35 (33.0) |
| rrs (WT1), MUT1 | AMr CMr | A1401G + WT | AMr CMr | 3 (2.8) |
| rrs ΔWT1 | AMs CMr | C1402T | AMr CMr | 1 (0.9) |
| rrs WT | AMs CMs | WT | AMr CMr | 6 (5.7) |
| rrs (MUT1) | AMr CMr | A1401G + WT | AMr CMs | 1 (0.9) |
| rrs WT | AMs CMs | WT | AMs CMs | 60 (56.6) |
| EMB | ||||
| embB ΔWT, MUT1B | EMBr | M306V (ATG-GTG) | EMBr | 26 (24.5) |
| embB ΔWT1, MUT1A | EMBr | M306I (ATG-ATA) | EMBr | 7 (6.6) |
| embB MUT1A | EMBr | M306I (ATG-ATA) + WT | EMBr | 1 (0.9) |
| embB ΔWT1 | EMBr | M306I (ATG-ATT) | EMBr | 1 (0.9) |
| embB ΔWT1 | EMBr | M306I (ATG-ATC) | EMBr | 1 (0.9) |
| embB WT | EMBs | WT | EMBr | 16 (15.1) |
| embB WT | EMBs | WT | EMBs | 54 (50.9) |
r, resistant; s, susceptible.
WT, wild-type pattern with all respective bands; ΔWT, omission of the respective band; (WT) or ([WT]), only a weak or very weak band, respectively.
Drug resistance was determined by conventional DST on LJ or MGIT, and in the case of a discrepancy, the DST was repeated with a second method and by determination of the MIC on LJ.
Comparison of the MTBDRsl assay and the DNA sequencing results of the gyrA gene showed no discrepancies. All 74 isolates phenotypically susceptible to OFL showed the wild-type pattern in the MTBDRsl assay. Of the 32 OFL-resistant isolates, 3 (9.4%) had wild-type patterns and were falsely classified as susceptible by the assay. The remaining 29 isolates (90.6%) were correctly identified as FLQ resistant. Of these, 22 had single patterns such as an absent wild-type band combined with a mutant band. The most prevalent mutation was the D94G (GAC-GGC) exchange indicated by the omission of the WT3 band together with the appearance of the MUT3C band (13 [40.6%] of 32), followed by the D94A (GAC-GCC) exchange in the same codon (4 [12.5%] of 32), and the A90V (GCG-GTG) mutation (3 [9.4%] of 32). Noteworthy is the high rate of mixtures showing the wild-type pattern and one or two mutations (7 [21.9%] of 32). In two cases, wild-type and mutant bands were both visible; in four other cases, weak wild-type bands and different combinations of mutant bands came up; and one had two mutant bands visible. A mixture of wild-type and mutant bands indicates heteroresistance. This observation was further supported by a second MTBDRsl test of colonies taken after growth in OFL-containing medium, since the wild-type bands disappeared and the mutant band(s) became stronger. All of these patterns could be unambiguously confirmed by the sequencing results, although in contrast to the hybridization pattern, revealing the actual existing sequence mixtures, this result could not be achieved in some cases from the DNA sequence alone.
With the MTBDRsl assay, all 60 AMs CMs strains showed wild-type patterns related to AM and CM resistance. In 39 (86.7%) of the 45 AMr CMr strains, a difference from the wild-type pattern could be detected, indicating resistance; in only 6 isolates (13.3%) was no mutation found. The A1401G exchange was the most prevalent (35 [77.8%] of 45), followed by mixtures of the wild-type band and the A1401G exchange (6.7%) in three isolates. One isolate showed a mixture of the wild-type band and the A1401G exchange, but only a weak band. With the conventional DST, the result was AMr but CMs. The proportion of predominant wild-type over mutated bacteria could be the reason for the CMs result with the conventional DST, although other reasons cannot be excluded. One isolate showed the omission of the WT1 band which could be specified by DNA sequencing as a C1402T exchange. According to Maus et al. (11), it was classified as AMs CMr, but with the conventional DST, the strain was AMr CMr.
EMB resistance was detected in 36 of 52 resistant isolates (69.2%), in which the M306V (ATG-GTG) exchange was the most prevalent (26 [50.0%] of 52), followed by the M306I (ATG-ATA) exchange in seven cases (13.5%). Two isolates were detected as resistant by the omission of the wild-type band, which could be specified by DNA sequencing showing a different mutation which does not correspond to any mutant band included in the assay. One isolate (1.9%) harbored a mixture showing a wild-type and a mutant band.
Results of the GenoType MTBDRsl obtained with sputum specimens.
A total of 64 specimens (42 smear positive, 12 scanty, and 10 smear negative) were chosen retrospectively for being either smear positive or smear negative but culture positive. The quality of the test strip results using DNA from strains or specimens was comparable, since no difference in the intensity of the hybridization bands could be observed. For the majority of specimens, the MTBDRsl assay produced interpretable results and the MTBC-specific control band appeared accurately (Fig. 1; Table 3). Only three of the gyrA and embB patterns (4.7%) and four of the rrs patterns (6.3%) were not analyzable, and therefore four specimens were excluded from the analysis. The reason for the failure is likely the long storage time of the frozen specimens. For specimens that were stored for more than 1 to 2 months at −20°C, the intensity of the bands became weaker. The sensitivity of the assay (in the sense of getting readable results) could also be a reason for the failure of the test. However, this is unlikely, since the strips of all 12 scanty and 10 smear-negative specimens were clearly interpretable.
TABLE 3.
GenoType MTBDRsl test results for the detection of FLQ, AM-CM, and EMB resistance in 64 smear-positive and smear-negative sputum specimensa
| Resistance and MTBDRsl pattern (gyrA, rrs, embB)b | MTBDRsl result | DST resultc | No. (%) of strains |
|---|---|---|---|
| FLQ | |||
| gyrA ΔWT3, MUT3C | FlQr | OFLr | 4 (6.3) |
| gyrA MUT1 | FlQr | OFLr | 1 (1.6) |
| gyrA MUT1 + 3C | FlQr | OFLr | 1 (1.6) |
| gyrA MUT3A + 3B | FlQr | OFLr | 1 (1.6) |
| gyrA MUT3B + 3C | FlQr | OFLr | 1 (1.6) |
| gyrA WT | FlQs | OFLr | 1 (1.6) |
| gyrA WT | FlQs | OFLs | 51 (79.7) |
| Not analyzable/excluded | None | OFLs | 4 (6.3) |
| AM-CM | |||
| rrs ΔWT1, MUT1 | AMr CMr | AMr CMr | 4 (6.3) |
| rrs (WT1), MUT1 | AMr CMr | AMr CMr | 1 (1.6) |
| rrs ΔWT1, MUT1, MUT2 | AMr CMr | AMr CMr | 1 (1.6) |
| rrs ΔWT1 | AMs CMr | AMr CMr | 1 (1.6) |
| rrs WT | AMs CMs | AMr CMr | 1 (1.6) |
| rrs WT | AMs CMs | AMs CMs | 52 (81.3) |
| Not analyzable/excluded | None | AMs CMs | 3 (4.7) |
| Not analyzable/excluded | None | AMr CMr | 1 (1.6) |
| EMB | |||
| embB ΔWT1, MUT1B | EMBr | EMBr | 7 (10.9) |
| embB ΔWT1, MUT1A | EMBr | EMBr | 2 (3.1) |
| embB MUT1B | EMBr | EMBr | 1 (1.6) |
| embB WT | EMBs | EMBr | 16 (25.0) |
| embB WT | EMBs | EMBs | 34 (53.1) |
| Not analyzable/excluded | None | EMBr | 4 (6.3) |
r, resistant; s, susceptible.
WT, wild-type pattern with all respective bands; ΔWT, omission of the respective band; (WT), only a weak band visible.
Drug resistance was determined by conventional DST on LJ or MGIT, and in the case of a discrepancy, the DST was repeated by a second method and by determination of the MIC on LJ.
All 51 strains phenotypically susceptible to OFL showed the correct wild-type pattern in the MTBDRsl assay. Of nine OFL-resistant isolates, one (11.1%) had wild-type bands and was falsely classified as susceptible by the assay. The remaining eight isolates (88.9%) were correctly identified as FLQ resistant. The most prevalent mutation was the D94G (GAC-GGC) exchange (four [44.4%] of nine). Again noteworthy is the high rate of mixtures (four [44.4%] of nine) indicating heteroresistance. In all cases, both wild-type and mutant bands were visible; in three cases, more than one mutant band came up (Table 3).
With the MTBDRsl assay, all 52 susceptible strains showed wild-type patterns related to AM and CM resistance. In seven (87.5%) of the eight AMr CMr strains, a difference from the wild-type pattern could be detected, indicating resistance; only one isolate (12.5%) had no mutation. The A1401G exchange was the most prevalent (4 [50.0%] of 8). Two isolates harbored mixtures (25.0%): one had a mixture of the wild-type band (a weak band) and the A1401G exchange; another showed two mutations. With the conventional DST, the result was AMr CMr. One isolate was classified as AMs CMr due to the omission of the WT1 band, but with the conventional DST the strain was AMr CMr.
EMB resistance was detected in 10 (38.5%) of 26 resistant isolates, in which the M306V (ATG-GTG) exchange was the most prevalent (7 [70.0%] of 10), followed by the M306I (ATG-ATA) exchange in 2 cases (20.0%) and a mixture of the wild-type pattern and the M306V exchange in 1. The 34 strains phenotypically susceptible to EMB showed correct wild-type patterns in the MTBDRsl assay.
Sensitivity and specificity of the GenoType MTBDRsl assay.
To calculate the sensitivity and specificity of the MTBDRsl assay, the results obtained with culture isolates and sputum specimens were combined. The overall sensitivity for FLQ, AM, CM, and EMB was 90.2, 83.3, 86.8, and 59.0%, respectively. The specificity was 100% for FLQ, AM, and EMB and 99.1% for CM.
DISCUSSION
The present study has shown that the MTBDRsl assay has the ability to detect FLQ-, AM-CM-, and EMB-resistant M. tuberculosis with a single test. As previously shown for other DNA strip assays (3, 7, 8, 23), the MTBDRsl assay has been proven reliable when applied either to culture isolates or directly to smear-positive specimens.
Sensitivities to confirm phenotypic FLQ resistance by identification of the associated gyrA mutations were recently reported to range from 70 to 90% (2, 13, 24). This is in concordance with the high sensitivity of 90.2% observed for the MTBDRsl assay in our study. Furthermore, the frequency of the D94G (GAC-GGC) mutation in gyrA was the highest (17 [41.5%] of 41), which is also comparable to those shown in other reports (13). An interesting finding of our study was the high proportion of heteroresistant isolates, which is in agreement with observations described by other authors (13, 24). In certain isolates, a mixture of wild-type and multiple mutant alleles of gyrA was even observed. Whether specific treatment regimens, individual fitness of wild-type clones and different mutated strains, or mutator (hypermutable) alleles of the DNA repair genes are the reason for this phenomenon remains speculative. Heteroresistance of M. tuberculosis is considered a preliminary stage of full resistance, and the finding of a high rate of heteroresistance may also reflect this prestage in certain settings. In principle, line probe assays are a useful tool to identify hereroresistance, especially compared to sequencing and culture-based DST. But to verify these findings, patients have to be monitored during treatment and clinical data have to be used.
The molecular basis of FLQ resistance is well understood. In contrast, only few studies have investigated the genetic background of AM and CM resistance (1, 11, 12). Sensitivities for the detection of AM-CM resistance by rrs mutations were 87.0 and 88.6% with the MTBDRsl assay in our study, which are in an acceptable range. Future investigations will show whether similar sensitivities will be found in different regions of the world.
Of special interest was the finding that one heteroresistant isolate (wild-type pattern and a weak band indicating a A1401G exchange) showed AM resistance but CM susceptibility compared to conventional DST. Although other reasons cannot be excluded, an explanation might be that the proportion of predominant wild-type bacteria masks the prestage of CM resistance in the conventional DST based on the proportion method. Maus and coworkers suggest that the C1402T mutation is associated with CM resistance and AM susceptibility. This study included one single strain with a confirmed C1402T exchange which was also AM resistant, which does not support this suggestion. However, this can be due to a second, as-yet-unidentified, mutation.
The sensitivity of detection of EMB resistance on the basis of genetics is lower (59.0%) than that of FLQ and AM-CM resistance detection. Consequently, the presence of a wild-type pattern regarding embB codon 306 in the assay has a low predictive value for EMB susceptibility (73.3%). All of the strains included in this study with an embB codon 306 mutation were EMB resistant. This demonstrates the strong association of this mutation with drug resistance. Some authors also propose that embB codon 306 mutations are associated with resistance to other antituberculosis drugs and with MDR M. tuberculosis. This fact underlines a more general impact of this mutation on drug resistance (6, 14). More extensive and detailed studies in different settings are needed to clarify this.
The usefulness of the new assay for rapid FLQ, AM-CM, and EMB susceptibility testing of M. tuberculosis was demonstrated. Implementation of the test is facilitated if other line probe assays are already part of the diagnostic algorithm in laboratories and can easily be included in the routine work flow. However, the new MTBDRsl assay should be evaluated in different settings before routine use, since the prevalence of mutations associated with FLQ, AM-CM, and EMB resistance may vary in different locations. A limitation of the test is the low sensitivity for the detection of EMB resistance: gene loci other than embB codon 306 may be associated with EMB resistance. Only mutations covered by wild-type or mutant probes can be detected. Novel mutations with locations besides the probe regions or outside the amplified targets can lead to misinterpretation.
As already reported for the MTBDRplus assay, the new MTBDRsl assay can be applied directly to smear-positive specimens. Moreover, the new assay was used successfully when applied to smear-negative specimens known to be culture positive. With a turnaround time of approximately 6 h, the diagnosis of second-line drug resistance can be shortened from weeks (conventional DST) to 1 day. The new MTBDRsl assay is a major improvement in the routine detection of FLQ and AM-CM resistance and to a lesser extent to EMB-resistant M. tuberculosis strains. Especially in combination with the MTBDRplus assay for the detection of MDR M. tuberculosis, the detection of XDR M. tuberculosis is made possible within 1 or 2 days. This can be substantial progress regarding the diagnosis of drug resistance to strengthen both the management of patient therapy and the prevention of transmission.
Acknowledgments
We thank K. Ott, B. Schlüter, and A. Witt, Forschungszentrum Borstel, Borstel, Germany, for their excellent technical assistance and Hain Lifescience for providing the GenoType MTBDRsl test kits.
Footnotes
Published ahead of print on 22 April 2009.
REFERENCES
- 1.Alangaden, G. J., B. N. Kreiswirth, A. Aouad, M. Khetarpal, F. R. Igno, S. L. Moghazeh, E. K. Manavathu, and S. A. Lerner. 1998. Mechanism of resistance to amikacin and kanamycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 421295-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonova, O. V., D. A. Gryadunov, S. A. Lapa, A. V. Kuz'min, E. E. Larionova, T. G. Smirnova, E. Y. Nosova, O. I. Skotnikova, L. N. Chernousova, A. M. Moroz, A. S. Zasedatelev, and V. M. Mikhailovich. 2008. Detection of mutations in Mycobacterium tuberculosis genome determining resistance to fluoroquinolones by hybridization on biological microchips. Bull. Exp. Biol. Med. 145108-113. [DOI] [PubMed] [Google Scholar]
- 3.Bang, D., A. B. Andersen, and V. O. Thomsen. 2006. Rapid genotypic detection of rifampin- and isoniazid-resistant Mycobacterium tuberculosis directly in clinical specimens. J. Clin. Microbiol. 442605-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canetti, G., W. Fox, A. Khomenko, H. T. Mahler, M. K. Menon, D. A. Mitchison, N. Rist, and N. A. Smelov. 1969. Advances in techniques of testing mycobacterial drug sensitivity, and the use of sensitivity tests in tuberculosis control programmes. Bull. W. H. O. 4121-43. [PMC free article] [PubMed] [Google Scholar]
- 5.Deutsches Institut für Normung. 1986. Medical microbiology—diagnosis of tuberculosis. Part 3: Detection of mycobacteria by culture methods DIN 58943-3. Beuth Verlag, Berlin, Germany.
- 6.Hazbón, M. H., M. Bobadilla del Valle, M. I. Guerrero, M. Varma-Basil, I. Filliol, M. Cavatore, R. Colangeli, H. Safi, H. Billman-Jacobe, C. Lavender, J. Fyfe, L. Garcia-Garcia, A. Davidow, M. Brimacombe, C. I. Leon, T. Porras, M. Bose, F. Chaves, K. D. Eisenach, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, and D. Alland. 2005. Role of embB codon 306 mutations in Mycobacterium tuberculosis revisited: a novel association with broad drug resistance and IS6110 clustering rather than ethambutol resistance. Antimicrob. Agents Chemother. 493794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hillemann, D., M. Weizenegger, T. Kubica, E. Richter, and S. Niemann. 2005. Use of the GenoType MTBDR assay for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 433699-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hillemann, D., S. Rüsch-Gerdes, and E. Richter. 2006. Application of the GenoType MTBDR assay directly on sputum specimens. Int. J. Tuberc. Lung Dis. 101057-1059. [PubMed] [Google Scholar]
- 9.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology. A guide for a level III laboratory. Centers for Disease Control, Atlanta, GA.
- 10.Ling, D. I., A. A. Zwerling, and M. Pai. 2008. GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur. Respir. J. 321165-1174. [DOI] [PubMed] [Google Scholar]
- 11.Maus, C. E., B. B. Plikaytis, and T. M. Shinnick. 2005. Molecular analysis of cross resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 493192-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maus, C. E., B. B. Plikaytis, and T. M. Shinnick. 2005. Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mokrousov, I., T. Otten, O. Manicheva, Y. Potapova, B. Vishnevsky, O. Narvskaya, and N. Rastogi. 2008. Molecular characterization of ofloxacin-resistant Mycobacterium tuberculosis strains from Russia. Antimicrob. Agents Chemother. 522937-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons, L. M., M. Salfinger, A. Clobridge, J. Dormandy, L. Mirabello, V. L. Polletta, A. Sanic, O. Sinyavskiy, S. C. Larsen, J. Driscoll, G. Zickas, and H. W. Taber. 2005. Phenotypic and molecular characterization of Mycobacterium tuberculosis isolates resistant to both isoniazid and ethambutol. Antimicrob. Agents Chemother. 492218-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piersimoni, C., A. Olivieri, L. Benacchio, and C. Scarparo. 2006. Current perspectives on drug susceptibility testing of Mycobacterium tuberculosis complex: the automated nonradiometric systems. J. Clin. Microbiol. 4420-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plinke, C., S. Rüsch-Gerdes, and S. Niemann. 2006. Significance of mutations in embB codon 306 for prediction of ethambutol resistance in clinical Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 501900-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Safi, H., B. Sayers, M. H. Hazbon, and D. Alland. 2008. Transfer of embB codon 306 mutations into clinical Mycobacterium tuberculosis strains alters susceptibility to ethambutol, isoniazid, and rifampin. Antimicrob. Agents Chemother. 522027-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salfinger, M., and G. E. Pfyffer. 1994. The new diagnostic mycobacteriology laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 13961-979. [DOI] [PubMed] [Google Scholar]
- 19.Sekiguchi, J., T. Miyoshi-Akiyama, E. Augustynowicz-Kopec, Z. Zwolska, F. Kirikae, E. Toyota, I. Kobayashi, K. Morita, K. Kudo, S. Kato, T. Kuratsuji, T. Mori, and T. Kirikae. 2007. Detection of multidrug resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 45179-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah, N. S., A. Wright, G. H. Bai, L. Barrera, F. Boulahbal, N. Martin-Casabona, F. Drobniewski, C. Gilpin, M. Havelkova, R. Lepe, R. Lumb, B. Metchock, F. Portaels, M. F. Rodrigues, S. Rüsch-Gerdes, A. van Deun, V. Vincent, K. Laserson, C. Wells, and J. P. Cegielski. 2007. Worldwide emergence of extensively drug-resistant tuberculosis. Emerg. Infect. Dis. 13380-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somoskovi, A., J. Dormandy, D. Mitsani, J. Rivenburg, and M. Salfinger. 2006. Use of smear-positive samples to assess the PCR-based GenoType MTBDR assay for rapid, direct detection of the Mycobacterium tuberculosis complex as well as its resistance to isoniazid and rifampin. J. Clin. Microbiol. 444459-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takiff, H., L. Salazzar, C. Guerrero, W. Philipp, W. M. Huang, B. Kreiswirth, S. T. Cole, W. R. Jacobs, Jr., and A. Telenti. 1994. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob. Agents Chemother. 38773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Traore, H., A. van Deun, I. C. Shamputa, L. Rigouts, and F. Portaels. 2006. Direct detection of Mycobacterium tuberculosis complex DNA and rifampin resistance in clinical specimens from tuberculosis patients by line probe assay. J. Clin. Microbiol. 444384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Doorn, H. R., D. D. An, M. D. de Jong, N. T. Lan, D. V. Hoa, H. T. Quy, N. V. Chau, P. M. Duy, D. Q. Tho, N. T. Chinh, J. J. Farrar, and M. Caws. 2008. Fluoroquinolone resistance detection in Mycobacterium tuberculosis with locked nucleic acid probe real-time PCR. Int. J. Tuberc. Lung Dis. 12736-742. [PubMed] [Google Scholar]
- 25.World Health Organization. 2008. Anti-tuberculosis drug resistance in the world. Fourth global report. WHO/HTM/TB/2008.394. World Health Organization, Geneva, Switzerland.
- 26.Zhang, Y., and A. Telenti. 2000. Genetics of drug resistance in Mycobacterium tuberculosis, p. 235-251. In G. F. Harfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, DC.