Abstract
Neurons are thought to use diverse families of cell-surface molecules for cell recognition during circuit assembly. In Drosophila, alternative splicing of the Down syndrome cell adhesion molecule (Dscam) gene potentially generates 38,016 closely related trans-membrane proteins of the immunoglobulin superfamily, each comprising one of 19,008 alternative ectodomains linked to one of two alternative transmembrane segments1. These ectodomains show isoform-specific homophilic binding, leading to speculation that Dscam proteins mediate cell recognition2. Genetic studies have established that Dscam is required for neural circuit assembly1,3-10, but the extent to which isoform diversity contributes to this process is not known. Here we provide conclusive evidence that Dscam diversity is essential for circuit assembly. Using homologous recombination, we reduced the entire repertoire of Dscam ectodo-mains to just a single isoform. Neural circuits in these mutants are severely disorganized. Furthermore, we show that it is crucial for neighbouring neurons to express distinct isoforms, but that the specific identity of the isoforms expressed in an individual neuron is unimportant. We conclude that Dscam diversity provides each neuron with a unique identity by which it can distinguish its own processes from those of other neurons, and that this self-recognition is essential for wiring the Drosophila brain.
The complexity and specificity of neuronal wiring implies the existence of a cellular recognition code that allows neurons to distinguish between one another11. It has been speculated that families of highly diverse cell-surface molecules could provide this function, such as the vertebrate neurexins12, cadherins13 and cadherin-related neuronal receptors14, and the insect Dscams1,15. However, it remains unclear to what extent the molecular diversity of such proteins is essential for wiring specificity, and how their diversity contributes to neuronal recognition. In Drosophila melanogaster, as many as 38,016 Dscam isoforms are generated by alternative splicing1. Each isoform consists of an ectodomain containing a unique combination of three different variable immunoglobulin-like domains linked to one of two alternative transmembrane segments (Fig. 1a). The variable ectodomain segments are encoded by 12, 48 and 33 alternatives for exons 4, 6 and 9, respectively, whereas the transmembrane domain is encoded by two versions of alternative exon 17 (Fig. 1a). A given ectodomain isoform binds strongly to itself, but only weakly, if at all, to other isoforms2. Thus, Dscam diversity could provide a molecular mechanism for selective recognition among neurons.
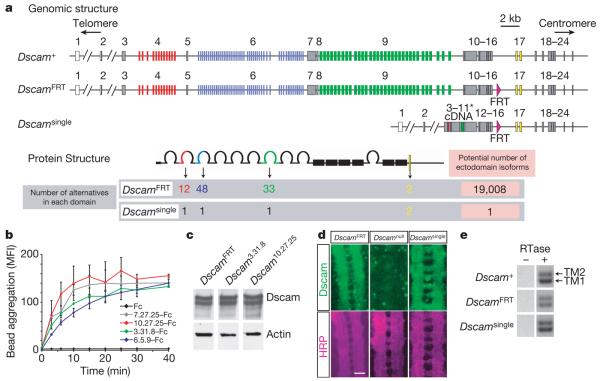
Figure 1. Generation and molecular characterization of Dscamsingle alleles.
a, Schematic representation of the genomic organization and proteins encoded by Dscam alleles used in this paper. Numbers above bars in the genomic structure indicate exons. Alternatively spliced exons are shown in colour. cDNA encoding a single isoform in the Dscamsingle includes exons 3–11 (asterisk). Pink triangles indicate the FRT site between exons 16 and 17. b, Using a bead-aggregation assay, Dscam10.27.25, Dscam3.31.8, and Dscam6.5.9 ectodomains show homophilic binding similar to the Dscam7.27.25 control2. Aggregation of fluorescent beads decorated with ectodomain–Fc fusion proteins was measured as an increase in the mean fluorescence intensity (MFI) of each particle. Binding experiments were performed twice. Error bars represent±1 s.d. c, Dscamsingle alleles express Dscam protein at wild-type levels. The level of Dscam protein in extracts of the larval central nervous system was assessed by immunoblotting. Actin was used as a loading control. d, Dscam protein expression pattern (green) is normal in Dscamsingle embryonic ventral nerve cord (stage 16). Anti-HRP (horseradish peroxidase) staining (purple) was used to visualize the neuropil. Scale bar, 10 μm. e, Expression of two alternative transmembrane (TM) domains in Dscamsingle animals. RT-PCR across exons encoding each of the alternative TM domains was performed using RNA extracted from the third instar larval brains. Gel electrophoresis separates products encoding TM1 and TM2 as indicated.
The potential role of Dscam diversity in neuronal wiring has previously been approached using various deletion mutations that remove subsets of alternative versions of exon 4 from the genomic locus. These alleles reduce the potential ectodomain diversity at most from 19,008 to some 5,000 isoforms. All of these mutants develop to adulthood and are fertile, and their nervous system organization is largely normal8,16. In one study, reducing the potential ectodomain diversity to ∼11,000 isoforms resulted in an increase in the variability of axon branching and the appearance of some ectopic branches in an identified somatosensory neuron5, alluding to a specific role for subsets of isoforms. However, because different cells splice Dscam differently7,17, these genomic deletions may result in variable reduction of Dscam protein levels within different cells. For example, a specific isoform of N-cadherin is required for targeting of R7 neurons in the visual system, but this is due to cell-specific splicing rather than an isoform-specific function18. It is therefore unclear whether the defects observed in Dscam isoform deletion mutants reflect a functional requirement for specific isoforms, a mosaic Dscam loss-of-function, or both. Thus, whether Dscam diversity is essential for neural circuit assembly remains a crucial and unresolved issue. A definitive test to address the importance of ectodomain diversity would be to completely eliminate alternative splicing, reducing Dscam ectodomain diversity to just a single isoform expressed from the endogenous locus.
We used homologous recombination to replace the genomic region encoding the variable ectodomains with a complementary DNA encoding a single isoform (Fig. 1a and Supplementary Fig. 1). Three distinct ectodomains were arbitrarily selected. These were shown to exhibit homophilic binding (Fig. 1b), a property shared with all other isoforms we have studied (>100) (refs 2; 19). We refer to these mutant alleles collectively as Dscamsingle, as the phenotypes of all three alleles were similar, and individually by indicating the selected exon variants in the allele name (for example, Dscam3.31.8 contains the variable exons 4.3, 6.31, and 9.8). All of these alleles carry an FRT (FLP recombinase target) sequence inserted in the intron between exons 16 and 17. Accordingly, we also generated a control allele, DscamFRT, which has an FRT insertion in the same location but retains the full complement of alternative exons (Fig. 1a). We verified the intended genomic rearrangements by sequencing 14 kb from the Dscam locus in each of these alleles. Sequencing of cDNAs confirmed that each Dscamsingle allele only expresses the designated ectodomain isoform, whereas DscamFRT expresses many different isoforms. Protein levels (Fig. 1c) and localization (Fig. 1d), as well as the relative use of the two alternative transmembrane exons (Fig. 1e) were similar among the Dscamsingle alleles and the DscamFRT and wild-type controls.
Like Dscamnull alleles1, all three Dscamsingle alleles are recessive lethal. Thus, not only is Dscam itself essential, but so is its diversity. To assess more carefully the viability of these mutants, newly hatched mutant larvae were isolated and grown without potential competition from their siblings. Under these conditions, 17–22% of Dscamnull homozygous animals survived until late pupal stages (Fig. 2a). In contrast, as many as 76–89% of Dscamsingle/Dscamnull animals survived to the pupal stages, suggesting that at least some Dscam function is diversity-independent. Notably, Dscamsingle homozygotes died in early larval development, regardless of the isoform encoded (Fig. 2a). Axonal pathways were disrupted in the embryonic ventral nerve cord in all of these mutant combinations, and this phenotype too was most severe in the Dscamsingle homozygotes (Fig. 2b). The phenotypes in animals carrying two different Dscamsingle alleles (that is, transheterozygous animals) have a similar spectrum of phenotypes to Dscamsingle homozygotes (Fig. 2a, b).
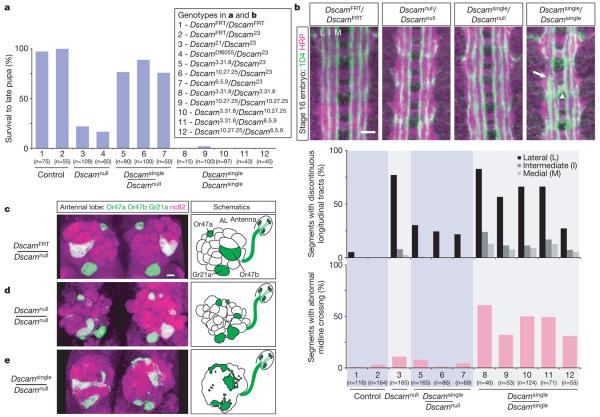
Figure 2. Viability and neuronal wiring defects in Dscamsingle mutants.
a, Survival of different genotypes to late pupal stages. Dscam21, Dscam23 and DscamDf6055 are protein-null alleles. b, Dscamsingle embryos show defects in embryonic central nervous system organization. Stage-16 embryos were examined for neuropil structure (anti-HRP, purple) and for three distinct longitudinal axon tracts (monoclonal antibody 1D4, anti FasII, green). Dscamsingle/Dscamsingle embryos (>95%; n=41) show severely disrupted longitudinal tracts (arrow) and aberrant midline crossing (arrowhead). Scale bar, 10 μm. c–e, Dscam diversity is required in the olfactory system. Antennal lobes (AL) were visualized with the presynaptic marker nc82 (purple) and synaptotagmin-GFP (green fluorescent protein) expression (green) in three classes of olfactory receptor neurons (see schematic). c, DscamFRT/Dscamnull controls show normal organization (n=88 antennal lobes). Scale bar, 10 μm. d, Loss of Dscam (Dscamnull/Dscamnull) results in mistargeting of ORNs to multiple glomeruli. The position and morphology of glomeruli are also abnormal (n=28). e, Loss of diversity (Dscamsingle/Dscamnull) leads to loss of glomerular boundaries, as well as formation of ectopic ORN termini throughout the antennal lobe. Severe phenotypes were observed for all three Dscamsingle alleles (100% penetrance: n=28, 128 and 16 for Dscam3.31.8, Dscam10.27.25 and Dscam6.5.9, respectively).
To explore further the neuronal wiring in these mutants, we examined the synaptic organization of the olfactory system (Fig. 2c-e). Olfactory receptor neurons (ORNs) project axons into the antennal lobe, where they form synapses with the dendrites of projection neurons and local interneurons. Typically, neurons in a given ORN class all express the same odorant receptor, and form connections within a distinct synaptic module, called a glomerulus20. Dscam is required autonomously in ORNs, projection neurons and interneurons to establish this circuitry4,6. In Dscamnull homozygotes, we observed a highly characteristic phenotype in which axons of a given ORN class target more than one glomerulus in each antennal lobe (Fig. 2d). Discrete glomeruli still formed in these mutants, but their stereotyped arrangement within the lobe was highly disrupted. In marked contrast, the antennal lobe of all Dscamsingle/Dscamnull animals was highly disorganized with no distinct glomerular structure, and ORNs formed many ectopic termini throughout the antennal lobe (Fig. 2e). Taken together, these data establish that Dscam diversity is essential for neural circuit formation.
What is the role of Dscam diversity in circuit assembly? One possibility is that diversity is instructive, with distinct isoforms specifying distinct synaptic connections1. This might be an attractive model, but there is little evidence to support it. An alternative model21 posits that Dscam diversity might provide a molecular mechanism for `self-avoidance', that is, the propensity of multiple dendritic or axonal branches from the same neuron (sister branches) to avoid each other and thereby elaborate appropriate receptive or terminal fields, respectively22,23. This phenomenon requires recognition and repulsion between self-neurites, but not between neurites of different neurons. Dscam diversity might provide this function, because each neuron expresses a unique set of Dscam isoforms7,17, and isoform-specific homophilic binding mediates repulsion2,8. To test this model, we focused on the simple and well-characterized sister branch segregation of mushroom body neurons in the central brain.
Each mushroom body comprises thousands of neurons. During development, each of these neurons extends an axon in a fascicle called the peduncle, at the end of which it bifurcates to produce two sister branches, one segregating into the dorsal lobe and the other into the medial lobe (Fig. 3a). Because many axons bifurcate at the same time and in close proximity to each other, each branch must be able to distinguish its sister branch from branches of other neurons, to ensure a high fidelity of sister branch segregation. Dscam is essential for this process3, and each mushroom body neuron expresses a distinct array of Dscam isoforms7. To satisfy the model that Dscam diversity promotes self-recognition and self-avoidance of sister branches21, two critical conditions must be met. First, the specific set of isoforms expressed in a given mushroom body neuron should not be important. Second, and in contrast, whatever set of isoforms a given mushroom body neuron expresses, it should be essential that this set is different from that of its neighbours. Transgenic rescue experiments and deletion mutant analysis have confirmed the first condition7,16. However, this condition on its own is also consistent with the null hypothesis that Dscam diversity is not required at all in mushroom body neurons. Thus, the critical test of the self-recognition model is to establish whether sister branch segregation requires neighbouring mushroom body axons to express different sets of Dscam isoforms.
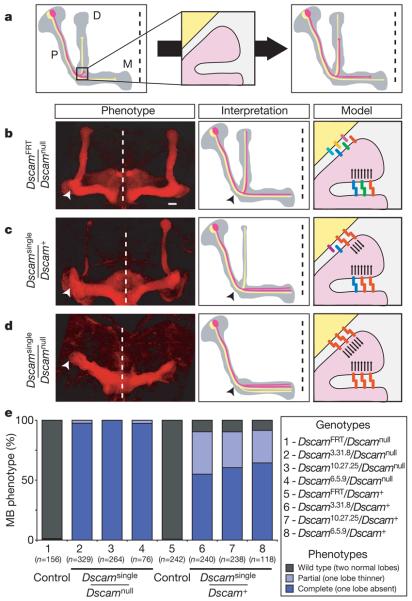
Figure 3. Dscam diversity is required for mushroom body development.
a, Schematic of mushroom body development (P, peduncle; D, dorsal lobe; M, medial lobe). Two representative mushroom body neurons are shown (yellow and pink). b–d, Left panels show mushroom body lobes from late pupae visualized by staining with monoclonal antibody 1D4 (anti-FasII). Only the lobe region is shown. Arrowhead indicates the location of the branch point. Dashed line, midline. Scale bar, 10 μm. Dscam isoforms expressed on sister branches (pink) and a non-self branch (yellow) are represented as coloured bars in panels on the right. Arrows indicate repulsive signals resulting from Dscam isoform-specific homophilic binding. b, Control animals (DscamFRT/Dscamnull). c, Dscamsingle/Dscam+ heterozygous animals. d, Dscamsingle/Dscamnull mutant animals. e, Quantification of mushroom body (MB) phenotypes.
To test this prediction, we examined mushroom body morphology in Dscamsingle and control animals (Fig. 3b-e). In DscamFRT controls, the normal bi-lobed structure of the mushroom body was observed, with two lobes representing populations of sister branches that had segregated correctly (Fig. 3b). In sharp contrast, this normal bi-lobed mushroom body morphology was never observed in Dscamsingle animals. In some 97% of Dscamsingle/Dscamnull mushroom bodies analysed, one of the two lobes was completely absent—typically the dorsal lobe (Fig. 3d). In the few remaining samples, one mushroom body lobe was significantly thinner than the other. We also observed a strong, but less severe, phenotype in ,∼90% of Dscamsingle/Dscam+ heterozygotes (Fig. 3c), with one mushroom body lobe either absent (∼60%) or thinner (∼30%), suggesting that some mushroom body sister branches segregate normally, but most do not. This dominant phenotype indicates that the mushroom body defects observed in Dscamsingle/Dscamnull animals do not result from the loss of any one isoform, but rather the presence of the same isoform on all axons. Thus, Dscam diversity is essential for mushroom body sister branch segregation, which is compromised even if only ∼50% of Dscam proteins in each neuron represent a single isoform shared by all neurons.
Although these data support a role for Dscam diversity in mushroom body branch segregation, it remains possible that the isoforms encoded in these Dscamsingle alleles are simply non-functional or even inhibitory for this process. To address this issue, we generated mosaic animals in which a single mushroom body neuron expresses only one Dscam isoform from a Dscamsingle allele, but all other neurons express the full complement of isoforms from the DscamFRT allele. This was achieved using an intragenic variation of the MARCM technique24, in which mitotic recombination was induced between FRT sites within two modified Dscam alleles (Fig. 4a and Supplementary Fig. 1). We tested two different Dscamsingle isoforms with this intragenic MARCM system, and found that the sister branches of isolated Dscamsingle mushroom body neurons segregated with high fidelity (Fig. 4b), but that sister branches of Dscamnull neurons did not3 (Fig. 4b). This experiment establishes that the single ectodomains we selected do indeed support sister branch segregation. Therefore, the lack of segregation in animals with a Dscamsingle allele must be due to the loss of diversity, not the loss of Dscam function. Taken together, these results demonstrate that Dscam diversity is dispensable within a single neuron, but essential within a population of neurons, supporting the notion that it provides each neuron with a unique cell-surface identity.
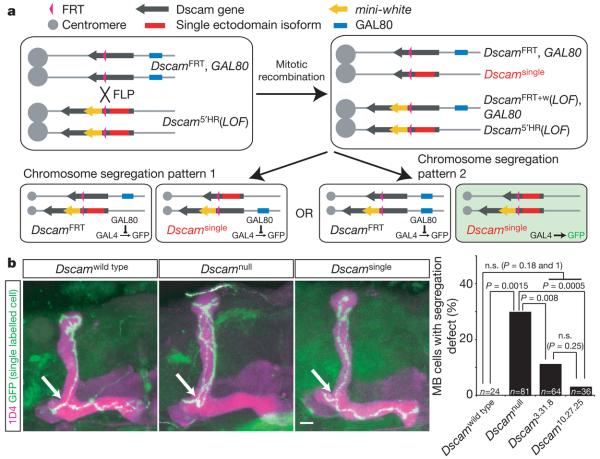
Figure 4. Dscamsingle is sufficient to promote branch segregation with high fidelity at the single-cell level.
a, Schematic of intragenic MARCM strategy to generate and label single Dscamsingle cells in an otherwise wild-type background (that is, transheterozygous with the DscamFRT and Dscam loss-of-function (DscamLOF) alleles). FLP recombinase induces mitotic recombination between FRT sites within the Dscam locus. Chromosomes can segregate in two ways. In one way (pattern 2), GFP-labelled (green) Dscamsingle/Dscam5′HR mutant cells are generated. The Dscam5′HR allele (Supplementary Fig. 1) is a loss-of-function allele. If chromosomes segregate in the alternative fashion (pattern 1), no labelled cells will be produced. One of these cells will carry an intact Dscamsingle allele. b, Branch segregation phenotypes. Labelled cells for Dscamwild type and Dscamsingle were generated using intragenic MARCM, and Dscamnull labelled cells were produced using conventional MARCM. The mushroom body was visualized by staining with monoclonal antibody 1D4, anti-FasII (purple), and the clones were labelled with membrane-targeted chimeric GFP (mCD8GFP, green)24. The genotype of each clone is indicated above the panel. Arrows indicate the branch point. Quantification is shown as a bar graph (using a two-tailed Fisher's exact test; n.s., not significant).
In conclusion, here we provide strong evidence that Dscam diversity is critical for neuronal wiring. We envision that Dscam diversity contributes to wiring specificity in many different ways. One of these is self-recognition and self-avoidance, as demonstrated here for mushroom body neurons. We propose that this is a central function for Dscam diversity in neural circuit formation.
METHODS SUMMARY
The strategy used to generate the Dscamsingle and DscamFRT alleles is indicated in Supplementary Fig. 1, and is based on the ends-in targeting strategy detailed in ref. 25. Three isoforms were selected essentially at random from sequenced cDNAs, with the only criterion being to select three distinct variants for each of exons 4, 6, and 9. Biochemical and molecular characterization of these alleles were peformed as described previously2,7,8. For survival analysis, each Dscam mutant animal was genotyped at first to second instar larval stages using kruppel–GFP, CyO balancer, and then transferred to fresh grape plates with a thin spread of yeast paste and raised at room temperature (∼22–25 °C). Independently isolated alleles harbouring the same isoform were used to generate homozygous Dscamsingle animals to avoid effects from potential second site mutations. Embryonic ventral nerve cords were immunostained as described previously7. For other analyses, animals that survived until late pupal stage were used. Immunostainings of pupal or adult brains were performed as described previously6,7. For intragenic MARCM analysis, clones were generated by inducing heat-shock-mediated expression of FLP recombinase at late larval to early pupal stages. Heat-shock was carried out at 37 °C for 1 h. Dscamnull mutant clones were generated using conventional MARCM as previously described7.
METHODS
Generation of Dscamsingle alleles
Homology regions were amplified by PCR from genomic DNA extracted from the reference strain for the Drosophila genome project26, and cloned into custom-built targeting vectors. For the single-ectodomain constructs, RT–PCR was performed on total RNA extracted from the heads of Canton-S adults, amplifying exons 3–11 and inserting this fragment in-frame into the flanking genomic region within the targeting vector, using an endogenous Asp 718 site in exon 3 and an NheI site engineered into exon 11, without altering the predicted amino acid sequence. Donor insertions on the X or 3rd chromosome were obtained by P-element-mediated transformation. These donor elements were excised and linearized with hsFLP and hsI-SceI, respectively25. The resulting virgin females were crossed to eyFLP males27, and reintegration of the targeting element was detected in the progeny by the presence of an eyFLP-resistant white+ marker (that is, flanked by a single FRT, rather than the two FRTs of the donor element). The final alleles were then obtained by FLP-induced recombination, as indicated in Supplementary Fig. 1, selecting for loss of the white+ marker. Two independently derived isolates were established for Dscam3.31.8 and Dscam10.27.25 alleles, obtained in turn from two independently derived intermediate alleles. One allele was generated for Dscam6.5.9.
Biochemical and molecular characterization
The bead aggregation assay was performed as previously described2. For immunoblots, approximately five brains dissected from the third instar larvae were homogenized directly in SDS sample buffer and western blots were performed using rabbit anti-Dscam antibody1 at 1:1,000 dilution. The membrane was re-probed with a 1:1,000 dilution of rabbit anti-actin antibody (Sigma). RT–PCR analysis to characterize use of alternative ectodomain isoforms was performed as previously described7. The use of exons 17.1 and 17.2, encoding alternative transmembrane domains, was characterized by RT–PCR with primers against exon 16 and exon 18, followed by gel electrophoresis separation of PCR products containing exon 17.1 or 17.2.
Immunohistology
The following antibodies were used for immunohistology: rabbit anti-Dscam (1:500; ref. 7), monoclonal antibody 1D4 (anti-FasII; 1:10), rabbit anti-GFP (1:1,000, Molecular Probes), monoclonal antibody nc82 (1:10; ref. 28), Cy5-conjugated goat anti-HRP (1:200, Jackson ImmunoResearch Laboratories) and Alexa488- or Alexa568-conjugated goat anti-mouse or anti-rabbit (1:200, Molecular Probes). Late pupal or adult brains were dissected, and immunostaining was performed essentially as previously described6,7. Stage-16 embryos were fixed and immunostained as previously described7. Immunofluorescent samples were analysed using an LSM 510 Meta (Zeiss).
Fly stocks
Dscam-null alleles (Dscam21, Dscam23 and DscamDf6055) have been described previously1,6. For antennal lobe phenotypic analyses, transgenic lines were generated, each containing a putative promoter of an Or gene (for example, Or47a) fused to a synaptotagmin–GFP marker (for example, Or47a–sytGFP). Three Or–sytGFP transgenes were used for this study (Or47a–sytGFP, Or47b–sytGFP and Gr21a–sytGFP) (X. L. Zhan, P. Cayirlioglu, I. C. Grunwald, D. Gunning and S.L.Z, unpublished reagents).
Supplementary Material
Acknowledgements
We thank members of the Zipursky and Dickson laboratories for critical comments on the manuscript. This work was supported by grants from the Austrian Science Fund (B.J.D) and NIH (S.L.Z.). Work at the Institute of Molecular Pathology is also supported by funds from Boehringer Ingelheim GmbH. S.L.Z. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Schmucker D, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 2.Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Zugates CT, Liang IH, Lee CH, Lee T. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002;33:559–571. doi: 10.1016/s0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- 4.Zhu H, et al. Dendritic patterning by Dscam and synaptic partner matching in the Drosophila antennal lobe. Nature Neurosci. 2006;9:349–355. doi: 10.1038/nn1652. [DOI] [PubMed] [Google Scholar]
- 5.Chen BE, et al. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. 2006;125:607–620. doi: 10.1016/j.cell.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 6.Hummel T, et al. Axonal targeting of olfactory receptor neurons in Drosophila is controlled by Dscam. Neuron. 2003;37:221–231. doi: 10.1016/s0896-6273(02)01183-2. [DOI] [PubMed] [Google Scholar]
- 7.Zhan XL, et al. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004;43:673–686. doi: 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 8.Matthews BJ, et al. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129:593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 9.Hughes ME, et al. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron. 2007;54:417–427. doi: 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soba P, et al. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron. 2007;54:403–416. doi: 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sperry RW. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc. Natl Acad. Sci. USA. 1963;50:703–710. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Missler M, Südhof TC. Neurexins: three genes and 1001 products. Trends Genet. 1998;14:20–26. doi: 10.1016/S0168-9525(97)01324-3. [DOI] [PubMed] [Google Scholar]
- 13.Takeichi M, et al. Cadherins in brain patterning and neural network formation. Cold Spring Harb. Symp. Quant. Biol. 1997;62:505–510. [PubMed] [Google Scholar]
- 14.Kohmura N, et al. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20:1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 15.Graveley BR, et al. The organization and evolution of the dipteran and hymenopteran Down syndrome cell adhesion molecule (Dscam) genes. RNA. 2004;10:1499–1506. doi: 10.1261/rna.7105504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, et al. Transmembrane/juxtamembrane domain-dependent Dscam distribution and function during mushroom body neuronal morphogenesis. Neuron. 2004;43:663–672. doi: 10.1016/j.neuron.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 17.Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nature Genet. 2004;36:240–246. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- 18.Nern A, et al. An isoform-specific allele of Drosophila N-cadherin disrupts a late step of R7 targeting. Proc. Natl Acad. Sci. USA. 2005;102:12944–12949. doi: 10.1073/pnas.0502888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wojtowicz WM, et al. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. doi: 10.1016/j.cell.2007.08.026. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jefferis GS, Hummel T. Wiring specificity in the olfactory system. Semin. Cell Dev. Biol. 2006;17:50–65. doi: 10.1016/j.semcdb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Zipursky SL, Wojtowicz WM, Hattori D. Got diversity? Wiring the fly brain with Dscam. Trends Biochem. Sci. 2006;31:581–588. doi: 10.1016/j.tibs.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Kramer AP, Kuwada JY. Formation of the receptive fields of leech mechanosensory neurons during embryonic development. J. Neurosci. 1983;3:2474–2486. doi: 10.1523/JNEUROSCI.03-12-02474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr. Biol. 2003;13:618–626. doi: 10.1016/s0960-9822(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 24.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 25.Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 26.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 27.Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- 28.Stortkuhl KF, Hofbauer A, Keller V, Gendre N, Stocker RF. Analysis of immunocytochemical staining patterns in the antennal system of Drosophila melanogaster. Cell Tissue Res. 1994;275:27–38. doi: 10.1007/BF00305373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.