Abstract
Background
Climatic changes during glacial periods have had a major influence on the recent evolutionary history of living organisms, even in temperate forests on islands, where the land was not covered with ice sheets. We investigated the phylogeographical patterns of the weevil Curculio sikkimensis (Curculionidae), a generalist seed predator of Fagaceae plants living in both deciduous oak and evergreen forests of Japan. Its genetic structure was compared to that of another host-specific seed predator, C. hilgendorfi, inhabiting only evergreen forests.
Results
We examined 921 bp of mitochondrial DNA for 115 individuals collected from 33 populations of C. sikkimensis from 11 plant species of three genera, Quercus, Lithocarpus, and Castanopsis. An analysis of molecular variance revealed that a large proportion (almost 50%, P < 0.001) of the total genetic variance could be explained by differences between two geographical regions, the southwestern and northeastern parts of the main islands of Japan. In contrast, no significant genetic differentiation of the weevil was observed among vegetation types of their utilized host plant species. The phylogeographical patterns of the generalist and the host-specific seed predator exhibited a congruent genetic boundary in the Chugoku-Shikoku region.
Conclusion
Our results suggest that geology and historical environment have contributed to shaping the present genetic structure of C. sikkimensis. The geographical patterns of genetic differentiation in the Chugoku-Shikoku region observed in the two types of Fagaceae-associated Curculio in this study have also been observed in several plant species growing in warm and cool temperate zones of Japan. The occurrence of this common pattern suggests that deciduous oak and evergreen forests of Japan survived together, or adjacent to each other, in small refugia during glacial ages, in the southwestern and northeastern parts of the main islands, although these two types of forests are presently distributed in cool and warm temperate zones of Japan, respectively.
Background
The current distribution of biological diversity cannot be understood without information about how organisms responded to historical geological and climatic conditions. In particular, the glacial-interglacial cycles of the Quaternary period may have played an important role in shaping the distribution of biodiversity among current populations, even in warm temperate zones, where the land was not covered with ice sheets [1,2]. The geological and geographical features of the Japanese Archipelago are comprised of several mountain ranges running largely parallel to a northeast to southwest axis. The mild coastal belt is close to these mountain ranges; thus the climate varies even in a narrow region. Consequently, a wide range of vegetation occurs, which accounts for various biogeographical factors that are not common on continental landmasses. Moreover, several landbridges between Japan and surrounding areas, which formed or disappeared in response to glacial-interglacial climatic changes, have also played important roles in determining the current distribution of Japanese biological diversity.
Phylogeographical patterns estimated from the extant genetic variation of organisms have proved highly informative in recovering the postglacial demographic histories of species [3,4]. Another powerful method is to compare intraspecific phylogeographical patterns among several taxa over the same area and to search for congruent geographic patterns of genetic variation, which would indicate the influence of common historical factors [5-17]. Comparing the intraspecific phylogeographical patterns among different species distributed in a single vegetation zone should be more informative because a group of species living together in the present environment likely responded in a similar or possibly the same manner to past geological or climatic events [5,18]. In the present study, we focused on the broadleaved deciduous and evergreen forest communities that characterize the biodiversity and endemism of the temperate zone, where the effects of climate change are particularly severe.
Palynological evidence indicates that broadleaved deciduous and evergreen forests in Japan were subjected to cold periods at least four times during the Quaternary [1,2]. During the glacial periods, climatic cooling caused southward and shifts to lower altitude in the geographic distribution of these forests. The pollen record indicates that refugial populations of broadleaved deciduous forests were sparsely distributed along the coasts of the Pacific Ocean and the Sea of Japan, while refugia of broadleaved evergreen forests were limited to southern areas mainly at the southern end of Kyushu. Fossil evidence also indicates that these populations migrated northward from refugia after the last glacial maximum [2,19,20]. These data suggest that populations of some forest-dependent insects, especially flightless or nonmigratory species, responded to the environmental changes associated with the glacial cycles in a similar fashion to that of their associated forests.
Most plant species growing in broadleaved deciduous and evergreen forests are parasitized by a diverse variety of herbivorous insects. We investigated and compared the mitochondrial DNA (mtDNA) genetic variation of two acorn weevils with different feeding types. These beetles are of particular interest because the females lay eggs in the host seeds. As a result, the fitness of the host and the weevil are intimately related [21]. The use of the rostrum during oviposition site preparation is considered the key adaptation that facilitates circumventing the physical defenses of the plant, such as the avoidance of attachment to the host [22]. All species of Curculio oviposit in seeds that are still on the tree. Females use the rostrum to excavate a hole in the seeds [23], after which they turn around and oviposit into the hole. The larvae feed on the seeds before leaving them to overwinter in the soil, where they pupate before emerging in spring or summer.
Five species of Curculio associated with Fagaceae seeds are distributed in Japan: C. sikkimensis, C. hilgendorfi, C. dentipes, C. robustus, and C. conjugalis. These include both generalist and host-specific seed predators of Fagaceae. Specifically, C. sikkimensis is a generalist associated with deciduous and evergreen trees [i.e., Quercus (deciduous and evergreen), Lithocarpus (evergreen), Castanopsis (evergreen), and Castanea (deciduous)], and C. hilgendorfi is a host-specific seed predator of Castanopsis. The other species are associated with several species of deciduous Quercus. Curculio sikkimensis and C. hilgendorfi are sometimes sympatrically distributed in evergreen forests.
In this study, we aimed to elucidate the biogeographical history of both types of forests by comparing the phylogeography of the two types (i.e., generalists and specialists) of acorn weevils. Our previous study showed that the mtDNA haplotypes found in the individuals of these two Curculio species, C. sikkimensis and C. hilgendorfi, validated each clade with high bootstrap support [24]. Moreover, our allozyme data showed that gene flow was disrupted between the two weevil species via reproductive isolation even in sympatric populations [24]. The two Curculio species exhibit similar adult morphologies, but can be identified by differences in the shape of the male genitalia [25]. Our previous study reported a phylogeographical and population demographic analysis of the host-specific seed predator C. hilgendorfi [24].
The purpose of this study was to assess the phylogeography of the generalist acorn weevil Curculio sikkimensis (HELLER) based on the current geographic distribution of mtDNA diversity. We report a phylogeographical pattern of C. sikkimensis collected from various Fagaceae seeds. This study addresses the following questions: 1] Do the mtDNA haplotypes of C. sikkimensis form host races? If C. sikkimensis exhibits some host specialization, the genetic differentiation of mtDNA in C. sikkimensis should be observable among their host Fagaceae species, among host plant genera, and between deciduous and evergreen host plants (i.e., deciduous and evergreen vegetation zones). 2] Do the phylogeographical patterns of Curculio species enable us to reconstruct the glacial and postglacial history of their associated temperate forests in Japan? Given that host plant species inhabit both broadleaved deciduous oak and evergreen forests, an investigation of the generalist predator C. sikkimensis provides an understanding of the biogeographical history of both deciduous oak and evergreen forests in Japan. However, the phylogeographical information of the specialist C. hilgendorfi reflects the history of evergreen forests. We compared the phylogeographical results of the generalist C. sikkimensis and the specialist C. hilgendorfi as well as those of the plant species growing to examine whether the evolutionary histories of their associated deciduous oak and evergreen forests are similar.
Methods
Insects
We sampled Fagaceae seeds of Quercus, Lithocarpus, and Castanopsis infested with C. sikkimensis from 33 localities in Japan (Table 1). Sampling was conducted during October and November 2002–2007, after larvae had grown up within the seeds. The seeds were kept in plastic bottles in the laboratory, where emerging larvae were collected. The larvae were preserved in 99% ethanol for DNA analysis.
Table 1.
Sample collection sites and host plant species of Curculio sikkimensis in Japan
| Localities | Host plants | Latitude | Longitude | No. samples | |
| 1 | Narita, Chiba | Quercus acuta | 35.47.00 | 140.19.15 | 3 |
| 2 | Mt. Takatsuka, Chiba | Quercus acuta | 34.56.10 | 139.56.35 | 4 |
| 3 | Higashizushi, Kanagawa | Quercus acuta | 35.18.10 | 139.36.15 | 4 |
| 4 | Uenohara, Yamanashi | Quercus myrsinaefolia | 35.37.55 | 139.06.50 | 2 |
| 5 | Ito, Shizuoka | Castanopsis sieboldii var.sieboldii | 34.52.35 | 139.05.45 | 2 |
| Quercus gilva | 1 | ||||
| 6 | Minamiizu, Shizuoka | Castanopsis sieboldii var.sieboldii | 34.39.00 | 138.51.10 | 7 |
| 7 | Minoshima, Wakayama | Quercus variabilis | 34.04.40 | 135.07.25 | 1 |
| 8 | Kitagawa, Kochi | Quercus gilva | 33.28.50 | 134.03.25 | 2 |
| 9 | Monobe, Kochi | Quercus crispula | 33.48.15 | 133.53.40 | 6 |
| 10 | Nangoku, Kochi | Quercus gilva | 33.37.10 | 133.35.40 | 2 |
| 11 | Kochi, Kochi | Quercus glauca | 33.33.02 | 133.28.50 | 2 |
| 12 | Susaki, Kochi | Quercus glauca | 33.24.55 | 133.18.35 | 1 |
| 13 | Himesaki, Sado Is., Niigata | Castanopsis sieboldii var.sieboldii | 38.05.05 | 138.33.45 | 1 |
| 14 | Futami, Sado Is., Niigata | Castanopsis sieboldii var.sieboldii | 37.58.45 | 138.15.25 | 2 |
| 15 | Hikone, Shiga | Castanopsis cuspidata | 35.17.05 | 136.16.00 | 1 |
| 16 | Mt. Hira, Shiga | Quercus crispula | 35.12.50 | 135.53.35 | 5 |
| 17 | Higashiyama, Kyoto | Castanopsis cuspidata | 35.00.10 | 135.47.15 | 4 |
| Quercus glauca | 1 | ||||
| 18 | Yoshida, Kyoto | Quercus serrata | 35.01.35 | 135.47.05 | 4 |
| Quercus glauca | 2 | ||||
| Castanopsis cuspidata | 1 | ||||
| 19 | Fushimi, Kyoto | Castanopsis cuspidata | 34.58.05 | 135.46.55 | 1 |
| 20 | Kasuga, Nara | Quercus gilva | 34.40.40 | 135.51.25 | 4 |
| Castanopsis cuspidata | 2 | ||||
| 21 | Oki Is., Shimane | Quercus serrata | 36.10.35 | 133.18.10 | 4 |
| 22 | Matsue, Shimane | Quercus acuta | 35.28.55 | 133.03.05 | 2 |
| 23 | Ooda, Shimane | Quercus myrsinaefolia | 35.11.55 | 132.28.05 | 4 |
| 24 | Kanayama, Okayama | Castanopsis sieboldii var.sieboldii | 34.44.20 | 133.56.45 | 2 |
| 25 | Kojima, Okayama | Castanopsis cuspidata | 34.30.20 | 133.50.55 | 1 |
| 26 | Niho, Yamaguchi | Quercus gilva | 34.12.60 | 131.32.45 | 4 |
| Castanopsis cuspidata | 2 | ||||
| 27 | Ogori, Yamaguchi | Castanopsis cuspidata | 34.06.35 | 131.23.55 | 2 |
| 28 | Kashii, Fukuoka | Quercus gilva | 33.39.10 | 130.27.10 | 2 |
| 29 | Kasuga, Fukuoka | Quercus gilva | 33.31.20 | 130.28.10 | 3 |
| 30 | Nobeoka, Miyazaki | Quercus gilva | 32.37.15 | 131.34.35 | 3 |
| 31 | Aya, Miyazaki | Quercus salicina | 32.01.40 | 131.10.10 | 8 |
| Quercus gilva | 5 | ||||
| Quercus acuta | 3 | ||||
| Lithocarpus edulis | 1 | ||||
| 32 | Issou, Yaku Is., Kagoshima | Lithocarpus edulis | 30.27.15 | 130.29.10 | 3 |
| 33 | Hanyama, Yaku Is., Kagoshima | Quercus salicina | 30.22.10 | 130.23.05 | 1 |
Sequencing of mtDNA
We analyzed sequence data for 115 C. sikkimensis individuals from 33 populations with 1–17 individuals per population (Table 1), from 11 plant species of the three genera, Quercus, Lithocarpus, and Castanopsis. Total DNA was extracted from the thoracic muscles of larvae using a standard proteinase K/SDS digestion and phenol-chloroform extraction. DNA fragments of the coding regions of mitochondrial cytochrome oxidase subunits I (COI) were amplified using the following PCR primers: COI-1729F, 5'-GGATCACCTGATATAGCATTCCC-3' and COI-2764R, 5'-CCTAAAAAATGTTGTGGGAAAAAGG-3' modified from [26]. Amplification of mtDNA was performed as follows: an initial 3 min denaturation (94°C) followed by 40 cycles of 0.5 min at 94°C, 0.5 min at 50°C, and 1 min at 70°C, and a final extension cycle of 10 min at 72°C. All PCRs were performed in a total volume of 20 μL using Ex-Taq polymerase (TaKaRa, Otsu, Japan), and the products were purified using ExoSAP-IT reagent (USB, Cleveland, OH, USA). Sequencing reactions were prepared using a BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) and analyzed on a PRISM 3100 sequencer (Applied Biosystems). Sequences [DDBJ accession nos. AB177451–AB177468, AB367602–AB367606, and AB457208–AB457233] were aligned using Sequence Navigator software (Applied Biosystems).
Data analyses
Haplotype diversity (h), nucleotide diversity (π), and Tajima's D [27] were calculated using Arlequin version 2.0 [28]. A statistical parsimony network [29] was constructed to investigate relationships among haplotypes within C. sikkimensis using the program TCS[30]. Ambiguous loops in the network were resolved following the recommendations of [31]. An analysis of molecular variance (AMOVA) was used to examine the amount of genetic variability partitioned within and among populations as well as among groups of populations [32] using Arlequin. To assess the significance of the various factors that could affect partitioning of genetic variability, populations were grouped according to a set of grouping criteria, including geographic distribution, host plant genera, and vegetation types. The significance of fixation indices was tested using a nonparametric approach [32]. When AMOVA analysis could not determine what number of partitioned groups is the most suitable, a spatial analysis of molecular variance (SAMOVA) based on a simulated annealing procedure was used to define the groups of populations [33]. The program SAMOVA 1.0 iteratively seeks the composition of a user-defined number (K) of groups of geographically adjacent populations that maximizes ΦCT. The program was run for 10,000 iterations for K = 2 to 4 from each of 100 random initial conditions.
Results
Haplotype diversity and the geographical distribution of C. sikkimensis
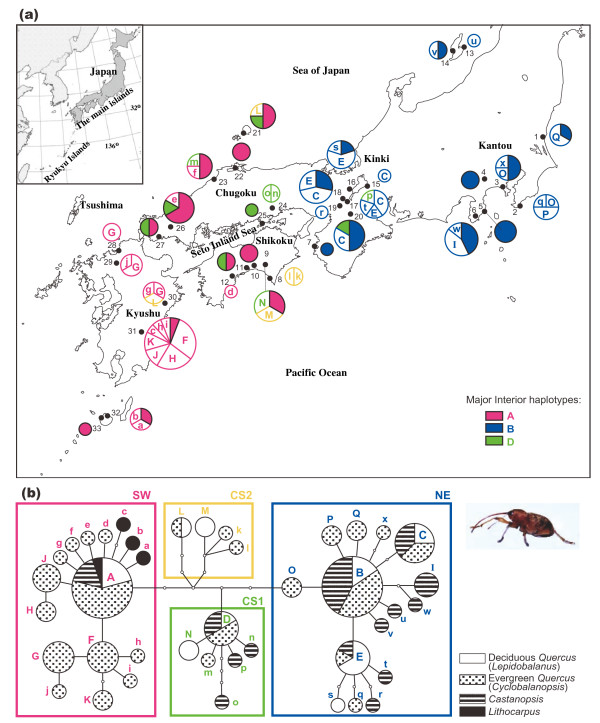
We identified 47 variable sites in the mtDNA regions and 41 haplotypes in total, of which 24 haplotypes were found only once. The calculated values of haplotype diversity and nucleotide diversity are provided in Table 2. Haplotypes found in more than two individuals are denoted by uppercase letters, with the A-haplotype being the most common. Haplotypes found only once are denoted in lowercase letters. The geographical distribution of the recognized mtDNA haplotypes of C. sikkimensis was highly structured (Fig. 1a).
Table 2.
Polymorphism summary statistics based on 921 bp of Curculio sikkimensis mtDNA
| No. samples | No. haplotypes | Haplotype diversity (h) | Nucleotide diversity (π) | Tajima's D |
| 115 | 41 | 0.9333 | 0.004654 | -1.60560 |
| P = 0.024 |
Figure 1.
Geographic distribution of mtDNA haplotypes of Curculio sikkimensis. (a) Geographic distribution of 41 mtDNA haplotypes found among 115 individuals of the generalist seed parasitic weevil, C. sikkimensis. Sample size ranged from 1 to 17 individuals per population. Circle sizes are proportional to haplotype frequencies. See Table 1 for the exact locations of populations and host plant species. (b) Relatedness among haplotypes detected in 921 bp of mtDNA (COI) of C. sikkimensis, represented in a statistical parsimony haplotype network. Each haplotype indicates the proportion of a host plant genus. Circle sizes are proportional to haplotype frequencies, and open dots indicate missing intermediate haplotypes.
The statistical parsimony haplotype network within C. sikkimensis with a 95% connection limit is shown in Fig 1b. Four main haplogroups (NE, CS1, CS2, and SW) were apparent in the network. Haplogroups NE and SW were found in the populations from northeastern (Kantou to Kinki district) and southwestern parts (Kyushu to the Chugoku-Shikoku region) of the main islands of Japan, respectively. Haplogroups CS1 and CS2 consisted of C. sikkimensis found in the Chugoku and Shikoku regions. The haplogroups NE, SW, and CS1 showed starlike shapes centered on haplotypes B, A, and D, respectively, which were the most frequently observed haplotypes.
Genetic structure observed in C. sikkimensis
No significant genetic structure of C. sikkimensis was observed when compared to the genera of the host plant species. Most haplotypes were found from more than two individuals, and they were shared among the individuals of C. sikkimensis collected from different host plant genera of Fagaceae (Fig. 1b). For example, weevil individuals with haplotype A were found in the seeds of all types of host plant genera. Moreover, the AMOVA revealed no genetic differentiation by the vegetation types that included host plants (-0.03%, P > 0.10; Table 3) or among the functional host plant groups (6%, P > 0.10; Table 3). The results suggest that the differences in living vegetation zones as well as in utilized host plant species have not contributed to shaping the present genetic structure of C. sikkimensis.
Table 3.
Analysis of molecular variance (AMOVA) of genetic variation in 921 bp of mtDNA of Curculio sikkimensis
| Grouping criteria and source of variation | df | Sum of squares | Variance components | % of total variation | Φ statistics with P-value |
| Geographic distribution I: [Kyushu, Chugoku to Shikoku] [Kinki to Kantou] | |||||
| Among groups | 1 | 91.7 | 1.56 | 52.8 | ΦCT = 0.52 |
| Among populations within groups | 31 | 64.5 | 0.29 | 10.0 | (P < 0.001) |
| Within populations | 82 | 89.6 | 1.09 | 37.0 | |
| Total | 114 | 245.9 | 2.95 | ||
| Geographic distribution II: [Kyushu] [Chugoku to Shikoku] [Kinki to Kantou] | |||||
| Among groups | 2 | 103.1 | 1.32 | 50.3 | ΦCT = 0.50 |
| Among populations within groups | 30 | 53.2 | 0.21 | 8.1 | (P < 0.001) |
| Within populations | 82 | 89.6 | 1.09 | 41.5 | |
| Total | 114 | 245.9 | 2.62 | ||
| Host plants: [Deciduous] [Evergreen] | |||||
| Among groups | 1 | 4.0 | -0.07 | -3.3 | ΦCT = -0.03 |
| Among populations within groups | 38 | 157.6 | 1.09 | 51.0 | (P > 0.10) |
| Within populations | 75 | 84.2 | 1.12 | 52.3 | |
| Total | 114 | 245.9 | 2.14 | ||
| Host plants: [Deciduous Quercus] [Evergreen Quercus] [Castanopsis] [Lithocarpus] | |||||
| Among groups | 3 | 25.5 | 0.15 | 6.8 | ΦCT = 0.06 |
| Among populations within groups | 36 | 136.0 | 0.97 | 43.2 | (P > 0.10) |
| Within populations | 75 | 84.2 | 1.12 | 49.8 | |
| Total | 114 | 245.9 | 2.25 | ||
Groupings of populations were examined based on their geographic distributions and host plants.
The geographical distributions of the recognized mtDNA haplotypes in C. sikkimensis were highly structured (Fig. 1a). The AMOVA showed strong genetic structure when grouping the populations according to their geographic distribution (Table 3). A large proportion (almost 50%, P < 0.001) of the total genetic variance was explained by differences among regions when comparing three geographical regions (i.e., Kyushu, locality numbers 28–33; Chugoku to Shikoku, locality numbers 21–27 and 8–12; and Kinki to Kantou, locality numbers 13–20 and 1–7) as well as two geographical regions (Kyushu to Chugoku-Shikoku, and Kinki to Kantou). The SAMOVA revealed that the value of ΦCT was larger at K = 2 than at K = 3, which indicated that partitioning into two groups was more appropriate. This result suggests that strong genetic differentiation exists between the southwestern and northeastern regions of the main islands of Japan.
Comparison of the phylogeographical pattern in C. sikkimensis and C. hilgendorfi
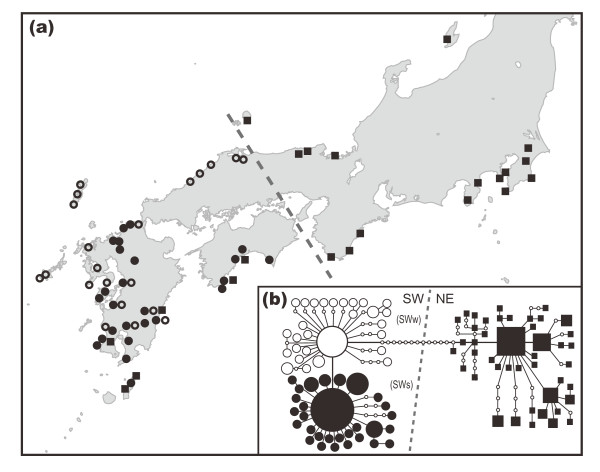
The haplotype and nucleotide diversities of the main islands of Japan showed similar values in C. sikkimensis (Table 2) and C. hilgendorfi (h = 0.964 and π = 0.00385, [24]). Congruent phylogeographical patterns were observed between C. sikkimensis (Fig. 1) and C. hilgendorfi (Fig. 2, [24]), with a deep gap between the SW and NE clades with respect to the mtDNA sequences. Two differences in the phylogeographical patterns were observed between C. sikkimensis and C. hilgendorfi. The first was the existence of CS clades in C. sikkimensis in the Chugoku and Shikoku regions, and the second was the lack of genetic differentiation in the SW clade of C. sikkimensis between the coasts of Pacific and the Sea of Japan.
Figure 2.
Geographic distribution of mtDNA haplotypes of Curculio hilgendorfi on the main islands of Japan (modified from [24]). (a) Geographic distribution of 102 mtDNA haplotypes found among 189 individuals of the host-specific seed parasitic weevil C. hilgendorfi on the main islands of Japan. (b) Relatedness among haplotypes detected in 2709 bp of mtDNA (COI, COII, and ND5) of C. hilgendorfi, represented in a statistical parsimony haplotype network. The values of haplotype diversity and nucleotide diversity of NE and SW clades are 0.964 and 0.00385, respectively.
Discussion
Phylogeographical patterns in C. sikkimensis
The observed gap between the SW and NE clades with respect to the mtDNA sequences of C. sikkimensis (Fig. 1) suggests that the southwestern and northeastern insect populations have been isolated for a long time, probably through several glacial and interglacial periods; the genetic groups have not been in contact for several thousands of years since the last glacial period; and long-distance migration of the weevil has been rare, resulting in negligible interpopulation genetic exchange events. This result suggests past fragmentation of this weevil species into at least two separate regions. Within the CS clade, the most common interior haplotype, D, was spread over the Chugoku and Shikoku regions. Genetic differentiation between the southwestern, northeastern, and Chugoku-Shikoku region was detected by the AMOVA. These results indicate the genetic uniqueness of the weevils distributed in the Chugoku-Shikoku region. Within the NE clade, a sub-haplogroup including the interior haplotype E was observed. These haplotypes also were often found in the Kinki region. Future studies examining more samples from this population may also reveal genetic uniqueness of the insects distributed in the Kinki region.
Molecular phylogenetic and ecomorphological studies on the genus Curculio in Europe and North America have suggested that the seed size of host plants is an important selective agent for changes in rostrum length and body size, and thus may be a key factor promoting morphological differentiation [21,34]. In our study, genetic differentiation of C. sikkimensis was not observed among the associated deciduous and evergreen forests or among host plant genera of Fagaceae (Table 3, Fig. 1b). These results suggest that genetic differentiation accompanied by host specialization among Fagaceae species is not occurring in C. sikkimensis, which is a uniquely polyphagous weevil species. Ecological and morphological studies of the weevils and their host plants in populations where various host plant species are sympatrically distributed may elucidate the evolution of Japanese acorn weevils.
Phylogeographical pattern of the generalist and the host-specific seed parasitic weevils
A greater genetic differentiation between NE and SW clades was found for the specialist seed predator C. hilgendorfi compared to the generalist predator C. sikkimensis (Figs. 1b, 2). This was likely the result of the more limited geographical distribution of the host plants (Castanopsis) of the specialist C. hilgendorfi. A geographic boundary of the mtDNA haplotypes was observed in the Chugoku-Shikoku region in the two different host types of Curculio. Such boundaries were also observed in the chloroplast DNA of several plant species growing in warm and cool temperate zones in Japan [[5,35], T. Iwasaki et al. unpublished data]. In Europe, the Alps and Pyrenees may have blocked the dispersal of many animal and tree species from Italian and Iberian refugia to the northern part of Europe [reviewed in [10,17]]. However, in the case of the geographical boundary in the Chugoku-Shikoku region in Japan, neither a north-south mountain range nor a temperature gap has been observed. Other geographical and climatic features of the Seto Inland Sea, such as a currently drier climate and limited on-shore wind, and the historical existence of a grassland landscape during dry cool climates in the glacial ages, might have served as barriers against recolonization of various species from their refugia.
Two differences in phylogeographical patterns were observed between C. sikkimensis and C. hilgendorfi. Castanopsis sieboldii var.sieboldii, which is the major host plant species of C. hilgendorfi, is rarely distributed around the Seto Inland Sea. In the case of the host-specific seed predator C. hilgendorfi, the lack of distribution of the main host plant species around the Seto Inland Sea may have prompted genetic differentiation between C. hilgendorfi on the Pacific Coast and those on the coast of the Sea of Japan. In contrast, C. sikkimensis is a generalist seed predator of Fagaceae plants and could migrate across the Seto Inland Sea using various Fagaceae plant species in the Chugoku and Shikoku regions. This polyphagous habit allowed C. sikkimensis to cross the Seto Inland Sea and also may have generated the intermediate CS clades. A more detailed history of the area around the Seto Inland Sea can be estimated by comparing the phylogeographical patterns of various organisms with diverse geographic distributions and life histories.
Insight into the glacial and postglacial history of temperate forests in Japan
Little phylogeographical data are available for the host plant species of these weevils due to the relatively slow rate of molecular evolution of plant chloroplast DNA [36-39], which is usually maternally inherited and thus often used in molecular phylogeographical studies. Extremely low levels of intraspecific variation in the chloroplast DNA of Japanese broadleaved evergreen species [40,41], including Castanopsis and Lithocarpus, as well as the possible introgression among Quercus species [41-43], have made it difficult to perform phylogeographical analyses of host plants. Among these host plant species, a phylogeographical pattern has been reported in deciduous Quercus; the geographic distribution of cpDNA haplotypes in deciduous Quercus indicated that one of the haplotypes is restricted to eastern Japan [44].
In this study C. sikkimensis living both in the broadleaved deciduous and evergreen forests and C. hilgendorfi living only in the broadleaved evergreen forests and in several plant species growing in these forests, shared a common phylogeographical pattern, with both exhibiting a genetic gap between the northeastern and southwestern parts of Japan. The geographic distributions of genetic diversity of plant species and those of their parasitic insects suggest that both were restricted to separate southwestern and northeastern refugia on the main islands of Japan during repeated glacial periods in the Quaternary. Moreover, these phytophagous insects, especially flightless or nonmigratory species, likely responded to the environmental changes associated with the glacial cycles in a fashion similar to that of their host plants. If the plant-insect association (i.e., the host range of the weevil) remained stable throughout the Quaternary, the presence of the phytophagous insects may be an indicator of the coverage of the temperate forests. Thus, incorporating phylogeographical information about herbivorous insect species with that of their host plant species could more precisely elucidate the phylogeographical patterns of their associated forests.
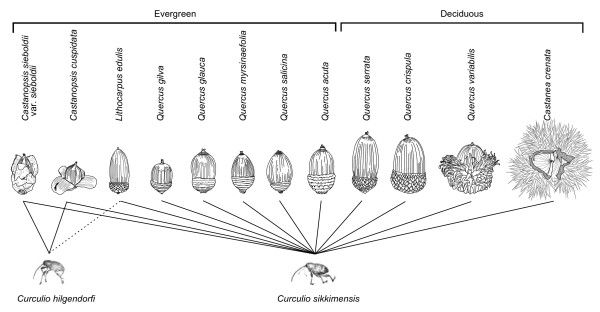
We did not find any genetic differentiation in the mtDNA of C. sikkimensis among their associated host plant species, which suggests that weevils occupying different host species do not shape host races. Although the size and shape of the utilized acorns varied among plant species (Fig. 3), C. sikkimensis can use almost all of these host plant species. Irrespective of the difference in host range between polyphagous C. sikkimensis and monophagous C. hilgendorfi, the phylogeographical patterns were very similar between the two species. This similarity is hypothesized to be due to (1) similar past distributions of refugia between deciduous oak and evergreen forests, or (2) similar past host ranges. That is, C. sikkimensis might have been restricted to evergreen Castanopsis species in the last glacial age and may have recently expanded its host range independently in each refugium after the last glacial maximum. In the former case, if some populations of C. sikkimensis survived during the glacial periods within deciduous oak forests and others in evergreen forests in different isolated refugia, some host specialization could have arisen or have been reinforced. The observation that the weevils inhabiting different acorn species do not shape host races would suggest that the populations of C. sikkimensis inhabiting deciduous oak forests and those in evergreen forests were not isolated during the long glacial periods. At the last glacial maximum, global sea level is estimated to have dropped to approximately 140 m lower than the present level [45]. The narrowing of the Tsushima Strait at that time caused a reduction in the Tsushima warm current into the Sea of Japan, resulting in further cooling and the aridification of the Japan Archipelago. Given this cool and arid climate, the areas covered by coniferous forests and cool-mixed forests expanded southwards [46]. This suggests that both deciduous oak and evergreen forests were restricted to the adjacent refugia at that time, although it is possible that the association between the insects and their host plant species could have established after the glacial period, as suggested by scenario 2 above. However, it is unlikely that C. sikkimensis expanded its host range to several deciduous and evergreen oak species independently from each refugium as recently as the last 20,000 years. In addition, in the European Curculio species, closely related weevils (e.g., C. humeralis, C. pardalis, and C. victoriensis) do not associate with common host plant species [21].
Figure 3.
Sizes and shapes of the acorns used by Curculio sikkimensis and C. hilgendorfi. The acorns and weevils have been drawn at an approximate scale of 1:2.
The coincident patterns of C. sikkimensis and C. hilgendorfi strongly support the hypothesis that both deciduous oak and evergreen forest types survived together or adjacent to one another in small refugia during glacial ages, although these forests are presently distributed in cool and warm temperate zones in Japan, respectively. A more detailed investigation of the pollen records or further genetic studies of organisms living in these types of forests will provide further insight into the evolutionary history of the broadleaved deciduous and evergreen forests of Japan.
Conclusion
Our data suggest that geology and historical environment have contributed to shaping the recent genetic structure of C. sikkimensis in temperate forests of Japan. Comparing the phylogeographical patterns of two types of phytophagous insects (i.e., a generalist seed predator living in both the cool and warm temperate zones and the host-specific seed predator living only in the warm temperate zone) and of several plant species growing in these temperate zones of Japan enabled us to specify the historical processes of these forests more precisely. We demonstrated that during glacial ages the cool and warm temperate forests in Japan survived together, or adjacent to each other, at least in the southwestern and northeastern parts of the main islands. Our analyses provide a foundation for studying the evolutionary history of the broadleaved deciduous and evergreen forests in Japan, which characterize the biodiversity and endemism of the cool and warm temperate zones, where the effects of climate change are particularly severe.
Authors' contributions
KA designed of the study, performed the molecular genetic studies, and drafted the manuscript. MK and NM helped in drafting the manuscript. All authors have read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank H. Toju for technical support in analyzing insect materials, and N. Tanaka, S. Kariyama, S. Morinaga, T. Sakura, T. Yumoto, R. Tsujino, A. Seo, S. Matsumura, S. Watanabe, Y. Kameda, A. Nagano, T. Hattori, A. Nakano, T. Suzuki, Y. Yoshida, K. Asami, and A. Nakagoshi for their assistance in the collection of seeds. We also thank the members of the Japanese Ferniest Club, T. Minamitani, Y. Omae, T. Kiuchi, H. Nishikawa, and M. Okubo, for sending us seed samples. This study was partly supported by Grants-in-Aid from the Japan Society for the Promotion of Science (no. 1701416 to K.A. and no. 16405014 to N.M).
Contributor Information
Kyoko Aoki, Email: aoki@sys.bot.kyoto-u.ac.jp.
Makoto Kato, Email: kato@zoo.zool.kyoto-u.ac.jp.
Noriaki Murakami, Email: nmurak@tmu.ac.jp.
References
- Minato M, Ijiri S. The Japanese archipelago (in Japanese) 3. Tokyo, Japan: Iwanamishoten; 1976. [Google Scholar]
- Tsukada M. Paleoecology. II. Synthesis (in Japanese) Tokyo, Japan: Kyoritsu; 1974. [Google Scholar]
- Avise JC. Phylogeography. London, UK: Harvard University Press; 2000. [Google Scholar]
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Aoki K, Suzuki T, Hsu T-W, Murakami N. Phylogeography of the component species of broad-leaved evergreen forests in Japan, based on chloroplast DNA. J Plant Res. 2004;117:77–94. doi: 10.1007/s10265-003-0132-4. [DOI] [PubMed] [Google Scholar]
- Arbogast BS, Kenagy GJ. Comparative phylogeography as an integrative approach to historical biogeography. J Biogeogr. 2001;28:819–825. doi: 10.1046/j.1365-2699.2001.00594.x. [DOI] [Google Scholar]
- Avise JC. Molecular population structure and the biogeographic history of a regional fauna: a case history with lessons for conservation biology. Oikos. 1992;63:62–76. doi: 10.2307/3545516. [DOI] [Google Scholar]
- Brochmann C, Gabrielsen TM, Nordal I, Landvik JY, Elven R. Glacial survival or tabula rasa? The history of North Atlantic biota revisited. Taxon. 2003;52:417–450. doi: 10.2307/3647444. [DOI] [Google Scholar]
- Fujii N, Ueda K, Watano Y, Shimizu T. Intraspecific sequence variation of chloroplast DNA in Pedicularis chamissonis Steven (Scrophulariaceae) and geographic structuring of the Japanese "alpine" plants. J Plant Res. 1997;110:195–207. doi: 10.1007/BF02509308. [DOI] [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc Lond, Ser B: Biol Sci. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Aguinagalde I, de Beaulieu J-L, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, et al. Glacial refugia: hotspots but not melting pots of genetic diversity. Science. 2003;300:1563–1565. doi: 10.1126/science.1083264. [DOI] [PubMed] [Google Scholar]
- Seo A, Watanabe M, Hotta M, Murakami N. Geographical patterns of allozyme variation in Angelica japonica (Umbelliferae) and Farfugium japonicum (Compositae) on the Ryukyu Islands, Japan. Acta Phytotax Geobot. 2004;55:29–44. [Google Scholar]
- Soltis DE, Gitzendanner MA, Strenge DD, Soltis PS. Chloroplast DNA intraspecific phylogeography of plants from the Pacific Northwest of North America. Plant Syst Evol. 1997;206:353–373. doi: 10.1007/BF00987957. [DOI] [Google Scholar]
- Soltis DE, Morris AB, McLachlan JS, Manos PS, Soltis PS. Comparative phylogeography of unglaciated eastern North America. Mol Ecol. 2006;15:4261–4293. doi: 10.1111/j.1365-294X.2006.03061.x. [DOI] [PubMed] [Google Scholar]
- Stehlik I. Resistance or emigration? Response of alpine plants to the ice ages. Taxon. 2003;52:499–510. doi: 10.2307/3647448. [DOI] [Google Scholar]
- Stewart JR, Lister AM. Cryptic northern refugia and the origins of the modern biota. Trends Ecol Evol. 2001;16:608–613. doi: 10.1016/S0169-5347(01)02338-2. [DOI] [Google Scholar]
- Taberlet P, Fumagalli L, Wust-Saucy A-G, Cosson J-F. Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- Griffin SR, Barrett SCH. Post-glacial history of Trillium grandiflorum (Melanthiaceae) in eastern North America: inferences from phylogeography. Am J Bot. 2004;91:465–473. doi: 10.3732/ajb.91.3.465. [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Miyoshi N. Chapter III-4 (in Japanese) In: Yasuda Y, Miyoshi N, editor. The illustrated vegetation history of the Japanese Archipelago. Tokyo, Japan: Asakura-shoten; 1998. pp. 224–236. [Google Scholar]
- Tsukada M. A vegetation map in the Japanese Archipelago approximately 20,000 years B.P. (in Japanese with English abstract) Jap J Ecol. 1984;34:203–208. [Google Scholar]
- Hughes J, Vogler AP. Ecomorphological adaptation of acorn weevils to their oviposition site. Evolution. 2004;58:1971–1983. doi: 10.1111/j.0014-3820.2004.tb00483.x. [DOI] [PubMed] [Google Scholar]
- Anderson RS. An evolutionary perspective on diversity in Curculionoidea. Mem Entomol Soc Wash. 1995;14:103–114. [Google Scholar]
- Gibson LP. Monograph of the genus Curculio in the New World (Coleoptera: Curculionidae). I. United States and Canada. Misc Publs Ent Soc Am. 1969;6:239–285. [Google Scholar]
- Aoki K, Kato M, Murakami N. Glacial bottleneck and postglacial recolonization of a seed parasitic weevil, Curculio hilgendorfi, inferred from mitochondrial DNA variation. Mol Ecol. 2008;17:3276–3289. doi: 10.1111/j.1365-294X.2008.03830.x. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Morimoto T, Kimoto S. The coleoptera of Japan in color, vol. IV (in Japanese) Higashiosaka, Japan: Hoikusha; 1984. [Google Scholar]
- Toju H, Sota T. Phylogeography and the geographic cline in the armament of a seed-predatory weevil: effects of historical events vs. natural selection from the host plant. Mol Ecol. 2006;15:4161–4173. doi: 10.1111/j.1365-294X.2006.03088.x. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L. ARLEQUIN, Version 2000: A software for population genetics data analysis. Geneva, Switzerland: University of Geneva; 2000. http://lgb.unige.ch/arlequin/ [Google Scholar]
- Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. http://darwin.uvigo.es/software/tcs.html [DOI] [PubMed] [Google Scholar]
- Pfenninger M, Posada D. Phylogeographic history of the land snail Candidula unifasciata (Helicellinae, Stylommatophora): Fragmentation, corridor migration, and secondary contact. Evolution. 2002;56:1776–1788. doi: 10.1111/j.0014-3820.2002.tb00191.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: applications to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupanloup I, Schneider S, Excoffier L. A simulated annealing approach to define the genetic structure of populations. Mol Ecol. 2002;11:2571–2581. doi: 10.1046/j.1365-294X.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- Hughes J, Vogler AP. The phylogeny of acorn weevils (genus Curculio) from mitochondrial and nuclear DNA sequences: the problem of incomplete data. Mol Phylogen Evol. 2004;32:601–615. doi: 10.1016/j.ympev.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Aoki K, Matsumura T, Hattori T, Murakami N. Chloroplast DNA phylogeography of Photinia glabra (Rosaceae) in Japan. Am J Bot. 2006;93:1852–1858. doi: 10.3732/ajb.93.12.1852. [DOI] [PubMed] [Google Scholar]
- Aoki K, Kato M, Murakami N. Mitochondrial DNA of phytophagous insects as a molecular tool for phylogeographic study of host plants. Acta Phytotax Geobot. 2005;56:55–69. [Google Scholar]
- Chase MW, Soltis DE, Olmstead RG, Morgan D, Les DH, Mishler BD, Duvall MR, Price RA, Hills HG, Qiu Y-L, et al. Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Ann Mo Bot Gard. 1993;80:528–580. doi: 10.2307/2399846. [DOI] [Google Scholar]
- Frascaria N, Maggia L, Michaud M, Bousquet J. The rbcL gene sequence from chestnut indicates a slow rate of evolution in the Fagaceae. Genome. 1993;36:668–671. doi: 10.1139/g93-089. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Li W-H, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Biol J Linn Soc. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Hattori T, Murakami N. Intraspecific sequence variation of chloroplast DNA among the component species of evergreen broad-leaved forests in Japan II. Acta Phytotax Geobot. 2004;55:125–128. [Google Scholar]
- Aoki K, Suzuki T, Murakami N. Intraspecific sequence variation of chloroplast DNA among the component species of evergreen broad-leaved forests in Japan. J Plant Res. 2003;116:337–344. doi: 10.1007/s10265-003-0103-9. [DOI] [PubMed] [Google Scholar]
- Dumolin-Lapègue S, Kremer A, Petit RJ. Are chloroplast and mitochondrial DNA variation species independent in oaks? Evolution. 1999;53:1406–1413. doi: 10.2307/2640887. [DOI] [PubMed] [Google Scholar]
- Whittemore AT, Schaal BA. Interspecific gene flow in sympatric oaks. Proc Natl Acad Sci USA. 1991;88:2540–2544. doi: 10.1073/pnas.88.6.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno M, Yokoyama J, Suyama Y, Ohyama M, Itoh T, Suzuki M. Geographical distribution of two haplotypes of chloroplast DNA in four oak species (Quercus) in Japan. J Plant Res. 2004;117:311–317. doi: 10.1007/s10265-004-0160-8. [DOI] [PubMed] [Google Scholar]
- Yokoyama Y, Kido Y, Tada R, Minami I, Finkel RC, Matsuzaki H. Japan Sea oxygen isotope stratigraphy and global sea-level changes for the last 50,000 years recorded in sediment cores from the Oki Ridge. Palaeogeogr, Palaeoclimatol, Palaeoecol. 2007;247:5–17. doi: 10.1016/j.palaeo.2006.11.018. [DOI] [Google Scholar]
- Gotanda K, Yasuda Y. Spatial biome changes in southwestern Japan since the Last Glacial Maximum. Quat Int. 2008;184:84–93. doi: 10.1016/j.quaint.2007.09.029. [DOI] [Google Scholar]