Abstract
The mitochondrial transcription factor A (mtTFA) is central to assembly and initiation of the mitochondrial transcription complex. Human mtTFA (h-mtTFA) is a dual high mobility group box (HMGB) protein that binds site-specifically to the mitochondrial genome and demarcates the promoters for recruitment of h-mtTFB1, h-mtTFB2 and the mitochondrial RNA polymerase. The stoichiometry of h-mtTFA was found to be a monomer in the absence of DNA, whereas it formed a dimer in the complex with the light strand promoter (LSP) DNA. Each of the HMG boxes and the C-terminal tail were evaluated for their ability to bind to the LSP DNA. Removal of the C-terminal tail only slightly decreased nonsequence specific DNA binding, and box A, but not box B, was capable of binding to the LSP DNA. The X-ray crystal structure of h-mtTFA box B, at 1.35 Å resolution, revealed the features of a noncanonical HMG box. Interactions of box B with other regions of h-mtTFA were observed. Together, these results provide an explanation for the unusual DNA-binding properties of box B and suggest possible roles for this domain in transcription complex assembly.
INTRODUCTION
The mitochondrion is unique among the organelles in eukaryotes because it contains its own genome independent of the nucleus and replicates this genome separately from the cell cycle (1). The human mitochondrial genome (mtDNA) is a circular double-stranded DNA molecule of 16 569 base pairs in length (2,3), which encodes 13 protein components of the oxidative phosphorylation pathway, 22 tRNAs and two rRNAs (2,4). The oxidative environment of the mitochondria and lack of an efficient DNA repair mechanism leave the mitochondrial genome highly susceptible to mutations, and such mutations have been implicated in a variety of disease processes (5,6).
Transcription of the mitochondrial genome initiates from three promoters: the light strand promoter (LSP) or two heavy strand promoters (HSP1 and HSP2) that are found in the 1.1 kb displacement loop region of the mtDNA (4,7). This generates long primary polycistronic transcripts that contain no introns. Transcription from the HSP1 produces the two rRNA species, whereas the tRNA and mRNA species are generated from transcripts initiating at the HSP2 and LSP (8). In contrast to transcription in the nucleus, only a small number of nuclear encoded protein components are required for mitochondrial transcription. These factors include the mitochondrial transcription factor A (h-mtTFA also known as TFAM), mitochondrial transcription factor B1 and B2 (h-mtTFB1 and h-mtTFB2), as well as the bacteriophage-related mitochondrial RNA polymerase (4,9).
Initiation of mitochondrial transcription requires specification of the mitochondrial promoter by h-mtTFA and subsequent recruitment of additional components of the transcription machinery. The first transcription factor of the mitochondrial transcription system to be discovered was h-mtTFA and DNaseI footprinting identified its binding sites in the LSP (10,11). The later back-to-back discovery and functional analyses of the factors h-mtTFB1 and h-mtTFB2 revealed that both proteins have roles in transcription activation (12,13). Interestingly, both h-mtTFB1 and h-mtTFB2 also have RNA methyltransferase activity (12–16). This led to the proposal that h-mtTFB1 and h-mtTFB2 have dual functions in the mitochondria, acting both as transcription factors and as methyltransferases that may aid in coupling transcription to translation in mitochondria (12,13,15,17). h-mtTFB1 and h-mtTFB2 interact with both the basic 25 amino acid residue tail of h-mtTFA and the mitochondrial RNA polymerase, and h-mtTFB1 has been shown to activate transcription independently of its methyltransferase activity (15).
mtTFA is a member of the high mobility group (HMGB) superfamily of DNA binding proteins defined by the HMG DNA binding domain (known as the HMG box). The HMG box is an L-shaped three-helix domain that binds to DNA in the minor groove and dramatically bends and unwinds DNA with the help of DNA intercalating residues. One group of HMGB proteins is associated with the maintenance and architecture of DNA in nuclear chromatin through nonsequence-specific DNA binding, and another is involved in the transcriptional activation of specific genes through site-specific DNA binding in gene promoter regions (18–20). The sequence-specific HMGB proteins typically contain one HMG domain, whereas the nonsequence-specific HMGB proteins usually have two HMG domains. mtTFA is unique in that it is site-specific, but contains tandem HMG-box DNA-binding domains. A 27 amino acid residue linker connects the HMG boxes, which are followed by a 25 residue carboxyl-terminal tail that is rich in basic amino acid residues. The C-terminal tail is essential for specific DNA recognition as well as transcription initiation and activation (21,22). h-mtTFA activates transcription by binding upstream of transcriptional start sites and induces conformational changes in the DNA such as bending and DNA unwinding (11). In contrast to other HMGB proteins, there is evidence that mtTFA can oligomerize. A dimer of h-mtTFA is needed to drive mtDNA into a compacted nucleoid structure (23), and in Xenopus laevis mtTFA (xl-mtTFA) tetramer formation is necessary for bidirectional transcriptional activation (24).
In addition to its role as a transcription factor, mtTFA has also been implicated as a primary structural factor in forming the architecture of the mitochondrial genome. The ability of mtTFA to influence mtDNA copy number is largely independent of its role in transcription (17,25,26). h-mtTFA has been shown to bind to DNA in a nonsequence-specific fashion, as indicated by DNA binding without a detectable DNaseI footprint (26,27). xl-mtTFA was identified as one of the main protein components in the nucleoprotein complex (28), indicating a role in xl-mtDNA packaging. Therefore, mtTFA shares a number of features with both the transcription factor-type and the chromosomal-type HMG proteins.
Several mechanisms of how h-mtTFA may initiate assembly of the transcription complex on mitochondrial DNA have been proposed (4). However, the precise molecular mechanism of assembly remains unclear. We investigated the contribution of the HMG domains of h-mtTFA to mitochondrial DNA binding to obtain a better understanding how h-mtTFA assembles on the mitochondrial promoter. We first determined the stoichiometry of h-mtTFA in the presence and absence of promoter DNA using a variety of complementary biophysical methods. Next, a closer examination of the DNA binding propensity of the individual domains of h-mtTFA suggested interesting functional differences, and the crystal structure at 1.35 Å resolution of box B of h-mtTFA provided insights into these observed functional differences.
MATERIALS AND METHODS
Protein expression and purification
The plasmids for expression of full-length and deletion constructs of h-mtTFA were created using Gateway Cloning Technology (Invitrogen) as described in the Supplementary Methods section. The h-mtTFA sequences with an N-terminal PreScission protease cleavage site were inserted into destination vectors that contained either an N-terminal His6 (pDEST527) or N-terminal GST (pDEST15) tag for protein expression in Escherichia coli strain BL21(DE3) + Codon (Stratagene). The constructs were verified by DNA sequencing.
The proteins were overexpressed in the E. coli strain BL21(DE3) + codon in LB supplemented with 100 μg/ml ampicillin and 7 μg/ml chloramphenical. A 1 l culture was inoculated from a glycerol stock and allowed to grow for ∼16 h at 37°C without the addition of IPTG, which allowed accumulation of protein due to leaky expression. The cell pellet was resuspended in 20 mM Tris-HCl pH 8.0, 1 M NaCl, 1 mM EDTA, 10% glycerol, protease inhibitors (Roche), and 50 mg of lysozyme prior to lysis by sonication. After DNaseI treatment and centrifugation (28 435g), the cleared lysate was incubated with Glutathione Sepharose (GE Healthcare) or Ni2+-NTA (Qiagen) resin. The resins were washed with 1 L of lysis buffer followed by an additional wash with 1 l of cleavage buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 1mM EDTA and 1 mM DTT). The proteins were either cleaved from the resin with PreScission Protease or eluted from the resin with glutathione, to give the intact GST-fusion proteins. The proteins were then dialyzed (50 mM HEPES–Na pH 7.0, 50 mM NaCl, 1 mM EDTA and 1 mM DTT) and further purified using cation exchange chromatography (Source S, GE Healthcare) and size-exclusion chromatography (Superdex 200, GE Healthcare). For the His6-tagged proteins, EDTA was omitted from buffers, and the proteins were eluted from the Ni2+-NTA resin using imidazole.
Size-exclusion chromatography of h-mtTFA proteins
A 16/60 Superdex™ 200 prep grade gel filtration column (GE Healthcare) was equilibrated in running buffer (50 mM HEPES–Na pH 7, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT) at a flow rate of 1 ml/min. A calibration curve was prepared using blue dextran (2000 kDa), amylase (158 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa) and RNase A (13.7 kDa) (GE Healthcare). The elution of these markers was monitored by UV absorption at 280 nm, and the elution volume for each protein was measured from the start of the sample application to the apex of the elution peak. The logarithm of molecular weight was plotted against Kav that was calculated for each protein as follows: Kav = Ve–Vo/Vt – Vo, where Ve = elution volume for the protein; Vo = column void volume = elution volume of blue dextran 2000; Vt = total column volume. The cleaved h-mtTFA proteins were concentrated to a volume of 2 ml at between 3 and 10 mg/ml using 5000 MWCO Vivaspin 20 centrifugal filter devices (Sartorius) before loading onto the column.
Analytical ultracentrifugation
Sedimentation velocity ultracentrifugation experiments were performed with a Beckman Optical XL-A analytical ultracentrifuge at 25°C in 50 mM HEPES–Na pH 7.0, 150 mM NaCl, 1 mM EDTA and 1 mM DTT with a 60 Ti rotor at 50 000 r.p.m. h-mtTFA was eluted from the gel filtration column and loaded into a Epon charcoal-filled two sector center piece, and the running buffer was used for the reference chamber. Samples were detected by UV absorbance at 280 nm. Sedimentation coefficients were corrected using SEDNTERP (29), which gave Vbar = 0.7365 and rho = 1.00461. The experimental data were analyzed with the program SEDFIT (30).
Electrophoretic mobility shift assays
Stoichiometric and quantitative electrophoretic mobility shift assays (EMSA) were used with the following linear duplex DNA fragment: 5′-GTGTTAGTTGGGGGGTGACTGTTAAAAGTG-3′. For competition experiments either the unlabeled 30 bp fragment of the LSP or a scrambled nonspecific 30 bp fragment 5′-ACTACTAACAGACCGCAACCTAAACACAAC-3′ was utilized. The 32P end-labeled DNA was prepared as described previously (31). The labeled DNA was then annealed with an excess of the complimentary strand of DNA and separated from any remaining ssDNA by gel purification. Protein dilutions of h-mtTFA were made in 50 mM HEPES–Na pH 7.5, 150 mM KCl, 1 mM EDTA, 10% glycerol and 1 mM DTT to give a final concentration range of 0.5 nM to 100 µM. Each 10 µl reaction mixture included 2 µg/ml bovine serum albumin, 1 µl of 10× binding buffer (500 mM HEPES–Na pH 7.5, 500 mM KCl, 20 mM MgCl2), 1 µl of the protein dilution, and <1 nM radiolabeled DNA. The reactions were allowed to incubate at 22°C for 30 min prior to loading on a pre-run 6% native polyacrylamide gel (30:1, acrylamide: bis-acrylamide in 0.33× TBE). After electrophoresis, the gel was dried and exposed to a phosphorimaging screen overnight. A Molecular Dynamics phosphorimaging system was used to digitize the gel images, and ImageQuant software (Molecular Dynamics, Inc.) was used to integrate band intensities corresponding to free and bound DNA at each protein concentration. The fraction of DNA bound was calculated and plotted for each protein concentration using Kaleidagraph (Synergy software). The resulting binding curves were fit to a single-site binding isotherm Y = ([P]/Kd)/(1 + [P]/Kd), where [P] is the total protein concentration, and Y is the fraction bound, under conditions where the DNA concentration is at least 10-fold lower than the expected Kd. The equilibrium dissociation constant (Kd) was obtained as a result of the curve fit of the mean of at least three independent experiments.
For unlabeled DNA duplexes equal amounts of the two strands were resuspended in 50 mM Tris–HCL (pH 7.4), 1 mM EDTA, annealed and further purified using a DEAE anion exchange column (Tosahaas) with an increasing salt gradient in 50 mM Tris–HCL (pH 7.4), 1 mM EDTA. Protein dilutions of mtTFA were made in 50 mM HEPES–Na (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol and 1 mM DTT to give a final concentration of 10 µM. Each 10 µl reaction mixture included 2 µg/ml BSA, 1 µl of 10× binding buffer [200 mM HEPES-Na (pH 7.5), 500 mM KCl, 20 mM MgCl2], 0.5–2 µl of the protein dilution, and 1-3 µM DNA. The reactions were allowed to incubate at 25°C prior to loading on a pre-run 8% native polyacrylamide gel (30:1, acrylamide: bis-acrylamide in 0.33× TBE). The samples were electrophoresed for 30 min at 145 V. The gels were then incubated with Sybr Green (Invitrogen) for >20 min and a Molecular Dynamics phosphorimaging system was used to digitize the gel images, and ImageQuant software (Molecular Dynamics, Inc.) was used for visualization.
Crystallization and structure determination
h-mtTFA box B (mtTFA110-179) was crystallized at 18°C from a 1:1 ratio of protein to well solution (0.1 M acetate-Na pH 5.0, 0.1 M CdCl2 and 21% PEG 4000) in a hanging drop format, after optimization from initial crystals found in commercial crystallization screens (Hampton 1 and 2; Hampton Research). The crystals were cryoprotected with glycerol and PEG 4000 and flash frozen prior to data collection. The protein crystallized in the space group C2221 with a = 30.28 Å; b = 99.35 Å; c = 51.90 Å, with one molecule per asymmetric unit.
Data were collected at the Molecular Biology Consortium at the Advanced Light Source, Lawrence Berkeley National Laboratory (Beamline 4.2.2) and were indexed, integrated, and scaled using d*trek (Rigaku) (Table 2). Anomalous data were collected at 0.98 Å, due to the presence of bound CdCl2 in the crystals (Table 2). Single wavelength anomalous diffraction (SAD) methods were used to obtain experimental phases for the structure of h-mtTFA110–179. The heavy atom positions were found and the structure was phased using the program SOLVE (32) and electron density was improved using CCP4 program DM and RESOLVE (32). Much of the initial model was built into a 1.35 Å SAD experimental map using the program COOT (33) prior to the first round of refinement. Refmac5 (CCP4 suite of programs) was used for maximum likelihood refinement with cross-validation using 10% of the reflections reserved for the RFree calculation. Water molecules were added over the process of several refinement cycles, and the structure was monitored for correct geometry and stereochemistry using COOT with a final validation using PROCHECK (34). The final protein model includes amino acids 111–177, with alternate conformations for residues Tyr120, Leu140, Val143, Lys155, Tyr158, Met173 and Ser175, but amino acids 130–134 were built as alanine due to poor electron density for this region.
Table 2.
Data collection and refinement statistics
| Characteristics | mtTFA110–179 |
|---|---|
| Wavelength (Å) | 0.98 |
| Space group | C2221 |
| Unit-cell parameters | 30.28 99.35 51.90 |
| a, b, c (Å) | |
| Resolution (Å) | 28.96–1.35 |
| Total number of refl. | 119 994 |
| Total of unique refl. | 17 556 |
| Multiplicity | 6.83 (5.17) |
| Rmergea | 4.9 (22.1) |
| I/σ (I) | 19.8 (5.7) |
| Overall completeness (%) | 99.2 |
| Resolution refinement (Å) | 28–1.35 |
| Reflections (working/test) | 15 765/1771 |
| Rcryst/Rfreeb | 18.8/21.4 |
| Nonhydrogen atoms | 674 |
| Water molecules | 51 |
| Bonds (Å) | 0.012 |
| Angles (°) | 1.524 |
| Mean B factor (Å) Protein | 20.38 |
| Ramachadran analysis: residues in the most-favored/allowed (%) | 57/100 |
aRmerge = ∑h∑i | Ihi − < Ih> |/∑h∑I|hi, were |hi is the i-th observation of the reflection h, while < Ih> is the mean intensity of reflection h.
bRfactor = ∑ ∥Fo| − ∥FC∥/|Fo|. Rfree was calculated with a set of randomly selected reflections (10%).
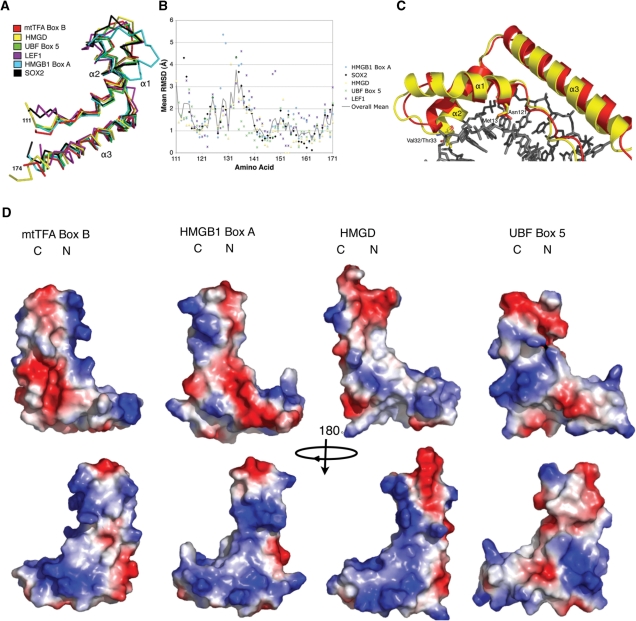
Root mean-squared deviation (RMSD) calculations were made between h-mtTFA box B and several other HMG protein structures that had been solved by either NMR or X-ray crystallography, and the overall backbone RMSD was calculated using the program Superpose (35). Overall RMSD values were calculated between each amino acid of the structures of HMGD [PDB ID 1qrv (36)], SOX2 HMG box A (PDB ID 1gt0, Remenyi, A., Scholer, H.R., Wilmanns, M. unpublished), HMGB1 box A [PDB ID 1ckt (37)], LEF1 [PDB ID 2lef (38)] and UBF5 [PDB ID 2hdz; Rong, H., Teng, M.K., and Niu, L., unpublished data)] and the X-ray crystal structure of h-mtTFA box B using the program COOT (33). Structural alignments were generated with using the program Chimera (39) and the structures were overlaid and images generated using the program PYMOL (40).
Western blot analysis
Deletion constructs of mtTFA were produced as previously described. A 1:1 Glutathione Sepharose 4B (GE Health Sciences) suspension was made and equilibrated in mtTFA buffer (50 mM HEPES-Na, (pH 7.0), 150 mM NaCl, 1 mM EDTA and 1mM DTT). GST-tagged mtTFA box B (mtTFA110–179) was added and equilibrated for 1 hour at 4°C. Beads were spun down and washed five times with mtTFA buffer prior to addition of His6-mtTFA deletion constructs. Beads were then washed with mtTFA buffer (50 mM HEPES–Na (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1mM DTT and 3% BSA). Samples were boiled in 2 × SDS–BME loading dye and loaded on a 15% SDS–PAGE. The gel was transferred for 1 h at 500 mA onto a PVDF membrane (Millipore) and blocked overnight in 3% BSA. Primary His antibody (Novagen) 1:1000 was added with 3% BSA for 1 h and washed in TBST buffer. Secondary HRP conjugated anti-mouse (GE Health Sciences) 1:100 000 was added in TBST for 1 h and washed in TBST. A 1:1 of HRP substrate was added and imaged by autoradiography. The membrane was stripped [25 mM glycine (pH 2) and 1% SDS] and reprobed with primary anti-GST antibody (UCDHSC Tissue Culture Core) with secondary antibody and imaged as described above.
RESULTS
Human mtTFA exists as a monomer in the absence of DNA
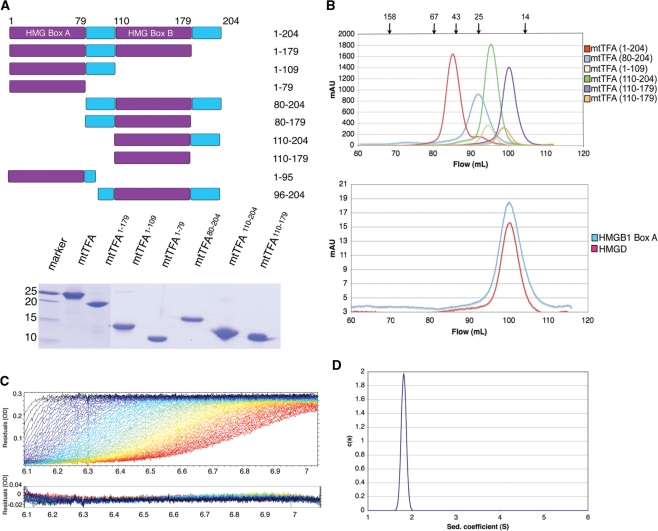
Although the function of h-mtTFA in mitochondrial transcription initiation has been investigated in a variety of species, the molecular mechanism of h-mtTFA assembly of the mitochondrial transcription complex is still unclear. In part, this is a result of disagreement about the stoichiometric state of h-mtTFA from the results of size-exclusion chromatography, which suggested that h-mtTFA is a dimer (23) and gradient and ultracentrifugation methods (10) which showed that it is a monomer or potentially an oligomer. To understand how h-mtTFA functions in promoter binding, it is important to know the stoichiometric state of h-mtTFA both in the presence and absence of DNA. Therefore, structural and biochemical information about HMG box structures was used to design a series of h-mtTFA deletion constructs (Figure 1A). The proteins were extensively purified so that they would be free of DNA for biophysical studies [Figure 1A, by previously established methods (36)]. Initially, we analyzed h-mtTFA using size-exclusion chromatography and observed that the full-length h-mtTFA, and all of the deletions of h-mtTFA, had elution volumes that were consistent with each protein having twice the expected molecular weight (Figure 1B and Table 1). However, two well-characterized HMG proteins that were known to be monomers in the absence of DNA, HMGD and HMGB1 box A (41,42), were also found to have elution volumes that indicated a size twice that of what was expected (Figure 1B and Table 1).
Figure 1.
Human mtTFA is an asymmetric monomer in the absence of DNA. (A) Deletion constructs of h-mtTFA. The upper panel shows a schematic diagram and the lower panel shows SDS–PAGE of h-mtTFA deletion constructs on a 15% polyacrylamide gel. (B) Size-exclusion chromatography (Superdex 200; GE Healthcare) elution profiles of h-mtTFA and h-mtTFA deletion constructs, mtTFA1–179, mtTFA1–109, mtTFA1–79, mtTFA80–204, mtTFA110–204 and mtTFA110–179 (top panel), and the single HMG domains, HMGB1 box A and HMGD (lower panel) in 50 mM HEPES–Na pH 7.0, 150 mM NaCl, 1 mM EDTA and 1 mM DTT. The position of each size standard is indicated by arrows above the top panel for amylase (158 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa) and RNase A (14 kDa). The void volume was at 45 ml and is not shown. (C) Sedimentation velocity profiles for the raw data acquired at different time points and the residuals after fittings had been performed using SEDFIT in 50 mM HEPES–Na pH 7.0, 150 mM NaCl, 1 mM EDTA and 1 mM DTT. (D) Calculated sedimentation coefficient distributions for the full-length h-mtTFA.
Table 1.
Mass estimates from SEC and AUC for human mtTFA and selected HMG boxes
| Protein | Theoretical MW | Est. SEC MW | Est. AUC MW |
|---|---|---|---|
| h-mtTFA | 24 775 | 48 977 | 25 958 |
| mtTFA1–109 | 13 229 | 22 998 | – |
| mtTFA1–79 | 9763 | 19 966 | – |
| mtTFA80–204 | 15 281 | 28 431 | – |
| mtTFA110–204 | 11 743 | 24 683 | – |
| mtTFA110–179 | 8753 | 16 151 | 9833 |
| HMGD | 8353 | 17 334 | – |
| HMGB1 box A | 8476 | 17 957 | – |
It is well known that asymmetric proteins have anomalous elution profiles in size exclusion chromatography (43). Therefore, we used analytical ultracentrifugation to unequivocally determine the stoichiometry of h-mtTFA. Three samples were subjected to sedimentation velocity analysis over a 10-fold concentration range. The homogeneity of the sample was determined using continuous size distribution analysis c(S) (Figure 1C and D). No evidence of concentration dependent self-association was seen (data not shown). The single and sharp peak in the c(S) analysis indicates a single homogeneous species with a Svedberg coefficient of 1.80 (Figure 1D) and the frictional ratio of 1.71 indicates that h-mtTFA exists in an asymmetric conformation. The calculated molecular weight from this analysis was 25 958 Da, which is within experimental error ( ± 10%) of the theoretical molecular weight for monomeric h-mtTFA of 24 384 Da. These results indicate that in the absence of DNA h-mtTFA is present as a monomer with an asymmetric shape in solution.
Human mtTFA assembles as a homodimer on the mitochondrial promoter
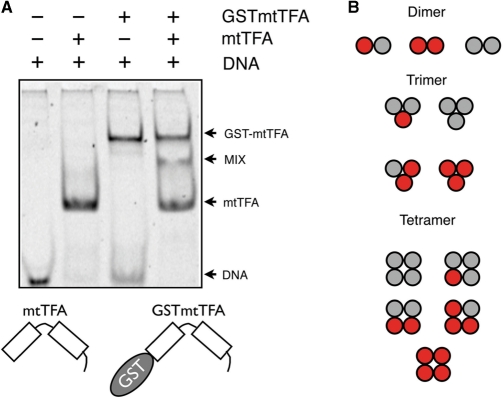
The LSP and HSP binding sites protected from DNaseI cleavage by h-mtTFA are approximately 30 bp in length (10,21). In contrast, the known HMGB binding site sizes are considerably shorter, from 8 bp for HMGD to 12 bp for LEF1 (36,38,44). To protect the observed length of DNA, we hypothesized that h-mtTFA assembles on the promoter DNA as either a dimer or an elongated monomer, with each HMG box protecting half of the binding site. The heterogeneous subunit assay introduced by Hope and Struhl (45) was used to determine the stoichiometry of h-mtTFA bound to the LSP promoter DNA (Figure 2). Since h-mtTFA has the ability to bind to DNA both sequence specifically and nonsequence specifically, we chose a minimal length LSP binding site of 30 bp that maintained high affinity h-mtTFA binding. h-mtTFA-DNA complexes appeared as a single band (Figure 2, lane 2). We also created a recombinant h-mtTFA fusion protein with an N-terminal GST tag (GST-h-mtTFA), and found that it also migrated as a single band but with a lower mobility than that of the h-mtTFA-DNA complexes (Figure 2, lane 3). When mixed together three bands appeared (Figure 2, lane 4). The top and bottom complexes could be attributed to DNA complexes with homodimers of mtTFA and GST-mtTFA, and the new band that migrates with an intermediate mobility between these corresponds to the mtTFA/GST-mtTFA heterodimer bound to DNA. The appearance of these bands is consistent with the dimerization of mtTFA on the LSP promoter DNA, as other oligomers would have give rise to additional bands in this experiment (Figure 2B).
Figure 2.
Human mtTFA binds to DNA as a dimer. (A) Heterogeneous subunit assay with mtTFA and GST-mtTFA was performed using 2 M mtTFA (lane 2), 2 M GST-mtTFA (lane 3), or both 1 M mtTFA and 1 M GST-mtTFA mixed together (lane 4), incubated with and loaded together with 1 M 30 bp unlabeled LSP DNA fragment (lanes 1 to 4). Arrows indicate the protein DNA complexes that form for mtTFA, GST-mtTFA, or the mixture. The appearance of three bands in lane 4 indicates that the binding species is a dimer of h-mtTFA, as illustrated in panel B. (B) The red and grey circles represent GST-mtTFA and mtTFA with the possible products of their assembly into dimers, trimers or tetramer forms.
Contribution of HMG boxes and the C-terminal tail of human mtTFA to DNA binding
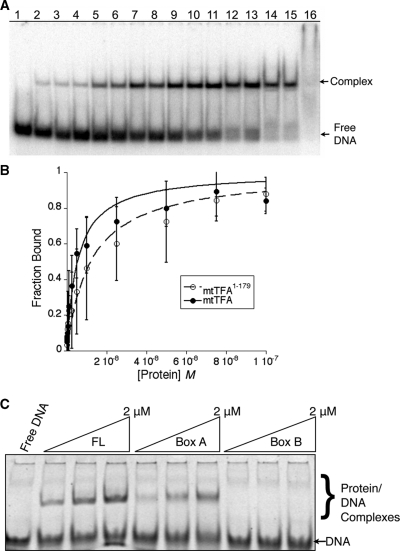
In order to understand the contribution of each part of h-mtTFA to DNA binding, we tested different constructs of h-mtTFA for their ability to bind to a 30 bp LSP DNA fragment. The C-terminal tail of h-mtTFA is important for its site-specific binding to the mitochondrial promoters and removal of this tail severally diminishes its ability to activate transcription (21). Additionally, deletion of the C-terminal tail reduced the DNA binding of h-mtTFA by three orders of magnitude (22). Based on these findings, we fully expected that deletion of the C-terminal tail would abolish the DNA binding of h-mtTFA. However, in quantitative EMSA (Figure 3A and B), we found that both h-mtTFA and the h-mtTFA1–179 proteins bound to the 30 bp LSP with a Kdapp, for h-mtTFA of 5.4 ± 0.6 nM and for h-mtTFA1–179 of 12.0 ± 1.7 nM. In addition, competition EMSA using the LSP DNA or nonspecific DNA (NS DNA) of the same length and composition showed that the NS DNA competed similarly to the LSP DNA for binding to mtTFA1–179, but that the LSP DNA competed better than NS DNA for binding to mtTFA (Supplementary Figure S1). These results suggest that the C-terminal tail of h-mtTFA is important for the DNA binding specificity, but that it does not contribute very much to the nonsequence-specific binding of h-mtTFA.
Figure 3.
Contributions of regions of human mtTFA to DNA binding. (A) Representation of typical EMSA used for binding curve analysis. Lane 1 contains no protein: lanes 2 through 16 contains increasing amounts of protein from 0.5 to 1000 nM. (B) Binding curves for mtTFA and mtTFA1–179 with error bars. (C) EMSA using 3 μM 30 bp LSP DNA with increasing concentrations of h-mtTFA, box A (mtTFA1–79) or box B (mtTFA110–179).
The region of h-mtTFA (mtTFA1–179) lacking the C-terminal tail binds to DNA with near wild type affinity and is composed of tandem HMG boxes separated by a linker region. In other HMGB proteins, with the possible exception of the upstream binding factor (UBF), all HMG boxes are thought to independently bind to DNA. To determine whether both of the HMG boxes of h-mtTFA contribute to LSP binding, we used stoichiometric EMSA with roughly equal amounts of protein and DNA at concentrations above the measured Kd. In this experiment, both the full-length protein h-mtTFA and HMG box A (h-mtTFA1–79) bound to the LSP DNA, whereas HMG box B (h-mtTFA110–179) failed to bind (Figure 3C). In case complexes of box B bound to DNA were just not sufficiently stable in electrophoresis, we tested the ability of box B to compete with mtTFA for DNA binding. We found that only at high concentrations of added box B (4 µM) was the complex of mtTFA–DNA disrupted (Supplementary Figure S2). This result could be due to weak binding of box B to either DNA or to other regions of mtTFA. Therefore, h-mtTFA box A had the ability to bind to DNA, but box B does not bind to duplex DNA at reasonable concentrations. These results support the conclusion that h-mtTFA binds to the LSP promoter DNA as a dimer, because the extent of protection is greater than that conferred by a single HMG-box, and box B does not bind to duplex DNA independent of box A.
Structure of human mtTFA box B
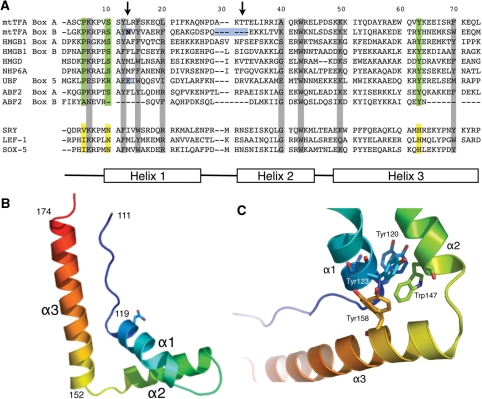
Sequence alignments of the HMG domains of h-mtTFA with other members of the HMG family suggest that h-mtTFA box B may not be a canonical HMG domain (Figure 4A). A variety of biophysical and structural analyses have defined the DNA binding modes of both sequence-specific and nonspecific HMGB proteins and has shown that these DNA binding modes are highly similar (20). The HMG box binds to the flattened, underwound and bent DNA minor groove using the large surface of the concave face of the protein, bending the DNA helix axis away from the site of contact (36,38,46,47). The binding interface includes electrostatic interactions, hydrogen bonds, and van der Waals contacts, as well as important partially intercalating nonpolar residues present in either helix α1 or α2 of the HMG box. The intercalating residue in helix α1 (Figure 4A, first arrow) is found in all of the sequence-specific HMGB proteins. In the nonsequence-specific HMGB proteins, there is also an additional intercalating residue at the beginning of helix α2 (Figure 4A, second arrow), and every known DNA binding HMG box has at least one nonpolar residue capable of intercalation at one or the other of these sites. Substitution of these intercalating residues with alanine or nonpolar amino acid residues significantly decreases the ability of the HMG domain to bind DNA (19,31). The presence of an asparagine residue at position 121 in h-mtTFA box B (Figure 4A, first arrow, blue box) suggests that this residue in box B would be unable to intercalate DNA (31). In addition, the other region that is known to make DNA contacts is completely absent from the N-terminus of helix α2 (Figure 4A, second arrow, blue box).
Figure 4.
X-ray crystal structure of human mtTFA box B. (A) Sequence alignments of box A and box B of h-mtTFA, box A and box B of HMG1, HMGD, NHP6A and Abf2. Global structure of an HMG protein is shown by diagram of helices α1, α3 and α3 at the bottom. Green boxes are residues that are conserved among all of the proteins. The arrows point to residues that are known to be involved in intercalating the DNA. (B) Ribbon diagram drawing of the backbone of box B showing the global fold of three helices stabilized into an L-shape configuration. The image was generated with PYMOL (40). (C) Hydrophobic core of box B with amino acids Tyr120, Tyr123, Trp147 and Tyr158 shown.
In order to determine whether the deficiency of h-mtTFA box B in DNA binding may be due to its unusual noncanonical form, we determined the structure using X-ray crystallography. Box B of h-mtTFA (amino acids 110–179) was crystallized using standard approaches, and the structure was solved using single-wavelength anomalous diffraction (SAD) methods (Table 2). The final refined structure is reported to 1.35 Å resolution (PDB ID 3fgh). Box B of h-mtTFA maintains the global fold of three helices stabilized in an L-shape configuration, which is stabilized by two hydrophobic cores similar to the other HMG proteins (Figures 4B and 5A). The hydrophobic core between helix α1 and helix α2 contains two amino acids (Tyr120 and Tyr158) that have multiple conformations suggesting a highly dynamic core that has not been seen in the nonsequence-specific HMG structures (Figure 4C), but is reminiscent of the less rigid hydrophobic cores seen in the sequence-specific transcription factors, Sox5 and SRY in the absence of DNA (46,48). The presence of Asn121 in helix α1 and the shortened length of helix α2 confirm that the intercalation residues in helix α1 and α2 that were predicted to be missing in this domain are in fact not there, and not compensated by other residues or structural changes. Interestingly, the yeast mitochondrial nucleoid protein Abf2 has similar anomalous features in its box B as human mtTFA (Figure 4A). Therefore, this analysis helped to explain why h-mtTFA box B fails to bind the LSP promoter DNA, and suggests that box B most likely evolved to play another role in h-mtTFA.
Figure 5.
Comparison of human mtTFA box B with other HMGB proteins. (A) Structural superposition of box B of h-mtTFA (3fgh; red) overlaid with HMGD (1qrv; yellow), UBF box 5 (2hdz; green), LEF1 (2lef; purple), HMGB1 box A (1ckt; cyan) and Sox2 (1gt0; black). (B) RMSD comparisons per amino acid were calculated using the program COOT, and were plotted against the amino acid sequence of h-mtTFA box B. The same color scheme was used as in (A). The solid line indicates the mean RMSD for the HMG boxes that were aligned. (C) Superposition of box B of h-mtTFA (red) on HMGD (yellow) in the HMGD–DNA complex and showing the shortened loop between helix α1 and α2 and the decreased size of helix α1. (D) Electrostatic surface potential plot of box B (mtTFA110–179), HMGB1 box A, HMGD, and UBF box 5 generated with PYMOL. Regions of positive potential are colored blue, and regions of negative potential are colored red.
To explore the possible roles of h-mtTFA box B in the function of h-mtTFA, we compared the box B structure to many HMG box structures that have been solved by crystallography and NMR. Table 3 lists the RMSDs of a representative group of HMG domains aligned with box B of h-mtTFA, and shows that there is a wide range of overall RMSD values between these structures. The structural alignment (Figure 5A) and per residue RMSD analysis (Figure 5B) illustrated that the N-terminal strand and helix α3 of the HMG boxes are the most conserved features. There were large RMSDs at the C-terminal end of helix α1, the linker connecting helix α1 and helix α2 between amino acids 131 and 141, and the N-terminal part of helix α2 (Figure 5B). The comparison of h-mtTFA box B to nonspecific HMG protein HMG boxes, such as HMGB1 box A bound to DNA (37) and HMGD bound to DNA (36), showed that helix α1 is shorter and there is a shorter linker connecting helix α1 and helix α2. This linker region is also significantly shorter than the linker in other HMG boxes, except for HMG box 5 of UBF, which was the most similar to box B of h-mtTFA in this region. Interestingly, this region is particularly important for duplex DNA binding and bending. As seen in an overlay diagram of box B with the HMGD-DNA complex (Figure 5C), this region of h-mtTFA box B does not approach the DNA as closely as HMGD does and it lacks the expected DNA intercalating residues. The presence of the polar amino acid Asn121 in the intercalating position normally occupied by a nonpolar residue in other HMG boxes suggests box B lacks the ability to intercalate the minor groove (Figure 5C).
Table 3.
Structural comparison of selected HMG boxes
| PDB | Protein | RMSD (Å) | Method |
|---|---|---|---|
| 2HDZ | UBF box 5 | 1.35 | X-Ray |
| 1J3C | HMGB2 box B | 1.40 | NMR |
| 1QRV | HMGD/DNA | 1.49 | X-Ray |
| 2LEF | LEF1/DNA | 1.50 | NMR |
| 1CKT | HMGB1 box A/DNA | 1.87 | X-Ray |
| 1J3X | HMGB2 box A | 2.04 | NMR |
| 1HMF | HMGB1 box B | 2.23 | NMR |
| 2YUL | SOX17 | 2.28 | NMR |
| 2YQI | HMGB3 box B | 2.33 | NMR |
| 2EZQ | HMGB3 box A | 2.36 | NMR |
| 1V63 | UBF box 6 | 2.38 | NMR |
| 1AAB | HMGB1 box A | 2.53 | NMR |
| 1GTO | SOX2/DNA | 2.93 | X-Ray |
| 1V64 | UBF box 3 | 3.10 | NMR |
| 1K99 | UBF box 1 | 3.13 | NMR |
| 1HRY | SRY/DNA | 3.73 | NMR |
| 1WGF | UBF box 4 | 5.42 | NMR |
Despite the differences in the tertiary structural features of box B of h-mtTFA compared to other HMG boxes, the surface electrostatic features of the HMG boxes are similar. The electrostatic surface map of h-mtTFA box B was compared to HMGB1 box A, HMGD, and UBF box 5, and showed clearly that the concave surface area that is typically involved in DNA binding was positively charged in all of the HMG boxes (Figure 5D). The presence of such a basic DNA binding surface most certainly suggests that box B may bind to DNA, although we suggest that it may not be in the canonical mode of binding seen for other HMG boxes.
Interactions of human mtTFA box B with other regions of mtTFA
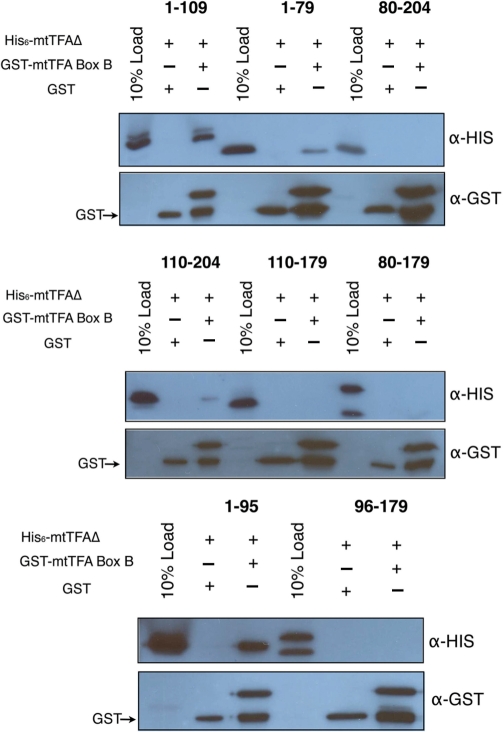
Although h-mtTFA is a monomer, the multiple domains within the protein may be important in the internal structuring of the protein. Kaufman et al. (23) previously suggested that box B played a role in dimerization, and this occurred through a coiled-coil mechanism. Although we showed that the full-length h-mtTFA is a monomer in solution, box B was seen to interact through a homomeric anti-parallel helical interaction involving helix α3 of symmetry related molecules in the crystal (Supplementary Figure S3). Therefore, we tested the ability of box B alone to dimerize using analytical ultracentrifugation and found that it also was present as a monomer in solution (Table 1 and Supplementary Figure S4). We next tried to detect interactions between box B and itself as well as a number of the h-mtTFA deletion proteins using pull-down assays (Figure 6) and found that box B interacted with box A and the linker region between box A and box B (Figure 6, top panel, lane 3). We truncated the linker region and confirmed that box B interacts with a region of amino acids between 80 and 95 (Figure 6, bottom panel, lane 3). Box B failed to interact with itself (Figure 6, middle panel, lane 6) but did show a weak interaction with the h-mtTFA80–204 protein (Figure 6, top panel, lane 9). Therefore, box B binds to box A and part of the linker included between 80-95 and likely is involved in some internal structuring of the protein. Although these experiments alone do not rule out that box B may play a role in DNA mediated dimerization they confirm that box B alone does not dimerize.
Figure 6.
Human mtTFA box B interacts with other regions of mtTFA. An N-terminal GST fusion with box B (mtTFA110–179) was tested for its ability to interact with the various deletion constructs of h-mtTFA (mtTFA1-109, mtTFA1–79, mtTFA1–80–204, mtTFA110–204, mtTFA110–179, mtTFA80–179, mtTFA1–95 and mtTFA96–179) in 50 mM HEPES–Na pH 7.0, 150 mM NaCl, 1 mM EDTA and 1 mM DTT. The reactions were electrophoresed on a 15% SDS–PAGE gel and transferred to nitrocellulose membrane (Millipore) and probed with anti-His and anti-GST antibodies, respectively.
DISCUSSION
This analysis of the structural features and DNA binding properties of the HMG boxes of h-mtTFA provides new insights into the first steps of mitochondrial transcription complex assembly. We first showed that h-mtTFA is a monomer in solution and assembles as a dimer on a single LSP binding site (Figures 1 and 2). The C-terminal tail of h-mtTFA was not needed for nonsequence-specific DNA binding (Figure 3A and B). Furthermore, the canonical HMG box h-mtTFA box A bound independently to the LSP DNA (Figure 3C), and as such appears to be the dominant DNA binding domain in h-mtTFA. In contrast, h-mtTFA box B did not bind to the LSP DNA by itself (Figure 3C), and as a variant HMG box (Figures 4 and 5) it has features that suggest why it may not be expected to bind to DNA in the canonical fashion. In addition, box B alone did not dimerize (Supplementary Figure S1), but it did interact with other regions of mtTFA (Figure 6). This suggests that box B may be involved in stabilizing interactions with h-mtTFA within the free protein and in the homodimeric protein DNA complex or potentially acting at a later step in transcriptional activation.
Implications of h-mtTFA stoichiometry in LSP recognition
In the absence of DNA, h-mtTFA is present as a monomer with a highly asymmetric shape (Figure 1). We attribute the anomalous elution profiles of a dimer seen in the studies by our lab and others using size-exclusion chromatography to the asymmetric shape of the protein (23). More difficult to explain is an additional peak that was seen in the presence of high salt (500 mM NaCl) in the size-exclusion experiments by Kaufman et al. (23,43). This peak was interpreted to be a monomer, although alternate explanations are possible, as the deoxycholate and high salt present in the chromatography buffer could have affected how h-mtTFA eluted due to potential micelle formation.
It has been widely thought that h-mtTFA minimally forms higher ordered structures on the mitochondrial promoters. Support for this model has come from the observation of higher ordered structures of dimers, trimers and tetramers with xl-mtTFA (24), and h-mtTFA has been shown to bind cooperatively as a dimer with nanomolar affinity (23). However, h-mtTFA has also been shown to function as a monomer when binding to Holliday Junctions (49). Here we clearly showed that in the absence of DNA, h-mtTFA exists as a monomer (Figure 1), and that it assembles as a dimer on a short segment of the mitochondrial promoter (Figure 2). This is a unique feature of mtTFA, as other HMG-box proteins bind DNA as monomers or in the case of UBF are obligate dimers (50,51). Thus, we conclude that a crucial step in setting up the preinitiation complex at the mitochondrial promoter is assembly and dimerization of h-mtTFA on the DNA.
Another step of transcription complex assembly involves the recruitment of h-mtTFB proteins and the mitochondrial polymerase. The LSP is an asymmetric sequence and initiation only occurs at one site that is located at a specific distance from the h-mtTFA binding site (21,52). Therefore, the fact that h-mtTFA is a monomer suggests that the sequence and structure of the DNA may also play a role in setting up an asymmetric complex. It was previously shown that the h-mtTFA C-terminal tail was required for footprinting the LSP and for transcriptional activity (21), as well as for recruitment of h-mtTFB1 and h-mtTFB2 (15). It is certainly possible that mtTFA and mtTFB can assemble together on the DNA, although this has not been observed potentially due to the dynamics of the complex. Interestingly, the ability of h-mtTFA to bind DNA was only slightly diminished by removal of the C-terminal tail (Figure 3A and B, Supplementary Figure S1), which suggests that it aids in the formation of the correct specific complex on the LSP. This could be because the tail contributes additional contacts that provide for the site-specificity for the mitochondrial promoter. Surprisingly, Kang and coworkers found that a similar C-terminal truncation in h-mtTFA decreased the binding affinity in surface plasmon resonance experiments by greater than 1000 fold, but in DNA unwinding in supercoiling assays this difference was only 8-fold (22). The difference in affinity that we measured for the deletion of the C-terminal tail is 2.4-fold. The discrepancies between these numbers could be due to the different protein preparations and the approaches that were used.
HMG box structural features and DNA binding
HMG boxes are found in proteins that are involved in numerous functions from signaling in inflammation to transcriptional regulation and the maintenance of DNA architectures (18,19,53). Although, it is thought that most HMG boxes bind and bend DNA in a similar fashion, many functions of HMG boxes are still not understood at the molecular level. h-mtTFA box B did not bind to DNA by itself (Figure 3C) and its sequence differs in regions of the protein that are known to be crucial for DNA binding in other HMGB proteins (Figures 4A and 5). This anomalous property of box B is most likely due to the unusual presence of a polar residue in the position typically occupied by a residue capable of intercalating into the minor groove of DNA (20,31). In addition, the lack of any putative intercalating residues in the region of the short helix α1 and the linker between helices α1 and α2 further explains it inability to bind DNA. These features suggest that box B of h-mtTFA would not bind DNA in the typical fashion seen for other HMG proteins. That is not to say that box B would fail to bind to either duplex or single stranded DNA under the correct conditions. For example, Ohno et al. (49) observed that both of the HMG boxes were needed for binding of h-mtTFA to Holliday Junction DNA. Therefore, depending on the context, box B may play a role in binding of certain types of DNA that may be generated in the course of promoter opening and stabilization of the transcription complex.
Box B of h-mtTFA interacts with other domains of h-mtTFA (Figure 6). Box B closely resembles UBF box 5, which is one of the HMG boxes in the multi-HMG-box organelle specific transcription factor UBF (Figure 5). UBF in mammals is involved in transcription of ribosomal DNA. It has six HMG boxes of which the first two are involved directly in DNA binding, but the function of the remaining HMG boxes is unknown (50,51,54,55). However, Moss and coworkers have proposed that HMG boxes 3-6 may be involved in formation of the ribosomal enhancesome through protein–protein interactions (50). HMGB proteins are known to interact with other proteins; for example HMGB1 interacts with P53, steroid receptors, and many others (18). Our results suggest that box B in h-mtTFA may be involved in interactions with other regions of h-mtTFA, including a portion of box A, a short putative helical segment that follows box A contained in the linker region (Figure 6), and potentially the C-terminal tail. The surface representation of box B also shows a deep acidic cavity in the core region between helix α1 and helix α3 that is not present in these other proteins (Figure 5D), and may be the site of additional protein–protein interactions, such as those that may occur with p53 (56). Therefore, we conclude that box B of h-mtTFA likely participates in an alternate mode of DNA binding and/or protein–protein interactions that occur during assembly of the transcription initiation complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The UCD Biomolecular X-ray Crystallography Center by HHMI (in part); the University of Colorado Cancer Center; National Institutes of Health; UMDF (05-101, #68859 to M.E.A.C). Funding for open access charge: MDA (#68859) and institutional funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Christopher Malarkey for critical reading of the article. We are very grateful to Gerald Shadel for providing the initial h-mtTFA plasmid construct and advice. We thank the staff at beamline 4.2.2 at the Advanced Light Source, Lawrence Berkeley National Lab, Brooke Hirsch at the UCD Biophysics Core Facility for collection of the mtTFA110–179 sedimentation velocity data, and Dominic Esposito at NCI, who generously donated the pDEST15 and pDEST 527 for the Gateway expression system.
REFERENCES
- 1.Clayton DA. Replication and transcription of vertebrate mitochondrial DNA. Annu. Rev. Cell Biol. 1991;7:453–478. doi: 10.1146/annurev.cb.07.110191.002321. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 3.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 5.Greaves LC, Taylor RW. Mitochondrial DNA mutations in human disease. IUBMB Life. 2006;58:143–151. doi: 10.1080/15216540600686888. [DOI] [PubMed] [Google Scholar]
- 6.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shadel GS, Clayton DA. Mitochondrial transcription initiation. Variation and conservation. J. Biol. Chem. 1993;268:16083–16086. [PubMed] [Google Scholar]
- 8.Shadel GS. Expression and maintenance of mitochondrial DNA: new insights into human disease pathology. Am. J. Pathol. 2008;172:1445–1456. doi: 10.2353/ajpath.2008.071163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoubridge EA. The ABCs of mitochondrial transcription. Nat. Genet. 2002;31:227–228. doi: 10.1038/ng0702-227. [DOI] [PubMed] [Google Scholar]
- 10.Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 12.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 13.McCulloch V, Seidel-Rogol BL, Shadel GS. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol. Cell Biol. 2002;22:1116–1125. doi: 10.1128/MCB.22.4.1116-1125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat. Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- 15.McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotney J, Shadel GS. Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J. Mol. Evol. 2006;63:707–717. doi: 10.1007/s00239-006-0075-1. [DOI] [PubMed] [Google Scholar]
- 17.Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol. Life Sci. 2007;64:2590–2606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas JO, Travers AA. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 20.Murphy FVt, Churchill ME. Nonsequence-specific DNA recognition: a structural perspective. Structure. 2000;8:R83–R89. doi: 10.1016/s0969-2126(00)00126-x. [DOI] [PubMed] [Google Scholar]
- 21.Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 22.Ohgaki K, Kanki T, Fukuoh A, Kurisaki H, Aoki Y, Ikeuchi M, Kim SH, Hamasaki N, Kang D. The C-terminal tail of mitochondrial transcription factor a markedly strengthens its general binding to DNA. J. Biochem. 2007;141:201–211. doi: 10.1093/jb/mvm020. [DOI] [PubMed] [Google Scholar]
- 23.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antoshechkin I, Bogenhagen DF, Mastrangelo IA. The HMG-box mitochondrial transcription factor xl-mtTFA binds DNA as a tetramer to activate bidirectional transcription. EMBO J. 1997;16:3198–3206. doi: 10.1093/emboj/16.11.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, Hamasaki N, Kang D. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol. Cell Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 27.Ghivizzani SC, Madsen CS, Hauswirth WW. In organello footprinting. Analysis of protein binding at regulatory regions in bovine mitochondrial DNA. J. Biol. Chem. 1993;268:8675–8682. [PubMed] [Google Scholar]
- 28.Bogenhagen DF, Wang Y, Shen EL, Kobayashi R. Protein components of mitochondrial DNA nucleoids in higher eukaryotes. Mol. Cell Proteomics. 2003;2:1205–1216. doi: 10.1074/mcp.M300035-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Lee KM, Hayes JJ. The N-terminal tail of histone H2A binds to two distinct sites within the nucleosome core. Proc. Natl Acad. Sci. USA. 1997;94:8959–8964. doi: 10.1073/pnas.94.17.8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys. J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klass J, Murphy FVt, Fouts S, Serenil M, Changela A, Siple J, Churchill ME. The role of intercalating residues in chromosomal high-mobility-group protein DNA binding, bending and specificity. Nucleic Acids Res. 2003;31:2852–2864. doi: 10.1093/nar/gkg389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55(Pt 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 35.Maiti R, Van Domselaar GH, Zhang H, Wishart DS. SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 2004;32:W590–W594. doi: 10.1093/nar/gkh477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy FV, IV, Sweet RM, Churchill ME. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 1999;18:6610–6618. doi: 10.1093/emboj/18.23.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399:708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 38.Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 39.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.DeLano WL. The PyMOL Molecular Graphics System (2002) on World Wide Web. 2002. http://www.pymol.org.
- 41.Jones DNM, Searles A, Shaw GL, Churchill MEA, Ner SS, Keeler J, Travers AA, Neuhaus D. The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster. Structure. 1994;2:609–627. doi: 10.1016/s0969-2126(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 42.Hardman CH, Broadhurst RW, Raine AR, Grasser KD, Thomas JO, Laue ED. Structure of the A-domain of HMG1 and its interaction with DNA as studied by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry. 1995;34:16596–16607. doi: 10.1021/bi00051a007. [DOI] [PubMed] [Google Scholar]
- 43.Tarvers RC, Church FC. Use of high-performance size-exclusion chromatography to measure protein molecular weight and hydrodynamic radius. An investigation of the properties of the TSK 3000 SW column. Int. J. Pept. Protein Res. 1985;26:539–549. doi: 10.1111/j.1399-3011.1985.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 44.Giese K, Pagel J, Grosschedl R. Functional analysis of DNA bending and unwinding by the high mobility group domain of LEF-1. Proc. Natl Acad. Sci. USA. 1997;94:12845–12850. doi: 10.1073/pnas.94.24.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hope IA, Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987;6:2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Werner MH, Huth JR, Gronenborn AM, Clore GM. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 47.Masse JE, Wong B, Yen YM, Allain FH, Johnson RC, Feigon J. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J. Mol. Biol. 2002;323:263–284. doi: 10.1016/s0022-2836(02)00938-5. [DOI] [PubMed] [Google Scholar]
- 48.Cary PD, Read CM, Davis B, Driscoll PC, Crane-Robinson C. Solution structure and backbone dynamics of the DNA-binding domain of mouse Sox-5. Protein Sci. 2001;10:83–98. doi: 10.1110/ps.32801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohno T, Umeda S, Hamasaki N, Kang D. Binding of human mitochondrial transcription factor A, an HMG box protein, to a four-way DNA junction. Biochem. Biophys. Res. Commun. 2000;271:492–498. doi: 10.1006/bbrc.2000.2656. [DOI] [PubMed] [Google Scholar]
- 50.Stefanovsky VY, Pelletier G, Bazett-Jones DP, Crane-Robinson C, Moss T. DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res. 2001;29:3241–3247. doi: 10.1093/nar/29.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bazett-Jones DP, Leblanc B, Herfort M, Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 52.Gaspari M, Falkenberg M, Larsson NG, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bianchi ME, Manfredi A. Chromatin and cell death. Biochim. Biophys. Acta. 2004;1677:181–186. doi: 10.1016/j.bbaexp.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 54.Stefanovsky VY, Moss T. The splice variants of UBF differentially regulate RNA polymerase I transcription elongation in response to ERK phosphorylation. Nucleic Acids Res. 2008;36:5093–5101. doi: 10.1093/nar/gkn484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stefanovsky VY, Bazett-Jones DP, Pelletier G, Moss T. The DNA supercoiling architecture induced by the transcription factor xUBF requires three of its five HMG-boxes. Nucleic Acids Res. 1996;24:3208–3215. doi: 10.1093/nar/24.16.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer Res. 2003;63:3729–3734. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.