Abstract
Translation initiation in eukaryotes is mediated by assembly of the eIF4F complex over the m7GTP cap structure at the 5′-end of mRNAs. This requires an interaction between eIF4E and eIF4G, two eIF4F subunits. The Leishmania orthologs of eIF4E are structurally diverged from their higher eukaryote counterparts, since they have evolved to bind the unique trypanosomatid cap-4 structure. Here, we characterize a key eIF4G candidate from Leishmania parasites (LeishIF4G-3) that contains a conserved MIF4G domain. LeishIF4G-3 was found to coelute with the parasite eIF4F subunits from an m7GTP-Sepharose column and to bind directly to LeishIF4E. In higher eukaryotes the eIF4E-eIF4G interaction is based on a conserved peptide signature [Y(X4)Lϕ], where X is any amino acid and Φ is a hydrophobic residue. A parallel eIF4E-binding peptide was identified in LeishIF4G-3 (20-YPGFSLDE-27). However, the binding motif varies extensively: in addition to Y20 and L25, binding strictly requires the presence of F23, whereas the hydrophobic amino acid (Φ) is dispensable. The LeishIF4E–LeishIF4G-3 interaction was also confirmed by nuclear magnetic resonance (NMR) studies. In view of these diversities, the characterization of the parasite eIF4E–eIF4G interaction may not only serve as a novel target for inhibiting Leishmaniasis but also provide important insight for future drug discovery.
INTRODUCTION
Leishmaniasis is caused by trypanosomatid parasites that cycle between invertebrate vectors and vertebrate hosts, transforming between the promastigote and amastigote life forms, respectively. Environmental conditions, such as temperature and pH switches, were shown to induce a stage-specific pattern of gene expression (1,2), which in some species is sufficient to produce the axenic form (amastigotes) observed in mammalian cells (3,4). Trypanosomatids are ancient eukaryotes that diverged early in evolution and therefore display a variety of unique molecular features. Transcription of protein-coding genes is polycistronic and the resulting pre-mRNAs are processed by trans-splicing and polyadenylation (5,6). During trans-splicing, a capped 39-nt exon of the spliced leader RNA is joined to the 5′-end of all transcripts. The 5′-cap of trypanosomatids consists of a highly modified structure, which in addition to the m7GTP cap found at the 5′-end of most eukaryotic mRNAs, contains 2′-O-methylations on the ribose moieties of the first four transcribed nucleotides and unusual base-methylations on the first adenine and fourth uridine (m7Gpppm36,6,2′Apm2′Apm2′Cpm23,2′U); this structure is thus denoted cap-4 (7).
Cap-dependent translation is a common mechanism in higher eukaryotes, which requires several factors. The cap-binding complex, eIF4F, consists of three subunits, with eIF4G serving as a scaffold protein for the whole complex (8,9). eIF4G holds the cap-binding protein, eIF4E, and the ATP-dependent RNA helicase, eIF4A. eIF4G also associates with eIF3 (10,11), a multisubunit factor that bridges the cap-binding complex and the small ribosomal subunit, as well as with the poly(A)-binding protein (PABP), which is found on the 3′-end of most eukaryotic mRNAs. These latter interactions allow the mRNA circularization, which is assumed to stimulate translation initiation (12,13). Assembly of the translation initiation complex on the 5′-mRNA cap structure (m7GTP) is rate-limiting and is therefore subject to strict regulation (9,14).
The human eIF4G (1600 amino acids) possesses three domains that are defined by viral protease cleavage patterns (10). The N-terminal segment includes eIF4E and PABP binding sites (15,16). This domain is initially unfolded, and partially folds upon binding to eIF4E in the human complex (17). In yeast it folds to a larger extent wrapping around eIF4E's N-terminus (18). The middle domain of eIF4G (MIF4G) is critical for assembly of the translation machinery, since it harbors the binding site for eIF3 and eIF4A (19,20). The MIF4G contains a single HEAT domain, whereas the human C-terminus of eIF4G contains two additional HEAT domains, which include the binding site for Mnk1, a kinase that phosphorylates eIF4E, and a second binding site for eIF4A (21). The large subunit of the nuclear cap-binding complex, CBP80, shares a common origin and domain structure with the eIF4G (22). Other eIF4G isoforms include the MIF4G domain but lack the N-terminal domain, such as the human 97 kDa protein DAP5 (23) or the 130 kDa splice-variant of the Caenorhabditis elegans eIF4G. They were shown to participate in cap-independent translation of transcripts encoding pro-apoptotic genes under stress conditions (24). DAP5 is also necessary for maintaining cell survival during mitosis by promoting cap-independent translation of at least two prosurvival proteins represented by Bcl2 and CDK1 (25).
The eIF4E–eIF4G interaction is a key target for translation control (26). This interaction relies on a short sequence of about 15 amino acids in the N-terminus of eIF4G, which contains a conserved motif Y(X4)LΦ, where X is any amino acid and Φ is a hydrophobic residue (17,27,28). This motif forms a short helical structure that binds conserved amino acids on the dorsal side of eIF4E (17). Several other proteins, such as the eIF4E-binding proteins (4E-BPs), contain this consensus sequence and thus compete with eIF4G for binding to eIF4E, depending upon its phosphorylation state. This regulates translation by affecting the 48S preinitiation complex formation (29).
Cap-binding proteins in trypanosomatids are expected to have gone through structural adaptations that enable them to interact with the unusual cap-4 structure. The biochemical and cellular features of four eIF4E isoforms in Leishmania major which were denoted LeishIF4E-1 through LeishIF4E-4, have been previously described (30–32). None of these isoforms could functionally complement a yeast eIF4E knockout strain, strongly supporting that they have a structurally diverged eIF4E/eIF4G interface. Furthermore, since there is yet no in vitro system for translation initiation for any of the trypanosomatids, the specific role of the different candidates was evaluated indirectly by biochemical and biophysical assays. Fluorescence titration assays showed that LeishIF4E-1 and LeishIF4E-4 bind m7GTP and cap-4 comparably well whereas LeishIF4E-2 binds mainly to the cap-4. LeishIF4E-3 was found to bind mainly to m7GTP, excluding its involvement in translation of cap-4 protected mRNAs (31). A bioinformatics search for eIF4G revealed five isoforms that contain the HEAT repeats of the MIF4G (32). Here we describe LeishIF4G-3, the most probable candidate for functioning as the Leishmania eIF4G, based on its ability to interact with the LeishIF4Es. Using yeast two-hybrid, site-directed mutagenesis and nuclear magnetic resonance (NMR) spectroscopy techniques, we further characterize the consensus peptide of LeishIF4G-3 (20-YPGFSLDE-27) that is responsible for this interaction and highlight variations between the Leishmania cap-binding complex and its counterpart from higher eukaryotic hosts.
MATERIALS AND METHODS
Organisms
Leishmania major (Friedlin strain) were cultured in Schneider's medium supplemented with 10% fetal calf serum (FCS), 4 mM l-glutamine and 25 µg/ml gentamycin.
Affinity purification of the LeishIF4F complex over m7GTP-Sepharose
Leishmania major promastigotes (5 × 108–1 × 109) were harvested, washed twice with PBS, and once with Column Buffer [CB: 20 mM HEPES pH 7.6, 2 mM EDTA, 1 mM DTT, 50 mM NaCl, 2 mM iododoacetamide and 1× protease Inhibitor cocktail (Sigma)]. The cells were resuspended in CB, incubated for 5 min on ice and lyzed by sonication, which was followed by centrifugation at 15 000g for 20 min at 4°C. The cellular extracts were loaded on an m7GTP-Sepharose column (Amersham) previously equilibrated with CB. The column was washed with CB containing 100 μM GTP. The proteins were eluted with CB containing 500 μM NaCl or with CB buffer containing 200 μM m7GTP, concentrated by Tri-chloroacetic acid (TCA) precipitation and separated over a sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (SDS–PAGE, 15%). Protein bands were identified by nano-liquid chromatography electrospray ionization tandem mass spectrometry (nano-LC-ESI-MS/MS) at the biological mass spectrometry facility of the Weizmann Institute. The different elution protocols yielded the same LeishIF4F subunits in the eluates.
The open reading frame (ORF) of LeishIF4G-3 was amplified by PCR using L. major genomic DNA as template. For details on the primers see Supplementary Data, Table 1. The amplified sequences were cloned into pGST or pHis expression vectors and the plasmids were used to transform Escherichia coli BL21 cells. Antibodies were raised against recombinant proteins excised from gels in New Zealand rabbits.
Polysome analysis over sucrose gradients
Leishmania major, which were grown at 26°C and incubated overnight at 35°C, were used for polysomal analysis over sucrose gradients (10–40%) as formerly described (31). Fractions were collected from the top of the gradients and their O.D260nm was measured. Proteins were recovered by TCA precipitation and separated by SDS–PAGE (15%) followed by immunoblotting using antibodies against the different translation initiation factors, as well as with antibodies against rpS6, marking the small ribosomal subunit.
GST in vitro pull-down assay
The GST-LeishIF4G-3 fusion protein or GST alone as control and LeishIF4Es were expressed in E. coli BL21 cells. Cells expressing each of these proteins were harvested and lyzed by sonication in Binding Buffer (BB: 20 mM MOPS pH 7.2, 50 mM KCl, 250 mM NaCl, 7 mM β-Mercaptoethanol, 2 mM MgCl2). GST-LeishIF4G-3 or GST alone was immobilized on Glutathione-Agarose beads (Sigma) previously equilibrated with BB. The beads were then washed with BB containing 0.1% Triton X-100 and incubated with supernatants of lyzed bacteria containing expressed LeishIF4Es. After extensive washing with BB containing 0.1% Triton X-100, the proteins were denatured with Laemmli's SDS–PAGE sample buffer, separated by SDS–PAGE (15%) and subjected to western blot analysis with appropriate antibodies.
In vivo pull-down analysis of LeishIF4F components
The coding regions of LeishIF4E-1 and LeishIF4E-4 lacking stop codons were amplified and cloned into pSNSAP1 (33), generating vectors pTAP-LeishIF4E-1 or LeishIF4E-4. The coding region of LeishIF4G-3 was amplified and tagged with the FLAG sequence (DYKDDDDK). The fragment was further cloned into pX, a Leishmania-specific expression vector (34), between two Hsp83 intergenic regions (IRs) that provide RNA processing signals, generating the pX-LeishIF4G-3-FLAG vector (primer sequences are provided in Supplementary Table 1). Leishmania major cells were transfected as previously described (35) with each of the vectors described above, and stable cell lines were selected in the presence of 200 μg/ml G-418. For pull-down assays, transgenic L. major cells (2 × 109) were harvested, washed, resuspended in CB and lyzed by sonication. For TAP-tagged protein purification, supernatants were agitated with streptavidin-Sepharose beads (GE Healthcare) for 3 h at 4°C. The beads were washed with CB containing 0.1% NP-40 and the proteins were eluted with CB supplemented with 2 mM biotin. For LeishIF4E-1, the eluate was loaded on IgG-Sepharose beads (GE Healthcare) and agitated for 3 h at 4°C. The beads were then washed with CB containing 0.1% NP-40 and the elution was performed by a short incubation in HAc pH 3.4, which was neutralized by the addition of saturated Tris buffer pH 7. For LeishIF4E-4 expressing cells, the eluate was loaded on an m7GTP-Sepharose column. The column was washed with CB containing 0.1% NP-40 and once with the same buffer containing 0.1 mM GTP. The proteins were eluted by 0.5 M NaCl in CB/0.1% NP-40. The eluted proteins were precipitated by TCA, fractionated by SDS–PAGE (12%) and subjected to western blot analysis with specific antibodies against the different LeishIF4F subunits. For the FLAG-tagged proteins the supernatant was rotated with prewashed anti-FLAG-Agarose beads (Sigma) for 3 h at 4°C. The beads were washed three times with CB containing 0.1% Triton X-100, and the bound proteins were analyzed by SDS–PAGE (12%).
Yeast two-hybrid assays
A yeast two-hybrid assay was performed using the commercial GAL4 Two-Hybrid Phagemid Vector Kit (Stratagene) following the manufacturer's instructions. The ORF of LeishIF4E-4 was fused to the GAL4-binding domain (pBD) whereas the ORF of LeishIF4G-3 was fused to the GAL4 activation domain (pAD) generating, respectively, pBD-LeishIF4E-4 and pAD-LeishIF4G-3. LeishIF4G-3 fragments corresponding to amino acids 1–49, 50–305, 306–635 and mutants of the amino terminal domain (1–49) were generated by PCR (see Supplementary Table 1 for primer details), using PFU-Turbo (Stratagene). All constructs were confirmed by DNA sequencing. The yeast strain YRG-2 (Mata ura352 his3-200 ade2-101 lys2-801 trp1-901 leu2-3 112 gal4-542 gal80-538 LYS2::UASGAL1-TATA GAL1-HIS3 URA3::UASGAL4 17mers(x3) TATACYC1-lacZ) was cotransformed with pBD-LeishIF4E-4 and pAD-LeishIF4G-3. Yeast transformants were cultured in Liquid SD-2 (-Trp/-Leu) medium at 30°C overnight, diluted to a final concentration of O.D600 = 0.15 and continued growth to O.D600 = 0.6. Yeast were spotted on SD-3 (-Trp/-Leu/-His) and SD-2 (-Trp/-Leu) plates, with or without 2.5 mM 3-amino-1,2,4-triazole (3-AT) (Sigma), respectively. Controls for the yeast two-hybrid assay are based by pAD-WT and pBD-WT (positive control) cotransformation, or pAD-WT and pLamin C plasmids (negative control), following the manufacturer's instructions.
Log phase yeast culture (O.D600 = 10) were harvested, washed with ddH2O and resuspended in 200 μl SDS–PAGE sample buffer. After boiling for 10 min, glass beads were added and the cells were vortexed for 5 min. The glass beads were then discarded and the extracts were centrifuged at 16 000g for 2 min. Samples (O.D600 = 1.0) were separated by SDS–PAGE (15%) and the gels were blotted and exposed to specific antibodies against LeishIF4E-4. The presence of recombinant pAD-LeishIF4G-3 fragments in transgenic yeasts was detected by PCR from yeast colonies (see Supplementary Figure 2) using primers derived from the pAD plasmid (see Supplementary Table 1).
LeishIF4E-1 NMR titration experiments with LeishIF4G-3
NMR spectra were recorded at 298K on a Varian Inova 600-MHz spectrometer equipped with a cryoprobe. NMR measurements were carried out on samples containing 250 µM 15N-labeled, partially deuterated sample of LeishIF4E-1 bound to m7GTP in buffer constituted of 50 mM Na2HPO4, 50 mM NaH2PO4, 0.5 mM EDTA, 2 mM DTT and 8% D2O at pH 6.5. LeishIF4G-3 peptide (NYLEPPYPGFSLDEVVRR synthesized at the Tufts University Core Facility) concentration was gradually increased in molar ratio ranging from 0.5 to 10 times relative to LeishIF4E-1 initial concentration. Trosy version of HSQC spectra (36) were recorded with 172 complex points in the indirect 15N dimension and 1024 complex points in the direct dimension.
RESULTS
LeishIF4G-3 is part of the parasite cap-binding complex and is purified on an m7GTP-Sepharose column
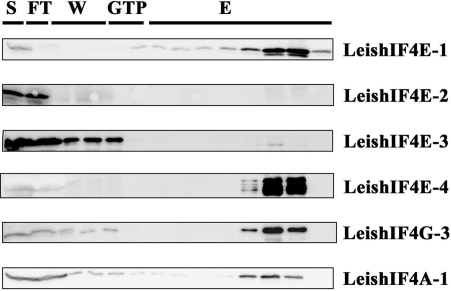
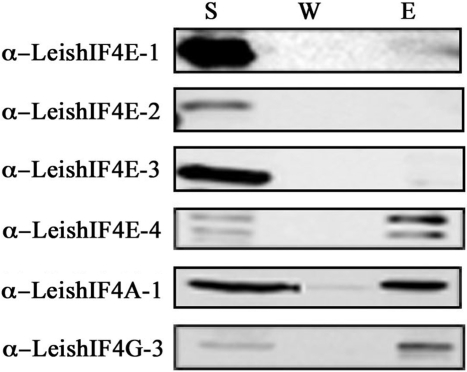
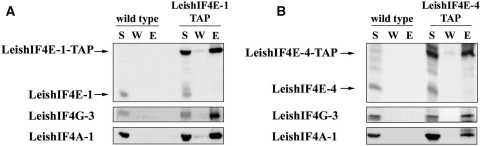
A bioinformatics search of the Leishmania and trypanosome genomes highlighted several proteins that share structural domains with eIF4G from higher eukaryotes. To confirm which of the candidates is indeed part of the translation initiation machinery, functional assays were pursued. In higher eukaryotes, components of the eIF4F complex can be eluted from an m7GTP-Sepharose column through the ability of eIF4E to interact with the m7GTP cap structure. Based on former results showing that specific recombinant LeishIF4E isoforms can interact with m7GTP, components of the endogenous cap-binding complex were also identified by affinity purification of proteins from whole cell extracts of L. major over an m7GTP-Sepharose column. The parasite cap-binding complexes were eluted and proteins were separated by SDS–PAGE. A ∼70 kDa excised band (Supplementary Figure 1A) was identified in two independent experiments by mass spectrometry as LmjF16.1600, a homolog of eIF4G that was denoted LeishIF4G-3 (Supplementary Figure 1B). This finding supports that LeishIF4G-3 is a potential candidate to act as a translational initiation factor and is in good agreement with a former report showing that this protein can interact with the parasite eIF4A helicase, which is one of the eIF4F subunits (32). Western blot analysis of the fractions that were eluted from the m7GTP-Sepharose column depicted the presence of LeishIF4E-1 and LeishIF4E-4, LeishIF4A as well as LeishIF4G-3 (Figure 1). LeishIF4E-2 and LeishIF4E-3 were not expected to be eluted from the m7GTP column. LeishIF4E-2 does not bind efficiently to m7GTP, and LeishIF4E-3 does not purify well from m7GTP-Sepharose, although it was shown to bind m7GTP using fluorescence titration assays (31,32). Thus, a probable form of the LeishIF4F complex which includes LeishIF4E-1 or LeishIF4E-4, LeishIF4A-1 and LeishIF4G-3 was purified over the m7GTP-Sepharose column.
Figure 1.
Leishmania major cap-binding complex purification. Leishmania major cells were lyzed and the soluble protein extract (S) was loaded over an m7GTP-Sepharose column. After collection of the flowthrough (FT) the column was washed (W) with CB (see Materials and Methods section) and with CB containing 100 μM GTP (GTP). The proteins were eluted (E) with CB containing 500 mM NaCl. Equal samples from the different fractions were separated by SDS–PAGE and analyzed by western blot using specific antibodies against the different subunits of the Leishmania cap-binding complex (LeishIF4E-1 through LeishIF4E-4, LeishIF4G-3, LeishIF4A-1). Similar results were obtained when the m7GTP-Sepharose column was eluted with free m7GTP.
LeishIF4G-3 comigrates with LeishIF4E-1 and LeishIF4E-4 on sucrose gradients
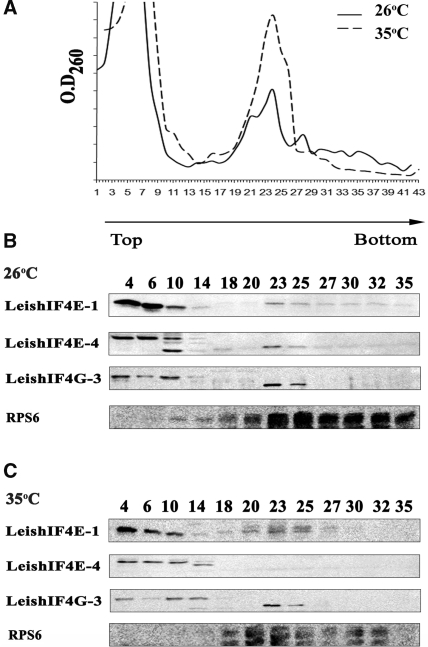
The LeishIF4G-3 ORF is smaller than the mammalian eIF4G-I and contains 635 compared with 1560 amino acids, respectively. It includes the middle domain of eIF4G (MIF4G), which is highly conserved among divergent organisms (see Supplementary Figure 2), but the other regions of LeishIF4G-3 bear no significant sequence similarity to the human eIF4GI, with a rather short N-terminus. In higher eukaryotes, eIF4G was shown to be mostly distributed in the cytoplasm (37), this was verified in Leishmania as well (data not shown). To investigate whether LeishIF4G-3 comigrates with LeishIF4E-1 and LeishIF4E-4 in particles of the same size, cell extracts of L. major cells that were grown at 26°C, a temperature used for growth of Leishmania promastigotes, were fractionated over sucrose gradients. The polysome profile was monitored by RNA absorbance at 260 nm (Figure 2A) and migration of the ribosomal small subunit was monitored by western analysis using antibodies directed against rpS6. Migration of the different LeishIF4F subunits were also monitored by western analysis. As shown in Figure 2B, LeishIF4G-3 comigrates with LeishIF4E-1 and LeishIF4E-4 in fractions that contain free RNPs at the top of the gradient (up to fraction 10), as well as in heavier fractions (10–14) that contain the 40S subunits, as demonstrated by their reaction with anti-rpS6 antibodies. In addition, for each of the proteins examined, a weaker reaction was observed in fractions that correspond to the 80S particles (fractions 23–25). However, this fraction highlighted lower bands of LeishIF4E-4 and LeishIF4G-3 as well as a higher band of LeishIF4E-1. Given that the antibodies against LeishIF4E-1 and LeishIF4E-4 do not cross-react with the other eIF4E paralogs in Leishmania, these bands may represent different modification forms of the protein, possibly caused by phosphorylation. A very weak association of LeishIF4E-1 and LeishIF4E-4 with polysomes was observed (fractions 27–35).
Figure 2.
LeishIF4G-3 comigrates with LeishIF4E-1 and LeishIF4E-4 by sucrose gradients analysis. Extracts of L. major cells grown at 26°C, and after exposure to 35°C for 18 h were loaded on 10–50% sucrose gradients. (A) Pattern of polysome migration on sucrose gradients. Fractions were collected from the top of the gradient and the O.D260nm was monitored. (26°C, continuous line; 35°C, broken line). Proteins were recovered by TCA precipitation, and immunoblotted with antibodies raised against LeishIF4E-1, LeishIF4E-4, LeishIF4G-3 as well as with anti-rpS6, for detection of fractions that contain complexes containing 40S particles. Western analysis was done on fractions of cells that were grown at 26°C (B) and 35°C (C).
Exposure of the parasites to elevated temperatures, which mimic the conditions in the human host (1), was shown to decrease the amount of polysomes in the cell, and as a result to increase the 80S fraction [(38) and Figure 2C]. This was reflected by the migration profile of rpS6. While during heat shock LeishIF4E-4 migrates only as RNPs, LeishIF4E-1 and LeishIF4G-3 maintain their migration profile with particles that contain the 40S small ribosomal subunit, including the 80S complex. It therefore appears that LeishIF4E-1 and LeishIF4G-3 most probably function at both life stages.
LeishIF4G-3 interacts with LeishIF4E-1 and LeishIF4E-4 in vitro
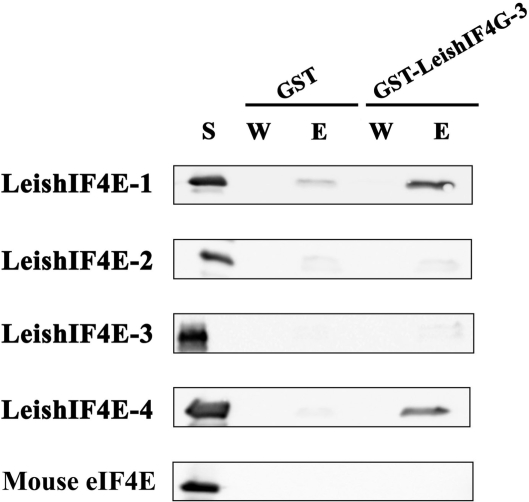
A common feature of the cap-binding complex in eukaryotes is the interaction between eIF4E and eIF4G. Copurification over m7GTP-Sepharose column and comigration on sucrose gradients strongly suggest the ability of the parasite proteins to physically interact, although being indirect. Therefore, pull-down assays using a GST-LeishIF4G-3 fusion protein, or GST alone were expressed in bacteria and immobilized onto Glutathione-Agarose beads. These beads were incubated with the soluble extracts of bacteria, each expressing one of the four LeishIF4Es, or the mouse eIF4E ortholog. The complexes were eluted and their components were identified by western blot analysis using specific antibodies. As shown in Figure 3, LeishIF4G-3 pulled-down LeishIF4E-1 and LeishIF4E-4, but not LeishIF4E-2 or LeishIF4E-3, when compared with the GST control. No interaction was observed between LeishIF4G-3 and the mouse eIF4E, supporting the observed divergence between LeishIF4G-3 and its mammalian ortholog.
Figure 3.
LeishIF4G-3 interacts with LeishIF4E-1 and LeishIF4E-4 in vitro. GST or GST-LeishIF4G-3 fusion protein were immobilized on Glutathione-Agarose beads. The beads were incubated with supernatant of bacteria that express the different LeishIF4E isoforms or the mouse eIF4E ortholog (S). After extensive washes (W) the protein complexes were eluted (E), and analyzed by western blot with specific antibodies against the different eIF4Es.
LeishIF4G-3 interacts with LeishIF4E-1 and LeishIF4E-4 in vivo
The ability of LeishIF4G-3 to interact with the different LeishIF4Es in vivo was investigated in transgenic L. major cell lines expressing a FLAG-tagged LeishIF4G-3. Cell extracts were incubated with anti-FLAG-Sepharose beads and the eluted fractions were analyzed with specific antibodies raised against the different LeishIF4F subunits. This analysis confirmed that LeishIF4G-3 could pull-down LeishIF4E-4, along with LeishIF4A-1 (Figure 4), and a very weak interaction between the tagged LeishIF4G-3 and LeishIF4E-1 was also detected.
Figure 4.
LeishIF4G-3-FLAG pulls down LeishIF4E-4. Leishmania major cells were transfected with pX-LeishIF4G-3-FLAG and cell lines expressing the FLAG tagged LeishIF4G-3 were selected in the presence of 200 μg/ml G418. The cells were lyzed by sonication and the soluble fractions (S) were incubated with anti-FLAG beads. After incubation for 3 h, the beads were washed (W), the proteins were eluted (E), separated by SDS–PAGE and analyzed by western blot.
In view of the different results obtained in the in vitro and in vivo tagging experiments that examine the interaction between LeishIF4G-3 and LeishIF4E-1, a reciprocal in vivo tagging approach was used. LeishIF4E-1 and LeishIF4E-4 were fused to the tandem affinity purification (TAP)-tag, using the pPNSAP1 vector (33), which adds two different in frame tags, a Protein A peptide and a streptavidin-binding peptide. The tagged LeishIF4E-1 was first bound to streptavidin beads, and the biotin-eluted fractions were further subjected to purification on an IgG-Sepharose column (Figure 5A). For the tagged LeishIF4E-4, the second round of purification was performed over m7GTP-Sepharose, taking advantage of the cap-binding activity of LeishIF4E-4 (Figure 5B). Western blot analysis of the purified complexes confirmed that LeishIF4G-3 was coprecipitated with LeishIF4E-1 and LeishIF4E-4. The pull-down of LeishIF4G-3 by the tagged LeishIF4E-1 was more efficient than the reciprocal pull-down of LeishIF4E-1 by tagged LeishIF4G-3, possibly due to over expression of the tagged LeishIF4E-1 in the transgenic parasites, which could serve to enhance a basically weaker interaction. The two TAP-tagged cap-binding complexes also contained LeishIF4A-1, the third components of the eIF4F complex.
Figure 5.
LeishIF4G-3 and LeishIF4A-1 copurifiy with TAP-tagged LeishIF4E-1 and LeishIF4E-4. Leishmania major wild-type or transgenic cells expressing TAP-tagged LeishIF4E-1 (A) and LeishIF4E-4 (B) were lyzed and the soluble supernatant (S) was loaded on streptavidin-Sepharose beads. The beads were washed, eluted with biotin and repurified in tandem over IgG-Sepharose beads (LeishIF4E-1) or over m7GTP-Sepharose beads (LeishIF4E-4). The corresponding beads were washed (W) and eluted (E, see Materials and Methods section). The W and E lanes were loaded with 20% of the wash and eluate volumes, respectively. All proteins were fractionated by SDS–PAGE (12%) and subjected to western blot analysis using antibodies specific against LeishIF4E-1, LeishIF4E-4, LeishIF4G-3 and LeishIF4A-1.
The interaction between LeishIF4G-3 and LeishIF4E-4 is mediated through a short peptide in its N-terminus
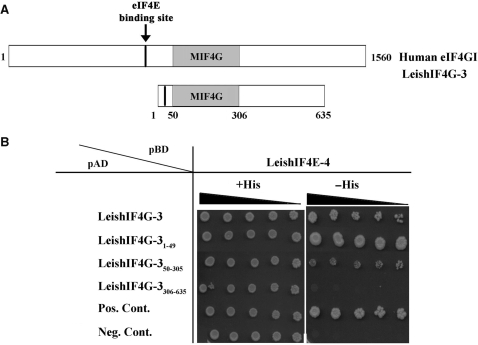
The GAL4 yeast two-hybrid system provides an alternative approach to monitor the interactions between LeishIF4G-3 and LeishIF4Es. This assay was used to map the LeishIF4G-3 fragment that is responsible for binding to LeishIF4E-4. LeishIF4E-4 was fused to the binding domain (BD) of GAL4 whereas LeishIF4G-3, or derivative fragments were fused to the GAL4 transcription activation domain (AD). The interaction between the tested proteins brings together the GAL4 DNA binding and transcriptional activation domains, resulting in transcription of the HIS3 gene, and growth of the cotransformed yeast cells on selective medium lacking histidine. The LeishIF4G-3 fragments that were investigated corresponded to the first 49 amino acids in the N-terminus (LeishIF4G-31-49), the MIF4G domain that comprises amino acids 50–305 (LeishIF4G-350–305) and the C-terminal domain that lies between amino acids 306–635 (LeishIF4G-3306–635) (Figure 6A). Growth in the absence of histidine was promoted only in yeast cells expressing either the full-length LeishIF4G-3 or its N-terminal domain, LeishIF4G-31–49, indicating that the amino terminus is indeed responsible for the interaction with LeishIF4E-4. The presence of the foreign genes (LeishIF4E-4 and the LeishIF4G-3 fragments) in the transgenic yeast was verified (see Supplementary Figure 3). While cells expressing the C-terminus LeishIF4G-3305–635 did not grow on medium-lacking histidine, the MIF4G domain of LeishIF4G-3 (LeishIF4G-350–305), showed a very weak interaction with LeishIF4E-4, which caused a feeble growth of yeast on the selective medium (Figure 6B).
Figure 6.
Monitoring the LeishIF4G-3 interaction with LeishIF4E-4 by the yeast two-hybrid assay. (A) Scheme of LeishIF4G-3 highlighting the location of its eIF4E-binding region and the MIF4G domain, as compared with the human eIF4GI. (B) Yeast cells were cotransformed with pBD-LeishIF4E-4 and pAD fused to different fragments of LeishIF4G-3 as indicated (1–49, 50–305, 306–635), or with positive or negative control plasmids (see Material and Methods section for details). The cells were cultured in Liquid SD-2 (-Trp/-Leu/+His), and 3-fold dilutions were spotted on SD-2 or SD-3 (-Trp/-Leu/-His) containing 2 mM 3-AT; the plates were incubated at 30°C for 10 days.
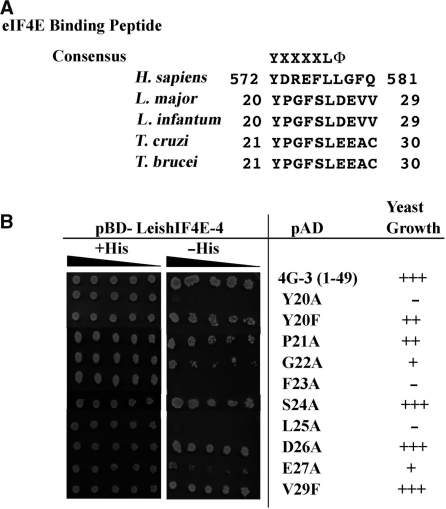
In higher eukaryotes the interaction of eIF4G with eIF4E is based on a short sequence motif Y(X4)LΦ, where X corresponds to any amino acid and Φ is a hydrophobic residue (27,28). A similar sequence element was found in the N-terminus of LeishIF4G-3 between positions 20 and 26 (YPGFSLD, Figure 7A). The peptide contains the consensus element 20-Y(X4)L-25, however, the conserved hydrophobic residue is replaced with an acidic amino acid, aspartic acid in Leishmania and glutamic acid in trypanosomes. Mutations were introduced in specific amino acids in the LeishIF4G-3 peptide, and their effect on binding to LeishIF4E-4 was examined using the yeast two-hybrid system (Figure 7B). The mutations were introduced into the N-terminal fragment of LeishIF4G-3 (amino acids 1–49) to avoid the masking of any negative binding by the presence of the MIF4G domain, which showed residual binding. Mutating the tyrosine at position 20 to alanine, but not to phenylalanine, eliminated the binding of LeishIF4G-31–49 to LeishIF4E-4. Similarly, the conserved leucine at position 25 is essential for binding, as expected. Unlike higher eukaryotes, where mutations in the amino acids that follow the tyrosine do not affect binding to eIF4E (27), in LeishIF4G-3 orthologs of trypanosomatids, these amino acids (21-PGFS-24) are conserved and some are also essential for binding. Replacement of phenylalanine 23 to alanine completely abrogates binding to LeishIF4E-4. Other mutations also affect yeast growth, though to a partial level: the exchange of glycine 22 with an alanine slowed down yeast growth rather efficiently whereas mutation of proline 21 had a much milder effect. In the flanking region, the only mutation that decreased binding to LeishIF4E-4 was the exchange of glutamic acid at position 27 with alanine, whereas exchange of aspartic acid 26 with alanine had no such effect. The valine residues at positions 28 and 29 are not conserved among trypanosomatids, suggesting that they are not required for the direct binding. Indeed, valine 29 is dispensable for yeast growth, as shown by its exchange to phenylalanine.
Figure 7.
Effect of mutations in the LeishIF4G-3 consensus peptide on binding to LeishIF4E-4. (A) Sequence conservation of the consensus peptide [Y(X4)LΦ] of LeishIF4G-3 in different trypanosomatids (L. major, L. infantum, T. cruzi, T. brucei), as compared with the human peptide (H. sapiens). (B) Yeast cells were cotransformed with pBD-LeishIF4E-4 and pAD fused to the LeishIF4G-31–49 fragment carrying different point mutations in the consensus peptide. The cells were cultured as described in the legend of Figure 6, and growth in the absence of histidine served as an indication for binding between LeishIF4G-3 and LeishIF4E-4.
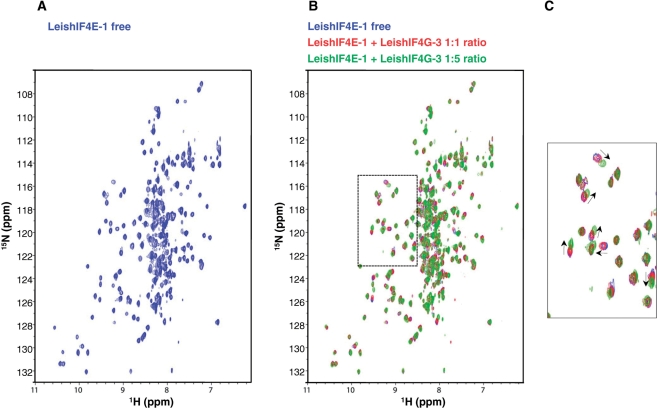
The binding to LeishIF4E-1 of a synthetic short peptide of 18 amino acids (14-NYLEPPYPGFSLDEVVRR-31) from the LeishIF4G-3 sequence, encompassing the consensus eIF4E-binding motif, was also investigated by NMR titration. LeishIF4E-1 was used for technical reasons, since the recombinant protein is more soluble than LeishIF4E-4 and stable at 25°C for a prolonged period. Further, LeishIF4E-1 binds m7GTP with an affinity similar to cap-4 (30). m7GTP was added to LeishIF4E-1 prior to recording the NMR spectra in order to stabilize the protein. A [15N,1H]TROSY-HSQC spectrum (36) was recorded using uniformly 15N-labeled LeishIF4E-1 (LeishIF4E-1 free) (Figure 8A). In subsequent experiments, unlabeled LeishIF4G-3 peptide was added in increasing molar ratios. This resulted in chemical shift changes in a select subset of resonances (Figure 8B and C) indicating that LeishIF4G-3 peptide binds to LeishIF4E-1.
Figure 8.
Binding of a LeishIF4G-3 peptide to LeishIF4E-1. (A) 15N, 1HTROSY-HSQC spectra of 15N-labeled LeishIF4E-1 (250 µM) in the presence of m7GTP (5 mM) for stabilization (LeishIF4E-1 free, in blue). (B) Overlays of 15N, 1HTROSY-HSQC spectra of 15N-LeishIF4E-1 free with increasing amounts of non-labeled LeishIF4G-3 peptide (NYLEPPYPGFSLDEVVRR). The concentrations of the LeishIF4G-3 added were 0 µM (blue, ratio 1:0), 250 µM (red, ratio 1:1) and 1.25 mM (green, ratio 1:5). (C) Enlargement of a region displaying the chemical shift differences between the spectra that is boxed in (B) in dashed lines. Peaks movements are indicated by arrows.
DISCUSSION
The cap-binding complex in Leishmania and trypanosomatids is of special interest in view of the unusual cap-4 structure, which is found at the 5′-end of all mRNAs. Components of the cap-binding complex, particularly LeishIF4E, must have gone through changes that enable efficient binding to, and assembly on this highly modified structure. This is reflected by the inability of the different LeishIF4Es to replace eIF4E from higher eukaryotes (31). A bioinformatics analysis highlighted the presence of several putative eIF4G homologs in Leishmania; in this study, we showed that one of these candidates, LeishIF4G-3 was eluted from an m7GTP-Sepharose column, along with LeishIF4E-1, LeishIF4E-4 and LeishIF4A-1, which are part of the eIF4F complex. This strongly suggests that LeishIF4G-3 is the general translation factor of trypanosomatids. However, proteins which were eluted from an m7GTP column are not restricted to translation, and could be associated with other RNA metabolism and transport processes between the nucleus and the cytoplasm. Thus, a more direct approach for monitoring the interactions between the putative LeishIF4G-3 and the Leishmania cap-binding translation initiation factors is required. We therefore characterized the interaction between LeishIF4G-3 and the different LeishIF4E homologs by pull-down assays. Only LeishIF4E-1 and LeishIF4E-4 were coprecipitated in vitro with the recombinant GST-LeishIF4G-3 fusion protein from Glutathione-Agarose beads, in agreement with our previous report on their ability to bind the human 4E-BP1. LeishIF4G-3 comigrates with LeishIF4E-1 and LeishIF4E-4 in sucrose gradients, supporting that these proteins can share the same sub-cellular particles.
The effect of heat shock on the migration profile of polysomes in Leishmania is in agreement with other reports in Trypanosoma brucei, showing that the relative amount of polysomes decreases whereas the 80S fraction is enlarged (38). In response to temperature elevation of Leishmania parasites, LeishIF4E-4 disintegrates from heavier complexes, and is found only at the top of the gradient, as free RNPs. However, LeishIF4E-1 which goes up in amastigotes (31), and LeishIF4G-3 continues to comigrate with the 80S particles, even when the general translation activity is low. This is in-line with the requirement that amastigotes resume their protein synthesis activity at elevated temperatures following differentiation to the life form that resides in mammals.
The interactions between recombinant proteins in vitro do not always provide a faithful reflection of the parallel in vivo processes, because they do not involve the complete subunit repertoire of their complexes. The interaction between LeishIF4G-3 and LeishIF4Es were therefore monitored in vivo. FLAG-tagged LeishIF4G-3 was able to pull-down only LeishIF4E-4. However, the reciprocal approach, where both LeishIF4E-1 and LeishIF4E-4 were TAP-tagged in vivo, verified the interaction between LeishIF4G-3 and LeishIF4E-1, as well as with LeishIF4E-4, in agreement with the results obtained from the in vitro pull-down assays. In the case of TAP-tagged LeishIF4E-1, the anti-LeishIF4G-3 antibodies identified a band that migrates slightly faster, suggesting perhaps that LeishIF4E-1 and LeishIF4E-4 interact with different phosphorylated forms of LeishIF4G-3 (Figure 5). This faster migrating band also cosediments with LeishIF4E-1 and LeishIF4E-4 in sucrose gradients (Figure 2).
In yeast eIF4G, the amino terminus is unfolded, and binding to eIF4E induces the folding of a region that encompasses 100 amino acids (positions 393–490) into a ‘molecular bracelet’ of α-helices that encircles the unstructured amino terminus of the eIF4E (18,39). In LeishIF4G-3 the amino terminus is much shorter, and consists of only 49 amino acids upstream of the first HEAT repeat. This region was found to be sufficient to promote the binding of LeishIF4G-3 to LeishIF4E-4, as indicated by the yeast two-hybrid assay.
An oligo-peptide in the N-terminal region of LeishIF4G-3 that shows partial conservation with the consensus signature peptide Y(X4)L of higher eukaryotes was verified by specific point mutations (Figure 7). The first tyrosine of the LeishIF4G-3 peptide is essential for binding to LeishIF4E-4. Its replacement by alanine abrogates the binding completely, whereas replacement by a phenylalanine residue indicates that the peptide is still functional, although growth of the yeast expressing the Y20F mutant is slower. A similar exchange in the human eIF4G peptide where tyrosine was replaced by phenylalanine had little, or no effect on its binding to eIF4E (27). Both amino acids are aromatic and a phenylalanine residue also occupies the parallel position in the consensus peptide of eIF4G from C. elegans (386-FGRDFMV-392). As expected, the leucine residue (L25) in the peptide is conserved and essential, and its replacement by alanine eliminates binding to LeishIF4E-4 completely. However, the subsequent residue in trypanosomatids is not hydrophobic, as in higher eukaryotes, and is replaced by an acidic residue, aspartic acid in L. major and L. infantum, or glutamic acid in T. brucei and T. cruzi (Figure 7A). This acidic residue is nonessential in Leishmania, and can be replaced by alanine without affecting the binding between LeishIF4G-3 and LeishIF4E-4. Another example where the hydrophobic residue is not conserved is the eIF4E-binding peptide (YERAFMK) of 4E-BP from Drosophila melanogaster (NP_477295), where the sixth position following the tyrosine in that peptide is occupied by a basic lysine residue, marked in bold. In contrast, the neighboring glutamic acid at position 27 from LeishIF4G-3 participates in the binding activity to LeishIF4E. We further highlighted additional deviations of the LeishIF4G-3 signature peptide as compared with its higher eukaryotic counterpart. The four amino acids that follow the essential tyrosine residue (Y20) are conserved among trypanosomatids, and our mutational analysis shows that the phenylalanine (F23) is essential, whereas the proline and glycine residues (P21 and G22) have a partial effect on the binding. In higher eukaryotes, replacement of amino acids at these positions has no effect on the binding to eIF4E (27). The different structural requirements in the peptide of LeishIF4G-3 are most probably required to promote assembly of the parasite eIF4F subunits on the highly modified cap-4 structure. The binding of the LeishIF4G-3 peptide to LeishIF4E-1 was also investigated by NMR spectroscopy. Following the addition of an 18 amino acids LeishIF4G-3 peptide to LeishIF4E-1, chemical shifts were observed, when compared with the LeishIF4E-1 NMR spectra recorded in the absence of the same peptide (Figure 8). The resonances that were most strongly affected mapped to amino acids in LeishIF4E-1 that are homologous (by sequence alignment) to the binding site in the mouse and yeast eIF4Es.
The deviations of the cap-4 binding complex of Leishmania from its higher eukaryote counterpart are further emphasized by the inability of LeishIF4G-3 to bind the murine eIF4E (Figure 3). This can be partly due to changes in the eIF4E binding peptide that are described above, and to the fact that the amino terminus of LeishIF4G-3 is rather short, questioning its ability to create the ‘molecular bracelet’ that has been shown for the yeast eIF4G. In higher eukaryotes, the amino terminus is also responsible for interacting with the PABP at the 3′-end of the mRNA. This is in agreement with our inability to show direct binding of LeishIF4G-3 to the Leishmania PABP (40) in the yeast two-hybrid assay (data not shown). At this stage we cannot exclude the possibility that the translation initiation complex in Leishmania may contain additional components that complement the function of the LeishIF4G-3 amino terminus in stabilizing the LeishIF4F complex.
Unlike most eIF4E homologs from higher eukaryotes, LeishIF4E-4 has a relatively long amino terminus of about 100 amino acids, and overall shows a low degree of conservation with eIF4E from higher eukaryotes. Furthermore, part of the residues that occupy positions which are expected to interact with eIF4G are not conserved (31). This includes exchange of the murine histidine 37 and proline 38 by methionine residues at both positions in LeishIF4E-4.
In conclusion, the variability in the amino termini of LeishIF4G-3 and LeishIF4E-4, as well as the specific changes of conserved amino acids that are expected to promote the interaction between them, could represent adaptations that occurred during evolution to enable the eIF4F assembly on the unique cap-4 moiety in trypanosomatids. In view of the above, we further speculate that the cap-4 binding complex could possibly serve as an intriguing target for the development of novel drugs, as shown for eukaryotes, where inhibitors of the interaction between eIF4E and eIF4G affect the growth of malignant cells in culture (41).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US-Israel Binational Foundation (grant number 2007287 to M.S. and G.W.); the UNICEF/UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (to M.S.); the Israel Ministry of Health (grant number 5440 to M.S.); the National Institutes of Health (grant numbers CA68262 and GM47467 to G.W.); a post-doctoral fellowship from the Fonds de la Recherche en Santé du Québec (to M.L.); the Howard Hughes Medical Institute (grant number 55005604 to E.D.); the National Science Support Project 2008-2010 No. PBZ-MNiSW-07/I/2007 (to E.D.). Funding for open access charge: US-Israel Binational Foundation (grant number 2007287 to M.S. and G.W.), National Institutes of Health (grant number CA075879 to G.W.) and Howard Hughes Medical Institute (grant number 55005604 to E.D.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr H. Arthanari for his help with NMR spectroscopy and for critical reading of the manuscript. We thank Prof. Orna Elroy-Stein for kindly providing us with anti-rpS6 antibodies.
REFERENCES
- 1.Shapira M, McEwen JG, Jaffe CL. Temperature effects on molecular processes which lead to stage differentiation in Leishmania. EMBO J. 1988;7:2895–2901. doi: 10.1002/j.1460-2075.1988.tb03147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zilberstein D, Shapira M. The role of pH and temperature on the development of Leishmania parasites. Annu. Rev. Microbiol. 1994;48:449–470. doi: 10.1146/annurev.mi.48.100194.002313. [DOI] [PubMed] [Google Scholar]
- 3.Bates P. Axenic culture of Leishmania amastigotes. Parasitol. Today. 1993;9:143–146. doi: 10.1016/0169-4758(93)90181-e. [DOI] [PubMed] [Google Scholar]
- 4.Barak E, Amin-Spector S, Gerliak E, Goyard S, Holland N, Zilberstein D. Differentiation of Leishmania donovani in host-free system: analysis of signal perception and response. Mol. Biochem. Parasitol. 2005;141:99–108. doi: 10.1016/j.molbiopara.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Liang XH, Haritan A, Uliel S, Michaeli S. Trans- and cis-splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Bangs JD, Crain PF, Hashizume T, Mccloskey JA, Boothroyd JC. Mass spectrometry of mRNA cap-4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 1992;267:9805–9815. [PubMed] [Google Scholar]
- 8.Hentze MW. eIF4G: a multipurpose ribosome adapter? Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. [DOI] [PubMed] [Google Scholar]
- 9.Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 10.Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 11.Korneeva NL, Lamphear BJ, Hennigan FL, Rhoads RE. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J. Biol. Chem. 2000;275:41369–41376. doi: 10.1074/jbc.M007525200. [DOI] [PubMed] [Google Scholar]
- 12.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 13.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q. Rev. Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 15.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tarun SZ, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 17.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell. 1999;6:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- 18.Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- 19.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcotrigiano J, Lomakin IB, Sonenberg N, Pestova TV, Hellen CU, Burley SK. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- 21.Bellsolell L, Cho-Park PF, Poulin F, Sonenberg N, Burley SK. Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1. Structure. 2006;14:913–923. doi: 10.1016/j.str.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Marintchev A, Wagner G. eIF4G and CBP80 share a common origin and similar domain organization: implications for the structure and function of eIF4G. Biochemistry. 2005;44:12265–12272. doi: 10.1021/bi051271v. [DOI] [PubMed] [Google Scholar]
- 23.Henis-Korenblit S, Shani G, Sines T, Marash L, Shohat G, Kimchi A. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc. Natl Acad. Sci. USA. 2002;99:5400–5405. doi: 10.1073/pnas.082102499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Contreras V, Richardson MA, Hao E, Keiper BD. Depletion of the cap-associated isoform of translation factor eIF4G induces germline apoptosis in C. elegans. Cell Death Differ. 2008;15:1232–1242. doi: 10.1038/cdd.2008.46. [DOI] [PubMed] [Google Scholar]
- 25.Marash L, Liberman N, Henis-Korenblit S, Sivan G, Reem E, Elroy-Stein O, Kimchi A. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol. Cell. 2008;30:447–459. doi: 10.1016/j.molcel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 27.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4G and the translational repressors 4E-binding proteins. Mol. Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altmann M, Schmitz N, Berset C, Trachsel H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997;16:1114–1121. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoffe Y, Zuberek J, Lewdorowicz M, Zeira Z, Keasar C, Orr-Dahan I, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Shapira M. Cap-binding activity of an eIF4E homolog from Leishmania. RNA. 2004;10:1764–1775. doi: 10.1261/rna.7520404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoffe Y, Zuberek J, Lerer A, Lewdorowicz M, Stepinski J, Altmann M, Darzynkiewicz E, Shapira M. Binding specificities and potential roles of isoforms of eukaryotic initiation factor 4E in Leishmania. Eukaryot. Cell. 2006;12:1969–1979. doi: 10.1128/EC.00230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhalia R, Reis CR, Freire ER, Rocha PO, Katz R, Muniz JR, Standart N, de Melo Neto OP. Translation initiation in Leishmania major: characterisation of multiple eIF4F subunit homologues. Mol. Biochem. Parasitol. 2005;140:23–41. doi: 10.1016/j.molbiopara.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeBowitz JH, Coburn CM, McMahon-Pratt D, Beverley SM. Development of a stable Leishmania expression vector and application to the study of parasite surface antigen genes. Proc. Natl Acad. Sci. USA. 1990;87:9736–9740. doi: 10.1073/pnas.87.24.9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laban A, Wirth DF. Transfection of Leishmania enriettii and expression of chloramphenicol acetyltransferase gene. Proc. Natl Acad. Sci. USA. 1989;86:9119–9123. doi: 10.1073/pnas.86.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pervushin KV, Wider G, Riek R, Wuthrich K. The 3D NOESY-[(1)H,(15)N,(1)H]-ZQ-TROSY NMR experiment with diagonal peak suppression. Proc. Natl Acad. Sci. USA. 1999;96:9607–9612. doi: 10.1073/pnas.96.17.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKendrick L, Thompson E, Ferreira J, Morley SJ, Lewis JD. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7)guanosine cap. Mol. Cell Biol. 2001;21:3632–3641. doi: 10.1128/MCB.21.11.3632-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel JD, Clayton C, Carrington M. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J. Cell Sci. 2008;121:3002–3014. doi: 10.1242/jcs.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hershey PE, McWhirter SM, Gross JD, Wagner G, Alber T, Sachs AB. The Cap-binding protein eIF4E promotes folding of a functional domain of yeast translation initiation factor eIF4GI. J. Biol. Chem. 1999;274:21297–21304. doi: 10.1074/jbc.274.30.21297. [DOI] [PubMed] [Google Scholar]
- 40.Bates EJ, Knuepfer E, Smith DF. Poly(A)-binding protein I of Leishmania: functional analysis and localisation in trypanosomatid parasites. Nucleic Acids Res. 2000;28:1211–1220. doi: 10.1093/nar/28.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.