Abstract
The vertebrate kinetochore complex assembles at the centromere on α-satellite DNA. In humans, α-satellite DNA has a repeat length of 171 bp slightly longer than the DNA in the chromatosome containing the linker histone H1. The centromere-binding protein CENP-B binds specifically to α-satellite DNA with properties of a centromeric-linker histone. Here, we analysed if linker histone H1 is present at or excluded from centromeric chromatin by CENP-B. By immunostaining we detected the presence, but no enrichment or depletion of five different H1 subtypes at centromeric chromatin. The binding dynamics of H1 at centromeric sites were similar to that at other locations in the genome. These dynamics did not change in CENP-B depleted cells, suggesting that CENP-B and H1 co-exist in centromeric chromatin with no or little functional overlap. By bimolecular fluorescence complementation (BiFC) and Förster resonance energy transfer (FRET), we revealed that the linker histone H1 subtypes H1° and H1.2 bind to centromeric chromatin in interphase nuclei in direct neighbourhood to inner kinetochore proteins.
INTRODUCTION
Centromeres are involved in faithful DNA segregation into daughter cells during mitosis. Centromeric chromatin consists of interspersed regions in which either histone H3 is present or, alternatively, both H3 histones are replaced by CENH3 (in humans: CENP-A). This centromeric chromatin region is framed by pericentromeric heterochromatin. During interphase, the kinetochores form a specialized chromatin of a roughly spherical structure (‘interphase pre-kinetochore’) distinct from the trilaminar structure of the kinetochores in mitosis after nuclear membrane break down (1). CENP-A forms a more compact complex with H4 compared to H3 resulting in a modified nucleosomal structure at the centromere (2,3). In addition to CENP-A, a larger number of inner kinetochore proteins are constitutively present at the centromeres during the whole cell cycle (4–9, recently reviewed by 10), although with cell cycle-dependent variations in their residence times (11,12). Essential for proper mitosis are CENP-A and CENP-C, which are found at all active centromeres including neo-centromeres (13,14), and depletion of CENP-A leads to the mislocalization of most but not all centromere proteins (15–17). Depletion of inner kinetochore proteins can result in chromosome missegregation and disruption of mitosis.
The 80 kDa centromere-binding protein CENP-B (18) not only binds to the centromere, but also to the pericentric heterochromatin domain distributed between sister kinetochores (19). It binds to a specific DNA sequence, the 17-bp ‘CENP-B box’ which is present in α-satellite repeats in human centromeres and in pericentromeric regions (20–23). The CENP-B/CENP-B box interaction (24) is crucial for the assembly of mammalian artificial chromosomes (25–27). CENP-B is dimeric and contains DNA-binding and dimerization domains at its N- and C-terminus, respectively (28–30). Binding of CENP-B to the CENP-B box bends the DNA by 59° which induces translational positioning of CENP-A containing nucleosomes on alphoid DNA in vitro (22,31). The length of alphoid DNA arrays and the density of CENP-B boxes have a strong effect on the CENP-A chromatin core and the formation of functional kinetochores (26). Thus, similar to the role of histone H1 in chromatin, binding of CENP-B to multiple adjacent CENP-B boxes arrayed in alphoid satellite DNA might promote assembly of a stable functional centromeric chromatin core with CENP-A nucleosomes in vivo. However, CENP-B seems neither be sufficient nor essential for functional kinetochore formation and maintenance in all cases: one of the two centromeres on human dicentric fusion chromosomes is frequently inactivated with essential kinetochore proteins missing (32,33) even though both centromers bind CENP-B (34,35). In mouse cells, functional kinetochores can be maintained without CENP-B (36) and CENP-B is neither detected on the Y chromosome centromere (37) nor on neo-centromeres. The active kinetochore state might be able to be maintained also in the absence of CENP-B, potentially by epigenetic mechanisms (27) and the contribution of linker histone H1. CENP-B might therefore serve as a centromeric linker histone, a question that we wished to address in this study.
CENP-C is an evolutionarily conserved essential kinetochore protein (38,39). It binds to centromeric DNA (40) adjacent to CENP-B, however in a sequence-independent manner (41,42). The requirement of CENP-A for CENP-C localization at the kinetochore (16) and the direct interaction between CENP-C and CENP-B (43) indicate that CENP-A, CENP-C and in most cases also CENP-B are tightly associated to form centromeric chromatin. We recently provided direct evidence for this model in in vivo Förster resonance energy transfer (FRET) studies (44).
Human α-satellite DNA is 4-bp longer than the canonical chromatosome DNA (45–49, reviewed in 50). Thus, human centromeric nucleosomes might be nearly identical to chromatosomes in its overall structure. Furthermore, CENP-A containing chromatin could be built from chromatosomes containing linker histone H1. In this case, the 24-bp long linker at centromeres is short compared to a mean value of ∼50 bp in a non-centromeric chromatin (51,52). However, the assembly of human centromeric nucleosomes and the structure of centromeric chromatin are unclear. In the yeast Saccharomyces cerevisiae, and the fruitfly Drosophila melanogaster centromeric nucleosomes are formed from hexamers or tetramers, respectively (52–56).
H1 represents a family of histone subtypes that are considered to stabilize the compaction of the chromatin into higher order structures (57–59). In higher organisms, linker histones have a conserved structure consisting of a central globular domain flanked by a long lysine-rich C-terminal tail and a shorter partly basic N-terminal extension. Specific subdomains of the C-terminal tail are crucial for H1 linker DNA binding and for stabilizing folded chromatin structures (60,61). Structural analysis revealed that the H1 central globular domain has two distinct binding sites (62) interacting with the DNA major groove near the dyad axis (63–65) and with the minor groove on the linker DNA about 15 bp away from the end of the nucleosomal core, respectively (66). H1 binds to nucleosomes without any known specificity of the underlying DNA sequence, protecting 15–20 bp of chromatosomal DNA (45,46). H1 exchange in chromatin of living cells is very rapid (60,66–68) indicating only transient interactions between H1 and nucleosomes, while core histones are stably incorporated into nucleosomes (69). H1.2 shows differential exchange dynamics at various stages of mitosis with the fastest rate at metaphase (70). H1 presence is able to suppress the ability of other proteins to bind to nucleosomal target sites (71); it might thus also interfere with CENP-B binding to chromatin.
Chromatin of different cell types varies in number and composition of histone H1 subtypes. Individual linker histone subtypes are supposed to fulfil different functions (reviewed in 72–74). There is a growing evidence for the participation of H1 histones in the regulation of gene activity (reviewed in 75, 76) and they are supposed to play a role in cell proliferation and differentiation (77,78), apoptosis (79), DNA repair (80) and aging (81). So far, eleven H1 homologous proteins have been described in mammals. These include five replication-dependent main class subtypes H1.1, H1.2, H1.3, H1.4 and H1.5 and the H1 replacement subtype H1° which is mainly restricted to cells which are arrested in proliferation. The seventh is the ubiquitous H1× and the last four subtypes are the developmental- and tissue-specific H1t, H1T2, HILS1 and H1Foo (reviewed in 74,82). The histone H1 subtypes in somatic cells in mammals share a highly conserved globular domain sequence while exhibiting variations in the N- and C-terminal tails (83). The different tissue- and developmental-specific H1 subtypes seem to be able to replace one another: in chicken DT40 cells (84) and in mice (85), deletion of one H1 subtype elevates the levels of the remaining subtypes, an indication that cells strive to maintain a constant level of H1. In mice, deletion of either the developmentally expressed H1°, the testis specific H1t, or any specific H1 somatic subtype did not affect survival and did not have any significant biological consequences. Thus, the correct overall amount of H1 rather than the correct relative amounts of the subtypes are crucial factors in H1 function (86,87). H1 subtypes might have a dual role: in addition to their general and exchangeable role, each individual subtype might also have a specific function (74). Inactive chromatin contains all subtypes, and centromeric heterochromatin and centromeres contain them in equal amounts (88).
Here, we analysed in living interphase cells if H1 is present in human centromeric chromatin and if it binds to linker DNA next to CENP-A containing nucleosomes, potentially in the direct vicinity also to CENP-B and -C.
MATERIALS AND METHODS
Immunofluorescence
For indirect immunofluorescence detection, HeLa cells (from the DSMZ, Braunschweig, Germany) were fixed with 3% paraformaldehyde (pFA) in phosphate buffered saline (PBS) for 15 min, permeabilized with 0.5% Triton X-100 in PBS for 10 min and blocked with 3% bovine serum albumin (BSA) in PBS. The following antibodies were used for indirect immunofluorescence labeling: affinity-purified anti-H1x antibody at a final concentration of 2 µg/ml (89), anti-H1.3 antibody (#ab24174, Abcam, Cambridge, UK) in a 1:50 dilution, H1.5 antibody (#ab24175, Abcam, Cambridge, UK) in a 1:50 dilution, a polyclonal anti-serum specific for H1.2 and H1.5 (Ab4112, 89) or a monoclonal anti-H1° antibody (kind gift of H. Zentgraf) in a 1:2 dilution. The following secondary antibodies were used in a 1:1000 dilution: Alexa Fluor 488-anti-mouse IgG (#A11017) and Alexa Fluor 488-anti-rabbit IgG (#11070) from Molecular Probes (distributed by MoBiTec, Göttingen, Germany). Anti-human IgG-Cy3 was used in a 1:400 dilution. The nuclei were visualized with the fluorochrome 4′-6-diamidine-2-phenyl indole (DAPI) using Vectashield Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA, USA).
Plasmids
The cloning of the vectors pCerulean-C1-CENP-A, pCerulean-N1-CENP-B and pCerulean-C2-CENP-C was described elsewhere (44). For the expression of human H1° fused to the N-terminus of Enhanced Yellow Fluorescent Protein (EYFP) in human cell lines, we used plasmid pSVH1°-EYFP (a kind gift of W. Waldeck, Heidelberg). For the expression of human H1° fused to the C-terminus of EYFP, we digested pSVH1°-EYFP with HindIII–BamHI and ligated the purified 608-bp fragment into HindIII–BamHI-treated pEYFP-C3 (Clontech, Palo Alto, CA, USA). This procedure left the amino acids RDPPDLDN at the C-terminus of H1°.
For bimolecular fluorescence complementation (BiFC) analysis, we cloned two fragments from cerulean (90,91): Cerf (cerulean first; aas M1 – E173) and Cerl (cerulean last; aas C155 – K239). They were used to construct several cerulean fragment carrying expression vectors, which are identical to the fluorescent protein vector system of BD Bioscience - Clontech, i.e. pCerf-C1, 2, 3 and pCerl-C1, 2, 3 for fusion to the C-termini of the cerulean fragments, as well as pCerf-N1 and pCerl-N1 for fusion to the N-termini of the cerulean fragments.
We constructed pCerf-C1-CENP-A by replacing the 722-bp EYFP AgeI-BsrGI fragment of pEYFP-C1-CENP-A (44) with the 545-bp Cerf containing AgeI-BsrGI fragment from pCerf-C1. For construction of pCerl-N1-CENP-B and pCerf-N1-CENP-B, we digested pEGFP-N1-CENP-B (44) with EcoRI and AgeI. The resulting 1799-bp CENP-B fragment was ligated into the 4237-bp EcoRI-AgeI fragment of pCerl-N1, respectively, into the 4519-bp EcoRI-AgeI fragment of pCerf-N1. pEYFP-C3-H1° was digested with HindIII and BamHI. The 608-bp H1° fragment was ligated into the 4512-bp HindIII–BamHI fragment of pCerf-C3 as well as into the 4230 bp HindIII–BamHI fragment of pCerl-C3 resulting in pCerf-C3-H1° and pCerl-C3-H1°.
For fluorescence lifetime imaging (FLIM) analysis, we constructed pmCherry-C1, 2, 3 and pmCherry-N1 vectors identically to the corresponding Clontech pEGFP (Enhanced Green Fluorescent Protein) vectors by replacing of the 722-bp AgeI-BsrGI fragment with the 713-bp AgeI-BsrGI mCherry carrying fragment from pmCherry-H2A. pEYFP-N1-CENP-B and pEYFP-C2-CENP-B (44) were digested with AgeI and BsrGI, and for both vectors the 722-bp fragments were replaced with the 722-bp AgeI-BsrGI EGFP harboring fragment from pEGFP-C2 resulting in pEGFP-N1-CENP-B and pEGFP-C2-CENP-B. For construction of pmCherry-C3-H1° and pmCherry-N1-H1°, the 608-bp HindIII–BamHI fragment of H1° (see above) was ligated into HindIII–BamHI digested pmCherry-C3, respectively, pmCherry-N1-H1°. After digestion of pEGFP-N1-H1.2 with HindIII and BamHI, the 647-bp H1.2 fragment was ligated into HindIII and BamHI opend pmCherry-C3 resulting in pmCherry-C3-H1.2. The linker sequence is YSDLELKL, and due to the cloning protocol the amino acid sequence DPPDLDN is fused to the C-terminal end of H1.2. pEGFP-C1-CENP-A resulted from the replacement of EYFP with EGFP in AgeI–BsrGI-digested pEYFP-C1-CENP-A. We further used vector pEGFP-C2-CENP-C (11). As a reference vector for positive FRET control measurements served a vector expressing the fusion pEGFP-mCherry (a kind gift of N. Audugé and M. Coppey-Moisan, Paris). The linker sequence between the N-terminal EGFP and mCherry is SGLRSRGDPAT.
All clones were verified by sequencing (MWG Biotech, Ebersberg, Germany). Full length protein expression of the fusion constructs was confirmed by western blots as described by Orthaus et al. (44).
Cell culture and transfection into HEp-2 cells
HEp-2 (HeLa derivative) cells were available in the laboratory. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM, PAA Laboratories, Pasching, Austria) supplemented with 10% fetal calf serum (PAA Laboratories, Pasching, Austria) in a 9.5% CO2 atmosphere at 37°C. For live cell imaging experiments, cells were washed with magnesium and calcium-containing PBS (Sigma-Aldrich, Taufkirchen, Germany) followed by detachment with trypsine/EDTA (ethylenediaminetetraacetic acid) (PAA Laboratories, Pasching, Austria) and re-seeding on 42-mm glass dishes (Saur Laborbedarf, Reutlingen, Germany) or into 35-mm glass bottom culture dishes (MatTek Corporation, Ashland, MA, USA) 48–72 h before experiments. Transfection with plasmid DNA was performed 24–48 h before analysis using FuGENE HD transfection reagent (Roche, Basel, Switzerland) according to the manufacturer's protocol. When expressing fluorescently tagged H1° and H1.2 in HEp-2 cells, we did not observe induction of cellular senescence or formation of senescence-associated heterochromatic foci (87).
Fluorescence recovery after photobleaching experiments
Fluorescence recovery after photobleaching (FRAP) experiments were performed on a Zeiss 510 confocal microscope with a ×63/1.40 oil immersion objective at 37°C and carried out as described by Hemmerich et al. (11). In short, bleaching was accomplished with a circular spot (at the centromere or at other nuclear sites) using the 488 nm (30 mW Argon laser) and 543 nm laser line (15 mW DPSS561-10). For each bleach-pulse 20–35 iterations were used. Fluorescence recovery was measured at lower laser intensity and 512 × 512 pixel resolution at 2.5 s intervals for 5–10 min. In seperate FRAP experiments, 10–20 cells with 1–2 locations each were analysed and averaged to a single FRAP curve.
BiFC
HEp-2 cells grown on coverslips in a six-well plate were transfected with 1 μg of the appropriate BiFC vectors (92) as indicated in each experiment using FuGene HD. For formation and maturation of the fluorescent fluorophore mediated by the interaction of the linked proteins, cells were incubated for 24–48 h. Afterwards, cells were fixed by 4% pFA for 10 min and permeabilized with 0.3% Triton X-100 for 5 min. The centromeres were stained with guinea pig serum against the N-terminal half of CENP-C (a kind gift of K. Yoda) and a species-specific secondary antibody linked to rhodamine (Jackson Immunoresearch, West Grove, USA), both at a dilution of 1:200. Cells were mounted in ProLong Gold (Molecular Probes, Eugene, USA) and analysed by confocal microscopy. As a control, each non-fluorescent half of the fluorophore was also expressed separately. In these control experiments and in cells expressing both parts of cerulean unlinked to any protein, no cerulean fluorescence could be detected.
Acceptor bleaching-based FRET measurements
FRET was measured using the acceptor photobleaching method (93,94) as described by Orthaus et al. (44). In brief, (co-)transfected cells grown on coverslips were analysed using a confocal laser scanning microscope (Axiovert 200M with LSM 510 Meta scanhead) and a C-Apochromat ×40/1.2 NA water immersion objective (Zeiss, Jena, Germany). Cerulean and EYFP were excited with the Ar 458 nm and 514 nm laser line, respectively. Bleaching of the acceptor EYFP was performed within a region of interest (ROI) including one centromere using the 514-nm laser line at 100% intensity and with 100-fold iteration. Two images were taken before and 8–10 images immediately after EYFP bleaching to assess changes in donor and acceptor fluorescence. Cerulean and EYFP intensities in the ROI were averaged and normalized relative to the highest acceptor value in the time series. The FRET efficiency was calculated by comparing the fluorescence intensity (IDA) before bleaching (e.g. in presence of the acceptor) with the intensity (ID) measured after bleaching (e.g. in the absence of the acceptor) according to:
| 1 |
When donor and acceptor fluorophores are in close vicinity, the intensity of the donor fluorescence increases after photobleaching of the acceptor.
FLIM-based FRET measurements
The donor fluorescence lifetime was determined by time-correlated single photon counting (TCSPC) in living human HEp-2 cells. For donor fluorescence excitation, a pulsed picosecond diode laser (LDH Series, PicoQuant, Berlin, Germany) with an output wavelength of 470 nm at a frequency of 20 MHz along with a dedicated driver (PDL Series, PicoQuant, Berlin, Germany) was used. Via a fibre coupling unit, the excitation light was guided into a confocal laser scanning microscope (Zeiss LSM 510 Meta, Carl Zeiss GmbH, Jena, Germany). Laser power was adjusted to give average photon counting rates not >104–105 photons/s (0.0001–0.001 photon counts per excitation event) to avoid pulse pile-up. Images of 256 × 256 pixels were acquired with a ×63 C-Apochromat water immersion objective (NA 1.20, Zeiss). Photons emitted by the sample were collected by the water immersion objective, passed through a sample-specific 520/40 BP filter and were detected by a single photon avalanche diode (PDM series, PicoQuant, Berlin, Germany). The data were acquired by the PicoHarp 300 TCSPC module (PicoQuant, Berlin, Germany) working in the TTTR mode (time-tagged time-resolved). To calculate the fluorescence lifetime, the SymPhoTime software package (v4.7, PicoQuant, Berlin, Germany) was used. Selected areas of the images corresponding to single centromeres (resulting in the fluorescence lifetime histograms) or the sum of all centromeric regions were fitted by maximum likelihood estimation (MLE). Depending on the quality of a fit indicated by the value of χ2, a mono- or bi-exponential fitting model including background was applied. A model was rejected when χ2 exceeded a value of 1.5. In this way, the presence of scattered light in few measurements could be identified and separated. However, due to low photon numbers and too close time constants, the simultaneous presence of two different donor fluorescence lifetimes for complexes with donor-only and donor plus acceptor in one centromere could not be separated by a bi-exponetial fit. Mean lifetimes τm for a series of control measurements are presented as mean ± SD. A donor fluorescence lifetime obtained from a centromere in a cell co-expressing donor and acceptor molecules was considered to be significantly different from the control measurement, when the lifetime differed from the mean of the control values by >3 SD. Thus, FRET was assumed to occur when the measured lifetime in centromeric regions of a cell τ (sample) was smaller than the mean −3 SD of the control measurements carried out in absence of an acceptor τm (control):
| 2 |
The FRET efficiency was calculated by comparing the donor fluorescence lifetime (τDA) in the presence of the acceptor with the respective fluorescence lifetimes (τD) of control measurements obtained in absence of an acceptor:
| 3 |
RNAi knock down
For knock down of CENP-B, the siGENOME SMARTpool (Dharmacon, Lafayette, USA) containing a set of four siRNAs against human CENP-B (GCACGAUCCUGAAGAACAA, GGAGGAGGGUGAUGUUGAU, CCGAAUGGCUGCAGAGUCU and CCAACAAGCUGUCUCCCUA) was used at a final concentration of 25 nM. HEp-2 cells were transfected using Hyperfect transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Repeatedly, after 24 h time steps the cells were harvested and counted (Beckman Coulter Counter, Fullerton, USA). Aliquots with equal cell numbers were lysed and analysed by western blotting in order to determine the time dependence of protein knock down as described by Wieland et al. (95). For the FRAP and FLIM experiments, cells grown in 35 mm glass bottom culture dishes (MatTek Corporation, Ashland, USA) were co-transfected with 5 μl siRNA pool against CENP-B and 1 μg of each plasmid DNA using 20 μl Hyperfect transfection reagent. Experiments were carried out after 4 days of incubation, when the protein level of CENP-B was decreased to 8%.
RESULTS
Nuclear H1 distribution with respect to centromeres
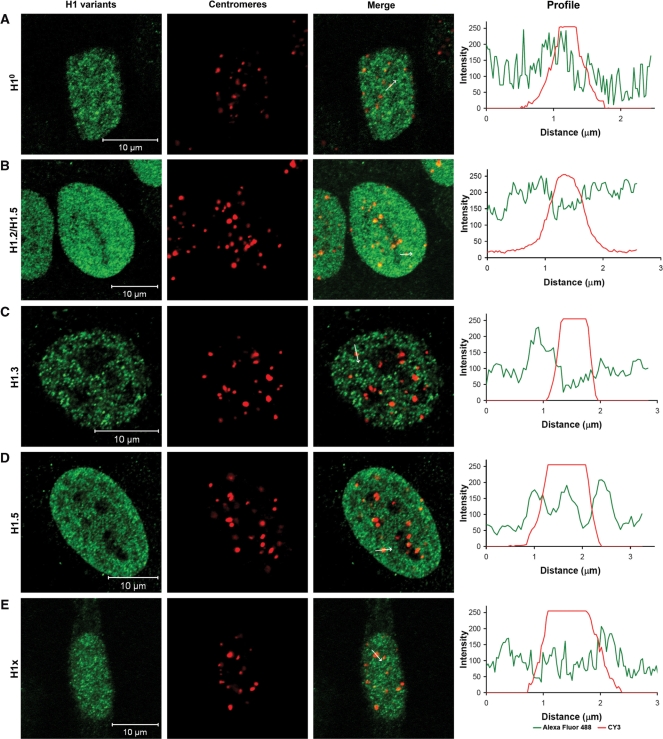
First, we asked if one of the H1 subtypes might be centromere-specific and if any enrichment or depletion of particular histone H1 subtypes can be detected at human centromeres. Therefore, human HEp-2 cells were stained with specific antibodies against the H1 subtypes H1°, H1.2, H1.3, H1.5 and H1x. Co-staining of centromeres with ACA (anti-centromere antibody) serum (containing antibodies against human CENP-A, -B and -C) revealed the presence of these H1 subtypes at interphase centromeres at levels similar to those at other locations in the nucleus. Furthermore, no specific association or exclusion of the analysed H1 subtypes at centromeres was found as indicated by the absence of any correlation in the fluorescence intensity overlay between the stained H1 subtypes and the centromeres. Images together with intensity profiles along one centromere are displayed in Figure 1 and a statistical analysis including a higher number of centromeres is presented in Supplementary Table 1. We conclude that these H1 proteins are neither enriched nor depleted at interphase centromeres. More detailed studies were carried out for H1° and H1.2 fusion proteins that have been ectopically expressed in human HEp-2 cells. The correct full length expression of each fusion protein was controlled by western blot analysis (Supplementary Figure 1). Microscopical analysis of the transfected interphase cells, co-stained with an H1° antibody, ensured that the fusion proteins showed a similar distribution as the endogeneous linker histones (Supplementary Figure 2). For life cell experiments, we selected these two subtypes for the following reasons: H1° is expressed during the complete cell cycle, mainly in terminally differentiated cells with an intermediate chromatin binding affinity, and H1.2 distributes throughout the nucleus, it is one of the most predominant subtypes in most human cells (96). H1.2 distribution correlates with DNA concentration (97), it seems to be a basic subtype being responsible for a ground level of chromatin compaction.
Figure 1.
Different H1 subtypes are present at centromeric chromatin, however, do not show any specific enrichment or reduction at human centromeres. HeLa cells were stained with (A) anti-H1°, (B) anti-H1.2/H1.5, (C) anti-H1.3, (D) anti-H1.5 and (E) anti-H1x antibodies and visualized by a species-specific secondary antibody fused to Alexa-Fluor 488 (green). Centromeres were stained with ACA (CY3, red). One confocal plane was examined per cell. Bars = 10 μm. The profiles (right) display the red and green fluorescence intensity along the indicated arrow in ‘Merge’.
H1 dynamics at centromeres
We next asked if H1 shows a particular binding mode to centromeric DNA. By FRAP, the dynamics of H1° exchange at interphase centromeres were studied in comparison to non-centromeric nuclear sites in human HEp-2 cells. For FRAP experiments, cells were co-transfected with mCherry-H1° and EGFP-CENP-A. At centromere positions, the mCherry fluorophore was irreversibly bleached by repetitive scanning at high laser intensity and fluorescence recovery at these spots was monitored over several minutes. Analysing 10–20 cells with 1–2 centromeres each, fast and complete recovery was observed (Supplementary Figure 3); our quantitative data are compatible with the absence of any immobile fraction. At centromeres, a recovery time t80% = 62 ± 15 s was measured, slightly smaller than, however, not significantly different from the recovery time at other nuclear sites (t80% = 76 ± 22 s) (Supplementary Figure 3). Our dynamic data are in the same order of magnitude as literature results (60,66) and consistent with their findings; quantitative differences might be due to different sizes of the bleached areas. Therefore, our FRAP measurements did not reveal a substantial difference in the dynamic behaviour of H1° at centromeres compared to non-centromeric nuclear sites. This would be the expected result for H1° binding similarly to centromeric compared to other chromatin.
To test whether human H1 and CENP-B can jointly bind to nucleosomal DNA linkers of CENP-A containing centromeric chromatin or if these two linker binding proteins exclude each other mutually, in the following we measured by BiFC, acceptor-bleaching FRET and FLIM the presence of histone H1° and H1.2 at the kinetochore in living human HEp-2 interphase cells.
Protein neighbourhoods at centromeres as analysed by BiFC
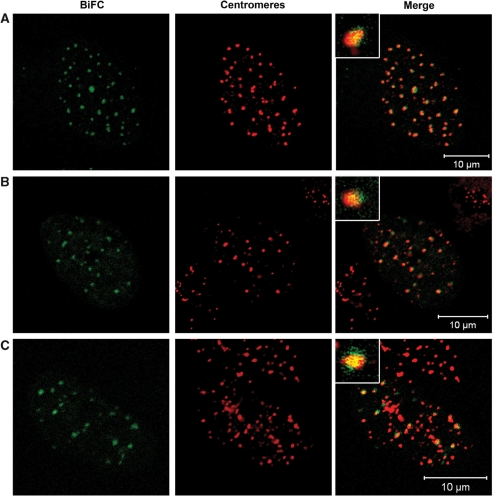
By BiFC, we determined if H1° and CENP-B are in intimate neighbourhood to one another. BiFC is based on the formation of a fluorescent complex formed by two fragments of a fluorophore (91,92,98). These fragments alone, the N- (‘Cerulean-first’) and C-terminus (‘Cerulean-last’) of cerulean are non-fluorescent; their association (and by this their fluorescence) is facilitated by the interaction or association between the proteins of interest which are fused to them. The kinetics of the association was measured by Hu et al. (99). As a positive control, CENP-A and CENP-B were used, both fused to the C- and the N-terminal fragment of cerulean, respectively. Recently, we could show that CENP-A and CENP-B are close to one another at human centromeres (44). Consistent with our previous observations, fluorescent cerulean foci within HEp-2 interphase cells could be detected (Figure 2A). Counterstaining with CENP-C revealed that these foci were formed at centromeres (Figure 2A). Outside centromeres only background fluorescence was observed suggesting that tight complex formation between CENP-A and CENP-B exclusively occurs at centromeres by local accumulation. This indicates that the fusion protein concentrations are too low to induce fluorescent complex formation by random contact, not mediated by the direct vicinity of the fusion partners.
Figure 2.
BiFC measurements revealed the presence of histone H1 next to CENP-B at centromic chromatin. HEp-2 cells were analysed expressing (A) Cerulean-first-CENP-A and CENP-B-Cerulean-last (positive control), (B) Cerulean-first-H1° and CENP-B-Cerulean-last and (C) Cerulean-last-H1° and CENP-B-Cerulean-first. The fluorescence of cerulean is shown in the first column ‘BiFC’ (green). Centromeres were stained with anti-CENP-C and rhodamine (red). Fluorescent complexes were formed at centromeric sites. The enlargements display this co-localization at one selected centromere. One confocal plane was examined per cell. Bars = 10 μm.
Next, human HEp-2 cells were co-transfected with plasmids containing either H1° or CENP-B fused to the fragments of cerulean. For both combinations, Cerulean-first-H1° and CENP-B-Cerulean-last as well as Cerulean-last-H1° and CENP-B-Cerulean-first, fluorescent foci were formed co-localizing with centromeres (Figure 2B and C). These results clearly show that CENP-B and H1° are in direct neighbourhood to one another in this region. Thus, we conclude that H1° is present in the human interphase pre-kinetochore next to CENP-B. Due to binding of the two parts of cerulean with slow dissociation kinetics, the protein complexes might accumulate and not display their true dynamics; the frequency of interaction is difficult to deduce from the BiFC data. Therefore, we next analysed the protein association by FRET in living cells. FRET is only observed during the period in which the proteins are close to one another.
Protein neighbourhoods at centromeres as analysed by acceptor-bleaching FRET and FLIM
As donor and acceptor fluorophors, the FRET pairs cerulean and EYFP as well as EGFP and mCherry were used. We constructed N-terminal fusion proteins of the inner kinetochore proteins CENP-A and CENP-C, and fusions at both termini of CENP-B, histones H1° and H1.2.
Upon expression in HEp-2 cells, the fluorescent-tagged kinetochore proteins localized in vivo at centromeric regions like the endogenous proteins (11, and data not shown). EGFP-tagged CENP-A is correctly incorporated into nucleosomes of centromeric heterochromatin (95) indicating that the fluorescent tag does not detectably alter the localization or other observed properties of the fusion protein in comparison to the endogenous protein. FRAP studies showed that EGFP-CENP–B and EGFP-CENP-C bind tightly to human centromeres (11) indicating that these tagged kinetochore proteins, like CENP-A, retained the centromere incorporation properties of the endogenous proteins. Also the fluorescently tagged histone H1° showed a similar distribution in the nucleus as the endogenous protein (Supplementary Figure 2).
For acceptor photobleaching methods (AB-FRET), cerulean and EYFP were used as donor and acceptor fluorophores, respectively. The cells containing the fluorescently tagged proteins were analysed in vivo as described by Orthaus et al. (44). In transiently transfected living HEp-2 cells, the fluorescence of EYFP was destroyed by bleaching single centromeres in interphase nuclei. Fluorescence intensity changes in the bleached area were recorded over time in the cerulean and EYFP channel. An increase of donor fluorescence intensity after acceptor bleaching is indicative for FRET. Basic control experiments for AB-FRET as described previously (44) were carried out. These control experiments allowed us to exclude photobleaching (100) or photo-conversion (101,102) of the donor fluorophore (data not shown). Also, during AB-FRET experiments, no negative influence of the bleaching procedure on cell morphology was observed.
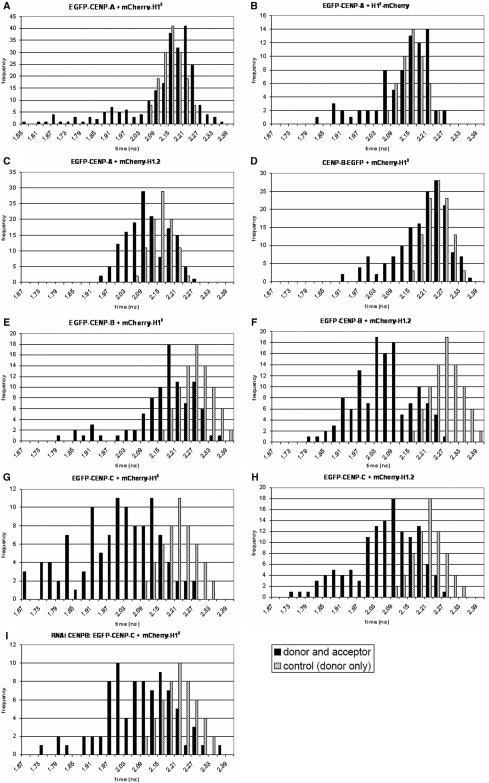
AB-FRET measurements have the disadvantage that the measured fluorescence intensities depend on the fluorophore concentrations. Therefore, energy transfer between the tagged proteins was further analysed by FLIM. Since EGFP has a fluorescence decay dominated by a single exponential (103), EGFP as donor with mCherry as acceptor fluorophore was used for the fluorescence lifetime measurements (104). The detection of FRET between EGFP and mCherry requires the fluorophores to be close to one another within a distance of 10 nm. FRET results in a shortening of the donor fluorescence lifetime. Unfused EGFP and mCherry, co-transfected in living human cells at similar expression levels, showed no FRET, allowing us to exclude that the FRET detected for the protein fusions might be caused by an incidental association of the fluorescent proteins (data not shown). Furthermore, the fluorescence lifetime of the donor EGFP was determined in living HEp-2 cells expressing the fluorophore alone or fused to either CENP-A, CENP-B or CENP-C. In cells expressing EGFP, EGFP-CENP-A, CENP-B-EGFP, EGFP-CENP-B or EGFP-CENP-C, the average fluorescence lifetimes were τm = 2.32 ± 0.04 ns (mean ± SD, n = 8 cells), τm = 2.17 ± 0.03 ns (n = 175 centromeres of six cells), τm = 2.23 ± 0.03 ns (n = 175 centromeres of five cells), τm = 2.28 ± 0.04 ns (n = 162 centromeres of seven cells) and τm = 2.23 ± 0.04 ns (n = 203 centromeres of nine cells), respectively. The donor-only lifetime distributions (Figure 4) also represent potential environmental influences on the donor fluorescence.
Figure 4.
Linker histone H1 binds to the centromere in direct vicinity to CENP-A, -B and -C. Displayed are the histograms of the donor fluorescence lifetimes evaluated at all centromeres by FLIM. HEp-2 cells were co-transfected with (A) EGFP-CENP–A, mCherry-H1°, (B) EGFP-CENP–A, H1°-mCherry, (C) EGFP-CENP–A, mCherry-H1.2, (D) CENP–B-EGFP, mCherry-H1°, (E) EGFP-CENP–B, mCherry-H1°, (F) EGFP-CENP–B, mCherry-H1.2, (G) EGFP-CENP–C, mCherry-H1°, (H) EGFP-CENP–C, mCherry-H1.2 and (I) EGFP-CENP–C, mCherry-H1° in CENP-B depleted cells. In these living cells, the donor fluorescence lifetimes were measured by TCSPC. The histograms display the fitted fluorescence lifetime values of all single centromeres (black bars). In addition, lifetime distributions of the corresponding donor-only control experiments are depicted as grey bars. The heights of the bars represent the numbers of centromeres (y-axis, frequency) whose lifetimes fall within the indicated 0.3 ns range (x-axis, time).
In order to assess the FRET efficiency between closely neighboured EGFP and mCherry, an EGFP-mCherry hybrid protein was analysed as a positive control, in which both fluorescent proteins are closely connected by a short linker (104). The mean fluorescence lifetime of EGFP within eight nuclei was significantly decreased to τm = 1.97 ± 0.01 ns, indicating that FRET occured between the two fluorophores with a FRET efficiency of 15%. We regarded a shortened donor lifetime as indicative of FRET when its value is 3 SDs smaller than the mean control value [see Equation (2)]. This value is in good quantitative agreement with the results of Tramier et al. (104). Under our experimental conditions, the fluorescence lifetime of EGFP did not shorten upon prolonged excitation.
In conclusion, our control experiments showed that a decrease of the donor fluorescence lifetime is indicative of a specific (direct or indirect) interaction between the proteins but not between the tags.
Since little to nothing is known about the orientation and rotational freedom of the fluorophores, we did not calculate molecular distances from our FRET data, but instead deduce from our results only the information that in case of FRET the fluorophores are close to one another in a range <∼10 nm.
CENP-A—H1
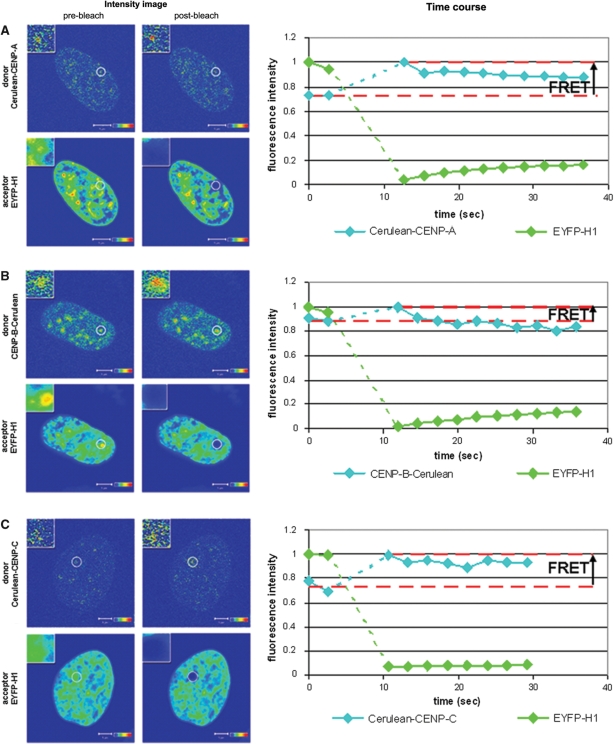
The flexible N-terminus of CENP-A sticks out of the nucleosome (47), expected to be close to linker binding proteins. We tested this hypothesis using AB-FRET and FLIM in interphase HEp-2 cells. EYFP-H1° showed a non-homogenous distribution within the cell nucleus which partially overlapped with Cerulean-CENP-A at centromeres (Figure 3A ‘Intensity image, pre-bleach’). After complete bleaching of the acceptor EYFP-H1° at centromeric regions, a fluorescence intensity increase of Cerulean-CENP-A was observed (Figure 3A ‘Intensity image, post-bleach’ and ‘Time course’). A FRET efficiency of 23 ± 3% was calculated indicating an association between both proteins (n = 11, single interphase kinetochores of 11 different cells). This result was confirmed by FLIM using the FRET pair EGFP-CENP-A and mCherry-H1°. The donor fluorescence lifetime at 241 centromeres in 10 different cells was measured and each single centromere was fitted by a mono-exponential decay. A large population of centromeres could be observed with a fluorescence decay time close to the control value (donor-only EGFP-CENP-A with τm = 2.17 ± 0.03 ns, see above). This indicates that most donors have no FRET acceptor in their close vicinity. However, a second population (23%) of centromeres was found with a donor fluorescence lifetime <2.09 ns (mean control value of 2.17 ns – 3 SD of 0.03 ns; Figure 4A).
Figure 3.
Acceptor bleaching FRET in vivo reveals the presence of linker histone H1° at human centromeres. The column ‘Intensity image’ shows confocal images that were acquired in the donor channel (upper panels) and the acceptor channel (lower panels) before (left panels) and after (right panels) bleaching of the acceptor. Intensities are encoded by colour ranging from blue (low intensty) to red (high intensity). The analysed centromere is shown in the inserts at a magnified scale. The column ‘Time course’ shows a plot of the time course of the fluorescence intensities that were recorded in the donor (cyan line) and the acceptor (green line) channel. Data were averaged over the indicated ROI and normalized to the maximum of the averaged intensities obtained in the respective channel. The dotted green and blue lines mark the bleaching period. The horizontal dashed red lines indicate the mean of the donor fluorescence intensities of the two measurements performed before photobleaching of the acceptor (IDA) and donor intensity recorded in the first image after acceptor photobleaching (ID). (A) Acceptor photobleaching of centromeres that contained Cerulean-CENP-A/EYFP–H1°, (B) CENP-B-Cerulean/EYFP-H1° or (C) Cerulean-CENP-C/EYFP-H1°. In all experiments, acceptor photobleaching resulted in an increase of the donor fluorescence intensity. This indicates that FRET occurs between the donor and acceptor fluorophores and that the respective proteins are in close vicinity to each other (<10 nm). Bars = 5 µm.
When analysing Cerulean-CENP-A and H1° now tagged at the C-terminus (H1°-EYFP), energy transfer at interphase kinetochores could not be found by AB-FRET (n = 11 interphase kinetochores). In the FLIM experiment using the FRET pair EGFP-CENP-A and H1°-mCherry, 77 centromeres in seven different cells were analysed as above. In this case, 26% of the centromeres showed a lifetime of EGFP-CENP-A <2.09 ns (Figure 4B). This indicates that N- as well as C-terminally tagged H1° was found at the interphase kinetochore next to the CENP-A N-terminus. Very high numbers (close to 100%) of centromeres with shortened donor lifetimes are not expected to be found since also untagged endogenous proteins are present in the cell which mix with the tagged proteins so that in a number of centromeres the tagged donor will have an untagged H1 as neighbour. This will be explained in more detail in the ‘Discussion’ section.
By FLIM, we additionally analysed a second H1 subtype H1.2. In cells co-expressing EGFP-CENP-A and mCherry-H1.2, 52 (35%) of 150 centromeres (in six cells) had shortened donor fluorescence lifetimes (<2.09 ns; Figure 4C). Thus, also H1.2 was found in CENP-A-containing centrometric chromatin.
CENP-B–H1
As a next step, we tested if CENP-B and H1 are located in close vicinity to each other. CENP-B binds via its N-terminus at the CENP-B box (28), which is supposed to be located on the linker DNA (31). Therefore, the distance between CENP-B and H1 is expected to be small.
To test this hypothesis, AB-FRET measurements with CENP-B, first tagged at its C-terminus (CENP-B-Cerulean), and EYFP-H1° were performed in interphase HEp-2 cell nuclei (Figure 3B). After bleaching of the acceptor EYFP-H1° at centromeric regions, the fluorescence intensity of CENP-B-Cerulean increased, indicating an FRET efficiency of 13 ± 1% (n = 8 interphase kinetochores). In agreement with these results, in the FLIM experiments using CENP-B-EGFP and mCherry-H1° donor, fluorescence lifetimes were significantly smaller than the control values (Figure 4D). In 158 centromeres of 11 cells, 21% of the donors had a shortened lifetime <2.14 ns (fluorescence lifetime of 2.23 ns – 3 SD of 0.03 ns). Then, centromeric regions with CENP-B tagged at its N-terminus (EGFP-CENP-B) and mCherry-H1° were analysed. Many centromeres did not show a decrease in the fluorescence lifetime (Figure 4E). However, 36 (40%) of 91 centromeres (six cells) had significantly smaller donor fluorescence lifetimes than the control (EGFP-CENP-B with τm = 2.28 ± 0.035 ns). When analysing EGFP-CENP-B and the other histone subtype mCherry-H1.2, also two populations of donor molecules were present. Of 129 centromeres in seven cells, 82% of the centromeres had shortened donor fluorescence lifetimes <2.18 ns (Figure 4F). Thus, different amounts of H1° and H1.2 were found with their N-termini close to the CENP-B N-terminus.
CENP-C–H1
Since CENP-C binds tightly to the inner kinetochore in direct association with CENP-A, -B and -I (11,44,105, Sandra Orthaus, unpublished data), it seems to be likely that CENP-C and H1 are in close vicinity to each other in human centromeres. To verify this hypothesis, the FRET efficiency between CENP-C and H1°, both tagged N-terminally, was determined (Figures 3C and 4G). In the AB-FRET experiments, the Cerulean-CENP-C fluorescence intensity increased when the acceptor EYFP-H1° was completely bleached indicating a FRET efficiency of 27 ± 2% (n = 3 interphase kinetochores, Figure 3C). In the FLIM experiment displayed in Figure 4G, seven cells were analysed that co-expressed EGFP-CENP-C and mCherry-H1°. In 70% of 112 centromeres, the donors showed a shortened lifetime <2.10 ns (donor-only EGFP-CENP-C with τm = 2.23 ± 0.04 ns − 3 SD of 0.04 ns). Also for histone mCherry-H1.2, 55% of the EGFP-CENP-C donors (72 centromeres in six cells) exhibit fluorescence lifetime values smaller than the control (Figure 4H). These results indicate that CENP-C and linker histone H1 are in close vicinity to each other within the human interphase kinetochore.
H1 tagged at its C-terminus was found to show shorter FRAP recovery times than when the tag is at the N-terminus (60). This can be explained by the fact that it is the C-terminal H1 domain which shows strongest binding to the linker DNA, and this binding may be distorted by the tag. On the other hand, the first half of the N-terminus of histone H1 is unstructured and does not bind to chromatin (106,107); this might enable it to tolerate the tag. Consistent with this view, we observed less C-terminally tagged H1 (H1-mCherry) in direct vicinity to tagged CENP-B and CENP-C than N-terminally tagged H1 (data not shown). Nevertheless, consistent with the FRAP data, also C-terminally tagged H1 is present at centromeric chromatin as indicated, for example, by our FRET data for EGFP-CENP-A and H1°-mCherry.
CENP-B RNAi
Our data show that H1 is present in centromeric chromatin in direct neighbourhood to inner kinetochore proteins CENP-A, -B and -C. We then asked if H1 is not only close to but also binds to these proteins, in particular to CENP-B. In addition, inner kinetochore proteins might form a compact structure which eventually could be altered when CENP-B is eliminated; and this might locally influence H1 dynamics. To elucidate these points, CENP-B was down-regulated by RNAi, and it was measured by FRAP in interphase HEp-2 cell nuclei if this influences the dynamics and residence time of H1 at centromeres. After 96 h of incubation with CENP-B siRNA, the protein level of CENP-B was diminished to 8% (Supplementary Figure 4). Measuring FRAP in these cells, we found t80% = 50 ± 18 s for mCherry-H1°, a value slightly smaller, within experimental error, however, not significantly different from the t80 value in untreated cells (t80% = 62 ± 15 s, Supplementary Figure 3). As in untreated cells, a complete fluorescence recovery could be observed also in CENP-B down-regulated cells. In our experiments, CENP-B down-regulation thus did not result in substantially different H1 dynamics. Furthermore, the close neighbourhood between CENP-C and H1 is not affected by CENP-B knock down: the donor lifetime distribution of EGFP-CENP-C in the presence of mCherry H1° was not significantly different in CENP-B depleted compared to untreated cells (Figure 4I). Thus, H1 association to CENP-C containing centromeric chromatin seems not to be dependent on the presence of CENP-B.
DISCUSSION
Within chromatin, adjacent nucleosomes are connected via DNA linkers that vary in length in a cell- and species-specific manner with mean linker lengths of 50 bp (51,52). Human centromeric DNA is built from 171 bp α-satellite repeats (108) which, for nucleosomes formed with protein octamers, would leave a short linker length of ∼24 bp. The α-satellite repeats contain a 17-bp CENP-B box, probably in the linker DNA (23), which is recognized by CENP-B (20,24,31). Nucleosome formation at centromeres and the centromeric chromatin structure are currently unclear. Centromeric nucleosomes might form either with octameric, hexameric or tetrameric protein complexes (52,54) each probably resulting in a different centromeric chromatin structure. Nucleosomes formed with protein octamers at centromeres would offer only limited space between adjacent nucleosomes (22). It is thus of interest if, in addition to CENP-B, linker histone H1 also contributes to centromeric chromatin structure.
H1 represents a family of histone subtypes. Several of these subtypes were analysed here for their presence at interphase centromeres. In general agreement with Parseghian et al. (88), our immunostaining experiments clearly showed the presence of the H1 subtypes H1°, H1.2, H1.3 H1.5 and H1x at centromeric chromatin at similar amounts as at non-centrometic sites. Fan et al. (85,109) quantified the amounts of H1 subtypes in different cell types revealing total levels of less than about one H1 per nucleosome. This may be due to the transient nature of H1 binding to chromatin indicating that linker DNA would not always be occupied with H1 proteins. Our results are consistent with chromatin immunoprecipitation (110), and multiple tandem affinity purification (8,9) experiments which presented hints that H1 might exist in CENP-A-containing nucleosomal protein complexes. We could neither detect an enrichment nor a depletion of any of these H1 subtypes at centromeres. H1x (111) seems to play a functional role in chromosome alignment and segregation (112) which seems not to be linked to centromeres: preferentially, H1x localizes to nucleoli in interphase and to the chromsome periphery during mitosis (112). Since H1 subtypes are able to replace one another, we continued our studies with a subselection of subtypes, H1° and H1.2.
A different binding mode of H1 to centromeric chromatin compared to other chromatin would influence its dynamics. By FRAP we measured the dynamics of H1° in interphase nuclei. We observed complete recovery in all cases and, within experimental error, no difference in recovery times at centromeres compared to other nuclear locations. These data support our immunostaining results that linker histone H1 is present in centromeric chromatin. They did not yield, however, any hint for a centromere-specific H1 binding mode.
In these measurements, the resolution is too limited to allow conclusions on a detailed (close to molecular) level. In order to find out if H1 binds to the linkers of CENP-A-containing nucleosomes in close neighbourhood to CENP-B, BiFC and FRET measurements were performed. The two non-fluorescent N- and C-terminal parts of cerulean were fused to CENP-B and H1°. These two proteins facilitated fluorescent complex formation at interphase centromeres, indicating that at this specific site CENP-B and H1° appear in close neighbourhood to one another. As a next step, energy transfer experiments were carried out between H1 and the inner kinetochore proteins CENP-A and -C as well as CENP-B in living human HEp-2 cells by two separate approaches, acceptor-bleaching FRET and FLIM. The data demonstrate a close neighbourhood of H1° and H1.2 with these three centromere binding proteins. In the transfected cells, the tagged proteins are present together with their unlabelled endogenous siblings. Therefore, the donor fusion proteins, when binding to the centromeres, have acceptor-tagged as well as untagged H1 proteins next to them. Our FRET value distributions thus showed long lifetimes of those donors without acceptor, as well as shorter lifetimes for donors with acceptor molecules in their vicinity. The number of donors in the one or the other situation is strongly influenced by the expression levels of the tagged and the endogenous proteins, as well as by the level of incorporation of the tagged kinetochore and H1 proteins into the complex. For our experiments, we always selected cells with low expression levels of the tagged proteins, however still allowing high quality data acquisition. As a consequence, donor-only and acceptor-only complexes might be frequent. Identifying donor molecules with shorter lifetimes thus indicates that a complex was formed in which the donor fusion is in close vicinity to the acceptor fusion. Our analysis of protein exchange dynamics revealed that the inner kinetochore complex is not stable over the cell cycle but shows fast exchange of some and stable incorporation of other proteins (11); kinetochore complex composition is thus flexible during the cell cycle. In our FRET analyses presented here, we did not distinguish different interphase cell cycle phases. Therefore, donors with different short lifetimes might represent different complex compositions or complex structures according to the cell cycle phase. We thus present the donor lifetime distributions, with lifetimes shorter than the control, as measured and did not calculate mean values. Lifetimes were observed even shorter than those of the positive control EGFP-mCherry. Such short lifetimes might be due to donors having more than one acceptor molecule next to them, as observed earlier (44). Most of the donor molecules linked to CENP-C were found to have an acceptor, either fused to H1° or H1.2, next to them, in normal as well as in CENP-B-depleted cells (Figure 4G, H and I). The situation is different for CENP-A: here, only a clearly smaller amount of donor molecules linked to CENP-A had an acceptor-H1 fusion next to them (Figure 4A, B and C). Both kinetochore proteins are centromere markers colocalizing with one another and observed by FRET to be in close vicinity (S. Orthaus, unpublished results). Thus, the FLIM results presented here seem to indicate that within the same complex, the donor fused to CENP-C, but less frequent the donor fused to CENP-A, is in the vicinity of the acceptor linked to H1. We have no indications that transfection with CENP-C is more efficient than with CENP-A (which would explain our observation); instead, CENP-C transfection efficiency seems to be lower. The higher number of centromeres showing FRET for donors fused to CENP-C could be due to CENP-C having a more flexible structure so that the donor fluorophore can probe a larger volume than those fused to CENP-A. We prefer the interpretation that the donor fluorophore fused to CENP-C might have more than one acceptor fluorophore fused to H1 next to it. Then, FRET could occur principally to several H1 molecules, and we would observe FRET also in the expected case when only a H1 subselection would carry an acceptor. Since the molecular architecture of the kinetochore complex is not determined yet, alternative explanations for the FLIM data are also conceivable. Different arrangements were observed for H1° compared to H1.2 next to CENP-B: while the donor linked to the N-terminus of CENP-B is in the neighbourhood of the acceptor fused to H1.2 in many cases, it is less frequent next to the acceptor linked to H1° (Figure 4D and F). CENP-B binds not only to CENP-A and -C containing centromeres, but also additionally to pericentromeric regions (19), so that our results might represent different complex architectures, potentially influencing H1° differently than H1.2.
In cells transiently transfected with tagged linker proteins, the total amount of these proteins is increased by additional 50–100% over all cells (see Supplementary Figure 1b and d) which might affect the H1 binding distribution to chromatin. In our experiments, we only selected cells showing moderate over-expression (still allowing for high quality data). The endogenous and the tagged linker proteins have similar chromatin binding properties (60,66) and a similar chromatin binding distribution (Supplementary Figure 2). Furthermore, endogenous H1 binds to CENP-A containing centromeric chromatin (Supplementary Table 1). The transient presence of additional tagged linker protein in the cell is thus expected to only influence the total amount of a stable combination of endogenous as well as tagged linker proteins at centromeric chromatin.
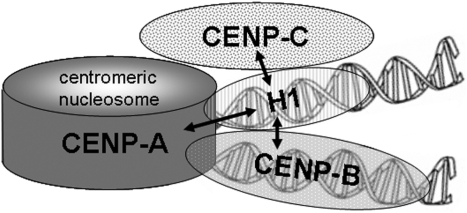
The impact of H1 on centromere structure and function is elusive. As CENP-B, also H1 might contribute to the positioning of successive nucleosomes and the pattern of nucleosome arrangement (49). CENP-B binds to the sequence-specific 17-bp boxes present on α-satellite DNA repeats and H1 is able to suppress the binding of other proteins to the same linker (71) suggesting that CENP-B and H1 might bind to neighbouring linkers (Figure 5). One could speculate that both linker DNA binding proteins, H1 and CENP-B, might cooperate leading to a more condensed and/or stable centromere-specific chromatin structure (113–115) which might contribute to function and identity of centromeric chromatin (113,115). In this case, the presence of CENP-B might influence H1 binding. This view, however, is not supported by our CENP-B knock down data. Within the experimental error range, H1 showed unchanged dynamics at centromeric chromatin in CENP-B down-regulated interphase cells. Furthermore, loss of CENP-B does not offer an additional H1 binding site since, due to its mode of interaction at the exit of both linker DNAs, only one H1 can be found per nucleosome. On the other hand, linker histone H1 might be able to partly take over CENP-B function in CENP-B-depleted centromeric chromatin. This would provide an explanation for the maintenance of an active kinetochore state despite the lack of CENP-B and/or CENP-B boxes. The molecular properties of CENP-B (18,116) are, however, distinct from those of H1: CENP-B, in contrast to H1, binds to a specific DNA binding site, the CENP-B box. Furthermore, CENP-B dimerizes tail-to-tail at its C-termini (29,30) also in vivo (S. Orthaus, unpublished results). We found no sequence homology and no subdomain similarities between CENP-B and H1. Thus, H1 is not expected to be able to fully replace CENP-B. If α-satellite chromatin forms a 30-nm fiber structure, CENP-B (and H1) might bind at the interior of the solenoid as discussed by Pluta et al. (28). This would be consistent with a tandem but not with a tail-to-tail dimerization of CENP-B (J. Sühnel and S. Diekmann, unpublished data). On the other hand, the negative charge of CENP-B is expected to destabilize this 30-nm structure (28) or might even induce another. Currently, the centromeric chromatin structure is unclear.
Figure 5.
Schematic representation of centromeric chromatin with a CENP-A containing nucleosome. In this model, the internucleosomal linker DNA is alternatively occupied by histone H1 and CENP-B. The arrows indicate the protein associations detected in this study.
A histone octamer together with the linker histone occupy the chromatosome DNA length of ∼167 bp. If centromeric nucleosomes form octamer complexes, the involvement of H1 in centromeric chromatin would thus determine a shorter limit for α-satellite repeat lengths: α-satellite repeats <167 bp would not allow for H1 binding. Human α-satellite repeats being slightly longer than chromatosome DNA are thus an additional hint that H1 might bind and contribute to centromeric chromatin formation.
We conclude that linker histone H1 is present in centromeric heterochromatin of human HEp-2 cells in direct vicinity to human inner kinetochore proteins (Figure 5).
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
FUNDING
German Bundesministerium fuer Bildung und Forschung (EXIST High-TEPP 03EXHT02 to S.O.). Funding for open access charge: EXIST High-TEPP 03EXHT02 to S.O.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Sühnel, W. Zuschratter, P. Hemmerich and C. Biskup for helpful discussions; D. Doenecke for critical reading of the manuscript; T. Zimmer, N. Klöcker, W. Waldeck, K. Sugimoto, M. Coppey-Moisan, N. Audugé and W. C. Earnshaw for the kind gift of plasmids; H. Zentgraf for providing the H1° antibody; K. Yoda for the CENP-C antibody; C. Weber for support; K. Hänecke for excellent assistance; and also both reviewers for very helpful comments.
REFERENCES
- 1.Roos UP. Light and electron microscopy of rat kangaroo cells in mitosis, II. Kinetochore structure and function. Chromosoma. 1973;41:195–220. doi: 10.1007/BF00319696. [DOI] [PubMed] [Google Scholar]
- 2.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 3.Conde e Silva NC, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 4.Earnshaw WC, Rothfield N. Identification of a familiy of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 5.Sugata N, Munekata E, Todokoro K. Characterisation of a novel kinetochore protein, CENP-H. J. Biol. Chem. 1999;274:27343–27346. doi: 10.1074/jbc.274.39.27343. [DOI] [PubMed] [Google Scholar]
- 6.Sugata N, Li S, Earnshaw WC, Yen TJ, Yoda K, Masumoto H, Munekata E, Warburton PE, Todokoro K. Human CENP-H multimers colocalise with CENP-A and CENP-C at active centromere-kinetochore complexes. Hum. Mol. Genet. 2000;9:2919–2926. doi: 10.1093/hmg/9.19.2919. [DOI] [PubMed] [Google Scholar]
- 7.Nishihashi A, Haraguchi T, Hiraoka Y, Ikemura T, Regnier V, Dodson H, Earnshaw WC, Fukagawa T. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell. 2002;2:463–476. doi: 10.1016/s1534-5807(02)00144-2. [DOI] [PubMed] [Google Scholar]
- 8.Foltz DR, Jansen LET, Black BE, Bailey AO, Yates JR, III, Cleveland DW. The human CENP-A centromeric complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 9.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, III, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 10.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 11.Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 2008;180:1101–1114. doi: 10.1083/jcb.200710052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellwig D, Münch S, Orthaus S, Hoischen C, Hemmerich P, Diekmann S. Live-cell imaging reveals sustained centromere binding of CENP-T via CENP-A and CENP-B. J. Biophoton. 2008;1:245–254. doi: 10.1002/jbio.200810014. [DOI] [PubMed] [Google Scholar]
- 13.van Hooser AA, Oki II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 14.Amor DJ, Kalitsis P, Sumer H, Choo KHA. Building the centromere: from foundation proteins to 3D organisation. Trends Cell Biol. 2004;14:359–368. doi: 10.1016/j.tcb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Sacharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 16.Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KHA. Early disruption of centromeric chromatin organisation in centromere protein A (CENP-A) null mice. Proc. Natl. Acad. Sci. USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progress and heterochromatin interactions. Nat. Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Earnshaw WC, Sullivan KF, Machlin PS, Cooke CA, Kaiser DA, Pollard TD, Rothfield NF, Cleveland DW. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooke CA, Bernat RL, Earnshaw WC. CENP-B: a major centromere protein located beneath the kinetochore. J. Cell Biol. 1990;110:1475–1488. doi: 10.1083/jcb.110.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeno M, Masumoto H, Okazaki T. Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range α-satellite DNA arrays of human chromosome 21. Hum. Mol. Genet. 1994;3:1245–1257. doi: 10.1093/hmg/3.8.1245. [DOI] [PubMed] [Google Scholar]
- 22.Yoda K, Ando S, Okuda A, Kikuchi A, Okazaki T. In vitro assembly of the CENP-B/α-satellite DNA/core histone complex: CENP-B causes nucleosome positioning. Genes Cells. 1998;3:533–548. doi: 10.1046/j.1365-2443.1998.00210.x. [DOI] [PubMed] [Google Scholar]
- 23.Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. Cenp-A, Cenp-B and Cenp-C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka Y, Nureki O, Kurumizaka H, Fukai S, Kawaguchi S, Ikuta M, Iwahara J, Okazaki T, Yokoyama S. Crystal structure of the CENP—B protein-DNA complex: the DNA-binding domains of CENP—B induce kinks in the CENP—B box DNA. EMBO J. 2001;20:6612–6618. doi: 10.1093/emboj/20.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohzeki J, Nakano M, Okada T, Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto Y, Nakano M, Ohzeki J, Larionov V, Masumoto H. A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere. EMBO J. 2007;26:1279–1291. doi: 10.1038/sj.emboj.7601584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okada T, Ohzeki JI, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 28.Pluta AF, Saitoh N, Goldberg I, Earnshaw WC. Identification of a subdomain of CENP-B that is necessary and sufficient for localisation to the human centromere. J. Cell Biol. 1992;116:1081–1093. doi: 10.1083/jcb.116.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoda K, Kitagawa K, Masumoto H, Muro Y, Okazaki T. A human centromere protein, CENP-B, has a DNA binding domain containing four potential alpha helices at the NH2 terminus, which is separable from dimerizing activity. J. Cell Biol. 1992;119:1413–1427. doi: 10.1083/jcb.119.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitagawa K, Masumoto H, Ikeda M, Okazaki T. Analysis of protein-DNA and protein-protein interactions of centromere protein B (CENP-B) and properties of the DNA-CENP-B complex in the cell cycle. Mol. Cell Biol. 1995;15:1602–1612. doi: 10.1128/mcb.15.3.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka Y, Tachiwana H, Yoda K, Masumoto H, Okazaki T. Human centromere protein B induces translational positioning of nucleosomes on α-satellite sequences. J. Biol. Chem. 2005;280:41609–41618. doi: 10.1074/jbc.M509666200. [DOI] [PubMed] [Google Scholar]
- 32.Merry DE, Pathak S, Hsu TC, Brinkley BR. Anti-kinetochore antibodies: use as probes for inactive centromeres. Am. J. Hum. Genet. 1985;37:425–430. [PMC free article] [PubMed] [Google Scholar]
- 33.Earnshaw WC, Migeon BR. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- 34.Earnshaw WC, Ratrie H, 3rd, Stetten G. Visualisation of centrometric proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- 35.Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- 36.Hudson DF, Fowler KJ, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wereford NG, de Kretser DM, Cancilla MR, et al. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Earnshaw WC, Bernat RL, Cooke CA, Rothfield NF. Role of the centromnere/kinetochore in cell cycle control. Cold Spring Harb. Symp. Quant. Biol. 1991;56:675–685. doi: 10.1101/sqb.1991.056.01.076. [DOI] [PubMed] [Google Scholar]
- 38.Saitoh H, Tomkiel J, Cooke CA, Ratrie H, 3rd, Maurer M, Rothfield NF, Earnshaw WC. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 39.Tomkiel J, Cooke CA, Saitoh H, Bernat RL, Earnshaw WC. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J. Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang CH, Tomkiel J, Saitoh H, Johnson DH, Earnshaw WC. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell Biol. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugimot K, Yata H, Muro Y, Himeno M. Human centromere protein C (CENP-C) is a DNA-binding protein which possesses a novel DNA-binding motif. J. Biochem. 1994;116:877–881. doi: 10.1093/oxfordjournals.jbchem.a124610. [DOI] [PubMed] [Google Scholar]
- 42.Politi V, Perini G, Trazzi S, Pliss A, Rask I, Earnshaw WC, Della Valle G. CENP-C binds the alpha-satellite DNA in vivo at specific centromere domains. J. Cell Sci. 2002;115:2317–2327. doi: 10.1242/jcs.115.11.2317. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki N, Nagano M, Nozaki N, Egashira S, Okazaki T, Masumoto H. CENP-B interacts with CENP-C domains containing Mif2 regions responsible for centromere localization. J. Biol. Chem. 2004;279:5934–5946. doi: 10.1074/jbc.M306477200. [DOI] [PubMed] [Google Scholar]
- 44.Orthaus S, Biskup C, Hoffmann B, Hoischen C, Ohndorf S, Benndorf K, Diekmann S. Assembly of the inner kinetochore proteins CENP-A and CENP-B in living human cells. Chembiochem. 2008;9:77–92. doi: 10.1002/cbic.200700358. [DOI] [PubMed] [Google Scholar]
- 45.Noll M, Kornberg RD. Action of micrococcal nuclease on chromatin and the location of histone H1. J. Mol. Biol. 1977;109:393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- 46.Simpson RT. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 47.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 48.Carruthers LM, Hansen JC. The core histone N-termini function independently of linker histones during chromatin condensation. J. Biol. Chem. 2000;275:37285–37290. doi: 10.1074/jbc.M006801200. [DOI] [PubMed] [Google Scholar]
- 49.Woodcock CL, Skoutchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 50.Catez F, Ueda T, Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells. Nat. Struct. Mol. Biol. 2006;13:305–310. doi: 10.1038/nsmb1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Widom J. A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc. Natl. Acad. Sci. USA. 1992;89:1095–1099. doi: 10.1073/pnas.89.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Camahort R, Li B, Florens L, Sanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 54.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 55.Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential S. cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dalal Y, Furuyama T, Vermaak D, Henikoff S. Structure, dynamics and evolution of centromeric nucleosomes. Proc. Natl. Acad. Sci. USA. 2007;104:15974–15981. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organisation of the nucleosome and of the salt dependent superstructure of chromatin. J. Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramakrishnan V. Histone structure and the organuisation of the nucleosome. Annu. Rev. Biophys. Biomol. Struct. 1997;26:83–112. doi: 10.1146/annurev.biophys.26.1.83. [DOI] [PubMed] [Google Scholar]
- 59.Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 60.Hendzel MJ, Lever MA, Crawford E, Th′ng JP. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- 61.Lu X, Hansen JC. Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J. Biol. Chem. 2004;279:8701–8707. doi: 10.1074/jbc.M311348200. [DOI] [PubMed] [Google Scholar]
- 62.Duggan MM, Thomas JO. Two DNA binding sites on the globular domain of histone H5 are required for binding to both bulk and 5S reconstituted nucleosomes. J. Mol. Biol. 2000;304:21–33. doi: 10.1006/jmbi.2000.4205. [DOI] [PubMed] [Google Scholar]
- 63.Thomas JO. Histone H1: location and role. Curr. Opin. Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 64.Travers AA. The location of linker histone in the nucleosome. Trends Biochem. Sci. 1999;24:4–7. doi: 10.1016/s0968-0004(98)01339-5. [DOI] [PubMed] [Google Scholar]
- 65.Sheng S, Czajkowsky DM, Shao Z. Localisation of linker histone in Chromatosomes by cryo-atomic force microscopy. Biophys. J. 2006;91:L35–L37. doi: 10.1529/biophysj.106.090423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown D, Izard T, Misteli T. Mapping the interaction surface of the linker H1 with the nucleosome of native chromatin in vivo. Nat. Struct. Mol. Biol. 2006;13:250–255. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lever MA, Th` ng JP, Sun X, Hendzel MJ. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- 68.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 69.Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen D, Dundr M, Wang C, Leung A, Lamond A, Misteli T, Huang S. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Juan L, Utley RT, Vignali M, Bohm L, Workman JL. H1-mediated repression of transcription factor binding to a stably positioned nucleosome. J. Biol. Chem. 1997;272:3635–3640. doi: 10.1074/jbc.272.6.3635. [DOI] [PubMed] [Google Scholar]
- 72.Parseghian MH, Hamkalo BA. A compendium of the histone H1 family of somatic subtypes: an elusive cast of characters and their characteristics. Biochem. Cell Biol. 2001;79:289–304. [PubMed] [Google Scholar]
- 73.Khochbin S. Histone H1 diversity: bridging regulatory signals to linker histone function. Gene. 2001;271:1–12. doi: 10.1016/s0378-1119(01)00495-4. [DOI] [PubMed] [Google Scholar]
- 74.Izzo A, Kamieniarz K, Schneider R. The histone H1 family: specific members, specific functions? Biol. Chem. 2008;389:333–343. doi: 10.1515/BC.2008.037. [DOI] [PubMed] [Google Scholar]
- 75.Brown DT. Histone H1 and the dynamic regulation of chromatin function. Biochem. Cell Biol. 2003;81:221–227. doi: 10.1139/o03-049. [DOI] [PubMed] [Google Scholar]
- 76.Harvey AC, Downs JA. What functions do linker histones provide? Mol. Microbiol. 2004;53:771–775. doi: 10.1111/j.1365-2958.2004.04195.x. [DOI] [PubMed] [Google Scholar]
- 77.Henriques JP, Casar JC, Fuentealba L, Carey DJ, Brandan E. Extracellular matrix histone H1 binds to perlecan, is present in regenerating skeletal muscle and stimulates myoblast proliferation. J. Cell Sci. 2002;115:2041–2051. doi: 10.1242/jcs.115.10.2041. [DOI] [PubMed] [Google Scholar]
- 78.Jedrusik MA, Vogt S, Claus P, Schulze E. A novel linker histone-like protein is associated with cytoplasmic filaments in C. elegans. J. Cell Sci. 2002;115:2881–2891. doi: 10.1242/jcs.115.14.2881. [DOI] [PubMed] [Google Scholar]
- 79.Konishi A, Shimizu S, Hirota J, Takao T, Fan Y, Matsuoka Y, Zhang L, Yoneda Y, Fujii Y, Skoultchi AI, et al. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell. 2003;114:673–688. doi: 10.1016/s0092-8674(03)00719-0. [DOI] [PubMed] [Google Scholar]
- 80.Downs JA, Kosmidou E, Morgan A, Jackson SP. Suppression of homologous recombination by the S. cerevisiae linker histone. Mol. Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 81.Happel N, Doenecke D, Sekeri-Pataryas KE, Sourlingas TG. H1 histone subtype constitution and phosphorylation state of the ageing cell system of human peripheral blood lymphocytes. Exp. Gerontol. 2008;43:184–199. doi: 10.1016/j.exger.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 82.Ausio J. Histone variants – the structure behind the function. Brief Funct. Genomic. Proteomic. 2006;5:228–243. doi: 10.1093/bfgp/ell020. [DOI] [PubMed] [Google Scholar]
- 83.Albig W, Meergans T, Doenecke D. Characterization of the H1.5 gene completes the set of human H1 subtype genes. Gene. 1997;184:141–148. doi: 10.1016/s0378-1119(96)00582-3. [DOI] [PubMed] [Google Scholar]
- 84.Takami Y, Nishi R, Nakayama T. Histone H1 variants play individual roles in transcription regulation in the DT40 chicken B cell line. Biochem. Biophys. Res. Commun. 2000;268:501–508. doi: 10.1006/bbrc.2000.2172. [DOI] [PubMed] [Google Scholar]
- 85.Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bustin M, Catez F, Lim J-H. The dynamics of histone H1 function in chromatin. Mol. Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 87.Funayama R, Saito M, Tanobe H, Ishikawa F. Loss of linker histone H1 in cellular senescence. J. Cell Biol. 2006;175:869–880. doi: 10.1083/jcb.200604005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Parseghian MH, Newcomb RL, Hamkalo BA. Distribution of somatic H1 subtypes is non-random on active vs. inactive chromatin II: distribution in human adult fibroblasts. J. Cell. Biochem. 2001;83:643–659. doi: 10.1002/jcb.1224. [DOI] [PubMed] [Google Scholar]
- 89.Stoldt S, Wenzel D, Schulze E, Doenecke D, Happel N. G1 phase-dependent nucleolar accumulation of human histone H1x. Biol. Cell. 2007;99:541–552. doi: 10.1042/bc20060117. [DOI] [PubMed] [Google Scholar]
- 90.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shyu YJ, Liu H, Deng X, Hu CD. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques. 2006;40:61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 92.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kenworthy A. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods. 2001;24:289–296. doi: 10.1006/meth.2001.1189. [DOI] [PubMed] [Google Scholar]
- 94.Karpova TS, Baumann CT, He L, Wu X, Grammer A, Lipsky P, Hager GL, McNally JG. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microscopy. 2003;209:56–70. doi: 10.1046/j.1365-2818.2003.01100.x. [DOI] [PubMed] [Google Scholar]
- 95.Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P. Functional complementation of human centromere Protein A (CENP-A) by Cse4 from S. cerevisiae. Mol. Cell Biol. 2004;24:6620–6630. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meergans T, Albig W, Doenecke D. Varied expression patterns of human H1 histone genes in different cell lines. DNA Cell Biol. 1997;16:1041–1049. doi: 10.1089/dna.1997.16.1041. [DOI] [PubMed] [Google Scholar]
- 97.Parseghian MH, Newcomb RL, Winokur ST, Hamkalo BA. The distribution of somatic H1 subtypes is non-random on active vs. inactive chromatin: distribution in human fetal fibroblasts. Chromosome Res. 2000;8:405–424. doi: 10.1023/a:1009262819961. [DOI] [PubMed] [Google Scholar]
- 98.Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 2006;1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hu CD, Chinenov Y, Kerppola TK. Visulisation of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 100.Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 1997;73:2782–2790. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Creemers TM, Lock AJ, Subramaniam V, Jovin TM, Völker S. Three photoconvertible forms of green fluorescent protein identified by spectral hole-burning. Nat. Struct. Biol. 1999;6:557–560. doi: 10.1038/9335. [DOI] [PubMed] [Google Scholar]
- 102.Malvezzi-Campeggi F, Jahnz M, Heinze KG, Dittrich P, Schwille P. Light-induced flickering of DsRed provides evidence for distinct and interconvertible fluorescent states. Biophys. J. 2001;81:882–887. doi: 10.1016/S0006-3495(01)75828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Volkmer A, Subramaniam V, Birch D, Jovin T. One- and two-photon excited fluorescence lifetimes and anisotropy decays of green fluorescent proteins. Biophys. J. 2000;78:1589–1598. doi: 10.1016/S0006-3495(00)76711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tramier M, Zahid M, Mevel JC, Masse MJ, Coppey-Moisan M. Sensitivity of CFP/YFP and GFP/mCherry pairs to donor photobleaching on FRET determination by fluorescence lifetime imaging microscopy in living cells. Microsc. Res. Tech. 2006;69:933–939. doi: 10.1002/jemt.20370. [DOI] [PubMed] [Google Scholar]
- 105.Cheeseman IM, Hori T, Fukagawa T, Desai A. KNL1 and the CENP-H/I/K comoplex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 2008;19:587–594. doi: 10.1091/mbc.E07-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vila R, Ponte I, Collado M, Arrondo JL, Jimenez MA, Rico M, Suau P. DNA-induced alpha-helical structure in the NH2-terminal domain of histone H1. J. Biol. Chem. 2001;276:46429–46435. doi: 10.1074/jbc.M106952200. [DOI] [PubMed] [Google Scholar]
- 107.Vila R, Ponte I, Jimenez MA, Rico M, Suau P. An inducible helix-Gly-Gly-helix motif in the N-terminal domain of histone H1e: a CD and NMR study. Protein Sci. 2002;11:214–220. doi: 10.1110/ps.29602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Alexandrov IA, Mashkova TD, Akopian TA, Medvedev LI, Kisselev LL, Mitkevich SP, Yurov YB. Chromosome-specific alpha satellites: two distinct families on human chromosome 18. Genomics. 1991;11:15–23. doi: 10.1016/0888-7543(91)90097-x. [DOI] [PubMed] [Google Scholar]
- 109.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Depletion of histone H1 in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1119–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 110.Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localised with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- 111.Happel N, Schulze E, Doenecke D. Characterisation of human histone H1x. Biol. Chem. 2005;386:541–551. doi: 10.1515/BC.2005.064. [DOI] [PubMed] [Google Scholar]
- 112.Takata H, Matsunaga S, Moriomoto A, Ono-Maniwa R, Uchiyama S, Fukui K. H1x with different properties from other linker histones is required for mitotic progression. FEBS Lett. 2007;581:3783–3788. doi: 10.1016/j.febslet.2007.06.076. [DOI] [PubMed] [Google Scholar]
- 113.Gilbert N, Allan J. Distinctive higher-order chromatin structure at mammalian centromeres. Proc. Natl. Acad. Sci. USA. 2001;98:11949–11954. doi: 10.1073/pnas.211322798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Segal E, Fondufe-Mittendorf Y, Chen L, Thastroem A, Field Y, Moore IK, Wang J-P, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Black BE, Jansen LET, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 116.Earnshaw WC. Anionic regions in nuclear proteins. J. Cell Biol. 1987;105:1479–1482. doi: 10.1083/jcb.105.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.