Abstract
The Wnt signaling pathway is necessary both for maintaining undifferentiated stem cells and for directing their differentiation. In mouse embryonic stem cells (ESCs), Wnt signaling preferentially maintains “stemness” under certain permissive conditions. T-cell factor 3 (Tcf3) is a component of the Wnt signaling and a dominant downstream effector in ESCs. Despite the wealth of knowledge regarding the importance of Wnt signaling underlying stem cells functions, the precise mechanistic explanation by which the effects are mediated is unknown. In this study, we identified new regulatory targets of Tcf3 using a whole-genome approach and found that Tcf3 transcriptionally represses many genes important for maintaining pluripotency and self-renewal, as well as those involved in lineage commitment and stem cell differentiation. This effect is in part mediated by the corepressors transducin-like enhancer of split 2 and C-terminal Binding Protein (CtBP). Notably, Tcf3 binds to and represses the Oct4 promoter, and this repressive effect requires both the Groucho and CtBP interacting domains of Tcf3. Interestingly, we find that in mouse preimplantation development embryos, Tcf3 expression is coregulated with Oct4 and Nanog and becomes localized to the inner cell mass of the blastocyst. These data demonstrate an important role for Tcf3 in modulating the appropriate level of gene transcription in ESCs and during embryonic development.
Keywords: Embryonic stem cells, Differentiation, T-cell factor 3, Transcription, Wnt signaling
INTRODUCTION
The ability of embryonic stem cells (ESCs) to grow in large numbers as pluripotent cells is dependent on the extrinsic sensing of the growth conditions coupled to the appropriate intrinsic cellular response. In mouse ESCs, key signaling pathways, including leukemia inhibitory factor (LIF)/gp130/Stat3, bone morphogenetic protein (BMP), and Wnt, have been found to be crucial for maintaining the undifferentiated state. In particularly, Wnt signaling is a highly conserved signaling pathway in metazoan animals that mediates cell-cell communications for the coordination of a variety of cellular processes, such as cell fate specification, cell proliferation, differentiation, survival, apoptosis, and migration [1].
During development, Wnt signaling has a dramatic influence on progenitor and stem cells in vivo. Inhibition of Wnt signaling via blocking of Wnt1 function is necessary for vertebrate forebrain specification [2], whereas activation of the signal is required in the entire central nervous system for expanding the progenitor cells by promoting proliferation and blocking apoptosis and differentiation [3]. In the neocortex of embryonic day (E) 11.5 embryos, Wnt activation promotes differentiation of neural stem cells [4]; in embryonic skin, Wnt is essential for fate decision of epidermal stem cells [5-7]. Wnt signaling has also been revealed to be important in the maintenance and differentiation of stem cells cultured in vitro. Elevated Wnt signaling blocks differentiation and promotes self-renewal and in vitro expansion of HSCs that can reconstitute the blood system of lethally irradiated mice [8, 9].
In ESCs, Wnt activation preferentially maintains “stemness” under certain permissive conditions. The overexpression of Wnt1 or stabilized β-catenin or abrogation of the APC complex leads to the inhibition of neural differentiation, mediated by the activation of downstream target genes [10, 11]. In addition, the use of a synthetic pharmacological inhibitor of GSK3β sustains ESC pluripotency and self-renewal via Wnt activation [12]. In spite of the wealth of knowledge regarding Wnt signaling underlying stem cells functions, the precise mechanistic explanation by which the effects are mediated is largely unknown. Although there are at least 19 Wnt ligands and 10 frizzled receptors, only four transcription factors have been known to directly mediate gene regulation in response to Wnt. However, the presence of numerous cofactors, such as β-catenin, Groucho/transducin-like enhancer of split (TLEs), and Hic5, adds tremendous diversity to the manner in which these factors can selectively regulate specific target genes [13-15]. The observations made through the activation of Wnt signaling via its ligands have not been informative of the transcriptional control of target genes by the Tcf/Lef factors. The direct genes bound by these factors and their interacting partners are key to better understanding the control of stemness versus differentiation.
We are particularly interested in the specific role of T-cell factor 3 (Tcf3) in the maintenance of mouse ESC pluripotency, as its loss delays the ability of these cells to differentiate via the relief of repression of Nanog [16]. Using whole genome analysis of Tcf3 regulatory sites in ESCs, we uncovered novel targets of Tcf3, many of which are implicated in stem cell pluripotency, developmental programs, signaling pathways, and oncogenesis. The cofactors TLE2 and CtBP (C-terminal binding protein) were found to be key partners of Tcf3 in mediating the repressive effect. Notably, Tcf3 transcriptionally represses Oct4 and is critical for maintaining the appropriate levels of Oct4 and Nanog in ESCs. Loss of Tcf3 by RNA interference (RNAi) knockdown blocks the ability of ESCs to differentiate. Developmentally, Tcf3 expression appears to be coregulated with Oct4 and Nanog during preimplantation development, and it becomes localized in the inner cell mass (ICM) of the blastocyst, similar to both Oct4 and Nanog. This suggests that Tcf3 might have important regulatory roles in ESCs and during development.
MATERIALS AND METHODS
Cell Culture and Transfection
All cell cultures were maintained at 37°C with 5% CO2. The culture of mouse ESCs, E14 (CRL-1821; American Type Culture Collection, Manassas, VA, http://www.atcc.org), either on feeders or under feeder-free conditions, was described previously [17]. HEK293T/17 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum and penicillin/streptomycin. Transfection of plasmids into mouse ESCs and HEK293 cells was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, http://www.invitrogen.com). Generation of stable overexpression ESC lines, Wnt stimulation, and differentiation of ESCs are described in supplemental online data.
Plasmid Construction
For RNAi design and construction of plasmids for short hairpin RNA (shRNA) synthesis, 19 base-pair gene-specific regions were designed on the basis of the algorithm of Reynolds et al. [18]. Oligonucleotides were cloned into pSuper.puro or pSuper.neo. retro.GFP (OligoEngine, Seattle, WA, http://www.oligoengine.com; sequences are given in supplemental online data). At least two shRNAs were designed to target each gene for screening on the basis of the Oct4 promoter-luciferase assay. All sequences were analyzed by BLAST to ensure specificity. Plasmids used in overexpression studies are given in supplemental online data.
RNA and Protein Analyses
Total RNA was extracted using Trizol (Invitrogen), column-purified with RNeasy kits (Qiagen, Hilden, Germany, http://www1.qiagen.com), and converted to cDNA, followed by real-time polymerase chain reaction (PCR) detection. Protein extraction and Western blotting were performed using standard molecular techniques and using antibodies described in the supplemental online data.
Chromatin Immunoprecipitation and DNA Microarray Analysis
Chromatin immunoprecipitation (ChIP) assays were carried out as described previously [17]. Quantitative PCR analyses were performed in real time using the ABI Prism 7900 Sequence Detection System (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com). Relative occupancy values were calculated by determining the apparent immunoprecipitation efficiency (ratios of the amount of immunoprecipitated DNA to that of the input sample) and normalized to the level observed. ChIP combined with DNA microarray was carried out according to the Agilent Mammalian ChIP-on-chip protocol (version 3; Agilent Technologies, Palo Alto, CA, http://www.agilent.com) [19] (supplemental online data).
Immunofluorescence Microscopy of Cells and Embryos
Cell cultures and embryos were fixed in 4% paraformaldehyde and permeabilized with 0.25% Triton X-100, followed by blocking with 1% bovine serum albumin in phosphate-buffered saline. ESCs and embryos were stained with primary antibodies (supplemental online data), followed by the appropriate secondary antibodies conjugated with Alexa Fluor 488, 568, 594, or 633 (Molecular Probes, Eugene, OR, http://probes.invitrogen.com). Images were captured with a confocal microscope (LSM 510 META; Carl Zeiss, Jena, Germany, http://www.zeiss.com).
Gene Expression Analysis
For the comparison of ESC versus embryoid body (EB) data, temporal gene expression data in triplicate for mouse ESC differentiation were downloaded from StemBase (http://www.stembase.ca) (experiment #:E113). For the comparison of ESC versus tissue data, the above mouse Affymetrix (Santa Clara, CA, http://www.affymetrix.com) gene expression data for ESCs at a time point of 0 hours were compared with a compendium of mouse gene expression data with 54 differentiated tissues and cell types downloaded from the Gene Atlas (http://symatlas.gnf.org) (supplemental online data).
Statistical Analysis
Student’s nonpaired t test was used to determine the statistical significance, where indicated.
RESULTS
Tcf3 Depletion Blocks ESC Differentiation
The transcription factors Tcf1, Tcf3, Tcf4, and Lef1 are downstream effectors of the Wnt signaling pathway and are direct mediators of gene transcription activity. The Tcf/Lef proteins contain amino-terminal β-catenin interaction domains and activate transcription of target genes when Wnt-stabilized β-catenin accumulates in the cell [20]. However, in the absence of stabilized β-catenin, Tcf/Lef proteins may function as transcription repressors through their interaction with corepressor proteins such as Groucho/TLEs, CtBP, and Hic5 [21].
All members of the Tcf/Lef protein family are expressed in mouse ESCs at different levels, with Tcf3 being the most abundant. It has previously been reported that Tcf3-/- ESCs delayed differentiation following LIF withdrawal [16]. To confirm whether this effect can be recapitulated in Tcf3-deficient ESCs mediated by RNAi, we performed both transient and stable knockdowns (supplemental online Figs. 1, 3A, 3B). All four shRNA designs elicited similar effects, and we used RNAi 3 for subsequent studies. Indeed, Tcf3 knockdown ESCs were resistant to differentiation upon LIF withdrawal, compared with control treated ESCs, which manifested extensive differentiation with the loss of colony forming property, as well as SSEA1 and alkaline phosphatase (AP) expression (Fig. 1A).
Figure 1.
Knockdown of Tcf3 in ESCs upregulates Oct4 and limits differentiation potential. (A): LIF was removed from culture media 24 h after control shRNA, vector, and Tcf3 shRNA transfection. Puromycin (1 μgml-1) was added to select for transfected cells. By d 7, Tcf3 shRNA-treated cells formed ESC colonies that could be propagated in the absence of LIF. Immunostaining showed that Tcf3 shRNA treatment in the absence of LIF maintained strong SSEA1 and AP expression. At least two different shRNA designs were used, and results were comparable (data not shown). Scale bars = 200 μm. (B): Control shRNA, vector, and Tcf3 shRNA-transfected stable ESC clones were generated, and one representative clone for each condition is presented. Tcf3 shRNA clones maintained spherical and intact EBs that stained positive for AP after 21 d, whereas control shRNA and vector-treated clones showed signs of disintegration with the appearance of cavities indicative of differentiation and negative for AP. Scale bars = 200 μm (left column) and 150 μm (right column). (C): Retinoic acid (RA) treatment of control shRNA ESCs showed extensive differentiation to flattened cells, with the loss of colony forming ability by d 5. Tcf3 shRNA ESCs maintained colony structures along with differentiated cells after 5 d of RA treatment. These cells could be propagated at least five times in the presence of RA, whereas control shRNA cells could not. During RA treatment, Tcf3 shRNA ESCs were able to maintain higher levels of Oct4 and Sox2 transcripts compared with control shRNA cells. Scale bar = 100 μm; error bars represent SEM. (D): Knockdown of Tcf3-upregulated Oct4, Utf1, Sox2, and Hist1h1b transcript levels after 3 d of transfection. *, significantly different from control shRNA control; p < .01; n = 3. Error bars represent SEM. (E): In both the presence and the absence of LIF, transient treatment of ESCs with Tcf3 shRNA upregulated the levels of Nanog and Oct4 proteins by 48 h. (F): Knockdown of Tcf3, but not Tcf2, Tcf4, or Lef1, upregulated Oct4 promoter activity in mouse ESCs, relative to control shRNA. Oct4 RNAi served as a positive control, whereas vector transfection served as a negative control. Four shRNAs were constructed for each gene to ensure specificity. *, significantly different from control shRNA control; p < .01; n = 3. Error bars represent SEM. Abbreviations: AP, alkaline phosphatase; d, day(s); h, hour(s); LIF, leukemia inhibitory factor; RNAi, RNA interference; shRNA, short hairpin; Tcf3, T-cell factor 3.
Since LIF is known to be essential for the propagation of mouse ESCs via activation of Stat3 in the Jak/Stat signaling pathway [22], we examined whether Tcf3-deficient ESCs could bypass the LIF receptor and directly activate Stat3, which is sufficient for sustaining self-renewal, or whether these cells use other strategies. In control shRNA ESCs, there was a rapid decline of phosphorylation on Ser 727 of Stat3 24 hours following LIF removal (supplemental online Fig. 2), and phosphorylated Stat3 was completely abrogated by 96 hours. The same observation was made for Tcf3-deficient ESCs upon LIF removal, indicating that the LIF/Jak/Stat pathway was not absolutely required for self-renewal (supplemental online Fig. 2).
To further determine the extent to which Tcf3-deficient ESCs are limited in differentiation into various cell lineages, we performed long-term cultures of ESCs into EBs. Clonal populations were isolated from control shRNA-, vector-, and Tcf3 shRNA-transfected ESCs using puromycin drug selection (supplemental online Fig. 3A), and clonal data from each representative condition are shown. In control shRNA and vector clones, typical cavitating EBs appeared by day 18. The EBs continued to enlarge, lose the spherical structure, and manifest signs of disintegration by day 21 (Fig. 1B). AP staining was completely absent, indicating that no pluripotent ESCs were present. In Tcf3-knockdown stable clones, the EBs maintained a tight and spherical morphology and stained positive for AP, suggesting the existence of undifferentiated pluripotent cells in the EBs.
Next, we examined whether Tcf3-deficient ESCs were resistant to the more potent retinoic acid (RA)-induced differentiation. After 5 days of RA treatment, control shRNA ESCs showed extensive differentiation with the loss of colonies, and Oct4 and Sox2 levels were downregulated (Fig. 1C). In contrast, Tcf3-deficient ESCs displayed modest signs of differentiation, with the persistence of undifferentiated ESC colonies (Fig. 1C). The levels of Oct4 and Sox2 diminished slightly but were comparatively higher than those of control shRNA cells. This mixture of undifferentiated and differentiated cells can be propagated in the presence of RA for at least five passages (data not shown). At this stage, removal of RA and reculturing in the presence of LIF allowed the recovery of fully undifferentiated ESC colonies and restoration of normal Oct4, Sox2, and Nanog mRNA levels (supplemental online Fig. 3C). One important assessment of ESC pluripotency is their ability to form teratomas composed of all germ lineages when transplanted subcutaneously into severe combined immunodeficient mice. With control shRNA ESCs, large teratomas with a variety of differentiated tissue types were obtained (supplemental online Fig. 3D). Tcf3-deficient ESCs produced smaller tumors consisting of homogeneous, undifferentiated cell types, indicating that in vivo differentiation was also blocked.
As it has been shown that Tcf3-/- ESCs had elevated Nanog protein levels, we tested whether Tcf3 RNAi knockdown of wild-type ESCs could also relieve the suppression of Nanog. Both Nanog mRNA and protein levels were markedly upregulated in Tcf3 shRNA-treated ESCs within 48 hours (Fig. 1D, 1E). Interestingly, we found that many other pluripotency-associated genes, such as Oct4, Sox2, and Utf1, as well as Hist1h1b, an indicator of cell cycling rate, were also increased (Fig. 1D, 1E). As it was previously shown that Oct4 levels were unaffected in Tcf3-/- ESCs, we sought to further confirm our results by using a combination of shRNA expression and an Oct4 promoter luciferase reporter. We saw that depletion of Tcf3 indeed resulted in a significant upregulation of the Oct4 promoter activity, whereas knockdown of other Tcf/Lef family members did not (Fig. 1F; shRNA knockdown efficiency is given in supplemental online Fig. 1). These results strongly suggested that Tcf3 directly regulates Oct4 activity in ESCs.
Tcf3 Occupancy in ESCs
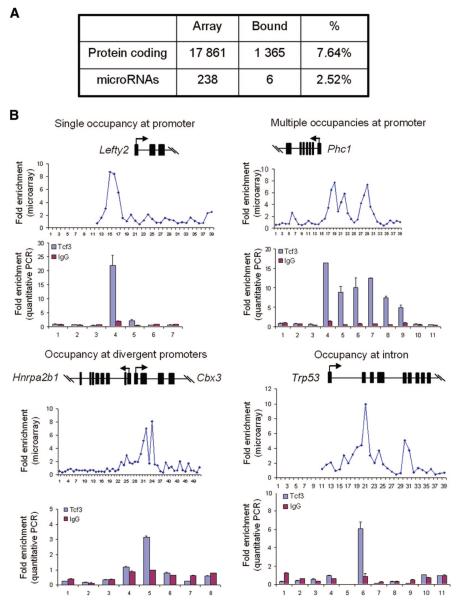
The depletion of Tcf3 profoundly prevented ESC differentiation and altered their self-renewing capacity. These changes in ESCs are unlikely to be caused by Nanog alone, as we demonstrated the effects of Tcf3 depletion on Oct4 as well. We speculated that Tcf3 may be regulating many key genes central to the pluripotency and developmental networks. Thus, we sought to identify the set of genes that Tcf3 regulates in mouse ESCs by using ChIP combined with DNA microarrays. We first generated stable Tcf3-overexpressing ESCs tagged with 2 × HA epitope, followed by immunoprecipitating for Tcf3-bound genes using a Tcf3-specific antibody. Immunoprecipitation for HA and analysis of a small representation of Tcf3 targets showed that the same genes were bound (data not shown). Tcf3 was found to be associated with 1,365 (7.64%) promoter regions for known protein encoding genes and 6 (2.52%) promoters for known microRNA genes (Fig. 2A; supplemental online data; supplemental online Fig. 4; supplemental online Table 1). The occupancy configurations of Tcf3 on some of the genes are depicted in Figure 2B. Although Tcf3 most commonly binds to a single site within a promoter (e.g., Lefty2), it may occupy more than one location, as illustrated on the genomic contig of Phc1. Tcf3 was also able to bind to intronic regions of genes such as Trp53, as noted previously [23], and occupies divergent promoters that may possibly control the transcription of two different genes in both directions of the plus and minus strand of DNA, such as that shown for Hnrrpa2b and Cbx3.
Figure 2.
Genome-wide location analysis of Tcf3 in mouse ESCs. (A): Tcf3-bound DNA fragments were immunoprecipitated, amplified, dye-labeled, and hybridized to DNA microarrays containing 45–60-mer probes that span -5.5 to +2.5 kilobases for 17,861 annotated mouse genes, relative to the transcription start sites. The Whitehead Neighborhood Model, P(Xbar) < 0.001, with intra-array median normalization was performed to identify bound target genes. It was found that 7.64% of protein-coding and 2.52% of microRNA genes were occupied by Tcf3. (B): Examples of Tcf3-bound genes. Microarray plots display normalized ChIP-enrichment ratios over input DNA for all probes within the genomic region. Quantitative real-time PCR scanning of the promoters was performed to confirm the enriched probe locations that corresponded to the microarray plot. Tcf3 bound to promoters at several configurations: single occupancy at the promoter (Lefty2), multiple sites on a promoter (Phc1), single occupancy of divergent promoters of two genes (Hnrpa2b1 and Cbx3), and occupancy at the intronic region (Trp53). Control immunoprecipitation was carried out with IgG antibody. Abbreviations: PCR, polymerase chain reaction; Tcf3, T-cell factor 3.
To determine the quality of the protein-DNA interactions in our study, we performed quantitative polymerase chain reaction (qPCR) on selected Tcf3 target genes. Eighty-four bound sequences were randomly selected, and qPCR was performed to determine the enriched targets. Seventy-eight of 84 targets were confirmed as positively bound, setting the false-positive rate at approximately 7% (supplemental online Fig. 5). To ensure that overexpressed Tcf3 was bound to specific targets in a manner similar to endogenous Tcf3, we performed ChIP for Tcf3 in wild-type ESCs. We confirmed that a representative sample of these target genes was indeed occupied by Tcf3 (supplemental online Fig. 6).
Co-Ocupancy of Tcf3, Oct4, and Nanog
Examination of the targets of Tcf3 in mouse ESCs revealed that many were pluripotency-associated factors. These included Oct4, Trp53, Mycn, Phc1, Eed, Gdf3, Fg4, Dppa5, and Dnmt3b, which are highly expressed in ESCs but become downregulated upon differentiation (Fig. 4D). Many of these factors have a detrimental effect on the ESC phenotype when disrupted [19, 24, 25]. We next identified genes occupied by Tcf3 that were also bound by both Oct4 and Nanog in mouse ESCs. We intersected our Tcf3 ChIP-on-chip data set with that obtained from the ChIP-PET analysis of Oct4 and Nanog [23] (Fig. 3A). It was immediately evident that many of the target genes shared by Oct4 and Nanog were occupied by Tcf3 as well. Together, Oct4, Nanog, and Tcf3 co-occupy at least 52 genes (Fig. 3A), and some examples are shown in Figure 3B. Notable targets include transcription factors (e.g., Oct4, Nanog, Tcf3, and Tbx3) and components of epigenetic regulators (e.g., Jarid2, Phc1, and Msh2). Significantly, Oct4, Nanog, and Tcf3 all co-occupied the promoters of one another. Although Oct4 and Nanog promote pluripotency and self-renewal through positive regulation of their target genes, including their own genes, the misexpression of pluripotency-associated genes can also alter the ESC phenotype. Tcf3 may serve as a modulator of pluripotency-associated gene expression in ESCs, as transient depletion of Tcf3 led to a relief of suppression (Fig. 3C). We further observed that Tcf3 bound in close proximity with Oct4 and Nanog in some cases (Fig. 3B). Hence, the observation that Tcf3 also occupies the targets of Oct4 and Nanog indicates that the repressive function of Tcf3 is essential for careful modulation of such genes.
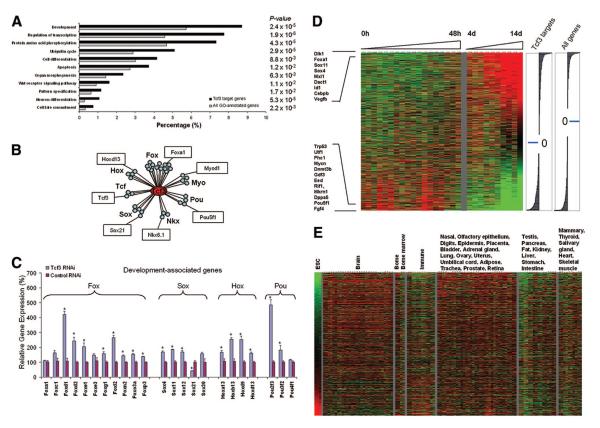
Figure 4.
Tcf3 regulates developmentally associated genes that are upregulated during ESC differentiation. (A): Gene Ontology (GO) analysis of Tcf3 target genes. Black bars represent the percentage of Tcf3 target genes in a particular GO category, and gray bars represent the percentage expected of all GO-annotated mouse genes on the microarray. The p value of this representation is based on a hypergeometric distribution. Top selected representations based on p value are shown. The complete list is given in supplemental online Table 2. (B): Examples of developmental transcription factor families bound by Tcf3. Tcf3 is represented by the red oval; individual transcription factors are represented by circles and grouped by families as indicated (supplemental online Table 3). Examples of transcription factors with defined roles in development are labeled. Transcription factor families include Hox, Fox, Tcf, Sox, Myo, Tbx, Nkx, Pax, and Pou. (C): Developmentally associated target genes belonging to the Fox, Sox, Hox, and Pou families are predominantly upregulated after Tcf3 RNAi depletion in ESCs. Two of the Fox members (Foxo6 and Foxf12) bound were not detected by quantitative polymerase chain reaction. (D): Tcf3 target genes were dynamically regulated during ESC differentiation. Expression profiling data sets during differentiation of ESCs into EBs were obtained [30]. Only Tcf3 target genes are shown. Green and red represent lower-than-average and higher-than-average expression intensities, respectively (supplemental online Table 4). The right panels show the ratio of the average signal value for undifferentiated ESCs and ESCs differentiated for 14 d for probes representing Tcf3 target genes analyzed and for all probes on the array, represented as the distribution of log2-transformed ratios for transcripts that are up- or downregulated by more than 1.5-fold in both experimental groups. (E): Expression of inactive Tcf3 target genes was upregulated in differentiated tissue types compared with undifferentiated ESCs. Gene expression data for ESCs were compared with a compendium of mouse expression data representing 54 other differentiated tissues and cell types [31] (supplemental online data; supplemental online Table 5). Abbreviations: d, day(s); Fox, forkhead box; h, hour(s); Hox, homeobox protein; Myo, myogenic basic domain; Nkx, NK transcription factor-related; Pax, paired box and paired-like; Pou, Pou domain-containing; RNAi, RNA interference; Sox, Sry box; Tbx, T-box; Tcf, T-cell factor.
Figure 3.
Tcf3 regulates pluripotency-associated genes. (A): Genes co-occupied by Tcf3, Oct4, and Nanog. From Loh et al. [23], 324 genes out of 356 genes cobound by Oct4 and Nanog could be mapped to the mouse promoter microarray. Of these, 52 genes were also bound by Tcf3. Notable examples implicated in ESC pluripotency were listed. (B): Different configurations of Tcf3, Oct4, and Nanog binding to genes. Exons are depicted in black boxes, and the arrows indicate the direction of each gene. The numbers on the right indicate the span from the first to the last exon. Note that the complete Tcf3 binding profile may not be represented, as the promoter microarrays do not cover the entire genomic region depicted. The binding of Tcf3 on Nanog was based on [16]. (C): Upon Tcf3 RNAi knockdown, most pluripotency-associated targets of Tcf3 became upregulated, relative to control RNAi. *, significantly different from control; p < .01; n = 4. Error bars represent SEM. Abbreviations: bp, base pairs; RNAi, RNA interference; Tcf3, T-cell factor 3.
Key Developmental Regulators Are Targets of Tcf3
To gather insights into other biological functions of Tcf3 in ESCs, we determined using Gene Ontology terms the categories of genes that were enriched. Genes involved in transcription and developmental hierarchies, such as cell differentiation, organ morphogenesis, pattern specification, neuron development, and cell fate commitment, among others, were overrepresented (Fig. 4A; a complete list is given in supplemental online Table 2).
Further analysis of the targets revealed that these were enriched for several families of homeotic genes that have wide-spread developmental roles, including the Hox clusters with highly conserved functions in patterning of the body axis (Fig. 4B; supplemental online Table 3). Tcf3-bound homeodomain genes also included members of the Fox, Tbx, and Sox gene families that are known to regulate early developmental programs. The forkhead family of Fox genes is involved in embryonic development, organogenesis, patterning of the germ layers, and immunity [26]. The Tbx family of genes regulates a wide variety of developmental processes, such as somite patterning, heart development, and limb formation [27, 28]. Members of the Sox family are indispensable in multiple aspects of development, including sex determination and neurogenesis [29]. We showed that in ESCs, many of these genes are repressed by Tcf3, as knockdown of this factor upregulated most members of development-associated gene families (Fig. 4C). The genes preferentially bound by Tcf3, when expressed, may mediate differentiation, explaining in part why Tcf3 is essential for maintaining ESC pluripotency.
Activation of Tcf3 Target Genes During Differentiation and Development
Since Tcf3 is associated with an important set of developmental regulators that must be silent in ESCs, we hypothesized that at least some of these genes must be activated during differentiation. We examined differential gene expression in differentiating ESCs [30]. Of the genes bound by Tcf3, the majority were expressed at low levels or absent in undifferentiated ESCs and became upregulated during EB differentiation (Fig. 4D). These include Dlk1, Foxa, Sox11, Sox4, Mxi1, Dact1, Cebpb, and Vegfb, which have been implicated in stem cell differentiation programs. Genes bound by Tcf3 also included ESC-associated transcription factors (Oct4, Sox2, Mycn, and Trp53), epigenetic regulators (Eed, Dnmt3b, and Phc1), signaling genes (Nodal, Spry2, and Fgf4), telomerase-associated factors (Rif1 and Mkrn1), and an RNA-binding protein, Dppa5/Esg1. These genes were highly expressed in undifferentiated ESCs and gradually downregulated upon 14 days of EB formation (Fig. 4D), supporting the notion that Tcf3 regulates the appropriate levels of ESC-associated factors required for pluripotency.
The observation that many Tcf3 targets have roles in development led us to further examine whether these are preferentially expressed in differentiated tissues and cell types. We compared the expression levels of Tcf3-occupied genes in ESCs with the expression level of these genes in 54 differentiated cell and tissue types [31], which are late terminal differentiation and developmental programs compared with EB formation. Tcf3-bound genes that were underexpressed in ESCs were generally upregulated in the various cell and tissue types, particularly in the brain, thyroid, heart, muscle, bladder, uterus, ovary, prostate, epidermis, and retina (Fig. 4E). By contrast, genes that were overexpressed in ESCs were downregulated in the terminally differentiated tissue and cell types, as expected. These data suggest that Tcf3 represses target genes that are involved in lineage specification in undifferentiated ESCs and that are primed to be activated transcriptionally during tissue or cell type-specific differentiation. Therefore, Tcf3 occupies both transcriptionally active genes that have roles in pluripotency and transcriptionally inactive genes that promote development upon activation. In Tcf3-knockdown ESCs, transcriptionally active genes are further upregulated, indicating that a regulated degree of repression control of such genes is important (Figs. 3C, 4C; supplemental online Fig. 7).
Mediation of Transcriptional Repression by Groucho/TLEs
The control of Wnt target gene expression requires regulated assembly of transcription repression and activation complexes. The binding of Tcf/Lef factors to DNA for transcriptional control is dependent upon their interaction with either β-catenin, Groucho/TLEs, or other coregulators [32, 33]. Several studies have demonstrated that Tcf/Lef-mediated transcriptional activation can be repressed by the addition of Groucho/TLEs [21, 34]. Although Tcf/Lef proteins can mediate either transcriptional activation or repression by interacting with different cofactors, our observation that most of the Tcf3 target genes in ESCs are upregulated upon knocking down Tcf3 suggests that it functions mainly as a repressor in ESCs. Indeed, many target genes that were bound by Tcf3 were similarly occupied by the TLE2 corepressor (Fig. 5A). We found that CtBP, another corepressor, also colocalized to a small number of Tcf3 target genes (supplemental online Fig. 8). In contrast, we did not observe any colocalization of β-catenin with Tcf3 on these target genes, and immunostaining revealed that β-catenin was largely restricted to the cytoplasm with very little nuclear localization (Fig. 5A; supplemental online Fig. 9). When we examined with greater resolution the occupancies of Tcf3, TLE2, CtBP, and β-catenin along the genomic locus of selected genes, we found that Tcf3 colocalized with TLE2 on Lefty2, Oct4, and Trp53 (Fig. 5B). Only CtBP appeared to bind Oct4 at one of the two Tcf3 bound loci, whereas β-catenin was not found to be bound in all instances. Upon activation of Wnt signaling by the addition of Wnt3A-conditioned medium to ESC for 96 hours, β-catenin was observed to bind cyclin D1, a reported target in several cell types (supplemental online Fig. 10).
Figure 5.
TLE2 and Tcf3 co-occupy the same target genes. (A): Tcf3 target genes were bound by TLE2 but not β-catenin. TLE2 immunoprecipitation (IP) showed strong enrichment for the same bound genes as Tcf3 in all instances examined. β-Catenin IP did not show appreciable enrichment. Hgf, Gpr154, and Spata19 were used as negative controls not bound by Tcf3. (B): Occupancy of Tcf3 and TLE2 occurred at the same genomic locus of Lefty2, Oct4, and Trp53. CtBP was observed to bind only Oct4, whereas β-catenin was not enriched at any of the repressed genes. All measurements were performed using quantitative polymerase chain reaction. (C): Sequential IP of Tcf3 followed by TLE2 showed enrichment of Tcf3 target genes. (D): Sequential IP of TLE2 followed by Tcf3 similarly showed enrichment of Tcf3 target genes. Abbreviation: Tcf3, T-cell factor 3.
As Tcf3-bound genes were tightly associated with TLE2 at the same loci, we determined whether both proteins were physically interacting with the same piece of DNA fragment. We performed sequential ChIP using Tcf3 antibody first, followed by immunoprecipitation with TLE2 antibody, and also the reverse. Remarkably, all DNA fragments associated with Tcf3 were similarly bound by TLE2 for the target genes tested, and the converse was also true (Fig. 5C, 5D). Control genes that were not targets of Tcf3 were not enriched. These data confirmed that the action of Tcf3 is largely inhibitory via interaction with the corepressor TLE2 and/or CtBP, and its trans-activation function through β-catenin appears to be less important.
Tcf3 Represses Oct4 Transcription
Our study indicates that Tcf3 might play crucial roles in ESCs by repressing genes involved in maintaining self-renewal and pluripotency. One of the key genes identified in our ChIP-chip data was Oct4. We sought to further characterize the regulation of Oct4 expression in ESCs by Tcf3. To show that Tcf3 directly represses the Oct4 promoter, we overexpressed Tcf3 in HEK293 cells that were cotransfected with an Oct4 promoter-luciferase construct. Overexpression of Tcf3 downregulated the activity of the Oct4 promoter by more than 50% within 48 hours (Fig. 6C, WT).
Figure 6.
Tcf3 represses Oct4 promoter. (A): Illustration of the mouse 3-kb Oct4 promoter with PE and DE. CR1—CR4 represent conserved regions of the promoter shared with human and rat. Seven putative Tcf/Lef binding sites are shown. M1—M4 represent each of the four constructs containing directed mutagenesis on the Tcf/Lef binding motif. (B): Chromatin immunoprecipitation analysis showed Tcf3 occupancy at two sites on the Oct4 promoter, corresponding to M1 and M2 near CR4, as well as M3 and M4 near CR2. Control IP was performed with IgG antibody and did not show any enrichment. Ten primer pairs were designed to scan the region of analysis. (C): Tcf3 overexpression repressed firefly luciferase activity driven by the WT Oct4 promoter to less than 50%, relative to vector control. Mutation of either site M2 or M4 did not affect Tcf3 repression of the mutant promoter. However, mutation of site M1 or M3 prevented Tcf3-mediated repression. Luciferase measurements were performed 48 hours post-transfection into HEK293 cells. *, significantly different from control short hairpin (shRNA); p < .01; n = 3 in three independent experiments. Error bars represent SEM. (D): Tcf3 mutant proteins were generated as depicted in the diagram. WT Tcf3 contains β-catenin, Groucho/TLE, and CtBP interaction domains and an HMG DNA binding domain. Tcf3 Δβ 48aa: 48-aa deletion from N terminus. Tcf3 Δβ 71aa: 71-aa deletion from N terminus. Tcf3 ΔGrg: 174-aa deletion of the Groucho/TLE interacting domain. Tcf3 ΔCtBP: 106-aa deletion from the C terminus. (E): Tcf3 Δβ 48aa and Tcf3 Δβ 71aa were able to mediate repression of the WT Oct4 promoter as with WT Tcf3. Disruption of either Groucho/TLE or CtBP interacting blocks the ability of mutant Tcf3 to repress Oct4 promoter. A noticeable upregulation of the luciferase activity was observed instead. Luciferase measurements were performed 48 hours post-transfection into HEK293 cells. Firefly luciferase measurements were relative to vector-transfected control. *, significantly different from control shRNA control; p < .01; n = 3 in three independent experiments. Error bars represent SEM. (F): The repression of the Oct4 promoter by Tcf3 overexpression was relieved by the addition of Wnt3A CM, whereas the enhanced activation of the promoter was observed with Oct4 overexpression. *, significantly different from —Wnt3A control; p < .01; n = 3 in three independent experiments. Error bars represent SEM. Abbreviations: aa, amino acid(s); CM, conditioned medium; DE, distal enhancer; HMG, high-mobility group; IP, immunoprecipitation; kb, kilobase; PE, proximal enhancer; Tcf3, T-cell factor 3; WT, wild-type.
Sequence analysis of the Oct4 promoter region revealed seven potential Tcf/Lef binding sites with the conserved motif of CTTTG(A)(A) or ACCAAA [14] (Fig. 6A). To determine the actual binding sites of Tcf3 on the promoter, we performed a ChIP assay and demonstrated that two regions near the conserved regions CR2 and CR4 of the Oct4 promoter were bound by Tcf3 (Fig. 6B). To further pinpoint the exact location of Tcf3 binding, each of the four possible sites (M1 to M4) was mutagenized to abolish the Tcf3 binding motif (Fig. 6A). A Tcf3 overexpression construct was cotransfected with each of the mutagenized Oct4 promoters into HEK293 cells. Tcf3 was able to repress the promoter containing mutations in M2 and M4, but not M1 and M3, suggesting that Tcf3 repressed Oct4 transcription activity by binding either the M1 or the M3 site (Fig. 6C).
To delineate the specific interacting domain that confers on Tcf3 its repressive function, we made mutant Tcf3 proteins lacking the β-catenin, Groucho/TLE, or CtBP interacting region (Fig. 6D; supplemental online Fig. 11) and cotransfected these constructs with the wild-type Oct4 promoter luciferase reporter. When different lengths of the amino-terminal β-catenin domain were deleted, the repression of the Oct4 promoter by Tcf3 was unaffected. However, the deletion of either the Groucho/TLE or the CtBP interaction domain abolished the repressive function, with the promoter being slightly activated relative to the vector control (Fig. 6E). These data demonstrated that the repressive effect of Tcf3 is mediated largely through its interaction with Groucho/TLE and/or CtBP, consistent with the colocalization of these corepressors with Tcf3 on the Oct4 promoter (Fig. 5B). Interestingly, the repression of Tcf3 on the Oct4 promoter could be relieved by the stimulation of Wnt signaling through the addition of Wnt3A within 72 hours (Fig. 6F). This effect can be further enhanced in an autoregulatory manner when Oct4 is cotransfected with its own promoter-luciferase, followed by stimulation with Wnt3A. However, it remains unclear whether β-catenin replaces Groucho/TLE to associate with Tcf3, resulting in activation, or associates with other Tcf/Lef factors to bind and activate the Oct4 transcription at other Tcf/Lef motifs located along promoter, thereby overriding the effect of Tcf3.
It has been shown that Tcf3 represses Nanog [16]. In our studies, Tcf3 also directly repressed Oct4 expression in ESCs, which led us to hypothesize that the inability of Tcf3-deficient ESCs to differentiate could be in part a result of elevated levels of both Oct4 and Nanog. If this is the case, ablation of either Oct4 or Nanog would cause Tcf3-deficient ESCs to differentiate. We treated Tcf3-deficient ESCs with control, Oct4, or Nanog shRNA. Although Tcf3-knockdown ESCs treated with control shRNA remained undifferentiated, introduction of Oct4 or Nanog shRNA caused depletion of Oct4 and Nanog transcripts and proteins, with induction of differentiation (supplemental online Fig. 12). Thus, the phenotype of the Tcf3 knockdown cells was in part dependent on Oct4 and Nanog.
Having demonstrated the essential role of Tcf3 in ESCs, we sought to ascertain its importance during mouse preimplantation development. The expression of Oct4 and Nanog during early development and in germ cells has been described previously [35-38]. Oct4 expression was observed clearly from the four-cell stage onward, whereas Nanog was detectable by the morula stage; both Oct4 and Nanog finally localized to the ICM of E3.5 and E4.5 blastocysts (Fig. 7A; supplemental online Fig. 13). Interestingly, Tcf3 was weakly expressed during the four-cell stage and was clearly visible from the eight-cell stage onward. Tcf3 also appeared to be ICM-specific and absent in the trophectoderm. The coexpression of Tcf3 with Oct4 and Nanog suggests that Tcf3 may have a regulatory role during early embryo development, particularly after zygotic gene activation.
Figure 7.
The role of Tcf3 during embryonic development and in ESCs. (A): Expression profiling of Tcf3 and Nanog during mouse preimplantation development showed that Nanog protein appeared in the mouse embryo from the morula stage onward and became localized to the inner cell mass (ICM) of the blastocyst (E3.5 and E4.5) but not the trophectoderm. Tcf3 protein appeared in the four-cell-stage embryo, and expression was maintained at the eight-cell and morula stages and became restricted to the ICM but not trophectoderm. Scale bar = 20 μm. Oct4 expression is given in supplemental online Fig. 13. (B): Schematic representation of the role of Tcf3 in ESCs. Tcf3 represses certain signaling pathways and oncogene families, in addition to developmentally associated genes. This prevents inappropriate activation of differentiation programs in ESCs. Tcf3 also regulates pluripotency-associated genes. Oct4 and Nanog are key Tcf3 targets that can be activated by other factors, such as Oct4, Nanog, Sox2, Sall4, LRH1, and Tpt1. Excessive levels of Oct4 drive ESCs into the endodermal lineage, whereas high levels of Nanog limit differentiation potential. Tcf3 serves as a repressor, as does GCNF, to attenuate Oct4 and Nanog levels among other pluripotency-associated factors. Abbreviations: DAPI, 4,6-diamidino-2-phenylindole; E, embryonic day; Tcf3, T-cell factor 3.
DISCUSSION
The importance of Wnt signaling in stem cell biology has been underscored through its role in maintaining progenitor cells in various tissue niches, including epithelial crypts of the intestine, the hippocampus, hematopoietic stem cells, and skin [7, 9, 39, 40]. In ESCs, Wnt activation preferentially supports self-renewal [12]. Recent studies of the transcriptional networks in ESCs have shown that essential regulators, including Oct4, Sox2, Sall4, and Nanog, function in a feed-forward manner that maintains the levels of pluripotency-associated genes. However, the manner in which Wnt signaling converges with the transcriptional regulation of pluripotency is unclear. Few investigations have focused on the effect of having elevated levels of pluripotency factors, where overexpressing Oct4 promotes endoderm lineage formation and elevated levels of Nanog sustain ESC self-renewal in conditions that would normally promote differentiation [37, 41]. Recent work has further defined the roles of Oct4, Sox2, and Nanog through their regulation of a wide array of genes [23, 42-44].
Less is understood regarding the negative regulation of pluripotency-associated genes. The tumor suppressor p53, for example, suppresses transcription of Nanog and induces differentiation after DNA damage to maintain genetic stability [24], and the orphan nuclear receptor GCNF represses Oct4 and Nanog during RA-induced differentiation of ESCs [45]. The fine-tuning of appropriate levels of these pluripotency factors by repressors is often underappreciated. Ectopic misexpression of Oct4 in adult mice leads to dysplastic growth in epithelial tissues and blocks progenitor cell differentiation [46].
In this study, we intersected components of the Wnt pathway with the transcriptional control of pluripotency-associated genes and mapped out the regulatory targets of Tcf3 in mouse ESCs. We found Tcf3 to be a potent repressor of the Oct4 gene, and Tcf3-deficient ESCs can self-renew but not differentiate. Intriguingly, although we observed the relief of Tcf3 suppression on Oct4 and Nanog in wild-type ESCs after RNAi-induced knockdown, it has been reported that Tcf3-/- ESCs have levels of Oct4 comparable to those of wild-type parental cells [16]. It may be possible that in the process of Tcf3-/- ESCs derivation, these cells have adjusted the levels of Oct4 through various means, such as transcriptional alterations by other upstream Oct4 regulators. Another discrepancy was that Tcf3-/- ESCs showed a delay in differentiation upon LIF withdrawal, whereas we observed that Tcf3 stable knockdown clones were completely resistant to differentiation. This effect could be attributed to the elevated levels of Oct4, in addition to Nanog, which were not observed in the Tcf3-/- cells. Our data also suggest that the resistance to differentiation might also be attributed to the upregulation of a host of other ESC-associated genes. It has been demonstrated that the overexpression of key factors can reprogram a differentiated somatic cell to a pluripotent stem cell [47], suggesting that the elevation of critical molecules is sufficient to drive self-renewal and/or confer pluripotency.
Apart from transcriptional regulation, cells also respond to extracellular signals. The targets of Tcf3 were enriched in many components of signaling pathways, such as Wnt, BMP, MAPK, and fibroblast growth factor signaling (supplemental online Fig. 14), which are implicated in gastrulation and lineage differentiation of mouse embryo, along with an essential role in self-renewal and differentiation of ESCs [48, 49]. Using KEGG analysis, we found that family members of the Wnt and MAPK pathways were overrepresented (supplemental online data; supplemental online Fig. 15). As Wnt activation drives self-renewal in ESCs [12], our results suggest that specific family members of the Wnt pathway must be maintained in a repressed state (supplemental online Fig. 7).
Another significant finding is that Tcf3 was found to bind key developmental genes. It has been suggested that polycomb group (PcG) proteins act as general transcription repressors that block expression of a large cohort of developmental regulators in ESCs, and all the PRC1 and PRC2 co-occupied genes contained trimethylated Lys 27 on histone H3, which is indicative of repression [50, 51]. Upon depletion of Eed, which is a component of PRC2 in mouse ESCs, PcG target genes become derepressed [50]. These observations parallel that of Tcf3, which serves a broader role, as well as an ESC-specific role. Tcf3 binds developmental genes and ESC-specific genes in the same stage-specific context, that is, undifferentiated ESCs. Upon depletion, both groups of target genes become derepressed. However, it remains unclear why there is a tendency for Tcf3-deficient cells to self-renew rather than differentiate. This may be partially attributed to the dominance of ESC-associated genes over differentiation-associated genes, which is also observed during somatic cell reprogramming by fusion with ESCs [52, 53]. This contrasts interestingly with its role in adult and embryonic skin progenitors, where Tcf3 repression of differentiation program dominates [7]. During development, Tcf3-/- embryos exhibit defective neural patterning with multiple notochords and mesodermal duplications at an early stage [6], further highlighting the critical role of Tcf3 in regulating the proper expression and dosage of developmental patterning genes. Although many Wnt genes and their secreted antagonists are expressed in the preimplantation stage [54, 55], it was observed that the ablation of neither Wnt1, Wnt3a, nor Wnt7b led to preimplantation defects [56-58], suggesting a redundancy in either the role of Wnts or their dominant functions during postimplantation development. Importantly, β-catenin did not appear to be localized in the nuclei of ICM cells [59], thereby confirming that Tcf3 acts as the dominant downstream repressor.
CONCLUSION
Taken together, our study suggests that Tcf3 fulfills two major roles (Fig. 7B). First, it regulates the proper dosage of ESC-associated genes, which enable ESCs to self-renew yet retain pluripotency. Second, Tcf3 represses differentiation and developmental programs to retain the differentiation potential of ESCs. Although Tcf3 appears to be expressed in many tissues, its role in regulating differentiation versus stemness programs has largely not been studied. Our study on Tcf3 target genes in ESCs therefore provides a valuable resource for elucidating what other candidate genes Tcf3 may be regulating in different systems. That Tcf3 represses many stemness genes also suggests that it could be modulating downstream targets in ESCs, which may be involved in somatic cell reprogramming.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by the Agency for Science, Technology and Research (Singapore) and the Singapore Stem Cell Consortium grant (SSCC-06-003). W.-L.T. is a recipient of the A*STAR graduate scholarship. The work is also partially supported by NIH Grants DK04763 and AI54973 (to B.L.). We are grateful to Dr. Paul Robson, Dr. Wing-Kin Sung, Karrie Ko, and Yin Loon Lee in the Genome Institute of Singapore for helpful discussions and experimental assistance and Boon Seng Soh for providing Wnt3A-conditioned medium.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST The authors indicate no potential conflicts of interest.
REFERENCES
- 1.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 2.Lagutin OV, Zhu CC, Kobayashi D, et al. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003;17:368–379. doi: 10.1101/gad.1059403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zechner D, Fujita Y, Hulsken J, et al. beta-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev Biol. 2003;258:406–418. doi: 10.1016/s0012-1606(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 4.Hirabayashi Y, Itoh Y, Tabata H, et al. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004;131:2791–2801. doi: 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- 5.Huelsken J, Vogel R, Erdmann B, et al. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 6.Merrill BJ, Pasolli HA, Polak L, et al. Tcf3: A transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–274. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 8.Reya T, Duncan AW, Ailles L, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 9.Willert K, Brown JD, Danenberg E, et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 10.Aubert J, Dunstan H, Chambers I, et al. Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat Biotechnol. 2002;20:1240–1245. doi: 10.1038/nbt763. [DOI] [PubMed] [Google Scholar]
- 11.Haegele L, Ingold B, Naumann H, et al. Wnt signalling inhibits neural differentiation of embryonic stem cells by controlling bone morphogenetic protein expression. Mol Cell Neurosci. 2003;24:696–708. doi: 10.1016/s1044-7431(03)00232-x. [DOI] [PubMed] [Google Scholar]
- 12.Sato N, Meijer L, Skaltsounis L, et al. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 13.Eastman Q, Grosschedl R. Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr Opin Cell Biol. 1999;11:233–240. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 14.Roose J, Clevers H. TCF transcription factors: Molecular switches in carcinogenesis. Biochim Biophys Acta. 1999;1424:M23–37. doi: 10.1016/s0304-419x(99)00026-8. [DOI] [PubMed] [Google Scholar]
- 15.Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 16.Pereira L, Yi F, Merrill BJ. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol Cell Biol. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Tam WL, Tong GQ, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 18.Reynolds A, Leake D, Boese Q, et al. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 19.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kléber M, Sommer L. Wnt signaling and the regulation of stem cell function. Curr Opin Cell Biol. 2004;16:681–687. doi: 10.1016/j.ceb.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Brantjes H, Roose J, Van De Wetering M, et al. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29:1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niwa H, Burdon T, Chambers I, et al. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 24.Lin T, Chao C, Saito S, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 25.Levine AJ, Brivanlou AH. GDF3, a BMP inhibitor, regulates cell fate in stem cells and early embryos. Development. 2006;133:209–216. doi: 10.1242/dev.02192. [DOI] [PubMed] [Google Scholar]
- 26.Wijchers PJ, Burbach JP, Smidt MP. In control of biology: Of mice, men and Foxes. Biochem J. 2006;397:233–246. doi: 10.1042/BJ20060387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buckingham M, Bajard L, Chang T, et al. The formation of skeletal muscle: From somite to limb. J Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plageman TF, Jr, Yutzey KE. T-box genes and heart development: Putting the “T” in heart. Dev Dyn. 2005;232:11–20. doi: 10.1002/dvdy.20201. [DOI] [PubMed] [Google Scholar]
- 29.Kiefer JC. Back to basics: Sox genes. Dev Dyn. 2007;236:2356–2366. doi: 10.1002/dvdy.21218. [DOI] [PubMed] [Google Scholar]
- 30.Sato N, Sanjuan IM, Heke M, et al. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol. 2003;260:404–413. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- 31.Su AI, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurlstone A, Clevers H. T-cell factors: Turn-ons and turn-offs. EMBO J. 2002;21:2303–2311. doi: 10.1093/emboj/21.10.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 34.Roose J, Molenaar M, Peterson J, et al. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 35.Scholer HR, Dressler GR, Balling R, et al. Oct-4: A germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scholer HR, Ruppert S, Suzuki N, et al. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- 37.Chambers I, Colby D, Robertson M, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 38.Mitsui K, Tokuzawa Y, Itoh H, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 39.Korinek V, Barker N, Moerer P, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 40.Lie DC, Colamarino SA, Song HJ, et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 41.Nichols J, Zevnik B, Anastassiadis K, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 42.Tomioka M, Nishimoto M, Miyagi S, et al. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 2002;30:3202–3213. doi: 10.1093/nar/gkf435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuroda T, Tada M, Kubota H, et al. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol Cell Biol. 2005;25:2475–2485. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okumura-Nakanishi S, Saito M, Niwa H, et al. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J Biol Chem. 2005;280:5307–5317. doi: 10.1074/jbc.M410015200. [DOI] [PubMed] [Google Scholar]
- 45.Gu P, LeMenuet D, Chung AC, et al. Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. Mol Cell Biol. 2005;25:8507–8519. doi: 10.1128/MCB.25.19.8507-8519.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 46.Hochedlinger K, Yamada Y, Beard C, et al. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 48.Loebel DA, Watson CM, De Young RA, et al. Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev Biol. 2003;264:1–14. doi: 10.1016/s0012-1606(03)00390-7. [DOI] [PubMed] [Google Scholar]
- 49.Molofsky AV, Pardal R, Morrison SJ. Diverse mechanisms regulate stem cell self-renewal. Curr Opin Cell Biol. 2004;16:700–707. doi: 10.1016/j.ceb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 50.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 51.Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tada M, Takahama Y, Abe K, et al. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 53.Cowan CA, Atienza J, Melton DA, et al. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 54.Kemp C, Willems E, Abdo S, et al. Expression of all Wnt genes and their secreted antagonists during mouse blastocyst and postimplantation development. Dev Dyn. 2005;233:1064–1075. doi: 10.1002/dvdy.20408. [DOI] [PubMed] [Google Scholar]
- 55.Kemp CR, Willems E, Wawrzak D, et al. Expression of Frizzled5, Frizzled7, and Frizzled10 during early mouse development and interactions with canonical Wnt signaling. Dev Dyn. 2007;236:2011–2019. doi: 10.1002/dvdy.21198. [DOI] [PubMed] [Google Scholar]
- 56.McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- 57.Takada S, Stark KL, Shea MJ, et al. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8:174–189. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- 58.Parr BA, Cornish VA, Cybulsky MI, et al. Wnt7b regulates placental development in mice. Dev Biol. 2001;237:324–332. doi: 10.1006/dbio.2001.0373. [DOI] [PubMed] [Google Scholar]
- 59.Kemler R, Hierholzer A, Kanzler B, et al. Stabilization of beta-catenin in the mouse zygote leads to premature epithelial-mesenchymal transition in the epiblast. Development. 2004;131:5817–5824. doi: 10.1242/dev.01458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.