Abstract
Establishment of cellular memory and its faithful propagation is critical for successful development of multicellular organisms. As pluripotent cells differentiate, choices in cell fate are inherited and maintained by their progeny throughout the lifetime of the organism. A major factor in this process is the epigenetic inheritance of specific transcriptional states or transcriptional memory. In this review, we discuss chromatin transitions and mechanisms by which they are inherited by subsequent generations. We also discuss illuminating cases of cellular memory in budding yeast and evaluate whether transcriptional memory in yeast is nuclear or cytoplasmically inherited.
Keywords: transcription, chromatin, cytoplasmic inheritance, memory, cell fate
Introduction: Transcriptional choice and its inheritance
All organisms regulate their genetic repertoire in response to their environment as well as cell intrinsic cues. Single-celled organisms like yeast can coordinately induce and repress sets of genes as a result of stimuli like nutrient starvation, mating pheromones or DNA damage. In addition to responding to extracellular signals, multicellular organisms undergo cell differentiation. Cell differentiation is the culmination of numerous, highly regulated gene expression events that occur during embryonic development and throughout the life of an adult organism, where it controls growth, homeostasis and tissue repair. Some of these gene expression patterns or transcriptional choices become marked by epigenetic alterations of the genome, resulting in a transcriptional memory of gene expression profiles that are inherited by progeny.
Cell fate determination is an integral part of embryonic development in all multicellular organisms. A single-celled zygote undergoes many mitotic divisions till the blastocyst stage, where the inner cell mass (ICM) contains all the totipotent cells that will ultimately give rise to the embryo-proper [1, 2]. As the ICM cells further divide, they reorganize to form the three germ layers – ectoderm, mesoderm and endoderm, which are fated to form distinct tissues and organ systems [1, 3, 4]. During these events, genes that encode pluripotency markers are transcriptionally repressed, and gene products characteristic of particular cell fates begin to be expressed [2, 5, 6]. As these cells further divide and differentiate, such characteristic gene expression states get ‘locked in’ and are faithfully replicated in all progeny of fate-restricted cells [7, 8]. This transcriptional memory can persist through multiple rounds of cell division, and in many cases throughout the lifetime of the organism even in the absence of the initiation signals [9–14]. Such cellular memory posits the existence of one or more mechanisms that transmit information of the active or silent gene state from mother to daughter cells. Mechanisms for providing cellular memory can be divided into two broad classes: cytoplasmic inheritance and nuclear inheritance [15–19].
Cytoplasmic inheritance of transcriptional memory involves the presence of a protein or small molecule located in the cytoplasm of the cell that gains memory of a particular transcriptional event (See Figure 1). During cell division, such a protein or small molecule signal can be passed on to daughter cells by distribution of the cytoplasm of the mother cell [20]. It can be envisaged that such memory would be relatively short-lived – its duration or persistence is limited by dilution and/or half-life of the cytoplasmically inherited memory factor. This model will be revisited in a later section of this review.
Figure 1. Mechanisms for nuclear and cytoplasmic inheritance of transcriptional memory.
A multipotent progenitor cell can respond to a particular signal(s) by altering its transcriptional profile. This step can lead to cell-fate commitment. Memory of adopted cell fate can be transmitted by various epigenetic/chromatin based mechanisms or by cytoplasmic signals. Changes in chromatin state can involve DNA-cytosine methylation, histone modifications and/or histone variants. Cytoplasmic inheritance could involve a signal-induced peptide, RNA or small molecule that maintains target genes in an ON or OFF state. Persistence of cellular memory in each case depends on faithful transmission of the ‘memory mark’ to subsequent generations. See text for details.
The nuclear or epigenetic inheritance model involves changes in the chromatin state of target genes, changes that can persist through DNA replication and mitosis [21]. These changes can be covalent marks on DNA and/or histones, and therefore would not alter the genomic information of pluripotent cells or their differentiated progeny. Where mechanisms for faithful replication of these marks have evolved, such nuclear or epigenetic memory can persist through many generations and indeed throughout life [7]. The substrate for these long-term memory marks is chromatin, and chromatin structure participates directly in transcriptional activation or repression of gene loci. Below we review some chromatin basics, and then we discuss several examples of chromatin based mechanisms for transcriptional memory.
Chromatin basics
The large genome of eukaryotes is packaged into chromatin, a DNA and protein containing structure. This structure affords compaction of the genome, thereby fitting it into the small volume of a nucleus. Importantly, and in ways still being avidly studied and discovered, it is a critical component of gene regulatory activities. The basic component of chromatin is a nucleosome, generated by wrapping approximately 147 bp of DNA ~1.7 times around an octamer of core histone proteins [22–24]. The canonical histone octamer contains 2 copies each of histones H2A, H2B, H3 and H4. Structurally, these histones consist of a central globular or ‘histone-fold’ domain and flexible NH2-terminal and COOH-terminal tail domains. As DNA wraps around the histone octamer, it makes intimate contacts at 14 positions [22, 23, 25, 26]. These tight associations occlude potential transcription factor binding sites and leads to steric hindrance to DNA-binding by transcriptional activators, repressors, and the core transcriptional machinery [27–29]. This protection of DNA is also evidenced biochemically by restricted access to DNA cleavage agents like micrococcal nuclease (MNase) and restriction enzymes [30, 31]. Beyond this primary level of nucleosomal or ‘beads-on-a-string’ structure are successive higher order chromatin structures that ultimately compact DNA into thick chromatin fibers, with the highest degree of compaction seen in the classical metaphase chromosome [32–34].
In addition to core histones, most eukaryotes also have an unrelated histone called linker histone (e.g. histone H1). Mammals have at least eight variants of histone H1, five of which are expressed ubiquitously in somatic tissue and some studies indicate that deletion of individual variants can cause distinct phenotypes [35–37]. Like core histones, linker histones are also highly basic in amino acid composition. There is on average 1 linker histone per nucleosome, which protects an additional ~20bp of DNA and stabilizes higher order chromatin folding [38–42]. Like their core histone counterparts, linker histones are also composed of two functional domains – the globular and tail domains. While the globular domain interacts with nucleosomes, the NH2-terminal and COOH-terminal tails of linker histone contain sites for post translational modifications, and the C-terminal tail is required for stabilizing higher order chromatin folding [43–54]. Whereas removal of H1 has no effect on viability of unicellular organisms like Tetrahymena, Aspergillus nidulans and Saccharomyces cerevisiae, linker histones are essential for cellular differentiation in higher eukaryotes [51, 55–61]. Xenopus, mouse, and human linker histone variants control transcriptional regulation of specific genes and regulate nucleosome spacing [36, 62–65]. There is evidence that H1 subtype switching also occurs in Xenopus and mammalian embryos during development [66]. This suggests a mechanism that may provide memory of cell fate specification during embryonic development.
Regulation of chromatin structure and function
How can chromatin structure be modified to regulate access to transcriptional regulators and the core transcriptional machinery? Cells employ two general enzymatic strategies that regulate chromatin dynamics. In the first case, ATP-dependent chromatin remodeling enzymes use energy derived from ATP hydrolysis to mobilize nucleosomes, evict some or all of the histones from the nucleosome, or to exchange histone variants [67]. These enzymes all contain ATPases of the Swi2/Snf2 superfamily and are broadly classified into the SWI/SNF, ISWI, CHD and Ino80 subfamilies [67, 68]. Some, like the SWI/SNF complex, play key roles in transcription; in yeast there are two SWI/SNF family members – RSC and SWI/SNF [69–72]. Members of the SWI/SNF family are primarily involved in transcriptional activation. For instance, the yeast SWI/SNF complex is recruited during activation of many genes during late mitosis as well as some highly inducible genes like INO1, SUC2, GAL1-10, PHO5 and PHO8 [73–77]. Complex eukaryotes have several different SWI/SNF complexes (also called BAF complexes), with the catalytic subunit being Brg1 or Brm, and a variety of associated subunits [71, 78–81]. Brg1 is essential for mammalian development, as mice harboring a brg1 deletion show periimplantation lethality [82, 83]. Some of the associated subunits are also tissue restricted. For instance BAF60c is restricted to the myocardial lineage, while BAF53b is neuron-specific [84, 85]. Other ATP-dependent remodeling enzymes, like ISWI and Mi-2, usually function in transcriptional repression [86–88]. Mi-2 complexes, a prominent member of the CHD (chromodomain containing) subfamily play critical roles at different stages of hematopoiesis, and ISWI complexes are critical for normal development and differentiation as evidenced by periimplantation lethality of mice lacking the catalytic subunit, Snf2h [89, 90]. The Ino80 subfamily, which includes INO80 and SWR1, are characterized by a split ATPase domain, and the SWR1 complex is required for exchange of the histone variant H2A.Z into nucleosomes [91–97].
The second category of chromatin remodeling enzymes includes enzymes that mediate posttranslational modifications of histones [98, 99]. Nucleosomal histones can be extensively modified at their N- or C-terminal tail domains, or even at some internal sites. Histone modifications function primarily by influencing the binding of non-histone proteins, like transcription factors and other chromatin remodeling enzymes, to nucleosomes, although some marks (e.g. H4K16ac) directly impact chromatin structure [100]. A wide variety of enzymes have been identified in all organisms that catalyze diverse modifications such as methylation, acetylation, phosphorylation, ubiquitylation and SUMOylation [101]. Histone modifications are reversible since there are also demethylases and deacetylases dedicated to removal of these groups [102, 103]. These modifications often are found in combinations and act synergistically to recruit or occlude chromatin associated proteins and generate transcriptionally favorable or unfavorable chromatin domains [104]. Histone modifying enzymes and ATP-dependent remodelers often act in concert for regulating gene expression [87, 105–108].
Finally, the most stable and replicable of chromatin modifications are those on the DNA itself, i.e. cytosine methylation of CpG islands by DNA methyltransferases [109, 110]. Though yeast and nematodes lack DNA methylation, it is critical in other metazoans for development and differentiation [111]. In addition there are RNA based chromatin regulatory processes exemplified by X-inactivation, heterochromatin formation and position-effect variegation, which have been discussed in detail elsewhere ([112–114] and others).
Strategies to establish transcriptional memory by modifying chromatin state
Establishing cellular memory of a particular transcriptional state is essential in multicellular organisms to obtain fruitful cell fate specification during development. As fate-restricted cells multiply during organogenesis, various mechanisms have evolved that replicate the transcriptional state of the progenitor cell in its daughters. For example, genes that confer pluripotency or alternate cell fates must be stably repressed, while genes characteristic of the chosen cell-fate are maintained in a transcriptionally active or poised state. As a consequence, despite having identical genomes, cells of different tissues must maintain and propagate their distinct transcriptional profiles or in other words, have transcriptional memory of the expression status at multiple loci [8, 11, 13, 14]. Since chromatin remodeling is often required to silence or activate genes, transcriptional memory could involve propagation of differential chromatin states in different tissues. Distinct chromatin states may be established and inherited by using various chromatin modifying strategies introduced above. We will discuss some specific enzymatic activities and their roles below.
DNA methylation
DNA methylation, predominantly at symmetrical CpG dinucleotides, is usually associated with gene silencing [115, 116]. Genomic methylation patterns are very stable and heritable in somatic differentiated cells. Once established, they are faithfully replicated at every cell division by the ‘methylation maintenance’ enzyme DNMT1, which uses a hemimethylated DNA substrate to restore the symmetrical CpG methylation pattern [117, 118]. There are two developmental stages – germ cells and preimplantation embryos – where the genomewide methylation pattern is reprogrammed. The fertilized egg undergoes a wave of demethylation during preimplantation development which erases part of the inherited, parental methylation pattern [119]. After implantation, the embryo then undergoes de novo methylation to establish a new embryonic methylation pattern. DNMT3A/B are the primary de novo DNA methylases that establish new methylation patterns and are expressed in most dividing cell types along with DNMT1 [109, 110, 120, 121]. Another protein, DNMT3L, is similar to DNMT3A/B in amino acid sequence but lacks enzymatic activity. It is expressed only in germ cells during de novo methylation and is believed to regulate DNMT3A/B [122, 123]. There is also an oocyte-specific form of DNMT1 (DNMT1o) that accumulates to very high levels in the cytoplasm of oocytes and persists in preimplantation embryos. It enters nuclei at the eight-cell stage to maintain imprinted methylation patterns [124]. Overall, DNA methylation has key roles in epigenetic gene regulation and silencing, in particular in gametic imprinting, X chromosome inactivation, and silencing of retrotransposons [7, 125, 126]. This methylation pattern has also been proposed to inhibit transposition as well as recombination and expansion of repetitive elements [127–129].
In dividing cells, the maintenance methyltransferase, DNMT1 provides epigenetic memory of transcriptionally silenced loci. Maintenance of imprinted silencing by DNMT1 is linked to DNA replication by its association with PCNA and CAF1 [117, 130–133] This association provides the mechanism for propagation of epigenetically silent states, but not transcriptionally active states, through S-phase to subsequent generations. Experiments suggest that methylation by itself does not prevent transcription [134–136], but instead transcriptional silencing of methylated loci is due to methyl-CpG binding domain proteins (MBDs), that alter chromatin structure [110]. The primary MBDs are MBD1, MBD2, MBD3 and MeCP2 [111, 137, 138]. The MBD proteins interact with and recruit histone deacetylases (HDACs) such as HDAC1, HDAC2, Mi-2/NURD which deacetylate the associated chromatin to render it transcriptionally incompetent [107, 139, 140].
Histone modifications
The NH2- and COOH-terminal tails of core histones are subject to extensive and dynamic posttranslational modifications (PTMs) [101]. These PTMs can disrupt the structure of the chromatin fiber or alter its interactions with nonhistone proteins. Prominent among marks associated with the transcriptionally active state are methylation of H3K4, ubiquitylation of H2BK123, and acetylation of several lysines within the N-terminal domains of H2B, H3 and H4 (H2BK11,16ac; H3K9,14,18,23,27ac; H4K5,8,12,16ac) [101, 141, 142]. Similarly, methylation of H3K9 or H3K27 are some of the more common marks associated with repressed loci [143, 144]. Different enzymes catalyze these histone modifications and can be conserved across species. Histone modifying enzymes are also important for controlling expression of many developmentally regulated genes. At the same time, histone demethylase and deacetylase enzymes have also been discovered that reverse these marks [145–150]. As expected, at most loci in dividing or differentiated cells, certain histone modifications can co-exist or act together to reinforce repressed or accessible chromatin states [151]. This observation has led to speculation that particular patterns of histone modifications might predict transcriptional status of genes in different cell lineages [152–156]. Interestingly, adult and embryonic stem cells have been shown to possess domains of ‘bivalent’ chromatin, where positive and negative histone modifications coexist at certain gene loci, which may poise these genes for appropriate regulation when the stem cells differentiate [157, 158]. Such chromatin domains may be essential for pluripotency and are resolved into and active or repressed state during lineage specification.
A key question is the precise mechanism by which histone modifications are transmitted through cell divisions. For histone PTMs to function as “memory” markers, they must be propogated through replication. Many studies have shown that a replication fork disrupts parental nucleosomes in front of the fork, and that both old and new histone octamers are rapidly assembled on to the daughter chromatids behind the fork. Pioneering studies of Jackson demonstrated that as DNA is replicated, old (H3–H4)2 tetramers are distributed to the daughter strands while newly synthesized (H3–H4)2 tetramers are deposited to fill in the gaps [159, 160]. Old and newly synthesized H2A-H2B dimers are then added to complete nucleosome assembly. Thus it appears that while hybrid octamers of old tetramers and new dimers can form, (H3–H4)2 tetramers remain essentially intact through DNA replication and chromatin assembly. Since only the parental tetramers contain histone modifications representing active or repressed chromatin states, “memory marks” would need to recruit enzymatic activities that would interpret the existing marks and replicate them on the newly formed octamers. Such a model has been proposed to explain how H3 K9me marks might be propogated by the Swi6 and Clr4 proteins [161], and there also appears to be potential candidates that can ‘read’ and ‘write’ H3K4me marks [162, 163].
Recent studies have suggested a different view of chromatin replication that suggests an alternative model for contributions of histone PTMs to transcriptional memory [164]. According to this model, the replication fork splits a parental (H3–H4)2 tetramer into two H3–H4 dimers which are segregated to the two newly replicated DNA strands. Thus, in this case each sister chromatid inherits a parental pattern of H3–H4 PTM which can then be used as a template to replicate modifications on the newly assembled nucleosomes.
Trithorax and Polycomb groups: Developmental Memory factors
One of the classic examples of transcriptional memory involves the developmental expression of the homeotic (Hox) genes in Drosophila and other metazoans [10, 165]. Hox gene expression is controlled by the interplay of two antagonistic sets of gene products that regulate chromatin structure – the Polycomb group (PcG) and the Trithorax group (TrxG). Hox genes encode homeodomain transcription factors that specify cell fates and therefore their transcription must be precisely regulated [166–168]. Mis-expression can lead to severe developmental abnormalities, such as formation of appendages at wrong positions. In fact, genes encoding Polycomb group proteins were first discovered in Drosophila as mutants that led to transformation of body-segmentation patterns along the embryonic anterior-posterior axis, thus transforming the identity of one segment to another [169–171]. Subsequently, genes encoding TrxG proteins were isolated as suppressors of polycomb mutants, indicating that TrxG proteins functionally antagonize PcG proteins [172–174]. The initial patterns of homeotic gene transcription are established in response to positional information in the early embryo [175]. Polycomb and Trithorax group proteins repress and activate genes respectively, and together they maintain gene expression patterns through subsequent cell divisions even after the regulators that initially established the precise segmentation patterns have long disappeared [165]. Thus, PcG and TrG proteins provide a memory of early cell fate decisions [176, 177].
Biochemical studies reveal that many PcG and TrxG products are components of several large complexes that have many different functions, all aimed at epigenetic control of chromatin configuration at homeotic and imprinted genes [178–180]. Several PcG and TrX complexes bind to and act via Polycomb and Trithorax response elements (PREs/TREs) [19, 181, 182], and a subset of PcG and TrxG members possess histone methyltransferase activity. The PRC2 complex contains E(Z) (called EZH2 in mammals), which is an H3K27 methyltransferase that mediates silencing of HOX and other loci. H3K27me is recognized by the chromodomain of the Polycomb (Pc) subunit of the PRC1 complex, and binding of PRC1 via H3K27me can generate a stably repressive chromatin structure that is refractory to gene expression [183, 184]. In support of this, there is evidence that PRC1 inhibits remodeling by the SWI/SNF chromatin remodeling complex and transcription by RNA Polymerase II in vitro [179, 185–187]. Likewise, the TrxG members, TRX (MLL is the mammalian ortholog) and ASH1, are H3K4 and H3K36 methyltransferases that are associated with activated gene expression [188–193]. The trx protein contains a SET domain, which is very similar to the SET domain within S. cerevisiae Set1p, the first identified H3K4-methyltransferase [194–198]. Trx and Ash1 are believed to promote transcriptional elongation, and this may be stimulated in part by the H3K4me and H3K36me marks. Thus, differential H3 lysine methylation status of promoter and coding regions, such as observed at the Drosophila Ubx gene, may confer a transcriptional ON or OFF status [199]. Other TrxG members includes genes that encode subunits of the Drosophila SWI/SNF chromatin remodeling complex, Brahma (BRM), Moira (MOR) and Osa (OSA) [173, 200–202]. Thus TrxG employs both histone methylation and ATP-dependent chromatin remodeling to maintain a heritable, transcriptional ON state. Due to the critical role of PcG and TrxG proteins in maintaining transcriptional memory of developmentally regulated genes, these complexes are the focus of extensive studies to determine mechanisms of cell fate choice and stem cell pluripotency.
Histone variants
The canonical histones that make up a core nucleosome particle – H2A, H2B, H3 and H4, are expressed and incorporated into chromatin only during DNA replication [203]. In addition, organisms express variants of canonical histones H2A (H2A.Z, macroH2A, H2A.X and H2ABbd) and H3 (H3.1, H3.2, H3.3, H3.1t and Cenp-A/Cse4) which are expressed and deposited throughout the cell cycle, are thought to have specific properties, and may establish structurally distinct chromosomal domains [164, 204–208]. Emerging evidence indicates that correct distribution of some histone variants is important for the epigenetic control of gene expression and cell fate decisions. Though the precise mechanisms for targeted deposition are still being worked out, distinct and often dedicated multiprotein complexes have been discovered that target histone variants for deposition at active genes, centromeres and transcriptionally silent loci [203]. For example, the ATP-dependent chromatin remodeling complex SWR1 is targeted to many RNA polymerase II promoters where it exchanges canonical H2A-H2B dimers for H2A.Z-H2B dimers [93–95, 209]. Likewise, the histone chaperone, HIRA, binds to H3.3–H4 dimers and deposits them within transcriptionally active chromatin [164, 210].
What are the functions of these histone variants? In yeast, H2A.Z is found within 1–2 nucleosomes that flank all RNA Polymerase II transcribed genes that are both active and inactive [211–213]. In addition it also prevents ectopic spreading of heterochromatin [214, 215]. Thus H2A.Z seems to be important to maintain an open state of chromatin. This premise is supported by the observation that nucleosomes that harbor H2A.Z-H2B dimers are less stable [213, 216, 217]. Though paradoxically, H2A.Z also promotes formation of 30nm fibers [218] and FRET studies suggest that octamers containing H2A.Z-H2B dimers are more stable in high salt concentrations than those containing H2A–H2B dimers [219]. Surprisingly, loss of yeast HTZ1 has a very mild phenotype in nutrient-rich growth conditions, indicating that transcription of most genes is unperturbed even upon loss of H2A.Z [220]. In contrast, the importance of H2A.Z for regulating chromatin configuration is clearly seen during mammalian embryonic development, as loss of H2A.Z results in preimplantation lethality in mice [218, 221]. There is also an active genomewide displacement of H2A.Z from early mouse PGCs that correlates with the timing of genomewide DNA demethylation, suggesting a role of H2A.Z loss in chromatin decondensation and reprogramming [222]. A more recently discovered H2A variant, H2A.Bbd (Barr body-deficient) appears to be exclusively associated with transcriptionally active chromatin and was shown to be excluded from inactive X chromosomes in mammalian females [223]. It is significantly diverged from canonical H2A in amino acid sequence and structure, and its distribution overlaps with regions of histone H4 acetylation, suggesting that this histone variant has evolved to perform a specialized function of epigenetically marking active chromatin [224–226]. In contrast to the distribution of H2A.Bbd, the H2A variant, macroH2A is enriched on the silenced X chromosome and epigenetically marks inactive chromatin [223, 227]. Its enrichment on the inactive X chromosome coincides with initiation of silencing.
Similar to H2A.Z, H3.3 may also be also involved in maintenance of open, transcriptionally active chromatin. H3.3 differs from canonical H3 at only 4 amino acid positions. However, unlike H3, H3.3 gene has a chromosomal location outside the histone gene cluster and it is synthesized independent of S phase. Thus, unlike the canonical H3, H3.3 deposition is replication-independent and transcription-dependent [206, 228]. As a result, H3.3 is also enriched in ‘activating’ posttranslational modifications such as H3K4me and H3K9,14,18,23ac, whereas canonical H3 preferentially accumulates repressive modifications like H3K9me [229–233].
H3.3 deposition plays critical roles during embryonic development. It replaces canonical H3 during meiotic X chromosome inactivation in the mouse germline [234]. This could provide epigenetic memory of maternally expressed and paternally imprinted genes at a stage where DNA methylation is reversed. Also in early mouse zygotes, H3.3 is incorporated only into paternal chromatin coinciding with its decondensation, soon after gamete fusion [235]. Consequently, a null mutation of the H3.3 chaperone, HIRA, has gastrulation defects in mouse embryos and is early embryonic lethal [236]. Also in accordance with the role of H3.3 in decondensation of paternal chromatin, mutations of Drosophila HIRA lead to formation of haploid embryos with only maternal chromosomes, which die before hatching [237].
It is very tempting to envisage a scenario where histone variants could mark different chromatin domains. As pluripotent cells adopt cell fates during differentiation, inheritance of such chromatin domains via a ‘histone variant signature’ could faithfully reproduce the transcription profile of a differentiated cell type in its successive progeny. However, some questions still remain. Besides the yet unsolved question of how these histone variants (and their chaperones) are targeted to mark loci, it is also still unclear how information of histone variant occupancy is replicated during S-phase. This knowledge is crucial to understand the mechanism of inheriting memory of transcriptional state. As is the case with inheriting histone modifications, the two popular models of replicating histone variant occupancy are the conservative and semiconservative model of histone deposition during DNA replication [238, 239]. One intriguing possibility that could facilitate inheritance of transcriptionally active states, and may be especially pertinent to inheritance of H3.3, is a model that involves a combination of both replication-coupled and replication-independent nucleosome assembly [240]. As the replication fork crosses a transcriptionally active locus, parental H3 and H3.3 tetramers would be used for assembly of nucleosomes onto the sister chromatids by replication-coupled deposition. Nucleosomes that harbor parental H3.3 would be present in lower concentration within newly synthesized chromatin, as only canonical H3 would be incorporated into newly synthesized, nonparental nucleosomes. However, this lower density of H3.3-containing nucleosomes might be sufficient to promote an active chromatin state, and transcription would continue from the locus after replication. Once transcription resumes, replication independent and transcription dependent deposition of H3.3 would reconstitute high levels of nucleosomes containing H3.3.
RNA-based silencing
Over the past few years, non-coding RNAs have been implicated in long term, chromatin-based gene silencing. The first wave of imprinted X-chromosome inactivation in preimplantation mouse embryos is mediated by the Xist RNA. Once transcribed, Xist RNA spreads from its origin in cis to coat the X-chromosome and recruits other silencing factors [241, 242]. Not only does Xist form a chromosomal memory of X inactivation during differentiation, Xist coating recruits further repressive chromatin changes – incorporation of the histone variant macroH2A, DNA methylation and recruitment of PcG proteins [243, 244]. These chromatin changes allow the inactivated X-chromosome to be stably silenced at later stages of development, even in the absence of Xist [245].
RNA interference (RNAi) mediated heterochromatin formation has also emerged as a robust means of establishing epigenetic inheritance [246, 247]. RNAi regulates heterochromatin formation and spreading at the pericentric dg and dh repeats in Schizosaccharomyces pombe [248, 249]. The process initiates when either of these repeats is transcribed. These RNAs are fed into the RNAi pathway, eventually generating small siRNAs that get incorporated into the RITS (RNA-induced transcriptional silencing) complex. RITS activity recruits the H3K9 methyltransferase, Clr4, and eventually the heterochromatin protein Swi6 binds to H3K9me to form silenced heterochromatin at centromeres. Inheritance of centromeric silencing through successive generations appears to require a mechanism connecting RNAi and early replication of centromeric repeats [250, 251].
Cellular memory in budding yeast
Unlike multicellular organisms, unicellular yeasts like Saccharomyces cerevisiae and Candida albicans demonstrate no significant differentiation or functional asymmetry between ‘progenitor’ mother cells and their progeny. Nevertheless, these cells have complex transcriptional networks that lend themselves to considerable robustness and sensitivity when a growing population of yeast encounter and respond to environmental changes. Can activation of such transcriptional networks lead to a heritable memory of adaptive response in yeast cells? There appear to be at least two clear situations where cellular memory is formed. One is the frequency of ‘white-opaque’ cell type switching in Candida albicans [252–254], and another is the rapid reactivation of galactose induced transcription of the GAL gene cluster (GAL1, GAL10 and GAL7) following a period of transcriptional repression [255–257]. Interestingly, in both examples memory of the transcriptional response persists for at least several generations. What is the mechanism by which transcriptional memory in yeast is inherited by successive generations? Yeast lack DNA methylation and therefore this mode of epigenetic inheritance is absent. For both of the above examples of transcriptional memory, regulators of chromatin structure have been suggested. Studies on white-opaque switching of C. albicans indicate that the histone deacetylases HDAC2 and RPD3 are involved since deletion of these genes affects switching frequency [258, 259]. Yet there is still no clear mechanism of how the mark of histone deacetylation is inherited by subsequent generations. For transcriptional memory at GAL genes, chromatin transitions as well as cytoplasmic signaling networks have been proposed and these will be discussed further.
How does the chromatin structure of yeast cells compare to the genome organization of more complex eukarayotes, such as Drosophila or vertebrates? Yeast chromatin, like all eukaryotes is composed of linear arrays of canonical nucleosomes, and it is believed that yeast chromatin is organized as a dynamic 30 nm fiber. Yeast chromatin is on average hyperacetylated, and this high level of acetylation correlates well with the observation that most of the yeast genome is transcribed. Yeast contain an H2A.Z histone variant (Htz1p), but the linker histone homolog, Hho1p, does not appear to play a role in chromatin organization or general genome function. Yeast cells contain members of all four classes of ATP dependent chromatin remodeling enzymes (e.g. SWI/SNF, ISW1/2, INO80, and Chd1p), as well as many different histone modifying enzymes. Notably, budding yeast lack the enzymes that generate several methylated lysines, including those that modify H3 K9 and H4 K20. In contrast, the fission yeast, S. pombe, contain both of these methylases. Consequently, fission yeast contain more classical heterochromatin domains that harbor H3 K9me2 and an HP1 homolog, whereas budding yeast rely on the Sir2/3/4 complex to generate heterochromatin-like structures that repress transcription and recombination. Thus, yeast cells, like more complex eukaryotes, can use heterochromatin as a mechanism to maintain an inactive transcription state over many generations.
The yeast GAL system and models of transcriptional memory
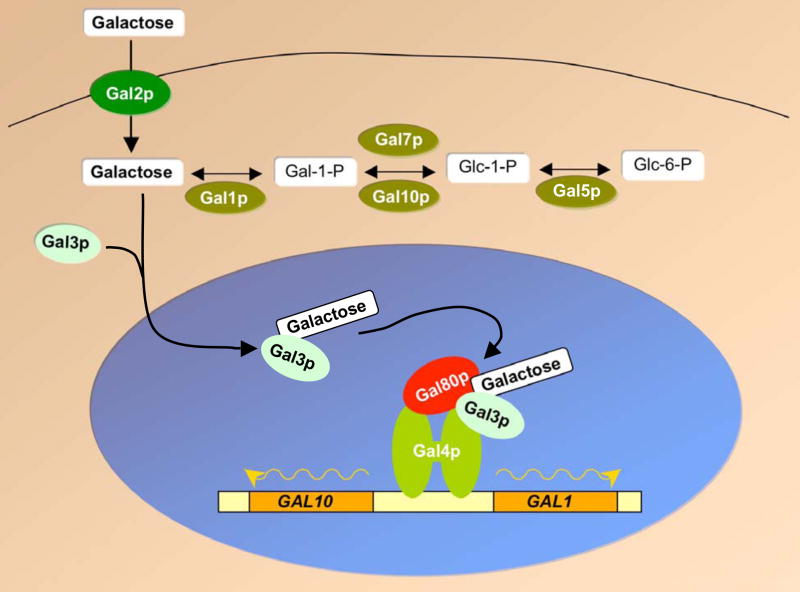
The GAL genes in budding yeast encode enzymes of the Leloir pathway which is activated by galactose and expresses proteins to internalize and metabolize this sugar. GAL genes can be broadly separated into two groups – the structural genes (GAL1, GAL5, GAL7, GAL10), which encode enzymes to metabolize galactose, and regulatory genes (GAL2, GAL3, GAL4, GAL80) that transport galactose and control expression of the structural genes. Expression of Gal1p (galactokinase), Gal7p (galactose-1-phosphate uridyl transferase) and Gal10p (uridine diphosphoglucose epimerase) are tightly regulated in the presence of different sugars and they are induced at least 1000-fold in the presence of galactose [260–263].
All GAL genes are activated by Gal4p, a zinc finger DNA binding protein that binds to target sequences upstream of each GAL gene. GAL1 and GAL10 are divergently transcribed on chromosome II, and their common intergenic regulatory region contains an upstream activating sequence (UASG) that contains 4 Gal4p binding sites (Figure 2)[261, 264]. When yeast cells are grown in glucose media, transcription from GAL1 and GAL10 is repressed by three mechanisms [262, 265]. Firstly, expression of the Gal4p activator is repressed in glucose media, limiting the amount of Gal4p that is bound to UASG. Secondly, the small amount of Gal4p that is bound to the UASG is inactivated by the Gal80p repressor which binds and masks the C-terminal activation domain of Gal4p. And thirdly, glucose-responsive repressor proteins Mig1p, Nrg1p and Nrg2p bind to UASG sequences located between UASG and the GAL1 or GAL10 promoter regions, and they actively repress transcription [266–269]. When cells are grown in ‘neutral’ carbon sources, such as raffinose or glycerol, glucose-repression mechanisms are alleviated, and thus Gal4p fully occupies its sites within UASG [261]. However, the Gal80p repressor remains bound to Gal4p, preventing transcriptional activation [261].
Figure 2. Schematic representation of activation of the GAL1-10 locus.
Transcriptional activation of GAL genes requires the presence of galactose sugar. Extracellular galactose is transported to the cytoplasm by the Gal2p permease. Intracellular galactose binds the Gal3p co-inducer protein and this complex inactivates Gal80p repressor. Inactivation of Gal80p leads to activation of the Gal4p activator. Gal4p is a transcription factor that binds to DNA sites upstream of the structural genes, like GAL1 and GAL10 and promotes their transcription (wavy arrows). GAL1 encodes galactokinase that converts galactose to galactose-1-phosphate. Other enzymes of the GAL regulon (Gal7p, Gal10p, Gal5p) further act on this substrate to metabolize galactose for energy production. See text for details.
When galactose is added to cells, it is transported to the cytoplasm by the Gal2p membrane-bound permease. Cytoplasmic galactose is signaled by the co-inducer protein, Gal3p, which binds galactose and translocates to the nucleus [270]. The Gal3p-galactose complex is believed to bind and inactivate the Gal80p repressor, which allows Gal4p to recruit the transcriptional machinery and chromatin remodeling and modification enzymes such as SWI/SNF and SAGA, respectively (Figure 2) [271–276]. Interestingly, transcriptional induction of GAL1 leads to translocation of the locus to the nuclear pore, a phenomenon that is believed to aid in the efficient export of mRNA into the cytoplasm [277–281].
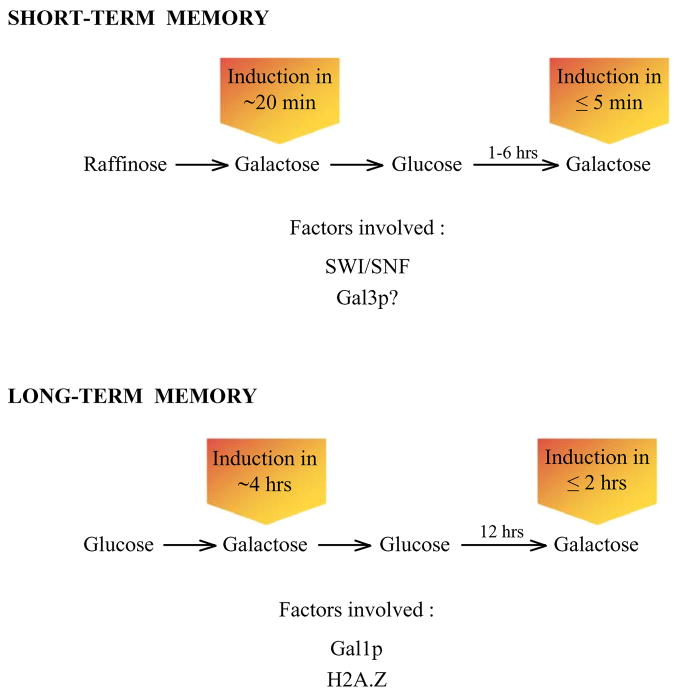
Brickner and colleagues described the first example of transcriptional memory involving the yeast GAL genes (see Figure 3) [257]. In their studies, cells were initially grown in glucose media and then shifted to galactose. In this case, transcriptional induction of GAL1 was quite slow (3–4 hours), as cells must eliminate glucose repression and synthesize Gal4p. Cells were then treated a second time with glucose, which repressed GAL1 expression, and following 12 hours (~6 cell divisions) of glucose growth, cells were once again switched to galactose medium. Remarkable, GAL1 induction was more rapid than the initial round of expression, with peak expression occurring in less than 2 hours. This phenomenon of rapid re-induction kinetics represents a form of transcriptional memory as the cells have “remembered” their previous exposure to galactose. Furthermore, the memory observed in this system appears to be a relatively long-term phenomenon as it is stable to at least 6 cell divisions.
Figure 3. Schematic to summarize the two forms of transcriptional memory observed at the GAL gene cluster.
Two different experimental regimens that lead to memory are shown. ‘Short-term’ and ‘long-term’ forms of memory differ in the required factors. See text for details.
To eliminate possible complications due to long term glucose repression, we performed similar “memory” experiments, but in our case naïve cells were propagated in media containing the neutral sugar, raffinose, prior to the transfer to galactose. In this regimen (Figure 3), the first round of GAL1 induction required only 20 minutes of galactose exposure for maximal expression. Cells were then treated with glucose for 1–6 hours to repress GAL1 expression, and then cells were transferred back to galactose medium. In this case, re-induction of GAL1 was almost instantaneous, with peak levels of expression occurring within 5 minutes of galactose expression [256]. But in contrast to the relatively long-term memory phenomenon described above, this memory was short-lived, with the rapid re-induction kinetics being lost when cultures were propagated for longer than three cell divisions (6 hours) in glucose media.
Interestingly, different factors have been implicated for ‘short-term’ and ‘long-term’ memory. Studies by Brickner and colleagues [257] suggest that formation and inheritance of the ‘long-term’ memory state requires the histone variant, H2A.Z, and memory correlated with the association of the GAL1 locus with the nuclear pore. The molecular role of H2A.Z is not clear, although Tzamarias and colleagues have suggested that it may also play in a role in the first round of expression by facilitating recruitment of the mediator complex [255]. It seems unlikely that the nuclear pore has a direct role in cellular memory, as a recent study indicates that parental nuclear pores are retained within the mother cell, while the daughter cell always receives newly synthesized and assembled pore complexes [282]. Thus, there does not appear to be a mechanism to faithfully propagate a GAL1-pore complex to daughter cells. In contrast to the long-term memory phenomenon, short-term memory does not require histone H2A.Z, post-translational histone modifications, or components of the nuclear pore, but instead requires the ATP-dependent chromatin remodeling enzyme, SWI/SNF [256 and our unpublished results].
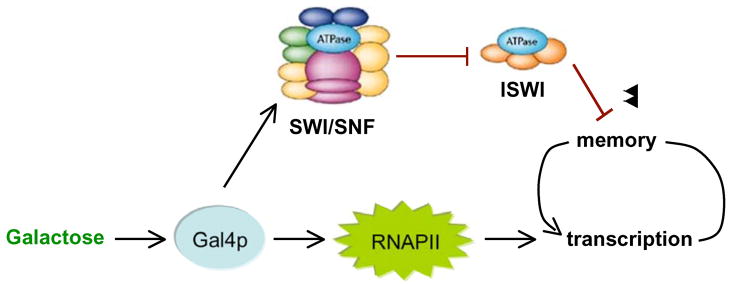
How could a chromatin remodeling enzyme promote transcriptional memory? One model proposes that SWI/SNF may influence the positioning of nucleosomes that are re-assembled at the promoter when the activated GAL1 gene is repressed by glucose. In this case, promoter elements might be located between positioned nucleosomes, poising the gene for a subsequent activation event. However, SWI/SNF is rapidly displaced from the GAL1 gene during glucose repression [256, 283], and thus the proposed nucleosome positions would need to be re-established after each round of DNA replication even in the absence of SWI/SNF. Interestingly, genetic analysis revealed that inactivation of either of the two ISWI complexes, ISW1 or ISW2, could rescue the defect in short-term transcriptional memory seen in the absence of SWI/SNF (Figure 4) [256]. This result seems to support a nucleosome positioning model, given that ISWI complexes typically function by establishing repressive nucleosome positions at RNAPII promoters [284].
Figure 4. Model for establishment of transcriptional memory through chromatin remodeling by SWI/SNF.
Intracellular galactose activates the Gal4p activator, which recruits RNA Polymerase II and other components of the transcriptional machinery. This leads to transcription of GAL1, GAL7 and GAL10 genes. Transcription leads to formation of memory, which can be inhibited by ISWI complexes. Activated Gal4p also recruits SWI/SNF, which antagonizes the repressive function of ISWI complexes at these loci, thereby contributing to establishment of transcriptional memory.
Like complex genetic networks of higher eukaryotes, yeast genetic networks are also controlled at different steps by feedback or feedforward loops. Such loops increase the responsiveness of the network to subtle environmental changes and reduce stochastic noise or cell heterogeneity in gene activity [285–287]. The GAL transcriptional network also incorporates signaling loops – positive signaling via Gal3p and negative signaling via Gal80p that can together enhance the robustness of the response of the population to galactose [288–291]. Thus, one simple model that might explain transcriptional memory of GAL genes posits that the initial induction of GAL genes alters the concentration of positive signaling molecules such that a second round of induction occurs with faster kinetics. Since memory is a heritable event, the increased concentration of signaling molecules would need to be inherited by daughter cells, presumably as a cytoplasmic component (Figure 1). Indeed, expression of the positive signaling factor, Gal3p, is induced 3–5 fold by growth in galactose. Likewise, the expression of Gal1p is induced >1000-fold by galactose. Gal1p and Gal3p are very closely related proteins that are believed to have diverged from a common ancestor. Gal1p can bind to galactose, and at high cellular concentrations, Gal1p can function as a galactose coinducer [292–295]. Interestingly, long-term memory at GAL genes requires Gal1p, but not Gal3p [255].
If transcriptional memory involves the cytoplasmic inheritance of the Gal1p signaling molecule, why is SWI/SNF required for short term memory? We favor a model in which the kinetics of GAL gene expression is influenced by two different rate determining steps. In the first round of expression, the kinetics of induction are determined by the rate at which galactose is sensed and an optimal level of signaling complex is generated (Figure 3). This step is quite slow compared to other steps of the transcription process, such as chromatin remodeling. In contrast, the signaling step is quite rapid during a second round of expression due to the high concentration of cytoplasmically inherited Gal3p and Gal1p. In this case, SWI/SNF-dependent chromatin remodeling at the promoter becomes the rate-limiting step for GAL induction. Thus, in this model, SWI/SNF does not play a direct role in transcriptional memory. A similar model may also explain why the histone variant H2A.Z influences long-term transcriptional memory.
Two questions arise – firstly, if Gal1p functions as a memory factor to robustly signal the presence of galactose in the environment, then why doesn’t the bona fide co-inducer, Gal3p, also provide long term memory? Secondly, do Gal1p or Gal3p also contribute to ‘short-term’ memory? As noted above, Gal3p levels only increase 3- to 5-fold when cells are switched to galactose media. Gal3p has a half-life of ~4 hours (2 cell divisions), so the increased levels of Gal3p may only survive for a few cell divisions. Thus, Gal3p may not facilitate long-term memory because the increased levels are too rapidly diluted. One would anticipate that Gal3p will contribute to short term memory, as the higher levels of Gal3p will still be available for the second round of induction. However, it is likely that Gal1p and Gal3p will function redundantly as “memory factors” since either factor should be able to enhance galactose signaling. Together, these results indicate that both short-term and long-term memory of GAL gene transcription is likely to be transmitted to future generations by the cytoplasmic distribution of signaling molecules. Whether chromatin-based mechanisms may also contribute to transcriptional memory at other loci in yeast remains to be elucidated.
Biography


Craig L. Peterson is a tenured Professor and Vice-Chair in the Program in Molecular Medicine at the University of Massachusetts Medical School. He received his B.S. in Molecular Biology from the University of Washington in 1983 and his Ph.D. in Molecular Biology from the University of California, Los Angeles in 1988. As a Helen Hay Whitney Foundation postdoctoral fellow with the late Ira Herskowitz at the University of California, San Francisco, Dr. Peterson initiated genetic studies that ultimately led to the discovery of the first chromatin remodeling enzyme, yeast SWI/SNF. He joined the faculty of the Program in Molecular Medicine in 1992 where he has pioneered genetic and biochemical analyses of chromatin remodeling enzymes, chromatin higher order folding, and the role of chromatin dynamics in transcription, DNA repair, and DNA replication. In addition to serving as an Executive Editor of BBA Gene Regulatory Mechanisms, he is also on the editorial boards of Molecular Cell, Current Biology, PLoS Biology, and Molecular and Cellular Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4. Garland Science; New York, NY: 2002. [Google Scholar]
- 2.Surani MA, Hayashi K, Hajkova P. Cell. 2007;128:747–62. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Gustafson T, Wolpert L. Int Rev Cytol. 1963;15:139–214. doi: 10.1016/s0074-7696(08)61117-1. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert SF. Developmental Biology. 6. Sinauer; Sunderland, MA: 2000. [Google Scholar]
- 5.Johnson BV, Rathjen J, Rathjen PD. Curr Opin Genet Dev. 2006;16:447–54. doi: 10.1016/j.gde.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Boyer LA, Mathur D, Jaenisch R. Curr Opin Genet Dev. 2006;16:455–62. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Reik W. Nature. 2007;447:425–32. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 8.Simon J. Curr Opin Cell Biol. 1995;7:376–85. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 9.Hagstrom K, Schedl P. Curr Opin Genet Dev. 1997;7:814–21. doi: 10.1016/s0959-437x(97)80045-7. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs JJ, van Lohuizen M. Semin Cell Dev Biol. 1999;10:227–35. doi: 10.1006/scdb.1999.0304. [DOI] [PubMed] [Google Scholar]
- 11.Francis NJ, Kingston RE. Nat Rev Mol Cell Biol. 2001;2:409–21. doi: 10.1038/35073039. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs JJ, van Lohuizen M. Biochim Biophys Acta. 2002;1602:151–61. doi: 10.1016/s0304-419x(02)00052-5. [DOI] [PubMed] [Google Scholar]
- 13.Fisher AG. Nat Rev Immunol. 2002;2:977–82. doi: 10.1038/nri958. [DOI] [PubMed] [Google Scholar]
- 14.Brock HW, Fisher CL. Dev Dyn. 2005;232:633–55. doi: 10.1002/dvdy.20298. [DOI] [PubMed] [Google Scholar]
- 15.Bantignies F, Cavalli G. Curr Opin Cell Biol. 2006;18:275–83. doi: 10.1016/j.ceb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Brown K. Crit Rev Eukaryot Gene Expr. 1999;9:203–12. doi: 10.1615/critreveukargeneexpr.v9.i3-4.50. [DOI] [PubMed] [Google Scholar]
- 17.Tuite MF, Lindquist SL. Trends Genet. 1996;12:467–71. doi: 10.1016/0168-9525(96)10045-7. [DOI] [PubMed] [Google Scholar]
- 18.Shoubridge EA. Hum Reprod. 2000;15(Suppl 2):229–34. doi: 10.1093/humrep/15.suppl_2.229. [DOI] [PubMed] [Google Scholar]
- 19.Ringrose L, Paro R. Annu Rev Genet. 2004;38:413–43. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 20.Austin KD, Hall JG. Pediatr Clin North Am. 1992;39:335–48. doi: 10.1016/s0031-3955(16)38298-0. [DOI] [PubMed] [Google Scholar]
- 21.Ng RK, Gurdon JB. Cell Cycle. 2008;7:1173–7. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- 22.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 23.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. J Mol Biol. 2002;319:1097–113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 24.Luger K, Richmond TJ. Curr Opin Struct Biol. 1998;8:33–40. doi: 10.1016/s0959-440x(98)80007-9. [DOI] [PubMed] [Google Scholar]
- 25.Suto RK, Edayathumangalam RS, White CL, Melander C, Gottesfeld JM, Dervan PB, Luger K. J Mol Biol. 2003;326:371–80. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]
- 26.Muthurajan UM, Park YJ, Edayathumangalam RS, Suto RK, Chakravarthy S, Dyer PN, Luger K. Biopolymers. 2003;68:547–56. doi: 10.1002/bip.10317. [DOI] [PubMed] [Google Scholar]
- 27.Luger K. Curr Opin Genet Dev. 2003;13:127–35. doi: 10.1016/s0959-437x(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 28.Workman JL, Kingston RE. Annu Rev Biochem. 1998;67:545–79. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 29.Hansen JC, Wolffe AP. Proc Natl Acad Sci U S A. 1994;91:2339–43. doi: 10.1073/pnas.91.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greil W, Igo-Kemenes T, Zachau HG. Nucleic Acids Res. 1976;3:2633–44. doi: 10.1093/nar/3.10.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sollner-Webb B, Melchior W, Jr, Felsenfeld G. Cell. 1978;14:611–27. doi: 10.1016/0092-8674(78)90246-5. [DOI] [PubMed] [Google Scholar]
- 32.Hansen JC. Annu Rev Biophys Biomol Struct. 2002;31:361–92. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 33.Horowitz-Scherer RA, Woodcock CL. Chromosoma. 2006;115:1–14. doi: 10.1007/s00412-005-0035-3. [DOI] [PubMed] [Google Scholar]
- 34.Horn PJ, Peterson CL. Science. 2002;297:1824–7. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 35.Alami R, Fan Y, Pack S, Sonbuchner TM, Besse A, Lin Q, Greally JM, Skoultchi AI, Bouhassira EE. Proc Natl Acad Sci U S A. 2003;100:5920–5. doi: 10.1073/pnas.0736105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Izzo A, Kamieniarz K, Schneider R. Biol Chem. 2008;389:333–43. doi: 10.1515/BC.2008.037. [DOI] [PubMed] [Google Scholar]
- 37.Sancho M, Diani E, Beato M, Jordan A. PLoS Genet. 2008;4:e1000227. doi: 10.1371/journal.pgen.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parseghian MH, Hamkalo BA. Biochem Cell Biol. 2001;79:289–304. [PubMed] [Google Scholar]
- 39.Hamiche A, Schultz P, Ramakrishnan V, Oudet P, Prunell A. J Mol Biol. 1996;257:30–42. doi: 10.1006/jmbi.1996.0144. [DOI] [PubMed] [Google Scholar]
- 40.Woodcock CL, Skoultchi AI, Fan Y. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 41.Zlatanova J, Caiafa P, Van Holde K. Faseb J. 2000;14:1697–704. doi: 10.1096/fj.99-0869rev. [DOI] [PubMed] [Google Scholar]
- 42.Zlatanova J, Seebart C, Tomschik M. Trends Biochem Sci. 2008;33:247–53. doi: 10.1016/j.tibs.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Thomas JO, Rees C, Finch JT. Nucleic Acids Res. 1992;20:187–94. doi: 10.1093/nar/20.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talasz H, Helliger W, Puschendorf B, Lindner H. Biochemistry. 1996;35:1761–7. doi: 10.1021/bi951914e. [DOI] [PubMed] [Google Scholar]
- 45.Wisniewski JR, Zougman A, Kruger S, Mann M. Mol Cell Proteomics. 2007;6:72–87. doi: 10.1074/mcp.M600255-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Dou Y, Mizzen CA, Abrams M, Allis CD, Gorovsky MA. Mol Cell. 1999;4:641–7. doi: 10.1016/s1097-2765(00)80215-4. [DOI] [PubMed] [Google Scholar]
- 47.Draves PH, Lowary PT, Widom J. J Mol Biol. 1992;225:1105–21. doi: 10.1016/0022-2836(92)90108-v. [DOI] [PubMed] [Google Scholar]
- 48.Bharath MM, Ramesh S, Chandra NR, Rao MR. Biochemistry. 2002;41:7617–27. doi: 10.1021/bi025773+. [DOI] [PubMed] [Google Scholar]
- 49.Ramakrishnan V. Crit Rev Eukaryot Gene Expr. 1997;7:215–30. doi: 10.1615/critreveukargeneexpr.v7.i3.20. [DOI] [PubMed] [Google Scholar]
- 50.Lu X, Hansen JC. J Biol Chem. 2004;279:8701–7. doi: 10.1074/jbc.M311348200. [DOI] [PubMed] [Google Scholar]
- 51.Vermaak D, Steinbach OC, Dimitrov S, Rupp RA, Wolffe AP. Curr Biol. 1998;8:533–6. doi: 10.1016/s0960-9822(98)70206-4. [DOI] [PubMed] [Google Scholar]
- 52.Villar-Garea A, Imhof A. PLoS ONE. 2008;3:e1553. doi: 10.1371/journal.pone.0001553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hale TK, Contreras A, Morrison AJ, Herrera RE. Mol Cell. 2006;22:693–9. doi: 10.1016/j.molcel.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 54.Garcia BA, Busby SA, Barber CM, Shabanowitz J, Allis CD, Hunt DF. J Proteome Res. 2004;3:1219–27. doi: 10.1021/pr0498887. [DOI] [PubMed] [Google Scholar]
- 55.Ushinsky SC, Bussey H, Ahmed AA, Wang Y, Friesen J, Williams BA, Storms RK. Yeast. 1997;13:151–61. doi: 10.1002/(SICI)1097-0061(199702)13:2<151::AID-YEA94>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 56.Shen X, Yu L, Weir JW, Gorovsky MA. Cell. 1995;82:47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 57.Steinbach OC, Wolffe AP, Rupp RA. Nature. 1997;389:395–9. doi: 10.1038/38755. [DOI] [PubMed] [Google Scholar]
- 58.Ramon A, Muro-Pastor MI, Scazzocchio C, Gonzalez R. Mol Microbiol. 2000;35:223–33. doi: 10.1046/j.1365-2958.2000.01702.x. [DOI] [PubMed] [Google Scholar]
- 59.Patterton HG, Landel CC, Landsman D, Peterson CL, Simpson RT. J Biol Chem. 1998;273:7268–76. doi: 10.1074/jbc.273.13.7268. [DOI] [PubMed] [Google Scholar]
- 60.Ner SS, Blank T, Perez-Paralle ML, Grigliatti TA, Becker PB, Travers AA. J Biol Chem. 2001;276:37569–76. doi: 10.1074/jbc.M105635200. [DOI] [PubMed] [Google Scholar]
- 61.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Cell. 2005;123:1199–212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 62.Bouvet P, Dimitrov S, Wolffe AP. Genes Dev. 1994;8:1147–59. doi: 10.1101/gad.8.10.1147. [DOI] [PubMed] [Google Scholar]
- 63.Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. Mol Cell Biol. 2003;23:4559–72. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kandolf H. Proc Natl Acad Sci U S A. 1994;91:7257–61. doi: 10.1073/pnas.91.15.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patterton D, Wolffe AP. Dev Biol. 1996;173:2–13. doi: 10.1006/dbio.1996.0002. [DOI] [PubMed] [Google Scholar]
- 66.Clarke HJ, McLay DW, Mohamed OA. Dev Genet. 1998;22:17–30. doi: 10.1002/(SICI)1520-6408(1998)22:1<17::AID-DVG3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 67.Smith CL, Peterson CL. Curr Top Dev Biol. 2005;65:115–48. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 68.Hogan C, Varga-Weisz P. Mutat Res. 2007;618:41–51. doi: 10.1016/j.mrfmmm.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 69.Peterson CL, Herskowitz I. Cell. 1992;68:573–83. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 70.Peterson CL, Dingwall A, Scott MP. Proc Natl Acad Sci U S A. 1994;91:2905–8. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nature. 1994;370:477–81. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 72.Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. Cell. 1996;87:1249–60. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 73.Krebs JE, Fry CJ, Samuels ML, Peterson CL. Cell. 2000;102:587–98. doi: 10.1016/s0092-8674(00)00081-7. [DOI] [PubMed] [Google Scholar]
- 74.Pollard KJ, Peterson CL. Mol Cell Biol. 1997;17:6212–22. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Winston F, Carlson M. Trends Genet. 1992;8:387–91. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 76.Dhasarathy A, Kladde MP. Mol Cell Biol. 2005;25:2698–707. doi: 10.1128/MCB.25.7.2698-2707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Adkins MW, Williams SK, Linger J, Tyler JK. Mol Cell Biol. 2007;27:6372–82. doi: 10.1128/MCB.00981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Imbalzano AN, Kwon H, Green MR, Kingston RE. Nature. 1994;370:481–5. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 79.Elfring LK, Deuring R, McCallum CM, Peterson CL, Tamkun JW. Mol Cell Biol. 1994;14:2225–34. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM. Genes Dev. 2000;14:2441–51. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Genes Dev. 1996;10:2117–30. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 82.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. Mol Cell. 2000;6:1287–95. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 83.Guidi CJ, Sands AT, Zambrowicz BP, Turner TK, Demers DA, Webster W, Smith TW, Imbalzano AN, Jones SN. Mol Cell Biol. 2001;21:3598–603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Nature. 2004;432:107–12. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 85.Olave I, Wang W, Xue Y, Kuo A, Crabtree GR. Genes Dev. 2002;16:2509–17. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Langst G, Becker PB. J Cell Sci. 2001;114:2561–8. doi: 10.1242/jcs.114.14.2561. [DOI] [PubMed] [Google Scholar]
- 87.Varga-Weisz P. Oncogene. 2001;20:3076–85. doi: 10.1038/sj.onc.1204332. [DOI] [PubMed] [Google Scholar]
- 88.Guschin D, Wade PA, Kikyo N, Wolffe AP. Biochemistry. 2000;39:5238–45. doi: 10.1021/bi000421t. [DOI] [PubMed] [Google Scholar]
- 89.Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K. Immunity. 2004;20:719–33. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 90.Stopka T, Skoultchi AI. Proc Natl Acad Sci U S A. 2003;100:14097–102. doi: 10.1073/pnas.2336105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Korber P, Horz W. Cell. 2004;117:5–7. doi: 10.1016/s0092-8674(04)00296-x. [DOI] [PubMed] [Google Scholar]
- 92.Bao Y, Shen X. Mutat Res. 2007;618:18–29. doi: 10.1016/j.mrfmmm.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. Science. 2004;303:343–8. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 94.Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, Richards DP, Wu X, Emili A, Hughes TR, Buratowski S, Greenblatt JF. Mol Cell. 2003;12:1565–76. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 95.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ebbert R, Birkmann A, Schuller HJ. Mol Microbiol. 1999;32:741–51. doi: 10.1046/j.1365-2958.1999.01390.x. [DOI] [PubMed] [Google Scholar]
- 97.Shen X, Mizuguchi G, Hamiche A, Wu C. Nature. 2000;406:541–4. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 98.Berger SL. Curr Opin Genet Dev. 2002;12:142–8. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 99.Couture JF, Trievel RC. Curr Opin Struct Biol. 2006;16:753–60. doi: 10.1016/j.sbi.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 100.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 101.Kouzarides T. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 102.Lan F, Nottke AC, Shi Y. Curr Opin Cell Biol. 2008;20:316–25. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shahbazian MD, Grunstein M. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 104.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Nat Rev Mol Cell Biol. 2007;8:983–94. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peterson CL, Logie C. J Cell Biochem. 2000;78:179–85. doi: 10.1002/(sici)1097-4644(20000801)78:2<179::aid-jcb1>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 106.Kingston RE, Narlikar GJ. Genes Dev. 1999;13:2339–52. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 107.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Nat Genet. 1999;23:62–6. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 108.Pollard KJ, Peterson CL. Bioessays. 1998;20:771–80. doi: 10.1002/(SICI)1521-1878(199809)20:9<771::AID-BIES10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 109.Goll MG, Bestor TH. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 110.Jaenisch R, Bird A. Nat Genet. 2003;33(Suppl):245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 111.Rountree MR, Bachman KE, Herman JG, Baylin SB. Oncogene. 2001;20:3156–65. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- 112.Riddle NC, Elgin SC. Curr Top Microbiol Immunol. 2008;320:185–209. doi: 10.1007/978-3-540-75157-1_9. [DOI] [PubMed] [Google Scholar]
- 113.White SA, Allshire RC. Curr Top Microbiol Immunol. 2008;320:157–83. doi: 10.1007/978-3-540-75157-1_8. [DOI] [PubMed] [Google Scholar]
- 114.Grewal SI, Elgin SC. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Klose RJ, Bird AP. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 116.Bird AP, Wolffe AP. Cell. 1999;99:451–4. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 117.Bestor TH. Hum Mol Genet. 2000;9:2395–402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 118.Pradhan S, Esteve PO. Biochemistry. 2003;42:5321–32. doi: 10.1021/bi034160+. [DOI] [PubMed] [Google Scholar]
- 119.Reik W, Dean W, Walter J. Science. 2001;293:1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 120.Li E, Bestor TH, Jaenisch R. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 121.Okano M, Bell DW, Haber DA, Li E. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 122.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. J Biol Chem. 2004;279:27816–23. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 123.Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Science. 2001;294:2536–9. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 124.Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. Cell. 2001;104:829–38. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- 125.Bird A. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 126.Li E. Nat Rev Genet. 2002;3:662–73. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 127.Yoder JA, Walsh CP, Bestor TH. Trends Genet. 1997;13:335–40. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 128.Walsh CP, Chaillet JR, Bestor TH. Nat Genet. 1998;20:116–7. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 129.Bird AP. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- 130.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 131.Leonhardt H, Page AW, Weier HU, Bestor TH. Cell. 1992;71:865–73. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 132.Rountree MR, Bachman KE, Baylin SB. Nat Genet. 2000;25:269–77. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 133.Sarraf SA, Stancheva I. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 134.Keshet I, Lieman-Hurwitz J, Cedar H. Cell. 1986;44:535–43. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 135.Kass SU, Landsberger N, Wolffe AP. Curr Biol. 1997;7:157–65. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 136.Buschhausen G, Wittig B, Graessmann M, Graessmann A. Proc Natl Acad Sci U S A. 1987;84:1177–81. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hendrich B, Bird A. Mol Cell Biol. 1998;18:6538–47. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 139.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Nature. 1998;393:386–9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 140.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Genes Dev. 1999;13:1924–35. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Roth SY, Denu JM, Allis CD. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 142.Peterson CL, Laniel MA. Curr Biol. 2004;14:R546–51. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 143.Martin C, Zhang Y. Curr Opin Cell Biol. 2007;19:266–72. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 144.Zhang Y. Genes Dev. 2003;17:2733–40. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- 145.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 146.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 147.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Mol Cell. 2005;19:857–64. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 148.Bradbury CA, Khanim FL, Hayden R, Bunce CM, White DA, Drayson MT, Craddock C, Turner BM. Leukemia. 2005;19:1751–9. doi: 10.1038/sj.leu.2403910. [DOI] [PubMed] [Google Scholar]
- 149.Agger K, Christensen J, Cloos PA, Helin K. Curr Opin Genet Dev. 2008;18:159–68. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 150.Lee MG, Wynder C, Cooch N, Shiekhattar R. Nature. 2005;437:432–5. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 151.Millar CB, Grunstein M. Nat Rev Mol Cell Biol. 2006;7:657–66. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 152.Nightingale KP, O’Neill LP, Turner BM. Curr Opin Genet Dev. 2006;16:125–36. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 153.Ng HH, Xu RM, Zhang Y, Struhl K. J Biol Chem. 2002;277:34655–7. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 154.Margueron R, Trojer P, Reinberg D. Curr Opin Genet Dev. 2005;15:163–76. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 155.Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Nat Genet. 2004;36:1291–5. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- 156.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. J Biol Chem. 2002;277:28368–71. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 157.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 158.Attema JL, Papathanasiou P, Forsberg EC, Xu J, Smale ST, Weissman IL. Proc Natl Acad Sci U S A. 2007;104:12371–6. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jackson V. Biochemistry. 1988;27:2109–20. doi: 10.1021/bi00406a044. [DOI] [PubMed] [Google Scholar]
- 160.Jackson V. Biochemistry. 1987;26:2325–34. doi: 10.1021/bi00382a038. [DOI] [PubMed] [Google Scholar]
- 161.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Science. 2002;297:2232–7. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 162.Ruthenburg AJ, Allis CD, Wysocka J. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 163.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. Cell. 2005;121:859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 164.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 165.Gould A. Curr Opin Genet Dev. 1997;7:488–94. doi: 10.1016/s0959-437x(97)80075-5. [DOI] [PubMed] [Google Scholar]
- 166.Fienberg AA, Utset MF, Bogarad LD, Hart CP, Awgulewitsch A, Ferguson-Smith A, Fainsod A, Rabin M, Ruddle FH. Curr Top Dev Biol. 1987;23:233–56. doi: 10.1016/s0070-2153(08)60627-4. [DOI] [PubMed] [Google Scholar]
- 167.Gaunt SJ. Bioessays. 1991;13:505–13. doi: 10.1002/bies.950131004. [DOI] [PubMed] [Google Scholar]
- 168.Krumlauf R. Bioessays. 1992;14:245–52. doi: 10.1002/bies.950140408. [DOI] [PubMed] [Google Scholar]
- 169.Simon J, Chiang A, Bender W. Development. 1992;114:493–505. doi: 10.1242/dev.114.2.493. [DOI] [PubMed] [Google Scholar]
- 170.Lewis EB. Nature. 1978;276:565–70. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 171.Kaufman TC, Lewis R, Wakimoto B. Genetics. 1980;94:115–133. doi: 10.1093/genetics/94.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. Mol Cell Biol. 2002;22:6070–8. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kennison JA, Tamkun JW. Proc Natl Acad Sci U S A. 1988;85:8136–40. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Daubresse G, Deuring R, Moore L, Papoulas O, Zakrajsek I, Waldrip WR, Scott MP, Kennison JA, Tamkun JW. Development. 1999;126:1175–87. doi: 10.1242/dev.126.6.1175. [DOI] [PubMed] [Google Scholar]
- 175.Wolpert L. Dev Genet. 1994;15:485–90. doi: 10.1002/dvg.1020150607. [DOI] [PubMed] [Google Scholar]
- 176.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Cell. 2007;128:735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 177.Grimaud C, Negre N, Cavalli G. Chromosome Res. 2006;14:363–75. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- 178.Francis NJ, Kingston RE, Woodcock CL. Science. 2004;306:1574–7. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 179.Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 180.Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH. Dev Cell. 2008;15:668–79. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 181.Ringrose L, Paro R. Development. 2007;134:223–32. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 182.Beisel C, Buness A, Roustan-Espinosa IM, Koch B, Schmitt S, Haas SA, Hild M, Katsuyama T, Paro R. Proc Natl Acad Sci U S A. 2007;104:16615–20. doi: 10.1073/pnas.0701538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Levine SS, King IF, Kingston RE. Trends Biochem Sci. 2004;29:478–85. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 184.Kohler C, Villar CB. Trends Cell Biol. 2008;18:236–43. doi: 10.1016/j.tcb.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 185.King IF, Francis NJ, Kingston RE. Mol Cell Biol. 2002;22:7919–28. doi: 10.1128/MCB.22.22.7919-7928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.King IF, Emmons RB, Francis NJ, Wild B, Muller J, Kingston RE, Wu CT. Mol Cell Biol. 2005;25:6578–91. doi: 10.1128/MCB.25.15.6578-6591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Otte AP, Kwaks TH. Curr Opin Genet Dev. 2003;13:448–54. doi: 10.1016/s0959-437x(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 188.Shearn A. Genetics. 1989;121:517–25. doi: 10.1093/genetics/121.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tanaka Y, Katagiri Z, Kawahashi K, Kioussis D, Kitajima S. Gene. 2007;397:161–8. doi: 10.1016/j.gene.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 190.Beisel C, Imhof A, Greene J, Kremmer E, Sauer F. Nature. 2002;419:857–62. doi: 10.1038/nature01126. [DOI] [PubMed] [Google Scholar]
- 191.Byrd KN, Shearn A. Proc Natl Acad Sci U S A. 2003;100:11535–40. doi: 10.1073/pnas.1933593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A. Science. 2001;294:1331–4. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- 193.Schwartz YB, Pirrotta V. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 194.Tripoulas N, LaJeunesse D, Gildea J, Shearn A. Genetics. 1996;143:913–28. doi: 10.1093/genetics/143.2.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce CM, Mazo A, Canaani E. Proc Natl Acad Sci U S A. 1998;95:4152–7. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. Proc Natl Acad Sci U S A. 2002;99:90–4. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Jenuwein T, Laible G, Dorn R, Reuter G. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Boa S, Coert C, Patterton HG. Yeast. 2003;20:827–35. doi: 10.1002/yea.995. [DOI] [PubMed] [Google Scholar]
- 199.Papp B, Muller J. Genes Dev. 2006;20:2041–54. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Collins RT, Furukawa T, Tanese N, Treisman JE. Embo J. 1999;18:7029–40. doi: 10.1093/emboj/18.24.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Crosby MA, Miller C, Alon T, Watson KL, Verrijzer CP, Goldman-Levi R, Zak NB. Mol Cell Biol. 1999;19:1159–70. doi: 10.1128/mcb.19.2.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Papoulas O, Beek SJ, Moseley SL, McCallum CM, Sarte M, Shearn A, Tamkun JW. Development. 1998;125:3955–66. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 203.Henikoff S, Ahmad K. Annu Rev Cell Dev Biol. 2005;21:133–53. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- 204.Wu RS, Bonner WM. Cell. 1981;27:321–30. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- 205.Park YJ, Luger K. Curr Opin Struct Biol. 2008;18:282–9. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Malik HS, Henikoff S. Nat Struct Biol. 2003;10:882–91. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 207.Chakravarthy S, Bao Y, Roberts VA, Tremethick D, Luger K. Cold Spring Harb Symp Quant Biol. 2004;69:227–34. doi: 10.1101/sqb.2004.69.227. [DOI] [PubMed] [Google Scholar]
- 208.Chakravarthy S, Luger K. J Biol Chem. 2006;281:25522–31. doi: 10.1074/jbc.M602258200. [DOI] [PubMed] [Google Scholar]
- 209.Luk E, Vu ND, Patteson K, Mizuguchi G, Wu WH, Ranjan A, Backus J, Sen S, Lewis M, Bai Y, Wu C. Mol Cell. 2007;25:357–68. doi: 10.1016/j.molcel.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 210.Ray-Gallet D, Quivy JP, Scamps C, Martini EM, Lipinski M, Almouzni G. Mol Cell. 2002;9:1091–100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- 211.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Raisner RM, Madhani HD. Curr Opin Genet Dev. 2006;16:119–24. doi: 10.1016/j.gde.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 213.Zhang H, Roberts DN, Cairns BR. Cell. 2005;123:219–31. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Stargell LA, Bowen J, Dadd CA, Dedon PC, Davis M, Cook RG, Allis CD, Gorovsky MA. Genes Dev. 1993;7:2641–51. doi: 10.1101/gad.7.12b.2641. [DOI] [PubMed] [Google Scholar]
- 215.Meneghini MD, Wu M, Madhani HD. Cell. 2003;112:725–36. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 216.Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausio J. J Biol Chem. 2001;276:41945–9. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- 217.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Nat Struct Biol. 2000;7:1121–4. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 218.Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ. Nat Struct Biol. 2002;9:172–6. doi: 10.1038/nsb767. [DOI] [PubMed] [Google Scholar]
- 219.Park YJ, Dyer PN, Tremethick DJ, Luger K. J Biol Chem. 2004;279:24274–82. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 220.Santisteban MS, Kalashnikova T, Smith MM. Cell. 2000;103:411–22. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 221.Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JR, Taylor H, Matthaei K, Rathjen PD, Tremethick DJ, Lyons I. Curr Biol. 2001;11:1183–7. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- 222.Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Nature. 2008;452:877–81. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Chadwick BP, Willard HF. J Cell Biol. 2001;152:375–84. doi: 10.1083/jcb.152.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bao Y, Konesky K, Park YJ, Rosu S, Dyer PN, Rangasamy D, Tremethick DJ, Laybourn PJ, Luger K. Embo J. 2004;23:3314–24. doi: 10.1038/sj.emboj.7600316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gautier T, Abbott DW, Molla A, Verdel A, Ausio J, Dimitrov S. EMBO Rep. 2004;5:715–20. doi: 10.1038/sj.embor.7400182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Gonzalez-Romero R, Mendez J, Ausio J, Eirin-Lopez JM. Gene. 2008;413:1–7. doi: 10.1016/j.gene.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 227.Costanzi C, Pehrson JR. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 228.Smith MM. Curr Opin Cell Biol. 2002;14:279–85. doi: 10.1016/s0955-0674(02)00331-9. [DOI] [PubMed] [Google Scholar]
- 229.Schwartz BE, Ahmad K. Genes Dev. 2005;19:804–14. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wirbelauer C, Bell O, Schubeler D. Genes Dev. 2005;19:1761–6. doi: 10.1101/gad.347705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Proc Natl Acad Sci U S A. 2004;101:1525–30. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Daury L, Chailleux C, Bonvallet J, Trouche D. EMBO Rep. 2006;7:66–71. doi: 10.1038/sj.embor.7400561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Chow CM, Georgiou A, Szutorisz H, Maia e Silva A, Pombo A, Barahona I, Dargelos E, Canzonetta C, Dillon N. EMBO Rep. 2005;6:354–60. doi: 10.1038/sj.embor.7400366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.van der Heijden GW, Derijck AA, Posfai E, Giele M, Pelczar P, Ramos L, Wansink DG, van der Vlag J, Peters AH, de Boer P. Nat Genet. 2007;39:251–8. doi: 10.1038/ng1949. [DOI] [PubMed] [Google Scholar]
- 235.van der Heijden GW, Dieker JW, Derijck AA, Muller S, Berden JH, Braat DD, van der Vlag J, de Boer P. Mech Dev. 2005;122:1008–22. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 236.Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ. Mol Cell Biol. 2002;22:2318–28. doi: 10.1128/MCB.22.7.2318-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P. Nature. 2005;437:1386–90. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- 238.Annunziato AT. J Biol Chem. 2005;280:12065–8. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- 239.Hake SB, Allis CD. Proc Natl Acad Sci U S A. 2006;103:6428–35. doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Henikoff S, Furuyama T, Ahmad K. Trends Genet. 2004;20:320–6. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 241.Chaumeil J, Le Baccon P, Wutz A, Heard E. Genes Dev. 2006;20:2223–37. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wutz A, Jaenisch R. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 243.Masui O, Heard E. Cold Spring Harb Symp Quant Biol. 2006;71:419–28. doi: 10.1101/sqb.2006.71.058. [DOI] [PubMed] [Google Scholar]
- 244.Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Nat Genet. 1999;22:323–4. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- 246.Moazed D, Buhler M, Buker SM, Colmenares SU, Gerace EL, Gerber SA, Hong EJ, Motamedi MR, Verdel A, Villen J, Gygi SP. Cold Spring Harb Symp Quant Biol. 2006;71:461–71. doi: 10.1101/sqb.2006.71.044. [DOI] [PubMed] [Google Scholar]
- 247.Zofall M, Grewal SI. Cold Spring Harb Symp Quant Biol. 2006;71:487–96. doi: 10.1101/sqb.2006.71.059. [DOI] [PubMed] [Google Scholar]
- 248.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 249.Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Science. 2006;313:1134–7. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- 250.Kloc A, Zaratiegui M, Nora E, Martienssen R. Curr Biol. 2008;18:490–5. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Nature. 2008;451:734–7. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 252.Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, Gerstein M, Yi S, Snyder M, Soll DR. Eukaryot Cell. 2006;5:1674–87. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zordan RE, Galgoczy DJ, Johnson AD. Proc Natl Acad Sci U S A. 2006;103:12807–12. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zacharioudakis I, Gligoris T, Tzamarias D. Curr Biol. 2007;17:2041–6. doi: 10.1016/j.cub.2007.10.044. [DOI] [PubMed] [Google Scholar]
- 256.Kundu S, Horn PJ, Peterson CL. Genes Dev. 2007;21:997–1004. doi: 10.1101/gad.1506607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Klar AJ, Srikantha T, Soll DR. Genetics. 2001;158:919–24. doi: 10.1093/genetics/158.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Srikantha T, Tsai L, Daniels K, Klar AJ, Soll DR. J Bacteriol. 2001;183:4614–25. doi: 10.1128/JB.183.15.4614-4625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Johnston M. Microbiol Rev. 1987;51:458–76. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lohr D, Venkov P, Zlatanova J. Faseb J. 1995;9:777–87. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 262.Johnston M, Flick JS, Pexton T. Mol Cell Biol. 1994;14:3834–41. doi: 10.1128/mcb.14.6.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Bhat PJ, Murthy TV. Mol Microbiol. 2001;40:1059–66. doi: 10.1046/j.1365-2958.2001.02421.x. [DOI] [PubMed] [Google Scholar]
- 264.Lohr D. Nucleic Acids Res. 1984;12:8457–74. doi: 10.1093/nar/12.22.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Griggs DW, Johnston M. Proc Natl Acad Sci U S A. 1991;88:8597–601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Frolova E, Johnston M, Majors J. Nucleic Acids Res. 1999;27:1350–8. doi: 10.1093/nar/27.5.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Treitel MA, Carlson M. Proc Natl Acad Sci U S A. 1995;92:3132–6. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Wu J, Trumbly RJ. Yeast. 1998;14:985–1000. doi: 10.1002/(SICI)1097-0061(199808)14:11<985::AID-YEA294>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 269.Zhou H, Winston F. BMC Genet. 2001;2:5. doi: 10.1186/1471-2156-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Wightman R, Bell R, Reece RJ. Eukaryot Cell. 2008 doi: 10.1128/EC.00261-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Bhaumik SR, Green MR. Genes Dev. 2001;15:1935–45. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Bhaumik SR, Raha T, Aiello DP, Green MR. Genes Dev. 2004;18:333–43. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Brandl CJ, Furlanetto AM, Martens JA, Hamilton KS. Embo J. 1993;12:5255–65. doi: 10.1002/j.1460-2075.1993.tb06221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Larschan E, Winston F. Genes Dev. 2001;15:1946–56. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Peng G, Hopper JE. Proc Natl Acad Sci U S A. 2002;99:8548–53. doi: 10.1073/pnas.142100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Platt A, Reece RJ. Embo J. 1998;17:4086–91. doi: 10.1093/emboj/17.14.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nature. 2006;441:774–8. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 278.Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Genes Dev. 2005;19:1188–98. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Cell. 2004;117:427–39. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 280.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, Nehrbass U. Nature. 2006;441:770–3. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 281.Abruzzi KC, Belostotsky DA, Chekanova JA, Dower K, Rosbash M. Embo J. 2006;25:4253–62. doi: 10.1038/sj.emboj.7601305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. Nature. 2008;454:728–34. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 283.Ng HH, Robert F, Young RA, Struhl K. Mol Cell. 2003;11:709–19. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 284.Mellor J, Morillon A. Biochim Biophys Acta. 2004;1677:100–12. doi: 10.1016/j.bbaexp.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 285.Kaern M, Elston TC, Blake WJ, Collins JJ. Nat Rev Genet. 2005;6:451–64. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 286.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 287.Stelling J, Sauer U, Szallasi Z, Doyle FJ, 3rd, Doyle J. Cell. 2004;118:675–85. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 288.Ronen M, Botstein D. Proc Natl Acad Sci U S A. 2006;103:389–94. doi: 10.1073/pnas.0509978103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Ramsey SA, Smith JJ, Orrell D, Marelli M, Petersen TW, de Atauri P, Bolouri H, Aitchison JD. Nat Genet. 2006;38:1082–7. doi: 10.1038/ng1869. [DOI] [PubMed] [Google Scholar]
- 290.Acar M, Becskei A, van Oudenaarden A. Nature. 2005;435:228–32. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- 291.Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EP, Landgraf D, Phillips I, Silver PA. Genes Dev. 2007;21:2271–6. doi: 10.1101/gad.1586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Hawkins KM, Smolke CD. J Biol Chem. 2006;281:13485–92. doi: 10.1074/jbc.M512317200. [DOI] [PubMed] [Google Scholar]
- 293.Hittinger CT, Carroll SB. Nature. 2007;449:677–81. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- 294.Meyer J, Walker-Jonah A, Hollenberg CP. Mol Cell Biol. 1991;11:5454–61. doi: 10.1128/mcb.11.11.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Platt A, Ross HC, Hankin S, Reece RJ. Proc Natl Acad Sci U S A. 2000;97:3154–9. doi: 10.1073/pnas.97.7.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]