Abstract
The functionally exchangeable L domains of HIV-1 and Rous sarcoma virus (RSV) Gag bind Tsg101 and Nedd4, respectively. Tsg101 and Nedd4 function in endocytic trafficking, and studies show that expression of Tsg101 or Nedd4 fragments interfere with release of HIV-1 or RSV Gag, respectively, as virus-like particles (VLPs). To determine whether functional exchangeability reflects use of the same trafficking pathway, we tested the effect on RSV Gag release of co-expression with mutated forms of Vps4, Nedd4 and Tsg101. A dominant-negative mutant of Vps4A, an AAA ATPase required for utilization of endosomal sorting proteins that was shown previously to interfere with HIV-1 budding, also inhibited RSV Gag release, indicating that RSV uses the endocytic trafficking machinery, as does HIV. Nedd4 and Tsg101 interacted in the presence or absence of Gag and, through its binding of Nedd4, RSV Gag interacted with Tsg101. Deletion of the N-terminal region of Tsg101 or the HECT domain of Nedd4 did not prevent interaction; however, three-dimensional spatial imaging suggested that the interaction of RSV Gag with full-length Tsg101 and N-terminally truncated Tsg101 was not the same. Co-expression of RSV Gag with the Tsg101 C-terminal fragment interfered with VLP release minimally; however, a significant fraction of the released VLPs was tethered to each other. The results suggest that, while Tsg101 is not required for RSV VLP release, alterations in the protein interfere with VLP budding/fission events. We conclude that RSV and HIV-1 Gag direct particle release through independent ESCRT-mediated pathways that are linked through Tsg101–Nedd4 interaction.
Keywords: Gag, HIV-1, L domain, Nedd4, RSV, Tsg101, Vps4
All retroviruses have in common three genes, gag, pol and env, which specify the structural and enzymatic functions of the virus (1). The Gag protein, encoded in gag, is sufficient for the formation and release of virus-like particles (VLPs) from Gag-expressing cells. The determinant of viral maturation and release through the plasma membrane is the late (L) domain, originally defined in Rous sarcoma virus (RSV) and subsequently shown to be functional in HIV type-1 (HIV-1) (2–5) (reviewed in 6). Although highly conserved within retroviral subgroups, the L domains in different families vary in amino acid sequence. HIV-1 has a Pro-Thr-Ala-Pro (PTAP) sequence as its L-domain core; equine infectious anemia virus (EIAV) has a Tyr-Pro-Asp-Leu sequence; RSV, Mason–Pfizer monkey virus (MPMV) and Moloney murine leukemia virus (Mo-MLV) have a Pro-Pro-Pro-Pro-Tyr (PY motif) sequence; and human T-cell leukemia virus (HTLV) and other retroviruses have both PY and PTAP motifs (4,7–12). These motifs are also found in the unrelated rhabdo-, filo- and herpes-virus groups, where they are also implicated in membrane trafficking (13–17). In addition, these motifs exist in plasma membrane- and endosomal membrane-associated cellular proteins, where they similarly serve as docking sites for cellular proteins involved in diverse functions including endocytic trafficking (18–20).
The retroviral L domains are functionally exchangeable (21–24). This observation suggests that they might direct an interaction with common cellular machinery, and recent studies support this notion. The prototypic PTAP, PY and YXXL motifs were found to interact with different cellular proteins (10,19,25–31). Tsg101, a protein involved in recognition and sorting of cellular endosomal cargo (32,33), binds to the PTAP motif in the p6 domain of the HIV-1 Gag protein (25,27,28) and is required for VLP release (27). Nedd4, an ubiquitin (Ub)-ligating (E3) enzyme, binds to the PY motif in the p2b region of the RSV Gag protein and is similarly required for VLP release (26). Another cellular protein, AIP-1, binds the YXXL motif in EIAV Gag and an LXXL motif downstream of the PTAP sequence in HIV-1 Gag (31,34–36). To date, approximately 20 additional cellular proteins have been found involved in endocytic sorting complexes required for trafficking [ESCRT 0, 1, 2 and 3; (33,37–41)] in a network that participates in the release of HIV-1 and other retroviruses (31,42–47, reviewed in 48). The cellular proteins are orthologs of the yeast class-E proteins, which are known to be required for vacuolar protein sorting (Vps proteins) (49). These proteins exist mainly as soluble proteins or complexes that are sequentially recruited from the cytosol to endosomal compartments. Cellular cargo destined for degradation, such as internalized ubiquitinated plasma membrane receptors undergoing downregulation, is transported to early ‘sorting’ endosomes through the action of Hrs (Vps27), a protein that binds the surface of early endosomes and recruits Tsg101 (50–55). Tsg101 induces the biogenesis of multivesicular bodies (MVBs) (40,56). Multivesicular bodies are carrier endosomes that contain the cargo within luminal vesicles due to the inward invagination of the regions on the endosomal membrane-bearing Hrs. The MVBs eventually fuse with lysosomes, resulting in exposure of cargo on the internal luminal vesicles to hydrolytic enzymes. Cargo destined to return to the plasma membrane (e.g. transferrin receptor) is transported to a different region on the early endosome and enters a recycling pathway. Afterwards, the cellular proteins involved in sorting are released from the endosome membrane into the cytoplasm for recycling through the action of members of the AAA family of ATPases, Vps4A and Vps4B (57).
While the linkage between Tsg101, the endocytic trafficking machinery and viruses with PTAP motifs is well documented, the mechanism that links Nedd4 and viruses with PY motifs to the machinery has been elusive. We previously reported that the L domain in RSV Gag interacts with endogenous Nedd4 proteins in primate COS-1 cells and human 293/E cells and with Nedd4-related proteins of avian origin, which we designated as L-domain-interacting proteins 1 and 2 (LDI-1, LDI-2; 26). L-domain-interacting protein-1 and LDI-2 lack the enzymatic [homologous to E6-AP carboxyl terminus (HECT)] domain but contain the N-terminal membrane-binding (C2) domain and the central PY-motif-binding WW domain. More recently, we identified the 3′-end of the gene encoding LDI-1 and determined that full-length (FL) LDI-1 has a C-terminal Ub ligase HECT domain similar to other Nedd4 family members (58). Some of the Gag proteins in the cytoplasm are modified, and mono- and di-ubiquitinylated forms of Gag are encapsi-dated. Modification is tightly correlated to efficient VLP release. We further demonstrated that a fragment of Nedd4 containing only the WW domain or a mutated FL Nedd4 protein containing an inactivating mutation in the catalytic site is a dominant-negative (DN) inhibitor of RSV Gag budding. The fact that Nedd4 proteins function as binding partners of the RSV L-domain, which is functionally exchangeable with the PTAP motif in HIV-1 Gag recognized by Tsg101, suggested that HIV-1 and RSV Gag might utilize the same trafficking pathway involving Tsg101 and Nedd4. Alternatively, the two Gag proteins might use Nedd4 and Tsg101 in pathways that are entirely separate but parallel, both pathways leading to particle release. Here, we show that release of VLPs assembled by RSV Gag was inhibited by a DN-interfering mutant of Vps4 that was previously shown to interfere with recycling of Tsg101 and to inhibit release of HIV-1, EIAV, Mo-MLV, MPMV and Ebola virus (10,12,17,27,41,47,59). We also show that Tsg101 interacted with both hemagglutinin (HA)-tagged LDI-1 and the endogenous Nedd4 proteins in COS-1 cells. Expression of HA-LDI-1 interfered with the Tsg101–Nedd4 interaction. Not surprisingly, HA-LDI-1 did not associate with HIV-1 Gag or interfere with its release. In contrast, Tsg101 and RSV Gag were found to co-localize, and the interaction required the Nedd4-binding site in Gag. Interestingly, co-expression with a Tsg101 protein lacking the N-terminal Ub E2-variant domain (UEV) in Tsg101 interfered with release of the Gag protein of Mo-MLV but had little or no effect on RSV Gag release. Intriguingly, however, the RSV VLPs were frequently detected in clusters that failed to detach from each other, suggesting that the Tsg101 mutant interfered with a Nedd4 function required for proper particle release. Thus, it appears that RSV and HIV-1 Gag direct particle release through ESCRT-mediated pathways that are independent but linked through Tsg101–Nedd4 interaction.
Results
A DN mutant of the AAA ATPase Vps4 inhibits release of RSV Gag
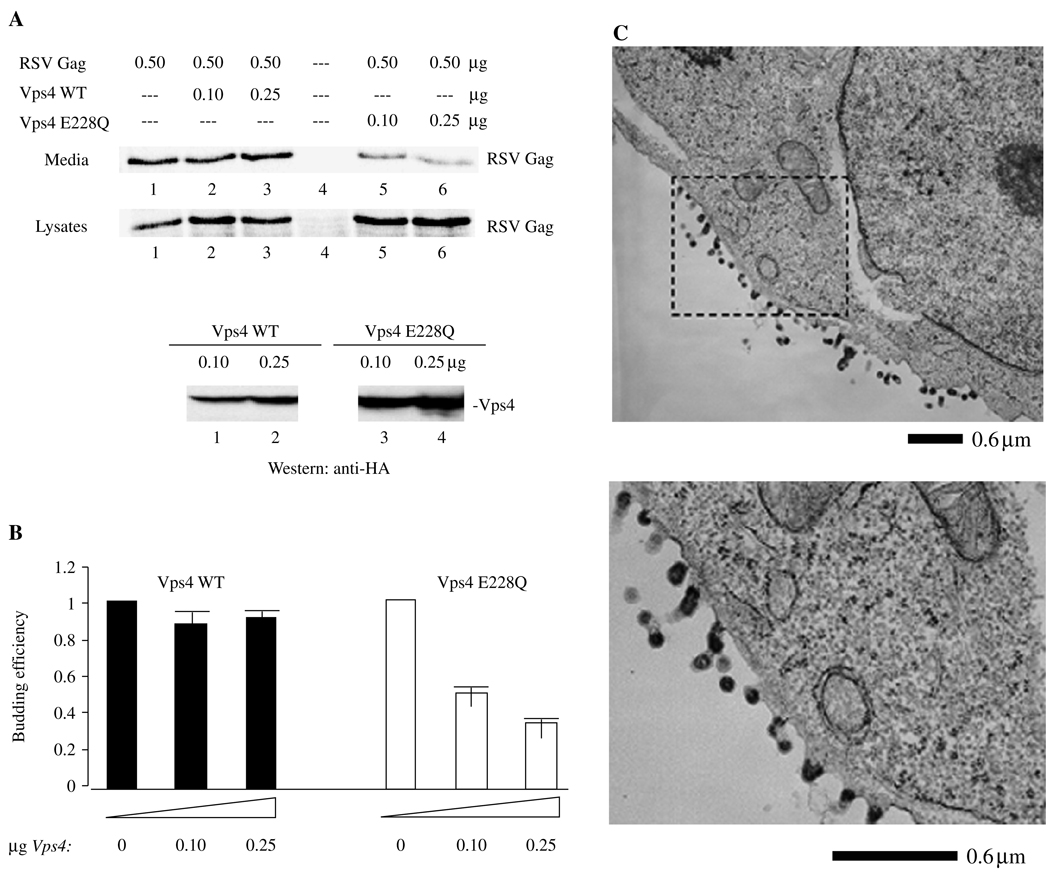
The Vps4 mutation used in this study contained an E228Q substitution, which prevents ATP hydrolysis (27). This mutant has been shown to suppress budding of HIV-1, Mo-MLV and other retroviruses (10,12,17,27,41,47,59), reflecting a requirement for a functional vacuolar-sorting pathway. To test the effect of the Vps4 mutant on RSV Gag release, we co-transfected different amounts of constructs encoding the wild-type (WT) Vps4 protein or the Vps4 substitution mutant with a constant amount of DNA-encoding RSV Gag containing a D37S mutation that inactivates PR (26). The use of the D37S mutant makes it easier to assess effects on release, as only the Gag precursor that directs productive particle formation is detected. Cell lysate and media fractions were prepared following labeling of the cells with 35S-Met and Cys for 2.5 h. Viral proteins were immunoprecipitated from the media and cell-lysate fractions and separated using SDS–PAGE. As shown in Figure 1A, co-expression of RSV Gag with WT Vps4 (lanes 2 and 3) did not interfere significantly with release of VLPs into the media (upper panel), as compared with control cells expressing Gag alone (lane 1). No non-specific signals were detected in a mock-treated sample (lane 4). In contrast, co-expression with Vps4 E228Q (lanes 5 and 6) reduced release of RSV VLPs into the media (upper panel); the amount of Pr76Gag detected in the cell lysate (lower panel) was comparable with, or greater than, control levels. Comparisons of the signal of Gag in the media and lysate fractions, which reflect the efficiency of Gag budding, indicated that the level of Gag release was approximately threefold lower in the presence of the Vps4 mutant as compared with Vps4 WT (Figure 1B). To confirm the results of the budding assay, we examined the co-transfected cells by electron microscopy (EM). No particles were detected in the vicinity of mock-transfected cells or cells transfected with DNA-encoding Vps4 WT (data not shown; cf. Figure 8A). In contrast, VLPs tethered to each other and to the cell were detected in samples cotransfected with RSV Gag and Vps4 E228Q (Figure 1C). This is similar to what is observed with RSV Gag containing an L-domain deletion; such budding structures were never observed following transfection of RSV Gag alone. The results indicate that release of RSV Gag requires proteins under the control of Vps4, as is the case for HIV-1 and other retroviral Gags, and suggest that, like them, RSV Gag uses components of the endocytic trafficking machinery.
Figure 1. Co-expression of Rous sarcoma virus (RSV) Gag and Vps4.
A) Effect on virus-like particle (VLP) release. Rous sarcoma virus Pr76Gag was expressed in 293/E cells (lane 1) in the presence of wild-type Vps4 (lanes 2 and 3) or Vps4 E228Q (lanes 5 and 6) and labeled with a mixture of 35S-Met and Cys for 2.5 h before harvesting at 48 h. Lane 4 is an untransfected control. The ratio of vps4: gag DNA (in µg) used for transfection were lanes 2 and 5, 0.2:1 and lanes 3 and 6, 0.5:1. Upper panel, VLP in media; Lower panel, Gag- and HA-tagged Vps4 in cell lysate. Proteins were detected by autoradiography. B) Semi-quantitative analysis of VLP release. The panel shows the ratio of the Gag signal in VLP isolated from the media to the Gag signal in cell lysates (VLP/cell lysate). C) Examination using electron microscopy (EM). Cells were prepared for thin section EM as described in Materials and Methods. The area outlined in the dashed rectangle is enlarged.
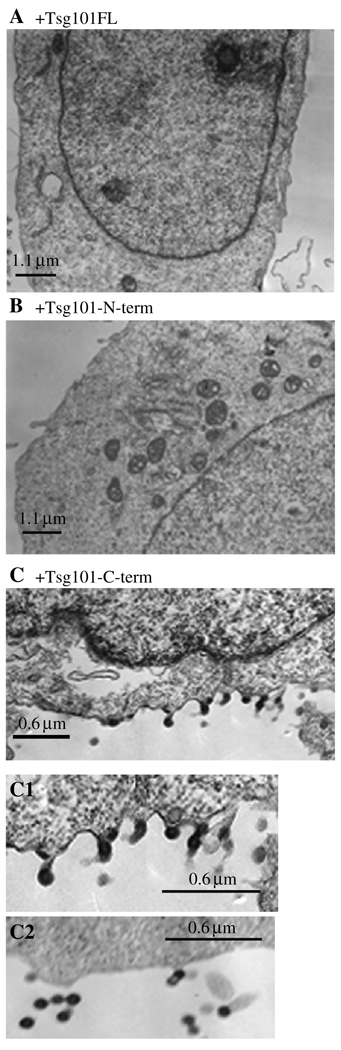
Figure 8. Electron microscopy (EM) of cells co-transfected with Rous sarcoma virus (RSV) gag and tsg101 full-length, tsg101 N-Term or tsg101 C-term.
Cells were prepared for thin section EM as described in Materials and Methods. Panel C1: enlargement of a region with doublet particles tethered to the cell; C2: released doublet particles in the extracellular region.
HA-LDI-1 co-localizes with RSV Gag and blocks its release but does not affect HIV-1 Gag
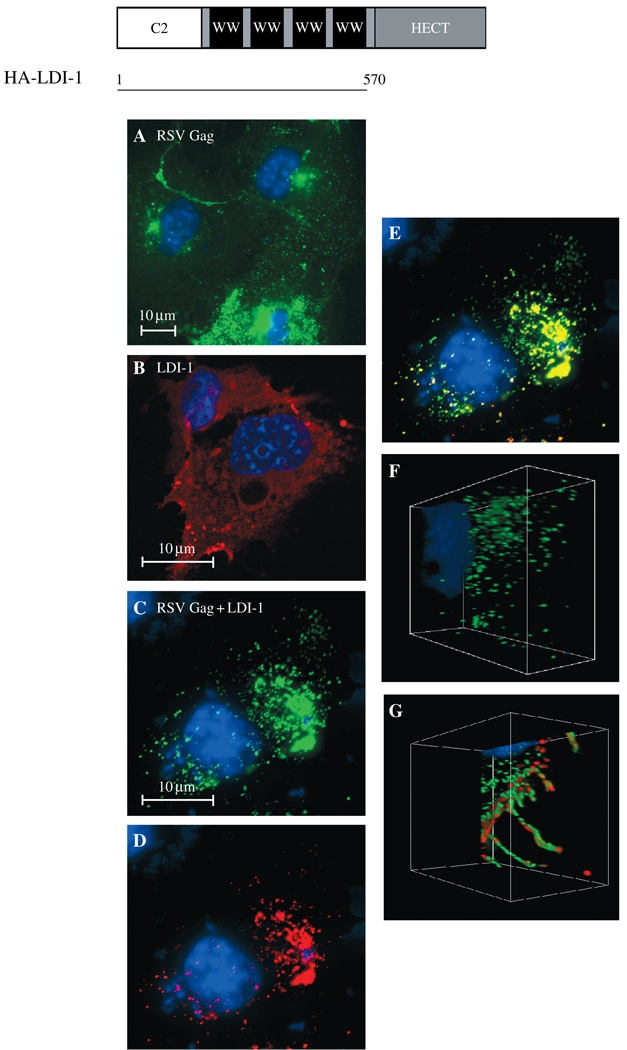
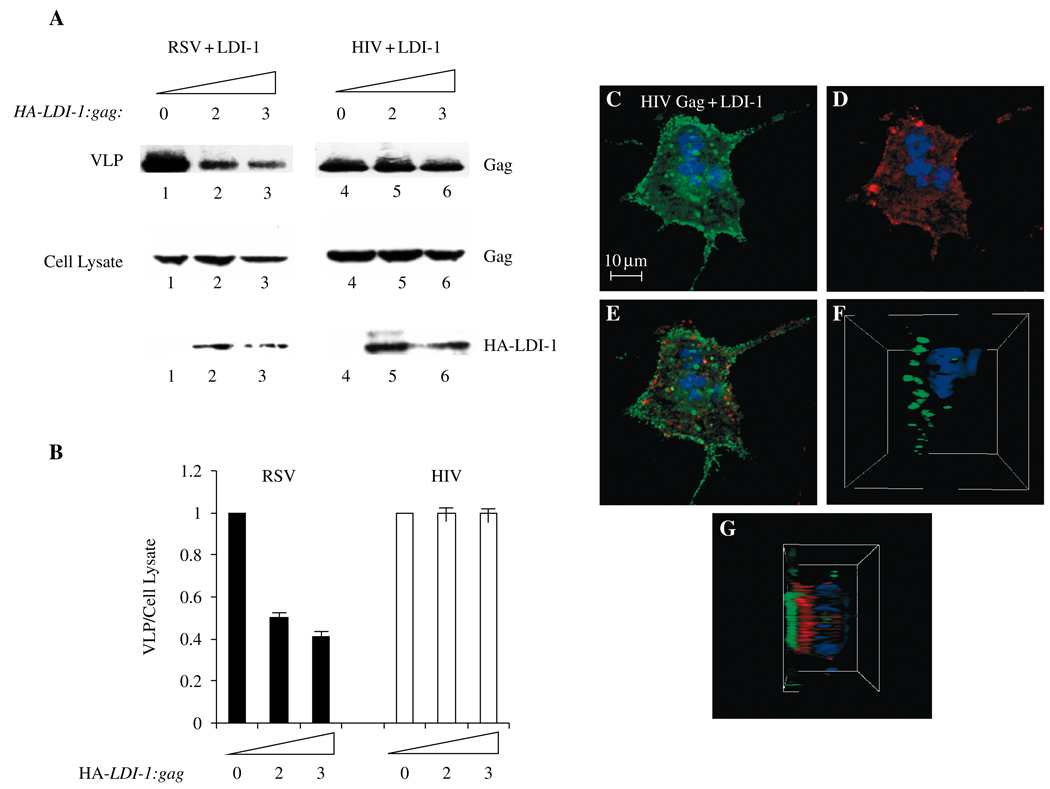
If RSV and HIV-1 Gag share the same pathway for budding, a possible convergent point could be a potential interaction between Nedd4 and Tsg101. As noted above, Nedd4 is an Ub-ligating (E3) enzyme (60). The primary sequence and the three-dimensional structure of the N-terminal domain of Tsg101 are homologous to that of Ub-conjugating (Ubc; E2) enzymes (61–63). The sequence in Tsg101 is highly conserved and most closely resembles the subgroup of E2 enzymes that the Nedd4 family specifically recognizes (61). Hemagglutinin-tagged LDI protein, comprised of the C2 and WW domains of avian Nedd4 with an N-terminal HA tag (26) (Figure 2, top), was first examined for association with RSV Gag in COS-1 cells using confocal microscopy (Figure 2, panels A–F). When expressed alone, RSV Gag-green fluorescent protein (GFP) exhibited dispersed, punctate fluorescence (panel A). Hemagglutinin-tagged LDI-1, expressed alone and detected by indirect immunofluorescence with mouse anti-HA antibody and a TRITC-tagged anti-mouse secondary antibody, was distributed throughout the cytoplasm (panel B). Following co-expression of Gag-GFP and HA-LDI-1 (panels C–E), the green (Gag, panel C) and red (HA-LDI-1, panel D) fluorescent signals were significantly mixed, as indicated by the yellow color (panel E). Similar results were obtained using 293 cells for fluorescence microscopy (not shown). The spatial relationship between RSV Gag-GFP and HA-LDI-1 was evident in a composite of 0.4-µ confocal images taken throughout the cell and rendered as a three-dimensional image (panels F and G). Here, co-localization is visualized as significant intermingling of the red and green fluorescent signals rather than the yellow color detected in the two-dimensional image. Rous sarcoma virus-green fluorescent protein fluorescence in the absence (panel F) and presence (panel G) of HA-LDI-1 expression is shown. This finding indicates that the change in Gag distribution evident in panels A and C reflects co-localization of Gag with HA-LDI-1. Expression of HA-LDI-1 interfered with release of RSV Gag-GFP in VLP assays (Figure 3, panels A and B, lanes 1–3). Inhibition was dependent on the amount of DNA encoding HA-LDI-1 that was transfected (twofold at an LDI: gag ratio of 2 and 2.5-fold at an LDI: gag ratio of 3). Electron microscopy studies of cells transfected with HA-LDI-1 (not shown) revealed VLPs in large inclusion bodies. Possibly, formation of these bodies interfered with antibody detection of the protein, as the expression level of HA-LDI-1 in the cell lysate did not appear as dose-dependent as the effect on VLP release. The level of HA-LDI-1 inhibition was similar to that obtained using FL LDI-1 protein containing an inactivating mutation in the catalytic domain (58). In contrast, HA-LDI-1 did not significantly interfere with HIV-1 Gag release (Figure 3, panel A, lanes 4–6; panel B, n = 10). Moreover, no co-localization with HA-LDI-1 was detected (panels C–E). The different spatial relationship between HIV-1 Gag-GFP and HA-LDI-1 as compared with RSV Gag-GFP and HA-LDI-1 was particularly evident in a three-dimensional composite of confocal images (panels F and G). The green fluorescence of HIV-1 GFP was detected as a defined layer in the absence (panel F) or presence (panel G) of HA-LDI-1. The red fluorescence attributable to HA-LDI-1 also was clearly defined and no co-localization was detected. Thus, HA-LDI-1 co-localized with RSV Gag and inhibited its release but did not associate with nor inhibit HIV-1 Gag release.
Figure 2. Confocal microscopy of cells co-expressing Rous sarcoma virus Gag-green fluorescent protein (RSV Gag-GFP) and hemagglutinin-L-domain-interacting protein (HA-LDI)-1.
Top, Schematic drawing of the Nedd4-related fragment used in this study. COS-1 cells were transfected with DNA encoding RSV-GFP alone (panel A), LDI-HA alone (panel B), or RSV Gag-GFP (green) and HA-LDI-1 (red) (panels C–G). Panel E shows the merged Gag-GFP (panel C) and LDI-1 (panel D) images. Panels F and G: three-dimensional renderings of stacked z sections through cells expressing RSV Gag alone (panel F) or LDI-1 and RSV Gag (panel G). The LDI-1 protein was detected by indirect immunofluorescence using mouse anti-HA as primary antibody and rabbit anti-mouse tagged with TRITC as secondary antibody.
Figure 3. Effect of L-domain-interacting protein (LDI)-1 expression on HIV-1-virus-like particle (VLP) release.
Panel A: Western analysis. COS-1 cells were transfected with DNA encoding Rous sarcoma virus (RSV) Gag-green fluorescent protein (GFP) (lanes 1–3) or HIV-1 Gag-green fluorescent protein (GFP) (lanes 4–6) alone (lanes 1 and 4) or with HA-LDI-1 (lanes 2, 3, 5 and 6) at HA-LDI-1 to gag ratios of 2:1 (lanes 2 and 5) or 3:1 (lanes 3 and 6). The Gag and LDI-1 proteins were detected using anti-GFP or anti-HA antibodies, respectively. Top panel: Gag-GFP in VLPs; center and lower panels: Gag-GFP (center) and HA-LDI-1 (lower) in cell lysates. Panel B: Semi-quantitative analysis of VLP release. The panel shows the ratio of the Gag signal in VLP isolated from the media to the Gag signal in cell lysates (VLP/cell lysate). Panels C–E: COS-1 cells were transfected with DNA encoding RSV Gag-GFP (green, panel C) and HA-LDI-1 (red, panel D). Panel E shows the merged images. Panels F and G: Three-dimensional renderings of stacked z sections through a cell expressing Gag and LDI-1. Left, HIV-1 Gag alone; right, LDI-1 co-expressed with HIV-1 Gag.
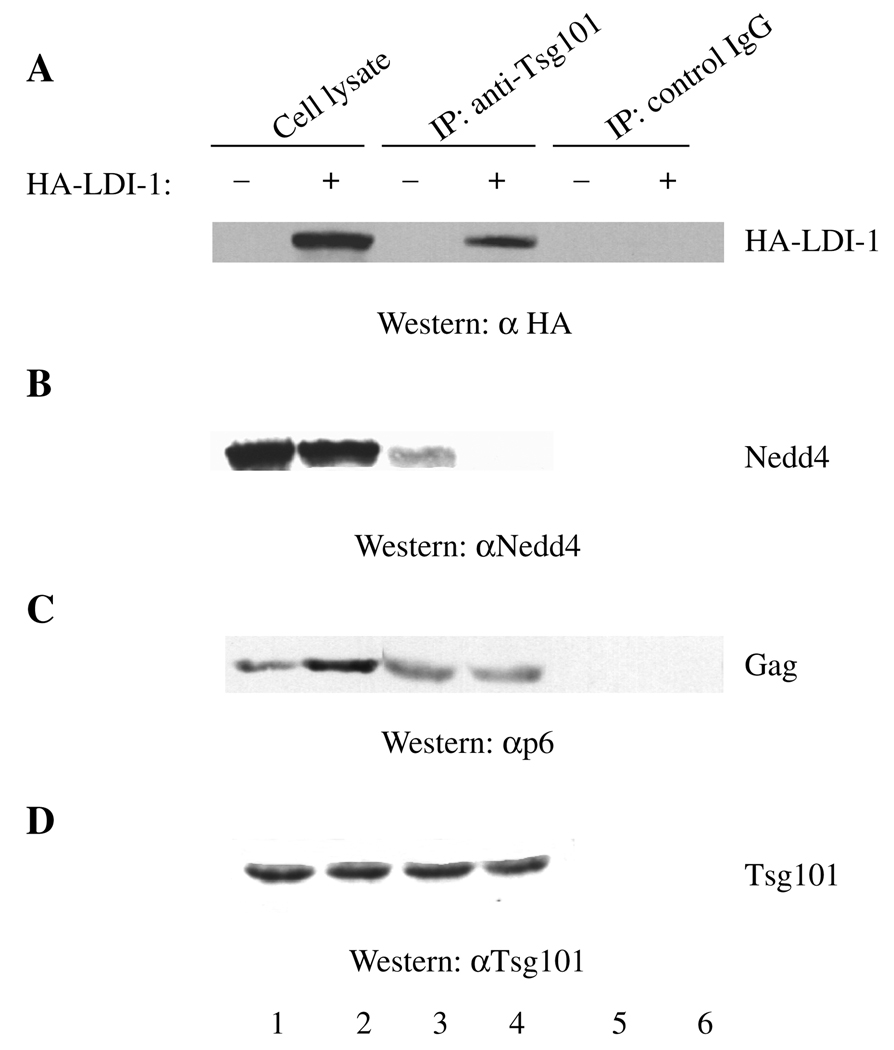
HA-LDI-1 associates with Tsg101 and competes with endogenous Nedd4 for interaction with Tsg101
The results above suggest that the RSV and HIV Gag-trafficking pathways are separate or, alternatively, that Nedd4 functions upstream of Tsg101 in the same pathway. To distinguish these possibilities, we next tested for Tsg101–LDI interaction. As shown in Figure 4A, the HA-LDI-1 expressed in transfected COS-1 cells (lane 2) was immunoprecipitated by antibody against Tsg101 (lane 4), as indicated by Western analysis using an antibody against the HA tag on the LDI-1 protein. Extracts prepared from cells that were not transfected with DNA encoding HA-LDI-1 (lanes 1, 3 and 5) contained no cross-reactive material and, in a control experiment using preimmune rabbit serum, no non-specific immunoprecipitation (Ip) was detected (lane 6), indicating that recognition of HA-LDI-1 was specific. Reprobing the blot with an anti-Tsg101 monoclonal antibody indicated that Tsg101 was present in the immune-precipitates (panel D). The results indicate that Tsg101 interacted with HA-LDI-1 specifically. As Tsg101 recognized both the endogenous Nedd4 protein in COS-1 cells and HA-LDI-1, we determined whether HA-LDI-1 could compete with the endogenous Nedd4 protein for interaction with Tsg101. To test this, we determined whether HA-LDI-1 interfered with Tsg101 binding to endogenous Nedd4 in cells expressing HIV-1 Gag, as measured by immune precipitation. Cells expressing Gag were used to provide a negative control in the cell lysate, as our previous studies indicated that LDI-1 does not bind the PTAP motif in HIV-1 Gag (26) and that HA-LDI-1 did not co-localize with HIV-1 Gag (Figure 3C). As shown in Figure 4, lanes 1 and 2, expression of HA-tagged LDI-1 (panel A) did not inhibit accumulation of endogenous Nedd4 (panel B, lanes 1 and 2), HIV-1 Gag (panel C, lanes 1 and 2) or endogenous Tsg101 (panel D, lanes 1 and 2). Although there appeared to be increased accumulation of Gag in the presence of HA-LDI-1 (panel C, lane 2), this was not reproducibly detected (n = 8). The polyclonal antibody against Tsg101 co-immunoprecipitated both Nedd4 (panel B, lane 3) and HIV-1 Gag (panel C, lane 3), as well as Tsg101 itself (panel D, lane 3). However, the presence of HA-LDI-1 in the extract (panel A, lane 4) reduced co-Ip of Nedd4 with Tsg101 approximately twofold to fourfold (panel B, compare lanes 3 and 4; n = 3). In contrast, expression of HA-LDI-1 did not detectably diminish the interaction of Tsg101 with HIV-1 Gag (panel C, lanes 3 and 4). As expected, Tsg101 was immunoprecipitated by the anti-Tsg101 antibody (panel D, lane 3), and LDI-1 expression did not prevent Tsg101 Ip (panel D, lane 4). Immunoprecipitation reactions using the preimmune control serum confirmed that co-immuno-precipitation of HA-LDI-1, Nedd4 and HIV-1 Gag required Tsg101 (panels A–D, lanes 5 and 6). The results indicate that the HA-tagged LDI-1 fragment can compete with endogenous Nedd4 for Tsg101 binding while not detectably affecting the interaction of Tsg101 with HIV-1 Gag. We do not know whether Tsg101 bound to HIV-1 Gag and Tsg101 linked to Nedd4 represent the same or different complexes.
Figure 4. Panels A–D, Effect of L-domain-interacting protein (LDI)-1 expression on Tsg101–Nedd4 interaction.
Extracts prepared from cells transfected with DNA encoding HIV-1 Gag (lane 1) or Gag and LDI-1 (lane 2) were incubated with buffer alone (lanes 3 and 5) or buffer containing antibody against Tsg101 (lane 4) or control IgG (lane 6). After washings, the samples were examined for immunoprecipitated proteins by SDS–PAGE and Western analysis. The panels show immunoblots of the same polyacrylamide gel that was successively probed for LDI-1, Nedd4, Gag and Tsg101. Panel A: HA-LDI-1 visualized by using antibody against the HA-tag on the protein. Panel B: Nedd4 detected by using the anti-Nedd4 antibody. Panel C: Gag detected by using antibody against the p6 domain. Panel D: Tsg101 detected by using the mouse monoclonal antibody against Tsg101.
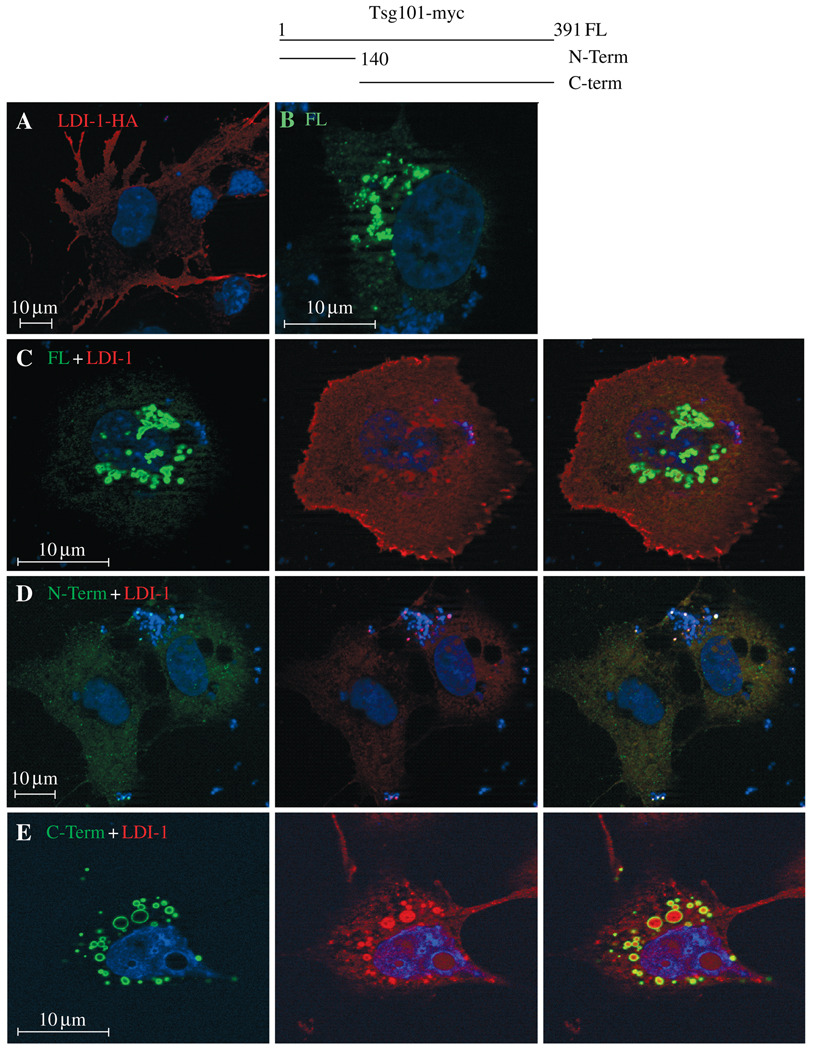
The biochemical interaction of Tsg101 with HA-LDI-1 detected by co-Ip was confirmed using confocal microscopy. As noted above, HA-tagged LDI-1, expressed alone, was detected throughout the cytoplasm (Figure 5A). Full-length myc-tagged Tsg101, in contrast, localized in the perinuclear region (panel B). However, following co-expression of the two proteins (panel C), some of the LDI-1 (red) appeared to localize to the perinuclear region that contained Tsg101 (green). Merged images revealed proximal red and green but not yellow fluorescence, suggesting that the proteins were in the same region but not directly associated. Both the perinuclear vesicular localization of Tsg101 FL and the proximity to HA-LDI-1 were abrogated by deletion of Tsg101 C-terminal residues 140–391 (panel D). Also, both HA-LDI-1 and Tsg101 fragments exhibited weak, diffuse fluorescence, similar to the pattern observed when they were expressed alone (panel A and data not shown). This suggested that the association of Tsg101 FL with HA-LDI-1 required determinants in the C-terminal region of Tsg101. Consistent with this notion, a significant fraction of HA-LDI-1 co-localized with a C-terminal fragment containing residues 140–391 in the perinuclear region (panel E). In contrast to the FL Tsg101 protein, LDI-1 and C-terminal fragment were directly associated, as indicated by the yellow fluorescence in the merged images. The results support the finding that Tsg101 and HA-LDI-1 associate and indicate that determinants in the C-terminal region of Tsg101 mediate the interaction.
Figure 5. Comparison of cells co-expressing L-domain-interacting protein (LDI)-1 and Tsg101.
Top center, schematic drawing of Tsg101-related proteins used in this study. COS-1 cells plated on slides for examination by confocal microscopy were transfected with (i) Panel A: DNA encoding HA-LDI-1 alone; (ii) Panel B: DNA encoding Tsg101 FL alone; (iii) Panel C: LDI-1 (red) and Tsg101-FL (green); (iv) Panel D: LDI-1 and Tsg101N-term; or (v) Panel E: LDI-1 and Tsg101 C-term. The HA-LDI-1 to tsg ratio was 1:1.
Tsg101 and RSV Gag co-localize
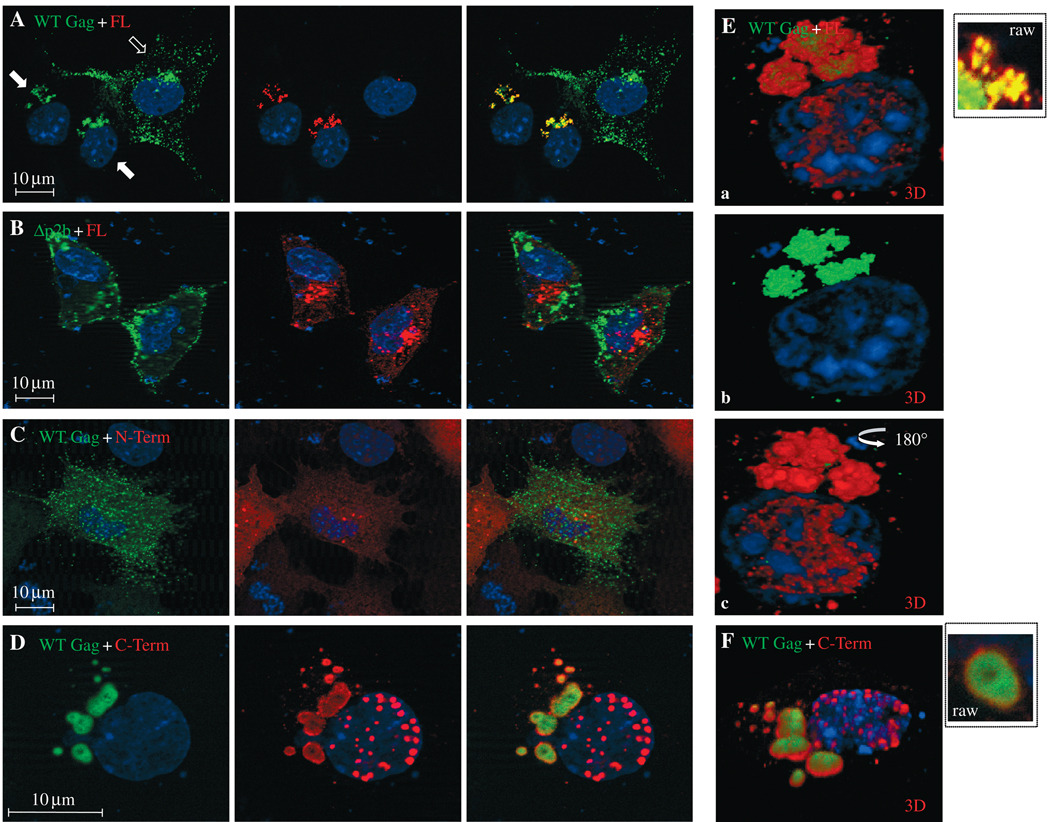
Because HA-LDI-1 interacted with Tsg101 and competed with endogenous Nedd4 for the protein, we determined whether RSV Gag and Tsg101 co-localized intracellularly (Figure 6). In the absence of Tsg101-myc expression, RSV Gag-GFP exhibited dispersed, punctate fluorescence as expected (panel A, open arrow). The same field shows cells that co-expressed Gag-GFP and Tsg101-myc (solid arrows). Under these conditions, both Gag (green) and Tsg-myc (red) accumulated in the perinuclear region and co-localized, as indicated by the yellow fluorescence. This co-localization required the p2b region of Gag that binds Nedd4 (26), as co-localization was abrogated by deletion of this region from the protein (panel B). Under these conditions, the subcellular distribution of Tsg101-FL was similar to that detected when Tsg101 was expressed alone (cf. Figure 5B). As was the case for HA-LDI-1, co-localization was not detected when Gag was co-expressed with the N-terminal fragment of Tsg101 (panel C). However, the C-terminal region that recognizes Tsg101’s ESCRT-1-binding partners (41,64) retained the ability to localize in proximity to Gag (panel D). Proximity was indicated by the closeness of the red and green fluorescent signals rather than the yellow fluorescence produced with Tsg101-FL. Most of the structures in which Gag accumulated in the presence of the C-terminal fragment were also larger (approximately 200–750 nm) than those formed by Tsg101-FL (<100 nm, panel A). Differences in the co-localization of Gag and Tsg101-FL versus Gag and the Tsg101 C-terminal fragment were evident in three-dimensional spatial images (panels E and F, respectively; insets show the Gag-Tsg101 clusters in panels A and D on the same scale). As noted above, co-localization is visualized as significant intermingling of the red and green fluorescent signals rather than the yellow color detected in the two-dimensional image. Co-localization was apparent in the three-dimensional spatial image of the merged Gag and Tsg101-FL (panel Ea). Removal of the red signal from merged images (panel Eb) and rotation of the merged image by 180 ° (panel Ec) suggested that Gag was ‘sandwiched’ in the small structures formed by Tsg101-FL. In contrast, in the larger structures formed by Tsg101 C-term, Gag was completely exposed (panel F). Taken together, the results indicate that RSV Gag co-localizes with Tsg101 and that co-localization is dependent on retention of the Nedd4-binding site in Gag. The results also suggest that determinants in the N-terminal region of Tsg101 influence the nature of the Tsg101-(Nedd4)– Gag interaction.
Figure 6. Comparison of cells co-expressing Rous sarcoma virus (RSV) Gag and Tsg101.
COS-1 cells were transfected with (i) Panel A: DNA encoding RSV wild-type (WT) Gag-green fluorescent protein (GFP) and Tsg101-FL (red); (ii) Panel B: δnp2b Gag-GFP and Tsg101-FL; (iii) Panel C: WT Gag and Tsg101N-term; or (iv) Panel D: WT Gag and Tsg101 C-term. Panel E (a, b, three-dimensional spatial images of a cell in panel A expressing Gag and Tsg101-FL; a) merged Tsg-FL/Gag images; b) Gag; and c) Tsg101-FL. Panel F: three-dimensional spatial image of cell in panel D. The tsg to gag ratio was 3:1. The open arrow in panel A indicates a cell in the field expressing Gag alone. Closed arrows indicated cells expressing Gag and Tsg101-FL. The image in panel D is enlarged ×2.5 relative to the magnification of the image in panel A. The insets in panels E and F show images of the Tsg101 Gag clusters in panels A and D on the same scale.
Effect of the C-terminal Tsg101 fragment on RSV Gag release
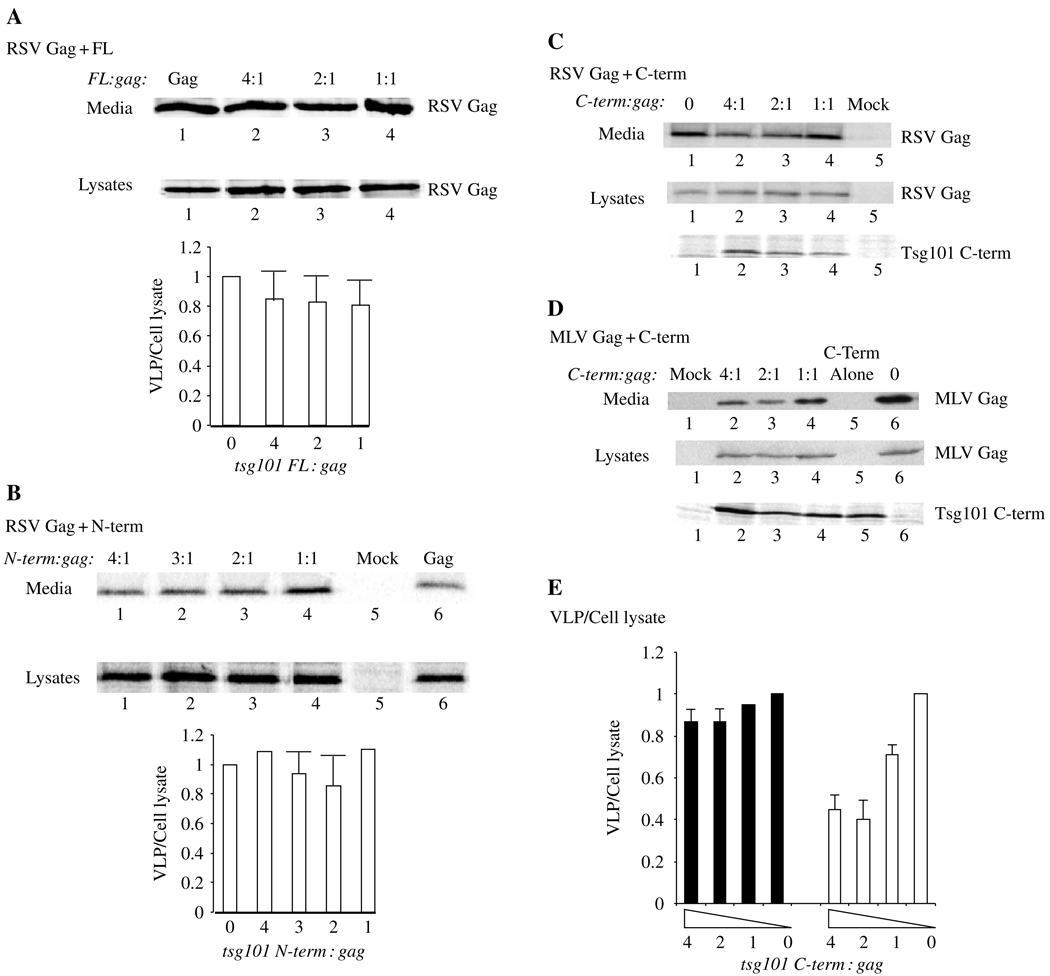
To determine whether the Tsg101–Gag interaction mediated by Nedd4 affected VLP release, we co-expressed RSV Gag with Tsg101-FL or fragments of the protein. The cells were metabolically labeled with 35S-Met and Cys as described above (cf. Figure 1), viral proteins were immunoprecipitated from the cell lysate, VLPs were isolated from the media, and the samples were analyzed by SDS–PAGE and autoradiography. Figure 7 shows the amount of WT Gag detected in the media (upper panels) or in the cell lysates (lower panels). No significant inhibition was observed with the FL protein or the N-terminal Tsg101 fragment (Figure 7, panels A and B; n = 8). Similar results were obtained with the C-terminal fragment (panel C). In a control experiment, the effect of the C-terminal fragment on Mo-MLV Gag release was determined (panel D). Our results confirmed previous observations that the C-terminal fragment of Tsg101 inhibits Mo-MLV Gag release (65) (compare lane 6 with lanes 2–4 in panel E). Panel E shows the efficiency of Gag release, expressed as the ratio of Gag in VLP to Gag in cell lysate (VLP/cell lysate). Although dose-dependent inhibition of Mo-MLV Gag release was observed (panel E, right; n = 2), RSV Gag release (left) was reproducibly more resistant (n = 8). To determine whether the released particles were assembled normally, the VLPs were recovered from the media fraction and subjected to sucrose density gradient centrifugation to equilibrium. The Pr76Gag protein in the samples banded at the density of VLPs (data not shown). Similar results were obtained with COS-1 cells; with DNA encoding a myc-tagged Tsg101 fragment (43) instead of a HA-tagged fragment (65); with RSV Gag expressed from pCMV-RSVgag that encodes Gag-GFP (66) or from p2036 (26); and when samples were probed by Western analysis (data not shown). The results indicate that despite the striking effects of Tsg101 on Gag localization, neither the FL nor the truncated Tsg101 protein interfered with VLP release. The fact that the Tsg101 C-terminal fragment detectably inhibited Mo-MLV Gag release suggests that the fragment interacts differently with the Gag-E3 ligase complexes formed by RSV and Mo-MLV.
Figure 7. Effect of the Tsg101 C-terminal fragment on Rous sarcoma virus (RSV) and Mo-MLV Gag release.
Panels A–C: Effect of Tsg101 full-length and Tsg101 fragment expression on RSV Gag release. Gag was expressed in 293/E cells alone or with the indicated Tsg101 protein at the indicated ratio of tsg: gag DNA. The cells were metabolically labeled with 35S-Met and Cys at 48 h after transfection, and media and cell lysate fractions were analyzed. The radiolabeled proteins in cell lysates were immunoprecipitated with polyclonal antibody against RSV antibody. An aliquot of the lysate in panel C was examined for Tsg101 by using antibody against the hemagglutinin (HA) tag on the protein in Western analysis. Panel D: Parallel study of the effect of Tsg101 C-term expression on Mo-MLV Gag release. Cells were harvested at 48 h post transfection and examined by Western analysis using antibody against the Mo-MLV CA protein. The blot was re-probed with antibody against HA to detect the Tsg101 protein. Proteins in cell lysates and virus-like particles (VLPs) isolated from media fractions were separated by SDS–PAGE, and autoradiograms were prepared. Panels A, B and E: semi-quantitative analysis of VLP release (VLP/cell lysate). The panels show the ratio of the Gag signal in VLP isolated from the media to the Gag signal in cell lysates.
Electron microscopy analysis of the 293/E cells incubated with Tsg101-FL (Figure 8, panel A) or the N-term fragment (panel B) revealed no VLPs, indicating that they were released and that they did not adhere to the cells. In contrast, examination of cultures incubated with the C-term fragment (panel C) typically revealed single or double particles adhered to the cell surface (enlarged in inset C1) or outside the cells (inset C2, taken from another cell). Examination of released VLPs in two independent experiments indicated that approximately 60% (35 of 58) of the particles were tethered doublets, indicating a defect in particle-particle detachment. Thus, although Tsg101 was not required for VLP release, it appears that proper Tsg101–Nedd4 interaction is necessary for efficient particle separation.
Discussion
A conserved protein network required for endocytic trafficking of cellular proteins has been identified in yeast and mammalian cells (31,33,37–39). The functioning of this network is controlled by Vps4 AAA ATPases and, based on their requirement for active Vps4, budding of HIV-1, EIAV, Mo-MLV, MPMV, HTLV-1, Ebola virus and RSV (10,12,17,47,59, this study) exploits this machinery. Previous studies showed that HIV-1 Gag binds Tsg101 and RSV Gag binds Nedd4 through their respective L domains (25–28). Here, we provide evidence that endogenous Tsg101 and Nedd4 proteins are stably associated in the cytoplasm, thereby providing a possible point where the functionally exchangeable Tsg101-binding and Nedd4-binding L domains might converge. L-domain-interacting protein-1, a Nedd4-related DN inhibitor of RSV Gag release, did not significantly inhibit HIV-1-VLP release (Figure 3). This finding is not surprising, as the HIV-1 L domain does not bind Nedd4-related proteins (26). Similarly, we found that a DN inhibitor of Tsg101 that affects HIV-1 Gag (43,65) and Mo-MLV Gag (65, this study) failed to block release of RSV Gag in our study. These observations indicate that Tsg101 and Nedd4 must function independently of each other in HIV-1 and RSV Gag trafficking. However, while intact Tsg101 was not required for efficient RSV Gag release in our studies, Nedd4 interaction with the FL Tsg101 protein was necessary for efficient particle–particle fission. This conclusion would appear to conflict with the results of a recent study where the C-terminal Tsg101 fragment was found to inhibit RSV Gag release (67). However, we suspect that the stronger inhibition observed in that study reflects differences in compartmentalization of the adventitiously expressed Tsg101 proteins. This notion is supported by the fact that Tsg101-Nedd4 co-localization was L-domain-dependent in our study (cf. Figure 6) but not in the aforementioned report. Thus, the RSV- and HIV Gag-trafficking pathways are linked through Tsg101–Nedd4 interaction even if both cellular proteins are not strictly required for the release per se.
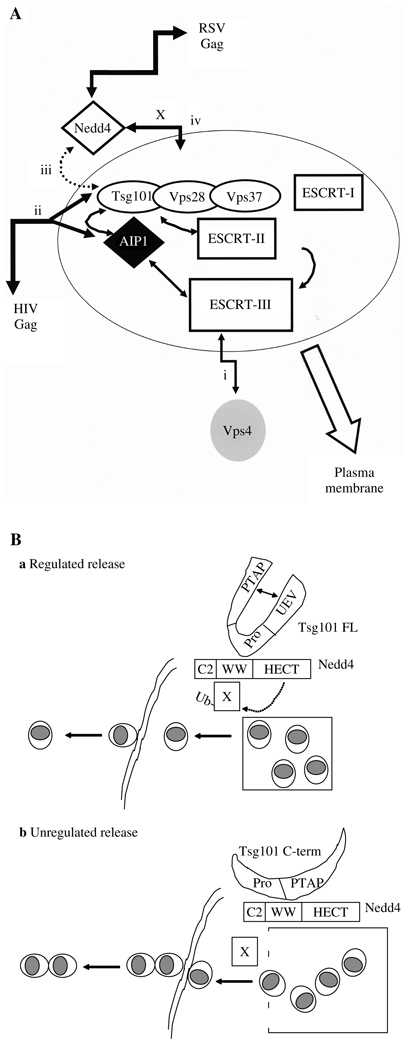
Figure 9A shows four aspects of HIV and RSV trafficking. (i) DN Vps4 regulates recycling of Vps proteins between cytosol and endocytic vesicles. Sensitivity to DN Vps4 indicates a requirement for functional endocytic trafficking machinery; both HIV and RSV Gag exhibit Vps4 DN sensitivity. (ii) HIV Gag accesses the machinery through interaction between its L-domain (PTAP) and the PT/SAP-binding pocket in the N-terminal UEV domain of Tsg101. Association with Tsg101 may allow HIV Gag to enter the early endosome and the MVB-sorting compartment, recruit ESCRT complexes and budding machinery and be transported with these to sites of release on the plasma membrane. (iii) Tsg101 interacts with Nedd4, but the Nedd4 component is not critical for HIV Gag release, perhaps because its contribution is redundant with other E3 ligases in the cell. Rous sarcoma virus and Mo-MLV Gag have both been shown to interact with more than one E3 ligase (26,68). Similarly, Tsg101 is not critical for RSV Gag release. By analogy with Mo-MLV, where siRNA-mediated depletion of Tsg101 did not inhibit Gag release (27), RSV release is not expected to require Tsg101: like Mo-MLV, it contains a PY-type L domain. Interestingly, in our study, the C-terminal fragment of Tsg101 had different effects on Mo-MLV and RSV Gag release. It has been reported that the PY motif of Mo-MLV Gag (DPPPYR) binds the E3 ligases WWP1 and WWP2 (68) and not Nedd4 (which binds the PPPPYL motif of RSV Gag) (26). This finding suggests that there are different E3 ligases for different PY motifs. The different manifestations of Tsg101 C-terminal-mediated inhibition might be explained if the fragment interacts with WWP1/WWP2 and Nedd4 differently and if other ligases can provide the release function for RSV Gag. (iv) In contrast to HIV, RSV Gag accesses the machinery through interaction with HECT domain E3 ligases. We speculate that a second factor, designated as X in Figure 9A, provides the link between RSV Gag, Nedd4 and ESCRT protein network.
Figure 9. A) Relationship between Rous sarcoma virus (RSV) and HIV-1 Gag-trafficking pathways.
B): a) Regulated RSV Gag release mediated by Tsg101–Nedd4 interaction and b) unregulated release resulting from Nedd4 interaction with the Tsg101 C-terminal fragment.
Although adventitious expression of the Tsg101 C-terminal fragment did not inhibit RSV Gag release in our studies, it nevertheless interfered with a step in the release process, i.e. budding of single particles. Because the Tsg101–Nedd4 interaction was altered by truncation of the N-terminal region, as revealed in both raw images and high-resolution three-dimensional images of the complex (Figure 6), we speculated that, in contrast to Tsg101-FL, the C-terminal Tsg101 fragment interfered with a Nedd4 function that regulates the timing of budding and fission events at the release site to ensure that bud fission occurs before new budding is initiated (Figure 9B). This may be the case for particles detected at the cell surface (Figure 8C) as well as for particles in the intracellular structures visualized using confocal microscopy (Figure 6D), where Gag appeared to be surface-exposed (Figure 6F). Possibly, this putative Nedd4 function involves the × factor proposed to link RSV Gag to the ESCRT machinery (cf. Figure 9A). Closely clustered extracellular RSV particles have been detected following treatment with proteasome inhibitors, which were suggested to limit the pool of free Ub (66). If an ubiquitination event is indeed required for particle-particle fission, our finding that the number of tethered particles increased significantly following co-expression of Gag with the Tsg101 C-term fragment suggests that interaction with the fragment inhibited Nedd4-dependent ubiquitination. Possibly, deletion of the UEV domain in Tsg101 exposed downstream Pro-rich sequences that are normally sequestered, and these, in turn, bound WW domains in Nedd4. These new interactions may have interfered with Nedd4 modification of factor × and, thereby, X-mediated regulation of particle release.
To date, Tsg101 has been shown to interact with the E3 enzymes Mdm2, p300 and Tsg101-associated ligase (69–72). Interestingly, however, although Tsg101 and Nedd4 are both involved in Ub-mediated functions in endocytic trafficking, the two proteins have not previously been linked. Nedd4-mediated monoubiquitinylation is a recognition signal for endocytic trafficking (60). As Tsg101 recognizes monoubiquitinylated cargo and regulates cargo sorting into MVBs (33,40), our findings potentially link these two cellular machineries. Similarly, Nedd4-mediated ubiquitination is a regulator of exocytosis in certain cell types (73). Perhaps our findings indicate that Tsg101 and Nedd4 participate in coordination of sorting and exocytosis events.
The functional exchangeability of the L domains combined with the ability of Tsg101 to interact with different E3 ligases may explain why retroviral trafficking mechanisms once appeared to be flexible and different. However, as all of the domains now appear to be under Vps4 control, there is ultimately no alternative release route for Gag that has entered the endocytic machinery, even if different endocytic pathways are used. Mutations to resistance against agents that target the viruses’ use of the machinery should thus be difficult to generate. However, HIV-1 exhibits variable degrees of L-domain-dependence under certain conditions (74,75). Also, as shown here and elsewhere (67), RSV Gag exhibits variable degrees of sensitivity to the Tsg101 C-terminal fragment. It will, thus, be crucial to identify factors that influence whether and how endocytic trafficking pathways are used in order to block alternative routes of virus release.
Materials and Methods
Constructs
HIV-1 and RSV Gag C-terminally tagged with GFP (Gag-GFP) were expressed from pCMV-HIV-1gag or pCMV-RSVgag (26,75). Constructs encoding RSV Pr76Gag or N-terminally HA-tagged LDI-1 (amino acids 1–570) in plasmid 2036 were described in Kikonyogo et al. (26). DNA encoding human vps4A or vps4A-E228Q was a gift of W Sundquist (University of Utah; 27) and was cloned with an HA-tag into the p2036 vector. DNA encoding Hrs-FLAG, Tsg101-myc or fragments thereof was described in Goff et al. (43). Constructs encoding HA-tagged N- or C-terminal fragments of the Tsg101 protein were gifts from E. Freed (60). A construct encoding MLV Gag was a gift from S. Goff (9).
Cell culture, transfection, preparation of cytoplasmic extracts and virus isolation
COS-1 cells were cultured in DMEM supplemented with fetal bovine serum and antibiotics to 60% confluency at 37 °C. Where indicated, the cells were transfected by using the FuGene 6 reagent (Roche, Indianapolis, IN, USA) according to the instructions of the manufacturer. At 48 h post transfection, the cells were harvested by scraping into cold PBS and collected by centrifugation. The pelleted cells were washed with cold PBS, allowed to swell in cold hypotonic buffer [10 mM Tris–HCl (pH 7.4) and 1 mM MgCl2] containing protease inhibitors and disrupted using a Dounce homogenizer with a type B pestle. The total lysate (1 mL) was spun for 10 min at 1000 × g at 4 °C to remove unbroken cells, nuclei and mitochondria, producing a total cytoplasmic fraction. For Ip experiments, the cytoplasmic extract was cleared of large particulate material by centrifugation at 10 000 × g. To isolate the VLPs assembled by Gag, we filtered the cell culture media (0.45 µm), applied to a cushion of 20% sucrose in a centrifuge tube and then spun at 274000 ×g for 90 min at 4 °C (Beckman SW41 rotor). The pelleted VLPs were suspended in 50 µL of PBS by gentle shaking at 4 °C. Samples were analyzed using SDS– PAGE and Western analysis. Semi-quantitative determinations of VLP release (VLP/cell lysate ratios) were made using a phosphorimager. Where indicated, 293/E cells were also examined. The 293/E cell line is an adenovirus-transformed human embryonic kidney cell line, which is stably expressing the EBNA1 protein of EBV. Transfection and metabolic labeling of cells were performed as previously described (26). Forty-eight hours post transfection, cells were washed twice with PBS and metabolically labeled with 35S-Met and Cys for 2.5 h. Cell lysate and media fractions were collected as described above, and RSV or Mo-MLV Gag was immunoprecipitated using specific polyclonal antibodies.
Ip assay
Protein A agarose beads (Pierce, Rockford, IL, USA) prewashed with immunoprecipitation (Ip) buffer [50 mM Tris–HCl (pH 7.5), 300 mM NaCl, 0.1% Triton-X-100 (TX-100)] containing protease inhibitors (1 mM, Roche), were incubated with the appropriate antibody, added to cytosolic extracts and rotated at 4 °C overnight. Immunoprecipitates were washed in Ip buffer, boiled in SDS sample buffer with 5% mercaptoethanol, resolved by SDS– PAGE and analyzed using Western blotting with the antibodies indicated in the text. Immunoprecipitated proteins were resolved using SDS–PAGE and analyzed using a phosphorimager.
Confocal microscopy
COS-1 cells were cultured in DMEM supplemented with fetal bovine serum and antibiotics to 60% confluency at 37 °C. Where indicated, the cells were transfected using the FuGene 6 reagent according to the instructions of the manufacturer. At 24 h post transfection, the cells were washed once in PBS and fixed in 4% formaldehyde (Fisher, Fairlawn, NJ, USA) in Ca2+-free, Mg2+-free PBS for 20 min. Samples were then washed three times for a total of 5 min with PBS, permeabilized with 0.1% TX-100 for 5 min and washed three times again with PBS. After blocking for 10 min in PBS containing 1% BSA, the cells were incubated with primary antibody for 1 h at 37 °C, rinsed with PBS and then incubated with TRITC-tagged secondary antibody for 30 min at 37 °C. The nuclear stain, Hoechst (Molecular Probes, Eugene, OR, USA), was added in the last 10 min. After rinsing, the cells were mounted using p-phenylenedia-mine and Immunomount. Confocal Images were captured with an inverted fluorescent/dic Zeiss Axiovert 200M microscope equipped with an AxioCam HRm camera (Zeiss, Thornwood, NY, USA) and mercury arc lamp light source using a ×63 Plan-Apochromat (NA 1.40) oil objective and operated using axiovision version 4.1 (Zeiss) software. More than 40 cell images were examined from duplicate samples in each experiment. Approximately 20 optical sections along the z-axis were acquired in increments of 0.4 µ. Figures show the central image or, where indicated, a composite of sections through the z plane of the cell rendered as a three-dimensional image. The fluorescent data sets were deconvolved using the constrained iterative method (axiovision version 4.1). The following excitation and emission wavelengths were used for imaging: Hoechst stain, λex360 ± 20/λem460 ± 25; fluorescein isothiocyanate (for GFP), λex480 ± 20/λem535 ± 25; Texas Red (for TRITC), λex560 ± 25/λem645 ± 35.
EM
Sixty-millimeter dishes of 293/E cells transfected with p2036-Gag were transfected with DNA encoding the protein indicated in the text above. Total DNA transfected per dish was normalized by using the p2036 vector. Forty-eight hours post transfection, cells were washed with PBS at room temperature and were fixed in 2.5% glutaraldehyde in 0.1 m sodium phosphate buffer (pH 7.4) at 4 °C for 30 min. Cells were scraped from the tissue culture dish and pelleted at 1000 × g for 10 min at 4 °C. The cell pellet was fixed for an additional 2 h in 2.5% glutaraldehyde and postfixed for 1 h with osmium tetroxide. The cell pellet was dehydrated in a series of alcohol washes and embedded in Epon. Ultrathin sections were stained with uranyl acetate and lead citrate and examined using a Zeiss 900 electron microscope.
Protein detection
Proteins were separated by electrophoresis through SDS-polyacrylamide gels. Following electrophoresis, the gels were transferred to nitrocellulose and were analyzed using Western blotting with the antibodies specified in the text above. Tsg101 was detected by using mouse monoclonal antibody or polyclonal antibodies (Santa Cruz, Santa Cruz, CA, USA). Anti-Nedd4 polyclonal antibodies were kind gifts obtained from D Rotin (77). Antiinfluenza HA epitope mouse monoclonal antibody was purchased from Covance, NJ, USA. Mouse monoclonal anti-CA antibody was purchased from NEN-DuPont (Newtown, CT, USA); monoclonal anti-FLAG antibody, anti-GFP and secondary antibodies were from Sigma (St. Louis, MO, USA). Anti-p6 rat monoclonal antibody was purchased from Advanced BioSciences (Dallas, TX, USA). Proteins were visualized by chemiluminescence using Lumi-Light reagents (Roche) or ECL (Amersham Biosciences, Piscataway, NJ, USA). The RSV polyclonal antibody used for Ip was prepared in rabbits immunized with detergent- and heat-denatured RSV.
Acknowledgments
We thank Dr E. Freed, Dr D. Rotin and Dr J. Wills for reagents. We are grateful to A. Goff and L.S. Ehrlich for helpful advice and discussions. This work was supported by National Institutes of Health grants GM 48294 (to C.C.) and CA 52047 (to J.L.). G.M. was partially supported by a W. Burghardt Turner Fellowship and by funding from NSF-Alliance for Graduate Education in the Professoriate (HRD 0450106).
References
- 1.Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 2.Wills JW, Cameron CE, Wilson CB, Xiang Y, Bennett RP, Leis J. An assembly domain of the Rous Sarcoma Virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiang Y, Cameron CE, Wills JW, Leis J. Fine mapping and characterization of the Rous Sarcoma Virus Pr76gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Göttlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 Gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang M, Orenstein JM, Martin MA, Freed EO. p6 Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demirov DG, Freed EO. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Puffer BA, Parent LJ, Wills JW, Montelaro RC. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasuda J, Hunter EA. Proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan B, Li X, Goff SP. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 1999;18:4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouamr F, Melillo JA, Wang MQ, Nagashima K, de Los Santos M, Rein A, Goff SP. PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J Virol. 2003;77:11882–11895. doi: 10.1128/JVI.77.22.11882-11895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Machesky NJ, Mansky LM. Both the PPPY and PTAP motifs are involved in human T-cell leukemia virus type 1 particle release. J Virol. 2004;78:1503–1512. doi: 10.1128/JVI.78.3.1503-1512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottwein E, Bodem J, Muller B, Schmechel A, Zentgraf H, Krausslich HG. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J Virol. 2003;77:9474–9485. doi: 10.1128/JVI.77.17.9474-9485.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harty RN, Paragas J, Sudol M, Palese PA. Proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J Virol. 1999;73:2921–2929. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FPA. PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implication for Filoviruses budding. Proc Natl Acad Sci USA. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda M, Ikeda A, Longan LC, Longnecker R. The Epstein-Barr virus latent membrane protein 2A PY motif recruits WW domain-containing ubiquitin-protein ligases. Virology. 2000;268:178–191. doi: 10.1006/viro.1999.0166. [DOI] [PubMed] [Google Scholar]
- 16.Jayakar HR, Murti KG, Whitt MA. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduces virus budding by inhibiting a late step in virion release. J Virol. 2000;74:9818–9827. doi: 10.1128/jvi.74.21.9818-9827.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Licata JM, Simpson-Holley M, Wright NT, Han Z, Paragus J, Harty RN. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins Tsg101 and Vps4. J Virol. 2003;77:1812–1819. doi: 10.1128/JVI.77.3.1812-1819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer BJ, Eck MJ. SH3 Domains. Minding your P’s and Q’s. Curr Biol. 1995;5:364–367. doi: 10.1016/s0960-9822(95)00073-x. [DOI] [PubMed] [Google Scholar]
- 19.Garnier L, Wills JW, Verderame MF, Sudol M. WW Domains and retrovirus budding. Nature. 1998;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 20.Katoh K, Shibata H, Susuki H, Nara A, Ishidoh K, Kominami E, Yoshimori T, Maki M. The ALG-2-interacting protein Alix associates with CHMP-4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J Biol Chem. 2003;278:39104–39113. doi: 10.1074/jbc.M301604200. [DOI] [PubMed] [Google Scholar]
- 21.Parent LJ, Bennett RP, Craven RC, Nelle TD, Krishna NK, Bowzard JB, Wilson CB, Puffer BA, Montelaro RC, Wills JW. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Craven RC, Harty RN, Paragas J, Palese P, Wills JW. Late domain function identified in the vesicular stomatitis virus M protein by use of Rhabdovirus-Retrovirus chimeras. J Virol. 1999;73:3359–3365. doi: 10.1128/jvi.73.4.3359-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan B, Campbell S, Bacharach E, Rein A, Goff SP. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J Virol. 2000;74:7250–7260. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F, Chen C, Puffer BA, Montelaro RC. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J Virol. 2002;76:1569–1577. doi: 10.1128/JVI.76.4.1569-1577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kikonyogo A, Bouamr F, Vana ML, Xiang Y, Aiyar A, Carter C, Leis J. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for Gag budding from cells. Proc Natl Acad Sci USA. 2001;98:11199–11204. doi: 10.1073/pnas.201268998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathways are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 28.Myers EL, Allen JF. Tsg101, an inactive homologue of ubiquitin ligase E2, interacts with human immunodeficiency virus type 2 Gag polyprotein and results in increased levels of ubiquitinated Gag. J Virol. 2002;76:11226–11235. doi: 10.1128/JVI.76.22.11226-11235.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puffer BA, Watkins SC, Montelaro RC. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1998;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pornillos O, Garrus JE, Sundquist WI. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 2002;12:569–579. doi: 10.1016/s0962-8924(02)02402-9. [DOI] [PubMed] [Google Scholar]
- 31.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DN, Scott A, Krausslich HG, Kaplan J, Morham SG, Sundquist WI. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 32.Babst M, Odorizzi G, Estepa EJ, Emr SD. Tumor susceptibility gene TSG101 and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1:242–258. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- 33.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-1. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 34.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP-1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 35.Martin-Serrano J, Bieniasz PD. A bipartite late budding domain in human immunodeficiency virus type 1. J Virol. 2003;77:12373–12377. doi: 10.1128/JVI.77.22.12373-12377.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA. 2003;100:12412–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katzmann DJ, Stefan CJ, Babst M, Emr SD. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: an endosomal-associated heterooligomeric protein complex required for MVB sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 39.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 40.Bache KG, Brech A, Mehlum A, Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol. 2003;162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bishop N, Woodman P. Tsg101/mammalian Vps23 and mammalian Vps28 interact directly and are recruited to Vps4-induced endosomes. J Biol Chem. 2001;276:11735–11742. doi: 10.1074/jbc.M009863200. [DOI] [PubMed] [Google Scholar]
- 42.Pornillos O, Higginson DS, Stray KM, Fisher RD, Garrus JE, Payne M, He G-P, Wang HE, Morham SG, Sundquist WI. HIV Gag mimics the Tsg101 recruiting activity of the human Hrs protein. J Cell Biol. 2003;162:425–434. doi: 10.1083/jcb.200302138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goff A, Ehrlich LS, Cohen SN, Carter CA. Tsg101 control of human immunodeficiency virus type 1 Gag trafficking and release. J Virol. 2003;77:9173–9182. doi: 10.1128/JVI.77.17.9173-9182.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scherer NM, Lehmann MJ, Jimenez-Soto LF, Ingmundson A, Horner SM, Cicchetti G, Alen PG, Pypaert M, Cunningham JM, Mothes W. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic. 2003;4:785–801. doi: 10.1034/j.1600-0854.2003.00135.x. [DOI] [PubMed] [Google Scholar]
- 45.Pelchen-Matthews A, Kramer B, Marsh M. Infectious HIV-1 assembles in late endosomes in primary macrophages. J Cell Biol. 2003;162:443–455. doi: 10.1083/jcb.200304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nydegger S, Foti M, Derdowski A, Spearman P, Thali M. HIV-1 egress is gated through late endosomal membranes. Traffic. 2003;4:902–910. doi: 10.1046/j.1600-0854.2003.00145.x. [DOI] [PubMed] [Google Scholar]
- 47.Tanzi GO, Piefer AJ, Bates P. Equine infectious anemia virus utilizes host vesicular protein sorting machinery during particle release. J Virol. 2003;77:8440–8447. doi: 10.1128/JVI.77.15.8440-8447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 49.Lemmon SK, Traub LM. Sorting in the endosomal system in yeast and animal cells. Curr Opin Cell Biol. 2000;12:457–466. doi: 10.1016/s0955-0674(00)00117-4. [DOI] [PubMed] [Google Scholar]
- 50.Komada M, Masaki R, Yamamoto A, Kitamura N. Hrs, a tyrosine kinase substrate with a conserved double zinc finger domain, is localized to the cytoplasmic surface of early endosomes. J Biol Chem. 1997;272:20538–20544. doi: 10.1074/jbc.272.33.20538. [DOI] [PubMed] [Google Scholar]
- 51.Raiborg C, Bache K, Gillody D, Madhust I, Stang E, Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- 52.Chin L-S, Raynor MC, Wei X, Chen HQ, Li L. Hrs interacts with sorting nexin 1 and regulates degradation of epidermal growth factor receptor. J Biol Chem. 2001;276:7069–7078. doi: 10.1074/jbc.M004129200. [DOI] [PubMed] [Google Scholar]
- 53.Bishop N, Horman A, Woodman P. Mammalian class E Vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J Cell Biol. 2002;157:91–101. doi: 10.1083/jcb.200112080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu Q, Hope LW, Brasch M, Reinhard C, Cohen SN. Tsg101 interaction with Hrs mediates endosomal trafficking and receptor downregulation. Proc Natl Acad Sci USA. 2003;100:7626–7631. doi: 10.1073/pnas.0932599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katz M, Shtiegman K, Tal-Or P, Yakir L, Mosesson Y, Harari D, Machluf Y, Asao H, Jovin T, Sugamura K, Yarden Y. Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic. 2002;3:740–751. doi: 10.1034/j.1600-0854.2002.31006.x. [DOI] [PubMed] [Google Scholar]
- 56.Lloyd T, Atkinson R, Wu M, Zhou Y, Penetta G, Bellen H. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell. 2002;108:261–269. doi: 10.1016/s0092-8674(02)00611-6. [DOI] [PubMed] [Google Scholar]
- 57.Fujita H, Yamanaka M, Imamura K, Tanaka Y, Nara A, Yoshimori T, Yolota S, Himeno MA. Dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E Vps phenotype in mammalian cells. J Cell Sci. 2003;116:401–414. doi: 10.1242/jcs.00213. [DOI] [PubMed] [Google Scholar]
- 58.Vana ML, Tang Y, Chen A, Medina G, Carter C, Leis J. Role of Nedd4 and ubiquitination of Rous sarcoma virus Gag in budding of virus-like particles from cells. J Virol. 2004;78:13943–13953. doi: 10.1128/JVI.78.24.13943-13953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shehu-Xhilaga Ablan S, Demirov DG, Chen C, Montelaro RC, Freed EO. Late domain-dependent inhibition of equine infectious anemia virus budding. J Virol. 2004;78:724–732. doi: 10.1128/JVI.78.2.724-732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rotin D, Staub O, Haguenaguer-Tsapis R. Ubiquitination and endocy-tosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J Membr Biol. 2000;176:1–17. doi: 10.1007/s00232001079. [DOI] [PubMed] [Google Scholar]
- 61.Koonin EV, Abagyan RA. Tsg101 may be the prototype of a class of dominant negative ubiquitin regulators. Nat Genet. 1997;16:330–331. doi: 10.1038/ng0897-330. [DOI] [PubMed] [Google Scholar]
- 62.Ponting CP, Cai Y-D, Bork P. The breast cancer gene product Tsg101: a regulator of ubiquitination? J Mol Med. 1997;75:467–469. [PubMed] [Google Scholar]
- 63.Pornillos O, Alam S, Rich RL, Myszka DG, Davis DR, Sundquist WI. Structural and functional interactions of the Tsg101 UEV domain. EMBO J. 2002;21:2397–2406. doi: 10.1093/emboj/21.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eastman SW, Martin-Serrano J, Chung W, Zang T, Bieniasz PD. Identification of human Vps37C, a component of ESCRT-1 important for viral budding. J Biol Chem. 2005;280:628–636. doi: 10.1074/jbc.M410384200. [DOI] [PubMed] [Google Scholar]
- 65.Goila-Gaur R, Demirov DG, Orenstein JM, Ono A, Freed EO. Defects in human immunodeficiency virus budding and endosomal sorting induced by Tsg101 over-expression. J Virol. 2003;77:9474–9485. doi: 10.1128/JVI.77.11.6507-6519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patniak A, Chau V, Wills JW. Ubiquitin is part of the retrovirus budding machinery. Proc Natl Acad Sci USA. 2000;97:13069–13074. doi: 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson MC, Spidel JL, Ako-Adjei D, Wills JW, Vogt VM. The C-terminal half of Tsg101 blocks Rous sarcoma virus budding and sequesters Gag into unique nonendosomal structures. J Virol. 2005;79:3775–3786. doi: 10.1128/JVI.79.6.3775-3786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT Ubiquitin ligases and virus release. J Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li L, Liao J, Ruland J, Mak T, Cohen SN. A Tsg101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control. Proc Natl Acad Sci USA. 2001;98:1616–1624. doi: 10.1073/pnas.98.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun Z, Pan J, Hope W, Cohen S, Balk S. Tumor susceptibility gene 101 protein represses androgen receptor transactivation and interacts with p300. Cancer. 1999;86:689–696. doi: 10.1002/(sici)1097-0142(19990815)86:4<689::aid-cncr19>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 71.Amit I, Yakir L, Katz M, Zwang Y, Marmor M, Citri A, Shtiegman K, Alroy I, Tuvia S, Reiss Y, Roubini E, Cohen M, Wides R, Bacharach E, Schubert U et al. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 2004;18:1737–1752. doi: 10.1101/gad.294904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grossman S, Deato M, Brignone C, Chan H, Kung A, Togami H, Nakatami Y, Livingston D. Polyubiquitination of p53 by a Ubiquitin Ligase activity of p300. Science. 2003;300:342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- 73.Hegde AN. Ubiquitin-proteasome-mediated local protein degradation and synaptic plasticity. Prog Neurobiol. 2004;73:311–357. doi: 10.1016/j.pneurobio.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 74.Ono A, Freed EO. Cell-type dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J Virol. 2004;78:1552–1563. doi: 10.1128/JVI.78.3.1552-1563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Demirov DG, Orenstein JM, Freed EO. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J Virol. 2002;76:105–117. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hermida-Matsumoto L, Resh MD. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J Virol. 2000;74:8670–8679. doi: 10.1128/jvi.74.18.8670-8679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Staub O, Yeger H, Plant PJ, Kim H, Ernst SA, Rotin D. Immunocolocalization of the ubiquitin-protein ligase Nedd4 in tissues expressing the epithelial Na+ channel (ENaC) Am J Physiol. 1997;272:1871–1880. doi: 10.1152/ajpcell.1997.272.6.C1871. [DOI] [PubMed] [Google Scholar]