Abstract
Biological polymers such as nucleic acids and proteins are ubiquitous in living systems, but their ability to address problems beyond those found in nature is constrained by factors such as chemical or biological instability, limited building-block functionality, bioavailability, and immunogenicity. In principle, sequence-defined synthetic polymers based on nonbiological monomers and backbones might overcome these constraints; however, identifying the sequence of a synthetic polymer that possesses a specific desired functional property remains a major challenge. Molecular evolution can rapidly generate functional polymers but requires a means of translating amplifiable templates such as nucleic acids into the polymer being evolved. This review covers recent advances in the enzymatic and nonenzymatic templated polymerization of nonnatural polymers and their potential applications in the directed evolution of sequence-defined synthetic polymers.
Biological Polymers: Evolvable but Structurally Narrow
Living systems must solve an enormous number of chemical challenges in order to sustain life. These challenges include the replication of genetic information and the translation of this information into functional molecules that mediate biological processes and maintain cellular structure. The molecular solutions to these challenges are predominantly sequence-defined biological polymers and the biosynthetic products that arise from their action.
The properties of DNA, RNA, and proteins are well suited to meet the diverse chemical needs of living systems. These biopolymers can exhibit a strong propensity to adopt stable three-dimensional structures in the aqueous, roughly neutral environment of most organisms. The folded state of these structures enables the precise positioning of functional groups in ways that might otherwise be enthalpically or entropically disfavored. The precise arrangement of functional groups, in turn, enables biological polymers to exhibit their remarkable binding and catalytic properties and allows them to interact with molecules of virtually every scale.
The ability to form well-folded structures, however, is not sufficient to explain the dominance of DNA, RNA, and proteins in biology. Among the many ways to construct a sequence-defined polymer, α-peptides and nucleotides are not the only solutions that can form higher-order structures. Many different varieties of foldable polymers (“foldamers”), including β-peptides and nucleic acid analogs, are known to adopt stable secondary, tertiary, and even quaternary structures (Hill et al., 2001; Horne and Gellman, 2008).
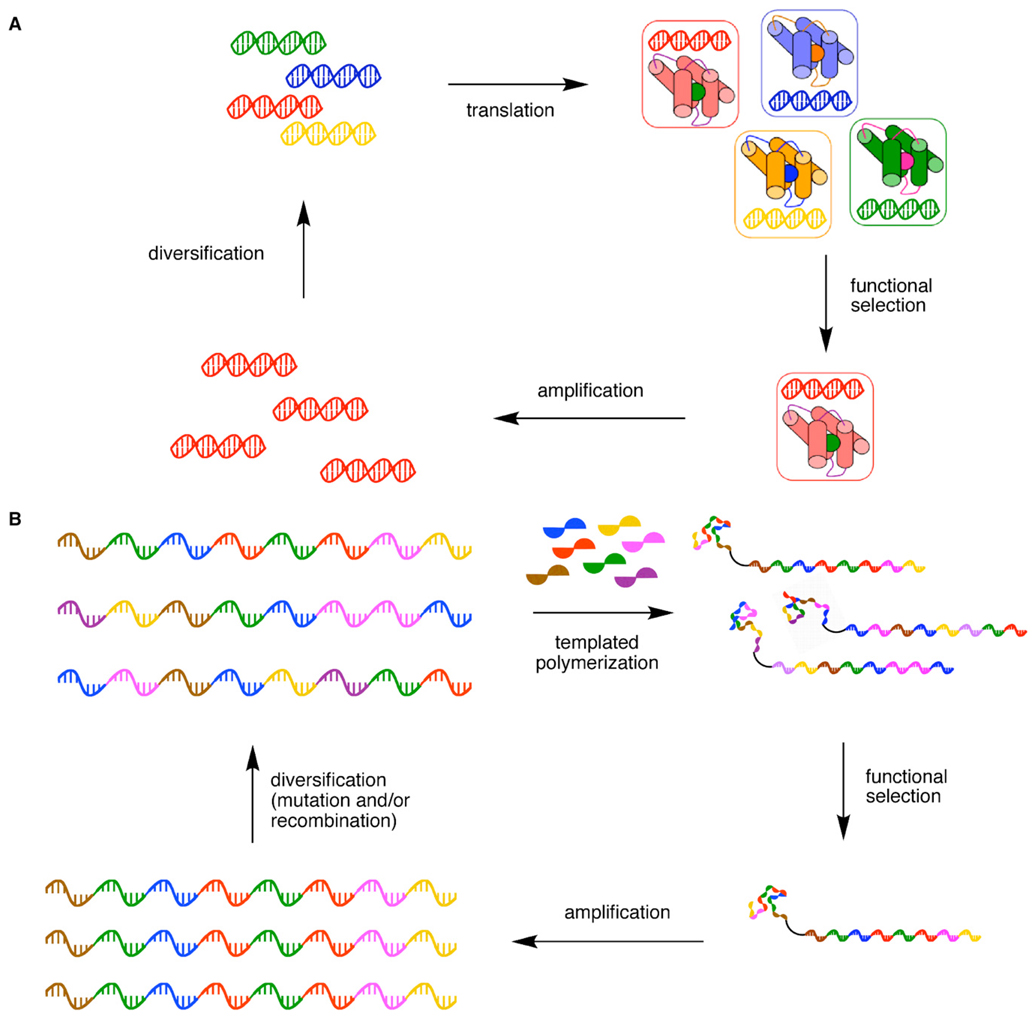
Instead, the dominance of DNA, RNA, and proteins among biological molecules with complex functional capabilities is almost certainly a consequence of their ability to evolve in the molecular context of biotic systems. A biological polymer with favorable functional properties, such as the ability to catalyze a reaction essential to life, increases the probability that the information carrier encoding its structure survives and replicates. This information is gradually diversified through mutation and recombination, then translated into a new generation of related polymer variants to complete the cycle of evolution. This ability of biopolymers to be translated from an information carrier (DNA or RNA) that can replicate and mutate (Figure 1A) allows them to explore sequence space in a manner guided by their functional capabilities. Compared with a random, nondirected exploration of sequence space, an evolution-directed search of sequence space can dramatically reduce the number of nonfunctional biopolymers that must be generated before a functional variant emerges (Sen et al., 2007; Joyce, 2004).
Figure 1. Key Components of the Evolution of Biological or Synthetic Polymers.
(A) Biopolymer evolution as it occurs in nature or in the laboratory. A polymer (such as a protein) is translated from and spatially associated with an information carrier (such as DNA). The resulting biopolymers undergo selection based on their functional properties. The information encoding surviving biopolymers replicates and mutates, resulting in a second generation of biopolymer variants related to those that survived selection.
(B) Synthetic polymers can in principle undergo a similar evolutionary process. Translation could be effected by nonenzymatic or enzymatic templated synthesis in a manner that associates each synthetic polymer with its information carrier. Following selection, the information carriers encoding surviving synthetic polymers are amplified and mutated to generate templates for a subsequent round of translation.
Despite these remarkable features, the structures of DNA, RNA, and proteins are constrained in ways that limit their usefulness in many therapeutic, diagnostic, and research applications. Biological polymers can be unstable to chemical conditions such as extreme pH or ion concentrations and solvent composition that lie outside of those typically found in living organisms. DNA, RNA, and proteins are also vulnerable to degradation by nuclease and protease enzymes. Moreover, therapeutic proteins and peptides introduced into a foreign host can provoke problematic immune responses. The therapeutic application of nucleic acids and proteins also suffers from delivery challenges, as most biological macromolecules are unable to cross the cell membrane and exhibit poor bioavailability.
Synthetic Polymers: Structurally Broad, but Not Evolvable
Sequence-defined polymers produced through chemical synthesis can overcome many of the constraints that limit the usefulness of biological polymers. Access to a much wider potential set of backbones and building blocks might allow researchers to create synthetic polymers that have enhanced mechanical strength or flexibility, have sophisticated optical properties, are able to conduct electricity, or are resistant to chemical or biological degradation. In addition, scientists could introduce building blocks into synthetic polymers with functional groups known to sense specific molecules (McQuade et al., 2000; Spangler et al., 2008; Miller and Chang, 2007), serve as biophysical probes (Spangler et al., 2008), or catalyze specific reactions (White et al., 2001). The ability of synthetic polymers to interact with biological systems can likewise be engineered to confer adhesion to certain tissues (Poncin-Epaillard and Legeay, 2003; Vasir et al., 2003; Woodley, 2001; Patil and Sawant, 2008), biodegradation (Alvarez-Lorenzo and Concheiro, 2008; Shoyele, 2008), or cytotoxicity (Hunter, 2006; Arnon et al., 1989; Galmarini et al., 2008; Kono et al., 2008); conversely, synthetic polymers can be made bio-orthogonal by the appropriate choice of building blocks to limit undesired interference with cellular processes (Yliperttula et al., 2008; Anderson et al., 2008).
A major challenge to fully realizing the potential of a given type of synthetic polymer is determining the appropriate building-block sequence that can confer a desired functional property. Significant recent advances have been achieved in the de novo design of RNA (Stormo, 2006), proteins (Das and Baker, 2008), and synthetic foldamers (Kirshenbaum et al., 1999; Kay et al., 2007; Patch and Barron, 2002). Some of the most successful recent examples of protein design (Jiang et al., 2008; Rothlisberger et al., 2008; Cochran et al., 2005), however, are based on models trained using empirical three-dimensional structural data that are not available for virtually all nonbiological polymers. The de novo design of sequence-defined polymers (synthetic or biological) with tailor-made structural or functional properties remains a difficult problem due to factors such as unpredictable conformational changes, unforeseen solvent interactions, and unknown stereoelectronic requirements (Tuchscherer et al., 1998; Ryadnov, 2007; Dill et al., 2007).
Evolvable Synthetic Polymers
The highly effective evolution-based approach to generating functional polymers in the laboratory has historically been limited to two types of molecules–proteins and nucleic acids–because for decades they were the only polymers that could be translated from the information in DNA or RNA. Methods to translate nucleic acids into nonnatural polymers would enable synthetic polymers to be diversified using mutagenesis and recombination, directly selected for desired binding or catalytic properties, and amplified by replicating the information encoding active molecules similar to the way that nature evolves proteins (Figure 1B).
In this article, we will review recent advances in the enzymatic and nonenzymatic synthesis of sequence-defined nonnatural polymers through templated polymerization. These developments represent promising ways to translate nucleic acids into polymers not limited to those used by living systems. The first half of this review focuses on nonnatural polymers created with the aid of DNA and RNA polymerases and the ribosome. The second half reviews efforts to translate information carriers into synthetic polymers in a sequence-specific manner without the aid of, and the constraints imposed by, biosynthetic enzymes.
Enzyme-Mediated Templated Synthesis of Nonnatural Polymers
Polymerase-Catalyzed Synthesis of Base-Modified Nucleic Acids
As nucleic acids have become increasingly important as sensors (O’Sullivan, 2002; Navani and Li, 2006; Lee et al., 2008), potential therapeutics (Eckstein, 2007; Kaur and Roy, 2008; Levy-Nissenbaum et al., 2008), and cellular probes (Phillips et al., 2008), numerous polymerase-compatible modifications have been investigated to increase chemical and biological stability and to confer other functional improvements.
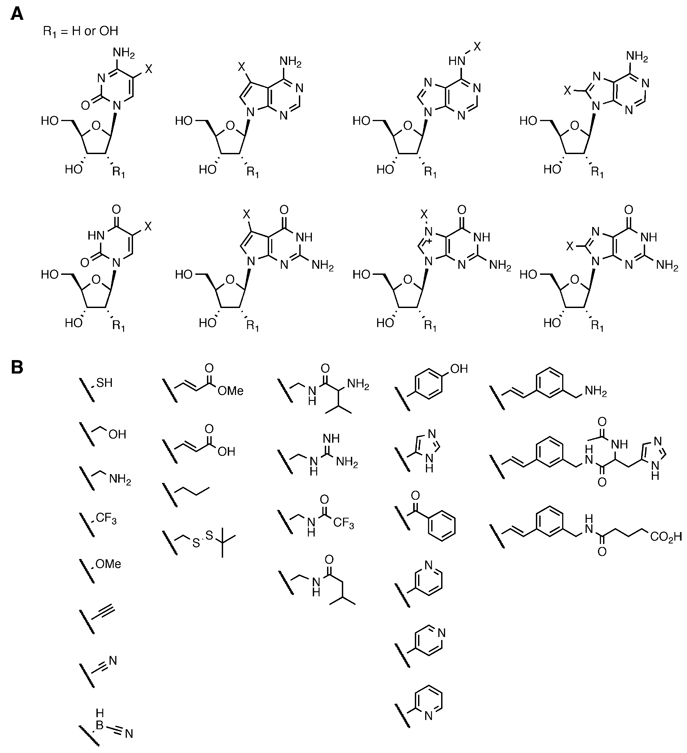
The enzyme-mediated polymerization of nucleotide triphosphates with altered base-pairing groups has been the subject of considerable research. The impetus for much of this work has been to study nucleic acid duplex stability and the function of DNA and RNA polymerases, and has been reviewed recently (Hwang and Romesberg, 2006; Henry and Romesberg, 2003; Hirao, 2006; Jung and Marx, 2005; Kool, 2001). The discovery of novel, polymerase-compatible nucleic acid bases has created interest in synthetic polymers that might contain higher densities of information than DNA or RNA, or ones that may even be used in orthogonal transcription systems. Scientists have also sought to increase the chemical diversity of oligonucleotides by incorporating chemical functionalities that are not present in natural RNA and DNA into nucleotide triphosphates. In this manner, more than 100 functionalized nucleotides have been incorporated into DNA and RNA, including those containing nucleophilic groups such as amines and thiols, electrophilic groups such as acrylates and aldehydes, proton donors and acceptors such as imidazole, pyridine, and guanidinium groups, and reactive groups such as cyanoborohydride. Many of these efforts are summarized in Figure 2; this area has also been reviewed recently (Jung and Marx, 2005; Porter et al., 1995; Hocek and Fojta, 2008; Krayevsky et al., 1996; Kuwahara et al., 2006; McGall, 2005).
Figure 2. Examples of Modified Nucleobases that Retain Compatibility with Polymerase Enzymes.
(A) Nucleic acid bases can be modified at several different positions (labeled X) without abolishing Watson-Crick base pairing or the ability of the resulting nucleotide to be incorporated into an oligonucleotide using a polymerase enzyme (Jung and Marx, 2005; Hocek and Fojta, 2008; Krayevsky et al., 1996; Kuwahara et al., 2006).
(B) More than 100 different base modifications have been introduced into nucleic acid polymers through the enzyme-mediated polymerization of base-modified nucleotide triphosphates. A representative set is shown here (Jung and Marx, 2005; Porter et al., 1995; Hocek and Fojta, 2008; Krayevsky et al., 1996; Kuwahara et al., 2006; McGall, 2005).
Polymerase-Catalyzed Synthesis of Backbone-Modified Nucleic Acids
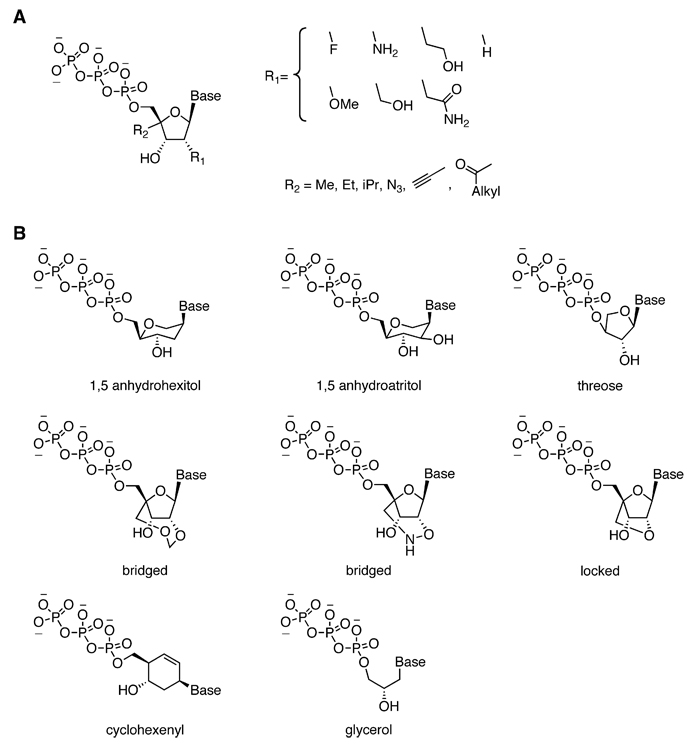
A more extreme form of polymer modification involves replacing or modifying the phosphate-ribose nucleic acid backbone. For example, modification of the 2′-hydroxyl group of RNA increases the stability of RNA and confers nuclease resistance. A number of different 2′ groups have been successfully incorporated in a sequence-specific manner using polymerase enzymes including fluoro-, amino-, methoxy-, and amido-ribonucleotides (Figure 3A). As a result of these advances, the discovery of aptamers from libraries of 2′-modified ribonucleotides has recently been achieved (Proske et al., 2002; White et al., 2008; Kubik et al., 1997).
Figure 3. Examples of Ribose-Modified or Non-Ribose-Based Nucleic Acids that Retain Compatibility with Polymerase Enzymes.
(A) Nucleic acid building blocks containing modified ribose groups that remain substrates for polymerase enzymes (Proske et al., 2002; White et al., 2008; Kubik et al., 1997; Summerer and Marx, 2004, 2005; Chen et al., 1993; Marx et al., 1998).
(B) Nonribose nucleic acid building blocks that retain the ability to form base pairs with DNA or RNA, and that can be incorporated into nucleic acids using polymerase enzymes (Vastmans et al., 2000, 2001, 2002; Pochet et al., 2003; Kempeneers et al., 2003, 2005; Chaput and Szostak, 2003; Horhota et al., 2005, 2006; Ichida et al., 2005a, 2005b; Veedu et al., 2007a, 2007b, 2008; Kuwahara et al., 2008; Chaput et al., 2002).
In their study into the mechanism of Klenow DNA polymerase, Marx and coworkers were able to incorporate building blocks containing alkyl groups at the 4′ position (Summerer and Marx, 2004). Several modifications at the 4′ position were studied as possible HIV reverse-transcriptase inhibitors, including the azide (Chen et al., 1993), alkyne (Summerer and Marx, 2005), and acyl (Marx et al., 1997, 1998) moieties (Figure 3A).
The polymerase-mediated incorporation of backbones that do not contain a ribose group has also been reported. The incorporation of anhydrohexitol nucleic acid (HNA) and anhydroaltritol (ANA) opposite DNA or RNA templates was studied by Herdewijn and coworkers (Figure 3B) (Vastmans et al., 2000, 2001; Pochet et al., 2003). They found that between four and six hexitol nucleotides could be incorporated selectively into an extending primer by various DNA and RNA polymerases, and as many as 15 hexitol monomers could be appended by the enzyme terminal transferase (Vastmans et al., 2002). Herdewijn and coworkers also studied the enzyme-mediated polymerization of cyclohexenyl nucleic acid purinotide triphosphates (Figure 3B, cyclohexenyl) and demonstrated the sequence-specific polymerization of these nucleotides opposite DNA templates containing T and C. The authors observed the polymerization of up to seven consecutive nonnatural nucleotides (Kempeneers et al., 2005).
The Szostak and Herdewijn groups studied the transcription of threose nucleic acids (TNA) from DNA templates (Figure 3B, threose) (Chaput and Szostak, 2003; Chaput et al., 2003; Kempeneers et al., 2003). TNA binds tightly to RNA and DNA and has a simpler backbone, leading some researchers to speculate that it may have served a role in the prebiotic world (Orgel, 2000). Szostak and coworkers investigated the kinetics of TNA synthesis and identified “Therminator” DNA polymerase as a particularly privileged enzyme for the efficient polymerization of TNA triphosphates (Horhota et al., 2005). Subsequent efforts have led to “transcribed” TNA oligomers of at least 80 consecutive nucleotides using this enzyme (Ichida et al., 2005a) and the demonstration of an in vitro selection system that in principle can be used to select for TNA aptamers and ribozymes (Ichida et al., 2005b). Meggers and coworkers simplified the phosphate-sugar backbone to its barest minimum through the synthesis and characterization of glycerol nucleic acids (GNA) (Zhang et al., 2005). Szostak and coworkers were able to show the sequence-specific enzyme-catalyzed incorporation of a single GNA nucleotide triphosphate (Figure 3B, glycerol) (Horhota et al., 2006).
Wengel, Kuwahara, and their respective coworkers recently described the enzymatic polymerization of locked nucleic acid (LNA) triphosphates on DNA and RNA templates (Figure 3B, locked) (Veedu et al., 2007a, 2007b, 2008; Kuwahara et al., 2008). LNAs are already routinely incorporated into DNA and RNA oligonucleotides through solid-phase synthesis and can increase the stability of double-stranded nucleic acids by preorganizing the DNA backbone (Crinelli et al., 2004). The discovery that LNA nucleotides can be incorporated enzymatically may enable the study of conformationally locked nucleic acids in large RNA molecules or the generation of aptamers with improved stability. Kuwahara and coworkers also studied the enzymatic polymerization of other 2′–4′ bridged nucleic acid triphosphates (Figure 3B, bridged) (Kuwahara et al., 2008).
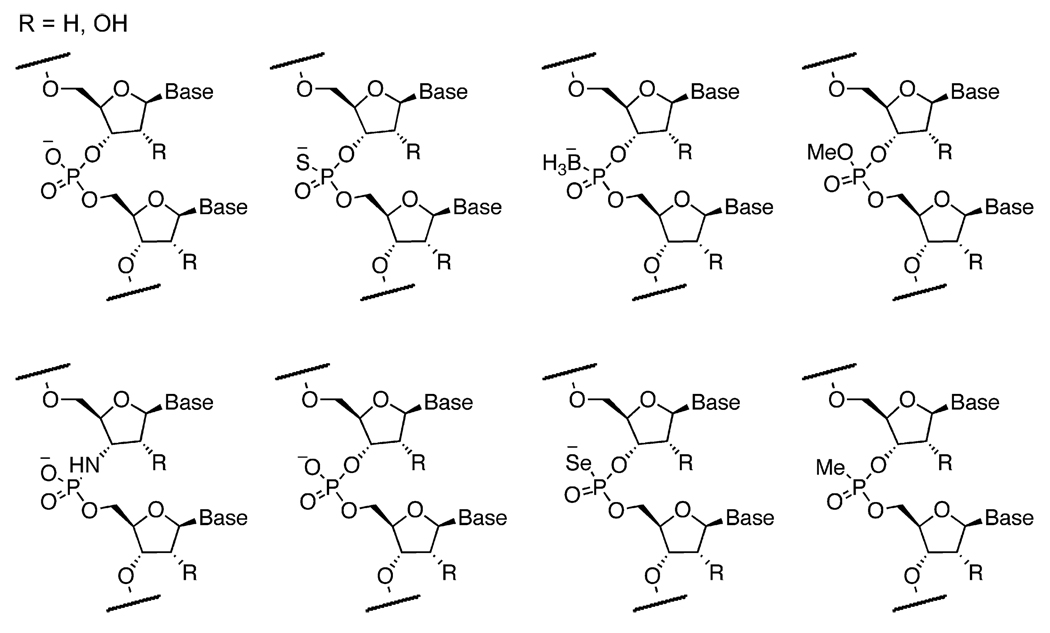
In addition to substituting the sugar group of the backbone, the phosphate group has also been the focus of recent efforts to generate sequence-defined nonnatural polymers using polymerase enzymes. As the site of most chemical and enzymatic degradation of oligonucleotides, the phosphodiester group has been replaced with a number of analogs that offer increased stability. Phosphate-backbone substitutions, in which one of the nonbridging oxygen atoms is replaced, can confer greater nuclease resistance, lipophilicity, and polarizability. Most polymerases will accept substitutions in only one of the two enantiotopic nonbridging oxygen atoms of the phosphate. All-phosphorothioate oligonucleotides have been generated by enzymatic transcription of DNA with sulfur-containing nucleotide triphosphates. Selections from enzyme-generated libraries containing phosphorothioate backbones have successfully yielded aptamers (Kang et al., 2007, 2008; King et al., 1998, 2002).
In a similar manner, an oxygen atom in the phosphate group can also be replaced with selenium to form phosphoroselenoate oligonucleotides (Carrasco and Huang, 2004). Phosphoroselenoates can aid in the analysis of nucleic acid crystallography (Wilds et al., 2002). Shaw and coworkers demonstrated that 5′(α-P-borano) nucleotide triphosphates are excellent substrates for DNA polymerases (Tomasz et al., 1992; Summers and Shaw, 2001; Li et al., 1995; He et al., 1999). Borane-containing oligonucleotides show increased resistance to nuclease degradation. In addition, aptamers and oligonucleotides containing boron may be used in combination with boron neutron capture therapy as activatable radiotherapeutics against tumors (Barth et al., 2005). Burke and coworkers recently reported a boron-containing aptamer to ATP that requires the borane group in order to function (Lato et al., 2002). Petkov and coworkers reported the incorporation of methylphosphono nucleotide triphosphate where one of the oxygens on the phosphate is replaced with a methyl group (Dineva et al., 1993). Escherichia coli DNA polymerase 1 was found to mediate the sequence-specific synthesis of methylphosphodiester nucleic acids. Phosphoramidates, in which one bridging oxygen is replaced with a nitrogen, have also been incorporated into DNA (Wolfe et al., 2002). Unlike phosphodiester bonds, the phosphate-amine bond in phosphoramidate oligonucleotides can be cleaved with mild acid (Letsinger and Wilkes, 1976), potentially opening the way for novel sequencing methods (Wolfe et al., 2002). Wolfe and coworkers have demonstrated a method in which the incorporation of a ribonucleotide before the phosphoramidate linkage allows for cleavage of an oligonucleotide only at specific dinucleotides (Wolfe et al., 2003). Representative examples of enzymatically synthesized nucleic acids containing phosphate replacements are shown in Figure 4.
Figure 4. Examples of Phosphate Analogs that Have Been Successfully Incorporated into Nucleic Acids Using Polymerase Enzymes.
Phosphate analogs have become increasingly common in alternative oligonucleotide backbones to improve the physicochemical properties of DNA and RNA or to introduce novel functional properties (Kuwahara et al., 2008; Crinelli et al.,2004; Kang et al., 2007, 2008; King et al., 1998, 2002; Carrasco and Huang, 2004; Wilds et al., 2002; Tomasz et al., 1992; Summers and Shaw, 2001; Li et al., 1995; He et al., 1999; Barth et al., 2005; Lato et al., 2002; Dineva et al., 1993; Wolfe et al., 2002, 2003; Letsinger and Wilkes, 1976).
Ribosome-Based Generation of Nonnatural Polymers
The ribosomal machinery creates oligomers through the RNA-templated coupling of activated amino acids. Several groups have taken advantage of this natural translation machinery to create novel biopolymers. Initial work pioneered by Schultz, Hecht, Chamberlain, and their respective coworkers (Bain et al., 1989, 1991; Noren et al., 1989; Ellman et al., 1992) focused on incorporating nonnatural amino acids site-specifically into existing proteins by using chemical (Hecht et al., 1978; Heckler et al., 1984) or enzymatic (Hartman et al., 2006) aminoacylation of amber suppressor tRNAs. These methods have been extensively reviewed (Wang et al., 2006; Xie and Schultz, 2005, 2006).
A limitation of nonsense suppression methods is the inability to simultaneously and independently direct the incorporation of more than two nonnatural amino acids. Reassignment of sense codons to nonnatural amino acids allows the incorporation of multiple amino acids in the same peptide. The PURE (protein synthesis using recombinant elements) system developed by Ueda and coworkers uses in vitro translation from purified recombinant factors derived from E. coli (Shimizu et al., 2001). As a result, the PURE system enables the withdrawal of aminoacyl-tRNA synthetases and amino acids from the reconstituted translation mixture and therefore the reassignment of multiple codons to nonnatural amino acids (Forster et al., 2003). The PURE system was used to prepare polypeptides containing between one and three different N-alkylated amino acids (Subtelny et al., 2008; Zhang et al., 2007; Frankel et al., 2003; Tan et al., 2004). Suga and coworkers integrated PURE with “flexizymes” (Murakami et al., 2006), ribozymes that acylate tRNAs, to enable the synthesis of more complex nonproteinogenic peptides. Using this combination of methods, Suga and coworkers demonstrated the single incorporation of 15 different N-methyl amino acids (Figure 5) and the multiple incorporation of 6 different N-methyl amino acids in response to arbitrarily chosen codons in a 12 or 15 amino acid peptide (Kawakami et al., 2007; 2008).
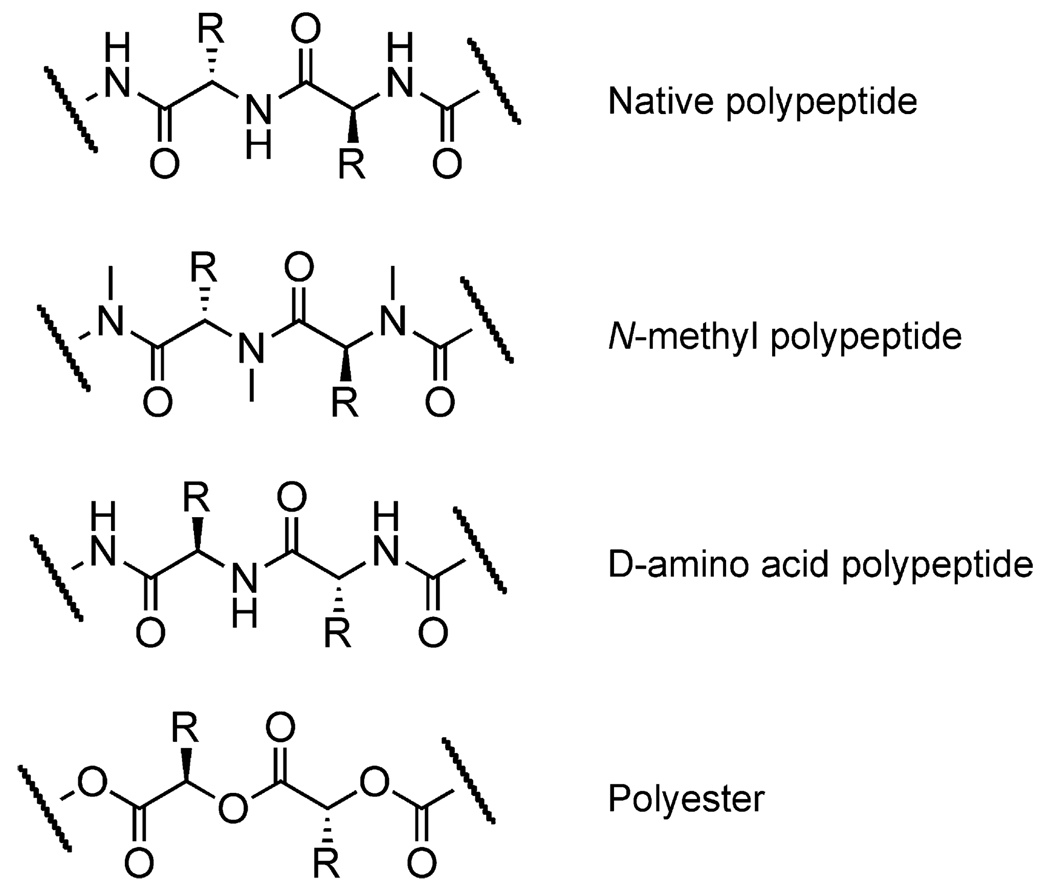
Figure 5. Nonnatural Polymers Created with Native or Engineered Ribosomes.
Due to the substrate requirements of the ribosome, all of the resulting backbones still resemble native polypeptides (Subtelny et al., 2008; Zhang et al., 2007; Frankel et al., 2003; Tan et al., 2004; Kawakami et al., 2007, 2008; Ohta et al., 2007, 2008a, 2008b; Dedkova et al., 2003, 2006).
The powerful combination of PURE and flexizymes allowed Suga and coworkers to polymerize 12 different α-hydroxy acids in combination to create polyesters up to 12 units long and containing up to three different side chains (Figure 5) (Ohta et al., 2007, 2008a). The authors reason that their inability to create longer polyesters might be due to the poor affinity of elongation factor Tu (EF-Tu) for α-hydroxy aminoacylated tRNAs, suggesting that modification of EF-Tu or the ribosome might allow the synthesis of longer polymers. This work and similar studies have recently been highlighted in an excellent review (Ohta et al., 2008b). Representative cases for ribosomally synthesized nonnatural polymers are shown in Figure 5.
Altering Natural Polymerase and Ribosomal Machinery to Accept Nonnatural Building Blocks
Although a variety of nonnatural polymers have been created using existing enzymes, progress using this approach can be hampered by the low efficiency with which natural polymerases or ribosomes incorporate nonnatural monomers that differ significantly from natural building blocks. To overcome this problem, some groups have increased the promiscuity of natural systems and improved the ability of polymerases and ribosomes to accept modified building blocks. Romesberg and coworkers have reviewed progress in evolving polymerases to accept a wide range of modified nucleotides (Henry and Romesberg, 2005; Holmberg et al., 2005).
Altering the building-block specificity of ribosomes appears to be a more difficult problem. Unlike polymerase enzymes, the ribosome consists of many large and intimately integrated RNA and protein molecules functioning together. Nevertheless, a few examples of ribosome modification for the creation of new materials have been reported. Hecht and coworkers have modified the peptidyl transferase center on the ribosome to permit the incorporation of (D)-amino acids into proteins (Dedkova et al., 2003, 2006) (Figure 5). This approach may one day enable the enzymatic synthesis of all-(D) proteins. Chin and coworkers evolved an orthogonal ribosome-termination factor pair that is independent of the E. coli translational machinery and shows an improved ability to incorporate p-benzoyl-(L)-phenylalanine into proteins (Wang et al., 2007).
Modifying nonribosomal proteins involved in ribosomal translation may also lead to improved nonnatural amino acid incorporation. Ohtsuki and coworkers rationally engineered an EF-Tu variant that enhanced the incorporation of amino acids containing bulky aromatic groups (Doi et al., 2007).
Nonenzymatic Translation of Nucleic Acids into Synthetic Polymers
Sequence-defined polymers generated through biosynthetic pathways are limited to those made of building blocks compatible with machinery such as the ribosome or polymerase enzymes. Despite significant advances that augment the structural diversity of biosynthetic proteins and nucleic acids (many of which are reviewed in this article), arbitrary building-block structures cannot in general be accommodated by the biosynthetic machinery or by known variants of these enzymes, limiting the structural and functional diversity of biosynthesized polymers.
An alternative approach to translating information carriers into nonnatural polymers does not use polymerase enzymes or the ribosome but instead relies on nonenzymatic template-directed polymerization. The impetus for the earliest studies in enzymefree templated polymerizations was to understand chemical routes by which information could be copied in a prebiotic world preceding the advent of proteins. In a series of seminal studies, Orgel and coworkers demonstrated nonenzymatic templatedirected synthesis of oligoribonucleotides on complementary oligonucleotides (Orgel, 1992; Sulston et al., 1969; Schneider-Bernloehr et al., 1970; Lohrmann and Orgel, 1976, 1977, 1979; Ninio and Orgel, 1978). These findings have been expanded to include nonnatural templates (Kozlov et al., 1999a, 1999b, 1999c, 2000; Chaput and Switzer, 2000; Schmidt et al., 1997a).
Nonenzymatic Synthesis of Polymers Containing Ribose Analogs
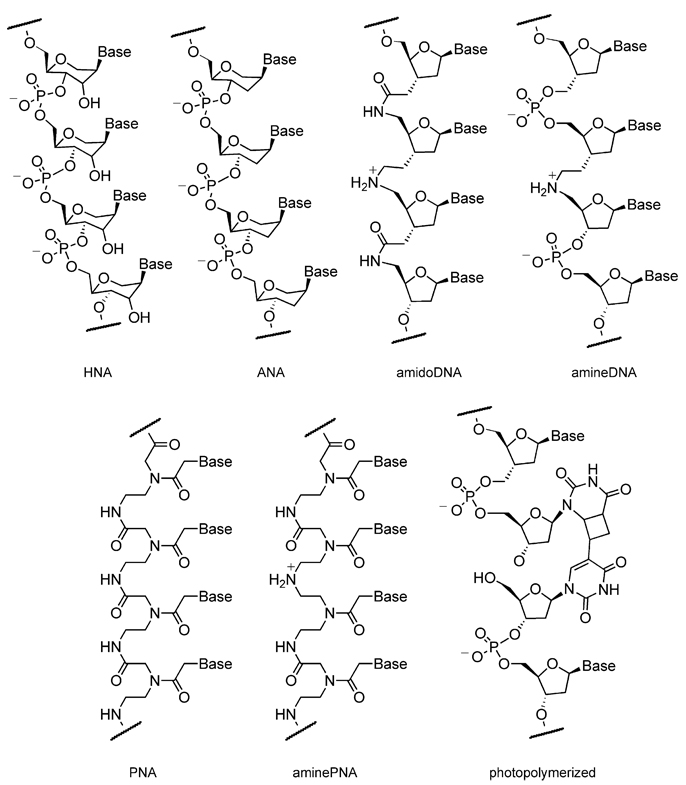
One notable challenge facing the RNA world hypothesis has been the prebiotic synthesis of β-ribofuranoside-5′-phosphates. This difficulty has led researchers to hypothesize that other information-carrying molecules, the components of which might be more easily formed than those of RNA, may have preceded RNA (Orgel, 2000; Anastasi et al., 2007). To understand the choice of ribose and deoxyribose in natural systems, Eschenmoser and coworkers systematically investigated structural alternatives to RNA (Eschenmoser, 1999). Their work led to the discovery of a set of nonnatural nucleic acids that efficiently form duplex structures with RNA and DNA and thereby demonstrated that natural nucleic acids are not unique in their ability to form stable, sequence-specific double-stranded structures. Orgel and coworkers reported the template-directed polymerization of some of these nonnatural nucleic acids using 2-methylimidazole-activated hexitol and altritol guanosine monomers (Kozlov et al., 2000) to make hexitol and altriol nucleic acids (HNAs and ANAs, respectively). HNA and ANA oligomers (Figure 6) form antiparallel duplexes with complementary DNA or RNA oligomers with structures that closely resemble that of A-form double-stranded nucleic acids (Hendrix et al., 1997a, 1997b; Wang et al., 2001; Allart et al., 1999). The authors found it difficult to create HNA or ANA oligomers longer than tetramers by template-directed polymerization, which suggests that not all monomers that form stable double-helical polymers necessarily undergo efficient templated oligomeriza-tion.
Figure 6. Synthetic Polymers Created by Nonenzymatic DNA- or RNA-Templated Polymerization of Synthetic Building Blocks.
See Anastasi et al. (2007); Eschenmoser (1999); Hendrix et al. (1997a); Zhan and Lynn (1997); Leitzel and Lynn (2001); Li et al. (2002); Li and Lynn (2002); Hud et al. (2007); Nielsen (1995); Dueholm et al. (1994); Englund and Appella (1995); and Nelson et al. (2000).
Orgel and coworkers discovered that an oligo-cytosine peptide nucleic acid (PNA) template facilitates the polymerization of activated guanosine RNA nucleotides, although the reaction is less regioselective for the formation of native 3′–5′ bonds than the templated polymerization using RNA templates (Bohler et al., 1995). The researchers extended the approach of PNA-templated RNA polymerization to adenine, cytosine, and thymine nucleotides and demonstrated that the polymerization proceeds in a sequence-specific manner (Schmidt et al., 1997a). Because the PNA template is achiral, it can in principle provide a route to access mirror-image nucleic acids. As expected, (L)-ribose-based activated nucleosides were shown to polymerize as readily as their (D)-ribose counterparts, although a racemic mixture of (D)- and (L)-ribose nucleotides showed considerable crossinhibition during polymerization (Schmidt et al., 1997b).
Reductive Amination-Based Polymerization of DNA and Amido-DNA
Lynn and coworkers developed a solution to the problem of information transfer between polymers that does not involve the formation of phosphodiester linkages. Goodwin and Lynn reasoned that, using a reaction that creates an equilibrium between transiently coupled and uncoupled substrates, the sequence-matched template-substrate complex would be thermodynamically favored and then could be locked in a subsequent irreversible step (Goodwin and Lynn, 1992). They incorporated an aldehyde and an amine group at the termini of nucleic acid building blocks and used reductive amination to ligate both DNA trimers (Figure 6, amineDNA) and nonnatural ribonucleotides in which the phosphate is replaced with an amide group (Figure 6, amidoDNA). The researchers found that this ligation reaction was not product inhibited, because the secondary amines formed from reductive amination exhibited reduced affinity for template compared with the imine-containing intermediates (Zhan and Lynn, 1997; Leitzel and Lynn, 2001). Lynn and coworkers extended this methodology to the DNA-catalyzed polymerization of synthetic mono-, di-, and tetra-nucleotides (Li et al., 2002; Li and Lynn, 2002) and made polymers of amidoDNA as long as 32 nucleotides (Hud et al., 2007).
Nonenzymatic Polymerization of PNA
Among synthetic polymers with the ability to recognize the information encoded in DNA or RNA, PNA (Figure 6, PNA) is an attractive backbone for several reasons. PNA resembles RNA in its ability to form double-helical complexes stabilized by Watson-Crick bonding between opposite strands. PNA also forms stable and selective duplexes with RNA, DNA, or itself (Nielsen, 1995). Side-chain-containing PNA building blocks can be readily accessed from α-amino acids and alcohols (Dueholm et al., 1994; Englund and Appella, 1995). Finally, the constituent components of PNA have been isolated under putative prebiotic conditions, raising the possibility that nonenzymatic PNA polymerizations may have relevance for primitive information-transfer events (Nelson et al., 2000).
Orgel, Nielsen, and coworkers used amine-carboxylic acid coupling to condense cytosine dimers of peptide nucleic acid into 10-mer PNAs (Bohler et al., 1995) in the presence, but not in the absence, of complementary G10 RNA templates. They also examined the sequence specificity of this polymerization reaction for PNA dimers (G2, A2, C2, and T2) on a decamer DNA template containing the sequence 5′-CCCCXXCCCC-3′, where X is any of the four nucleotides. The authors found that in each case, the building block complementary to X is preferentially incorporated, but found considerable (up to 35% in the worst case) misincorporation of the wrong building block as well as significant amounts of truncated products (Schmidt et al., 1997c).
Building on the work of Lynn, Nielsen, Orgel, and their respective coworkers, Rosenbaum and Liu sought to develop the sequence-specific DNA-templated polymerization of PNA aldehydes. Liu and coworkers previously reported the strong dependence of DNA-templated reductive amination reaction efficiency on the adjacency of template-bound reactants (Gartner et al., 2002). These observations together with Lynn’s successes with DNA-templated DNA and amidoDNA polymerization using reductive amination suggested an effective method to translate DNA sequences into corresponding peptide nucleic acids. Indeed, PNA tetramers containing aldehyde C termini exhibited highly efficient polymerization on DNA templates containing five or ten repeats (20 or 40 bases) of complementary nucleotides (Rosenbaum and Liu, 2003) (Figure 6, aminoPNA). The polymerization also proceeded in a sequence-specific manner, showing a preference to terminate rather than to incorporate a sequence-mismatched PNA building block.
By adorning the PNA building blocks with chemical functional groups not available to biological nucleic acids, Liu and coworkers increased the structural and functional potential of the resulting DNA-templated PNA polymers (Kleiner et al., 2008). In agreement with previous studies on the stability of modified PNA-DNA duplexes (Dueholm et al., 1994; Englund and Appella, 1995), they observed that the presence of a side chain at the γ position did not significantly impede polymerization provided that the (L) stereochemistry of the side chain was used, that both side-chain stereochemistries at the α position of the PNA building block are also tolerated, and that a side chain at the γ position with (D) stereochemistry results in a marked decrease in DNA-templated PNA polymerization efficiency. These findings raise the possibility of using DNA-templated PNA polymerization as a basis for the directed evolution of functionalized PNA polymers.
Photopolymerization
Extending previous studies describing the photoligation of DNA (Lewis and Hanawalt, 1982; Letsinger et al., 1997; Liu and Taylor, 1998), Saito and coworkers demonstrated the photo-induced oligomerization of DNA hexamers (Fujimoto et al., 2000) through the [2+2] photocyclization between vinyluracil and uracil (Figure 6, photopolymerized). Photopolymerizations offer several potential advantages as compared to chemical polymerization. The use of light avoids the need to add reagents, simplifies purification, allows spatial and temporal control of polymerization, and in some cases, such as this one, enables polymerization to be reversed. The building blocks contained 5-vinyldeoxyuridine as the 5′ nucleic base of a building block, which reacts with a 3′-uracil on neighboring building blocks under 366 nm light. The resulting joined DNA can be quantitatively reverted to starting building blocks by irradiation at 302 nm. This photoligation strategy may prove useful in the templated polymerization of a variety of nonnatural polymers, including many of the backbones mentioned in this review.
Conclusion
Recent examples of nonnatural sequence-defined polymers formed through the enzymatic or nonenzymatic polymerization of nucleic acids have provided a tantalizing taste of synthetic polymers that could one day be synthesized and evolved in a manner resembling the biosynthesis and evolution of biological polymers. The integration of the translation methods described above with in vitro selection would represent a powerful tool for the discovery of novel functional polymers with tailor-made properties that in principle could extend beyond those of DNA, RNA, and proteins.
In addition to the functional polymers that might arise from synthetic polymer evolution, research in this area will illuminate the relationship between building-block structure, backbone structure, and the functional potential of a polymer. These studies may even provide insights into why DNA, RNA, and proteins have come to play their special roles in biology. The transitions from a world based on a primitive polymer to a world based on RNA and then a world based on proteins were mediated by information-transfer steps that largely remain to be discovered. The translation of information-carrying polymers into a variety of other, more functional polymers could begin to generate plausible model systems for understanding these transitions. The continued development of key components toward the evolution of synthetic polymers may therefore give rise both to improved therapeutics, sensors, catalysts, or materials, as well as to new insights into life’s chemical origins.
ACKNOWLEDGMENTS
Y.B. and D.R.L. gratefully acknowledge support from the Office of Naval Research (N00014-03-1-0749), the National Institutes of Health (GM065865), and the Howard Hughes Medical Institute. We thank Elizaveta Freinkman and Kevin Esvelt for helpful discussions. Y.B. gratefully acknowledges the support of an NSF Graduate Research Fellowship.
REFERENCES
- Allart B, Khan K, Rosemeyer H, Schepers G, Hendrix C, Rothenbacher K, Seela F, Van Aerschot A, Herdewijn P. D-altritol nucleic acids (ANA): hybridisation properties, stability, and initial structural analysis. Chem. Eur. J. 1999;5:2424–2431. [Google Scholar]
- Alvarez-Lorenzo C, Concheiro A. Intelligent drug delivery systems: polymeric micelles and hydrogels. Mini Rev. Med. Chem. 2008;8:1065–1074. doi: 10.2174/138955708785909952. [DOI] [PubMed] [Google Scholar]
- Anastasi C, Buchet FF, Crowe MA, Parkes AL, Powner MW, Smith JM, Sutherland JD. RNA: prebiotic product, or biotic invention? Chem. Biodivers. 2007;4:721–739. doi: 10.1002/cbdv.200790060. [DOI] [PubMed] [Google Scholar]
- Anderson JM, Rodriguez A, Chang DT. Foreign body reaction to biomaterials. Semin. Immunol. 2008;20:86–100. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon R, Hurwitz E, Schechter B. The use of antibodies and polymers as carriers of cytotoxic drugs in the treatment of cancer. Horiz. Biochem. Biophys. 1989;9:33–56. [PubMed] [Google Scholar]
- Bain JD, Glabe CG, Dix TA, Chamberlin AR, Diala ES. Biosynthetic site-specific incorporation of a non-natural amino-acid into a polypeptide. J. Am. Chem. Soc. 1989;111:8013–8014. [Google Scholar]
- Bain JD, Diala ES, Glabe CG, Wacker DA, Lyttle MH, Dix TA, Chamberlin AR. Site-specific incorporation of nonnatural residues during in vitro protein biosynthesis with semisynthetic aminoacyl-transfer RNAs. Biochemistry. 1991;30:5411–5421. doi: 10.1021/bi00236a013. [DOI] [PubMed] [Google Scholar]
- Barth RF, Coderre JA, Vicente MG, Blue TE. Boron neutron capture therapy of cancer: current status and future prospects. Clin. Cancer Res. 2005;11:3987–4002. doi: 10.1158/1078-0432.CCR-05-0035. [DOI] [PubMed] [Google Scholar]
- Bohler C, Nielsen PE, Orgel LE. Template switching between PNA and RNA oligonucleotides. Nature. 1995;376:578–581. doi: 10.1038/376578a0. [DOI] [PubMed] [Google Scholar]
- Carrasco N, Huang Z. Enzymatic synthesis of phosphoroselenoate DNA using thymidine 5′-(α-P-seleno)triphosphate and DNA polymerase for X-ray crystallography via MAD. J. Am. Chem. Soc. 2004;126:448–449. doi: 10.1021/ja0383221. [DOI] [PubMed] [Google Scholar]
- Chaput JC, Switzer C. Nonenzymatic oligomerization on templates containing phosphodiester-linked acyclic glycerol nucleic acid analogues. J. Mol. Evol. 2000;51:464–470. doi: 10.1007/s002390010109. [DOI] [PubMed] [Google Scholar]
- Chaput JC, Szostak JW. TNA synthesis by DNA polymerases. J. Am. Chem. Soc. 2003;125:9274–9275. doi: 10.1021/ja035917n. [DOI] [PubMed] [Google Scholar]
- Chaput JC, Sinha S, Switzer C. 5-propynyluracil.diaminopurine: an efficient base-pair for non-enzymatic transcription of DNA. Chem. Commun. (Camb.) 2002:1568–1569. doi: 10.1039/b204535d. [DOI] [PubMed] [Google Scholar]
- Chaput JC, Ichida JK, Szostak JW. DNA polymerase-mediated DNA synthesis on a TNA template. J. Am. Chem. Soc. 2003;125:856–857. doi: 10.1021/ja028589k. [DOI] [PubMed] [Google Scholar]
- Chen MS, Suttmann RT, Papp E, Cannon PD, McRoberts MJ, Bach C, Copeland WC, Wang TS. Selective action of 4′-azidothymidine triphosphate on reverse transcriptase of human immunodeficiency virus type 1 and human DNA polymerases α and β. Biochemistry. 1993;32:6002–6010. doi: 10.1021/bi00074a011. [DOI] [PubMed] [Google Scholar]
- Cochran FV, Wu SP, Wang W, Nanda V, Saven JG, Therien MJ, DeGrado WF. Computational de novo design and characterization of a four-helix bundle protein that selectively binds a nonbiological cofactor. J. Am. Chem. Soc. 2005;127:1346–1347. doi: 10.1021/ja044129a. [DOI] [PubMed] [Google Scholar]
- Crinelli R, Bianchi M, Gentilini L, Palma L, Magnani M. Locked nucleic acids (LNA): versatile tools for designing oligonucleotide decoys with high stability and affinity. Curr. Drug Targets. 2004;5:745–752. doi: 10.2174/1389450043345083. [DOI] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with Rosetta. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Dedkova LM, Fahmi NE, Golovine SY, Hecht SM. Enhanced D-amino acid incorporation into protein by modified ribosomes. J. Am. Chem. Soc. 2003;125:6616–6617. doi: 10.1021/ja035141q. [DOI] [PubMed] [Google Scholar]
- Dedkova LM, Fahmi NE, Golovine SY, Hecht SM. Construction of modified ribosomes for incorporation of D-amino acids into proteins. Biochemistry. 2006;45:15541–15551. doi: 10.1021/bi060986a. [DOI] [PubMed] [Google Scholar]
- Dill KA, Ozkan SB, Weikl TR, Chodera JD, Voelz VA. The protein folding problem: when will it be solved? Curr. Opin. Struct. Biol. 2007;17:342–346. doi: 10.1016/j.sbi.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Dineva MA, Chakurov S, Bratovanova EK, Devedjiev I, Petkov DD. Complete template-directed enzymatic synthesis of a potential antisense DNA containing 42 methylphosphonodiester bonds. Bioorg. Med. Chem. 1993;1:411–414. doi: 10.1016/s0968-0896(00)82151-3. [DOI] [PubMed] [Google Scholar]
- Doi Y, Ohtsuki T, Shimizu Y, Ueda T, Sisido M. Elongation factor Tu mutants expand amino acid tolerance of protein biosynthesis system. J. Am. Chem. Soc. 2007;129:14458–14462. doi: 10.1021/ja075557u. [DOI] [PubMed] [Google Scholar]
- Dueholm KL, Petersen KH, Jensen DK, Egholm M, Nielsen PE, Buchardt O. Peptide nucleic-acid (PNA) with a chiral backbone based on alanine. Bioorg. Med. Chem. Lett. 1994;4:1077–1080. [Google Scholar]
- Eckstein F. The versatility of oligonucleotides as potential therapeutics. Expert Opin. Biol. Ther. 2007;7:1021–1034. doi: 10.1517/14712598.7.7.1021. [DOI] [PubMed] [Google Scholar]
- Ellman JA, Mendel D, Schultz PG. Site-specific incorporation of novel backbone structures into proteins. Science. 1992;255:197–200. doi: 10.1126/science.1553546. [DOI] [PubMed] [Google Scholar]
- Englund EA, Appella DH. Synthesis of γ-substituted peptide nucleic acids: a new place to attach fluorophores without affecting DNA binding. Org. Lett. 1995;7:3465–3467. doi: 10.1021/ol051143z. [DOI] [PubMed] [Google Scholar]
- Eschenmoser A. Chemical etiology of nucleic acid structure. Science. 1999;284:2118–2124. doi: 10.1126/science.284.5423.2118. [DOI] [PubMed] [Google Scholar]
- Forster AC, Tan Z, Nalam MN, Lin H, Qu H, Cornish VW, Blacklow SC. Programming peptidomimetic syntheses by translating genetic codes designed de novo. Proc. Natl. Acad. Sci. USA. 2003;100:6353–6357. doi: 10.1073/pnas.1132122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A, Millward SW, Roberts RW. Encodamers: unnatural peptide oligomers encoded in RNA. Chem. Biol. 2003;10:1043–1050. doi: 10.1016/j.chembiol.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Fujimoto K, Matsuda S, Takahashi N, Saito I. Template-directed photoreversible ligation of deoxyoligonucleotides via 5-vinyldeoxyuridine. J. Am. Chem. Soc. 2000;122:5646–5647. [Google Scholar]
- Galmarini CM, Warren G, Kohli E, Zeman A, Mitin A, Vinogradov SV. Polymeric nanogels containing the triphosphate form of cytotoxic nucleoside analogues show antitumor activity against breast and colorectal cancer cell lines. Mol. Cancer Ther. 2008;7:3373–3380. doi: 10.1158/1535-7163.MCT-08-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner ZJ, Kanan MW, Liu DR. Expanding the reaction scope of DNA-templated synthesis. Angew. Chem. Int. Ed. Engl. 2002;41:1796–1800. doi: 10.1002/1521-3773(20020517)41:10<1796::aid-anie1796>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Goodwin JT, Lynn DG. Template-directed synthesis: use of a reversible reaction. J. Am. Chem. Soc. 1992;114:9197–9198. [Google Scholar]
- Hartman MC, Josephson K, Szostak JW. Enzymatic aminoacylation of tRNA with unnatural amino acids. Proc. Natl. Acad. Sci. USA. 2006;103:4356–4361. doi: 10.1073/pnas.0509219103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Porter KW, Hasan A, Briley JD, Shaw BR. Synthesis of 5-substituted 2′-deoxycytidine 5′-(α-P-borano)triphosphates, their incorporation into DNA and effects on exonuclease. Nucleic Acids Res. 1999;27:1788–1794. doi: 10.1093/nar/27.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht SM, Alford BL, Kuroda Y, Kitano S. “Chemical aminoacylation” of tRNA’s. J. Biol. Chem. 1978;253:4517–4520. [PubMed] [Google Scholar]
- Heckler TG, Chang LH, Zama Y, Naka T, Hecht SM. Preparation of ′2,(′3)-O-acyl-pCpA derivatives as substrates for T4 RNA ligasemediated “chemical aminoacylation.”. Tetrahedron. 1984;40:87–94. [Google Scholar]
- Hendrix C, Rosemeyer H, DeBouvere B, VanAerschot A, Seela F, Herdewijn P. 1′,5′-anhydrohexitol oligonucleotides: hybridisation and strand displacement with oligoribonucleotides, interaction with RNase H and HIV reverse transcriptase. Chem. Eur. J. 1997a;3:1513–1520. [Google Scholar]
- Hendrix C, Rosemeyer H, Verheggen I, Seela F, VanAerschot A, Herdewijn P. 1′,5′-anhydrohexitol oligonucleotides: synthesis, base pairing and recognition by regular oligodeoxyribonucleotides and oligoribonucleotides. Chem. Eur. J. 1997b;3:110–120. [Google Scholar]
- Henry AA, Romesberg FE. Beyond A, C, G and T: augmenting nature’s alphabet. Curr. Opin. Chem. Biol. 2003;7:727–733. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Henry AA, Romesberg FE. The evolution of DNA polymerases with novel activities. Curr. Opin. Biotechnol. 2005;16:370–377. doi: 10.1016/j.copbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Hill DJ, Mio MJ, Prince RB, Hughes TS, Moore JS. A field guide to foldamers. Chem. Rev. 2001;101:3893–4012. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]
- Hirao I. Unnatural base pair systems for DNA/RNA-based biotechnology. Curr. Opin. Chem. Biol. 2006;10:622–627. doi: 10.1016/j.cbpa.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Hocek M, Fojta M. Cross-coupling reactions of nucleoside triphosphates followed by polymerase incorporation. Construction and applications of base-functionalized nucleic acids. Org. Biomol. Chem. 2008;6:2233–2241. doi: 10.1039/b803664k. [DOI] [PubMed] [Google Scholar]
- Holmberg RC, Henry AA, Romesberg FE. Directed evolution of novel polymerases. Biomol. Eng. 2005;22:39–49. doi: 10.1016/j.bioeng.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Horhota A, Zou KY, Ichida JK, Yu B, McLaughlin LW, Szostak JW, Chaput JC. Kinetic analysis of an efficient DNA-dependent TNA polymerase. J. Am. Chem. Soc. 2005;127:7427–7434. doi: 10.1021/ja0428255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horhota AT, Szostak JW, McLaughlin LW. Glycerol nucleoside triphosphates: synthesis and polymerase substrate activities. Org. Lett. 2006;8:5345–5347. doi: 10.1021/ol062232u. [DOI] [PubMed] [Google Scholar]
- Horne WS, Gellman SH. Foldamers with heterogeneous backbones. Acc. Chem. Res. 2008;41:1399–1408. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hud NV, Jain SS, Li XH, Lynn DG. Addressing the problems of base pairing and strand cyclization in template-directed synthesis—a case for the utility and necessity of ‘molecular midwives’ and reversible backbone linkages for the origin of proto-RNA. Chem. Biodivers. 2007;4:768–783. doi: 10.1002/cbdv.200790063. [DOI] [PubMed] [Google Scholar]
- Hunter AC. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Adv. Drug Deliv. Rev. 2006;58:1523–1531. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Hwang GT, Romesberg FE. Substituent effects on the pairing and polymerase recognition of simple unnatural base pairs. Nucleic Acids Res. 2006;34:2037–2045. doi: 10.1093/nar/gkl049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida JK, Horhota A, Zou KY, McLaughlin LW, Szostak JW. High fidelity TNA synthesis by Therminator polymerase. Nucleic Acids Res. 2005a;33:5219–5225. doi: 10.1093/nar/gki840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida JK, Zou K, Horhota A, Yu B, McLaughlin LW, Szostak JW. An in vitro selection system for TNA. J. Am. Chem. Soc. 2005b;127:2802–2803. doi: 10.1021/ja045364w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, III, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce GF. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- Jung KH, Marx A. Nucleotide analogues as probes for DNA polymerases. Cell. Mol. Life Sci. 2005;62:2080–2091. doi: 10.1007/s00018-005-5117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Lee MS, Watowich SJ, Gorenstein DG. Combinatorial selection of a RNA thioaptamer that binds to Venezuelan equine encephalitis virus capsid protein. FEBS Lett. 2007;581:2497–2502. doi: 10.1016/j.febslet.2007.04.072. [DOI] [PubMed] [Google Scholar]
- Kang J, Lee MS, Copland JA, III, Luxon BA, Gorenstein DG. Combinatorial selection of a single stranded DNA thioaptamer targeting TGF-β1 protein. Bioorg. Med. Chem. Lett. 2008;18:1835–1839. doi: 10.1016/j.bmcl.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G, Roy I. Therapeutic applications of aptamers. Expert Opin. Investig. Drugs. 2008;17:43–60. doi: 10.1517/13543784.17.1.43. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Murakami H, Suga H. Exploration of incorporation of Nα-methylated amino acids into peptides by sense-suppression method. Nucleic Acids Symp. Ser. (Oxf) 2007;51:361–362. doi: 10.1093/nass/nrm181. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Murakami H, Suga H. Messenger RNA-programmed incorporation of multiple N-methyl-amino acids into linear and cyclic peptides. Chem. Biol. 2008;15:32–42. doi: 10.1016/j.chembiol.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Kay ER, Leigh DA, Zerbetto F. Synthetic molecular motors and mechanical machines. Angew. Chem. Int. Ed. Engl. 2007;46:72–191. doi: 10.1002/anie.200504313. [DOI] [PubMed] [Google Scholar]
- Kempeneers V, Vastmans K, Rozenski J, Herdewijn P. Recognition of threosyl nucleotides by DNA and RNA polymerases. Nucleic Acids Res. 2003;31:6221–6226. doi: 10.1093/nar/gkg833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempeneers V, Renders M, Froeyen M, Herdewijn P. Investigation of the DNA-dependent cyclohexenyl nucleic acid polymerization and the cyclohexenyl nucleic acid-dependent DNA polymerization. Nucleic Acids Res. 2005;33:3828–3836. doi: 10.1093/nar/gki695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DJ, Ventura DA, Brasier AR, Gorenstein DG. Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry. 1998;37:16489–16493. doi: 10.1021/bi981780f. [DOI] [PubMed] [Google Scholar]
- King DJ, Bassett SE, Li X, Fennewald SA, Herzog NK, Luxon BA, Shope R, Gorenstein DG. Combinatorial selection and binding of phosphorothioate aptamers targeting human NF-κ B RelA(p65) and p50. Biochemistry. 2002;41:9696–9706. doi: 10.1021/bi020220k. [DOI] [PubMed] [Google Scholar]
- Kirshenbaum K, Zuckermann RN, Dill KA. Designing polymers that mimic biomolecules. Curr. Opin. Struct. Biol. 1999;9:530–535. doi: 10.1016/S0959-440X(99)80075-X. [DOI] [PubMed] [Google Scholar]
- Kleiner RE, Brudno Y, Birnbaum ME, Liu DR. DNA-templated polymerization of side-chain-functionalized peptide nucleic acid aldehydes. J. Am. Chem. Soc. 2008;130:4646–4659. doi: 10.1021/ja0753997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono K, Kojima C, Hayashi N, Nishisaka E, Kiura K, Watarai S, Harada A. Preparation and cytotoxic activity of poly(ethylene glycol)-modified poly(amidoamine) dendrimers bearing adriamycin. Biomaterials. 2008;29:1664–1675. doi: 10.1016/j.biomaterials.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Kool ET. Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu. Rev. Biophys. Biomol. Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- Kozlov IA, De Bouvere B, Van Aerschot A, Herdewijn P, Orgel LE. Efficient transfer of information from hexitol nucleic acids to RNA during nonenzymatic oligomerization. J. Am. Chem. Soc. 1999a;121:5856–5859. doi: 10.1021/ja990440u. [DOI] [PubMed] [Google Scholar]
- Kozlov IA, Politis PK, Pitsch S, Herdewijn P, Orgel LE. A highly enantio-selective hexitol nucleic acid template for nonenzymatic oligoguanylate synthesis. J. Am. Chem. Soc. 1999b;121:1108–1109. doi: 10.1021/ja9836489. [DOI] [PubMed] [Google Scholar]
- Kozlov IA, Politis PK, Van Aerschot A, Busson R, Herdewijn P, Orgel LE. Nonenzymatic synthesis of RNA and DNA oligomers on hexitol nucleic acid templates: the importance of the A structure. J. Am. Chem. Soc. 1999c;121:2653–2656. doi: 10.1021/ja983958r. [DOI] [PubMed] [Google Scholar]
- Kozlov IA, Zielinski M, Allart B, Kerremans L, Van Aerschot A, Busson R, Herdewijn P, Orgel LE. Nonenzymatic template-directed reactions on altritol oligomers, preorganized analogues of oligonucleotides. Chem. Eur. J. 2000;6:151–155. doi: 10.1002/(sici)1521-3765(20000103)6:1<151::aid-chem151>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Krayevsky AA, Alexandrova LA, Dyatkina NB, Kukhanova MK, Shirokova EA. Modified substrates of DNA polymerases and design of antivirals. Acta Biochim. Pol. 1996;43:125–132. [PubMed] [Google Scholar]
- Kubik MF, Bell C, Fitzwater T, Watson SR, Tasset DM. Isolation and characterization of 2′-fluoro-, 2′-amino-, and 2′-fluoro-/aminomodified RNA ligands to human IFN-γ that inhibit receptor binding. J. Immunol. 1997;159:259–267. [PubMed] [Google Scholar]
- Kuwahara M, Suto Y, Minezaki S, Kitagata R, Nagashima J, Sawai H. Substrate property and incorporation accuracy of various dATP analogs during enzymatic polymerization using thermostable DNA polymerases. Nucleic Acids Symp. Ser. (Oxf) 2006;50:31–32. doi: 10.1093/nass/nrl016. [DOI] [PubMed] [Google Scholar]
- Kuwahara M, Obika S, Nagashima J, Ohta Y, Suto Y, Ozaki H, Sawai H, Imanishi T. Systematic analysis of enzymatic DNA polymerization using oligo-DNA templates and triphosphate analogs involving 2′,4′-bridged nucleosides. Nucleic Acids Res. 2008;36:4257–4265. doi: 10.1093/nar/gkn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lato SM, Ozerova ND, He K, Sergueeva Z, Shaw BR, Burke DH. Boron-containing aptamersto ATP. Nucleic Acids Res. 2002;30:1401–1407. doi: 10.1093/nar/30.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JO, So HM, Jeon EK, Chang H, Won K, Kim YH. Aptamers as molecular recognition elements for electrical nanobiosensors. Anal. Bioanal. Chem. 2008;390:1023–1032. doi: 10.1007/s00216-007-1643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitzel JC, Lynn DG. Template-directed ligation: from DNA towards different versatile templates. Chem. Rec. 2001;1:53–62. doi: 10.1002/1528-0691(2001)1:1<53::AID-TCR8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Letsinger FL, Wilkes JS. Incorporation of 5′-amino-5′-deoxythymidine 5′-phosphate in polynucleotides by use of DNA polymerase I and a ϕX174 DNA template. Biochemistry. 1976;15:2810–2816. doi: 10.1021/bi00658a017. [DOI] [PubMed] [Google Scholar]
- Letsinger RL, Wu TF, Elghanian R. Chemical and photochemical ligation of oligonucleotide blocks. Nucleosides Nucleotides. 1997;16:643–652. [Google Scholar]
- Levy-Nissenbaum E, Radovic-Moreno AF, Wang AZ, Langer R, Farokhzad OC. Nanotechnology and aptamers: applications in drug delivery. Trends Biotechnol. 2008;26:442–449. doi: 10.1016/j.tibtech.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Lewis RJ, Hanawalt PC. Ligation of oligonucleotides by pyrimidine dimers—a missing link in the origin of life. Nature. 1982;298:393–396. doi: 10.1038/298393a0. [DOI] [PubMed] [Google Scholar]
- Li XY, Lynn DG. Polymerization on solid supports. Angew. Chem. Int. Ed. Engl. 2002;41:4567–4569. doi: 10.1002/1521-3773(20021202)41:23<4567::AID-ANIE4567>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Li H, Porter K, Huang F, Shaw BR. Boron-containing oligodeoxyribonucleotide 14mer duplexes: enzymatic synthesis and melting studies. Nucleic Acids Res. 1995;23:4495–4501. doi: 10.1093/nar/23.21.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhan ZYJ, Knipe R, Lynn DG. DNA-catalyzed polymerization. J. Am. Chem. Soc. 2002;124:746–747. doi: 10.1021/ja017319j. [DOI] [PubMed] [Google Scholar]
- Liu J, Taylor JS. Template-directed photoligation of oligodeoxyribonucleotides via 4-thiothymidine. Nucleic Acids Res. 1998;26:3300–3304. doi: 10.1093/nar/26.13.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann R, Orgel LE. Template-directed synthesis of high molecular weight polynucleotide analogues. Nature. 1976;261:342–344. doi: 10.1038/261342a0. [DOI] [PubMed] [Google Scholar]
- Lohrmann R, Orgel LE. Reactions of adenosine 5′phosphorimidazolide with adenosine analogs on a polyuridylic acid template. J. Mol. Biol. 1977;113:193–198. doi: 10.1016/0022-2836(77)90049-3. [DOI] [PubMed] [Google Scholar]
- Lohrmann R, Orgel LE. Studies of oligoadenylate formation on a poly (U) template. J. Mol. Evol. 1979;12:237–257. doi: 10.1007/BF01732341. [DOI] [PubMed] [Google Scholar]
- Marx A, MacxWilliams MP, Bickle TA, Schwitter U, Giese B. 4′-acylated thymidines: a new class of DNA chain terminators and photocleavable DNA building blocks. J. Am. Chem. Soc. 1997;119:1131–1132. [Google Scholar]
- Marx A, Amacker M, Stucki M, Hubscher U, Bickle TA, Giese B. 4′-acylated thymidine 5′-triphosphates: a tool to increase selectivity towards HIV-1 reverse transcriptase. Nucleic Acids Res. 1998;26:4063–4067. doi: 10.1093/nar/26.17.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGall GH. Nucleoside triphosphate analogs for nonradioactive labeling of nucleic acids. In: Vaghefi MM, editor. Nucleoside Triphosphates and Their Analogs: Chemistry, Biotechnology, and Biological Applications. Boca Raton, FL: CRC Press; 2005. pp. 269–327. [Google Scholar]
- McQuade DT, Pullen AE, Swager TM. Conjugated polymerbased chemical sensors. Chem. Rev. 2000;100:2537–2574. doi: 10.1021/cr9801014. [DOI] [PubMed] [Google Scholar]
- Miller EW, Chang CJ. Fluorescent probes for nitric oxide and hydrogen peroxide in cell signaling. Curr. Opin. Chem. Biol. 2007;11:620–625. doi: 10.1016/j.cbpa.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Ohta A, Goto Y, Sako Y, Suga H. Flexizyme as a versatile tRNA acylation catalyst and the application for translation. Nucleic Acids Symp. Ser. (Oxf) 2006;50:35–36. doi: 10.1093/nass/nrl018. [DOI] [PubMed] [Google Scholar]
- Navani NK, Li Y. Nucleic acid aptamers and enzymes as sensors. Curr. Opin. Chem. Biol. 2006;10:272–281. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Levy M, Miller SL. Peptide nucleic acids rather than RNA may have been the first genetic molecule. Proc. Natl. Acad. Sci. USA. 2000;97:3868–3871. doi: 10.1073/pnas.97.8.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PE. DNA analogues with nonphosphodiester backbones. Annu. Rev. Biophys. Biomol. Struct. 1995;24:167–183. doi: 10.1146/annurev.bb.24.060195.001123. [DOI] [PubMed] [Google Scholar]
- Ninio J, Orgel LE. Heteropolynucleotides as templates for non-enzymatic polymerizations. J. Mol. Evol. 1978;12:91–99. doi: 10.1007/BF01733260. [DOI] [PubMed] [Google Scholar]
- Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. A general method for site-specific incorporation of unnatural amino acids into proteins. Science. 1989;244:182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- Ohta A, Murakami H, Higashimura E, Suga H. Synthesis of polyester by means of genetic code reprogramming. Chem. Biol. 2007;14:1315–1322. doi: 10.1016/j.chembiol.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Ohta A, Murakami H, Suga H. Polymerization of α-hydroxy acids by ribosomes. ChemBioChem. 2008a;9:2773–2778. doi: 10.1002/cbic.200800439. [DOI] [PubMed] [Google Scholar]
- Ohta A, Yamagishi Y, Suga H. Synthesis of biopolymers using genetic code reprogramming. Curr. Opin. Chem. Biol. 2008b;12:159–167. doi: 10.1016/j.cbpa.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Orgel LE. Molecular replication. Nature. 1992;358:203–209. doi: 10.1038/358203a0. [DOI] [PubMed] [Google Scholar]
- Orgel L. Origin of life. A simpler nucleic acid. Science. 2000;290:1306–1307. doi: 10.1126/science.290.5495.1306. [DOI] [PubMed] [Google Scholar]
- O’Sullivan CK. Aptasensors—the future of biosensing? Anal. Bioanal. Chem. 2002;372:44–48. doi: 10.1007/s00216-001-1189-3. [DOI] [PubMed] [Google Scholar]
- Patch JA, Barron AE. Mimicry of bioactive peptides via non-natural, sequence-specific peptidomimetic oligomers. Curr. Opin. Chem. Biol. 2002;6:872–877. doi: 10.1016/s1367-5931(02)00385-x. [DOI] [PubMed] [Google Scholar]
- Patil SB, Sawant KK. Mucoadhesive microspheres: a promising tool in drug delivery. Curr. Drug Deliv. 2008;5:312–318. doi: 10.2174/156720108785914970. [DOI] [PubMed] [Google Scholar]
- Phillips JA, Lopez-Colon D, Zhu Z, Xu Y, Tan W. Applications of aptamers in cancer cell biology. Anal. Chim. Acta. 2008;621:101–108. doi: 10.1016/j.aca.2008.05.031. [DOI] [PubMed] [Google Scholar]
- Pochet S, Kaminski PA, Van Aerschot A, Herdewijn P, Marliere P. Replication of hexitol oligonucleotides as a prelude to the propagation of a third type of nucleic acid in vivo. C. R. Biol. 2003;326:1175–1184. doi: 10.1016/j.crvi.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Poncin-Epaillard F, Legeay G. Surface engineering of biomaterials with plasma techniques. J. Biomater. Sci. Polym. Ed. 2003;14:1005–1028. doi: 10.1163/156856203769231538. [DOI] [PubMed] [Google Scholar]
- Porter KW, Tomasz J, Huang F, Sood A, Shaw BR. N7-cyanoborane-2′-deoxyguanosine 5′-triphosphate is a good substrate for DNA polymerase. Biochemistry. 1995;34:11963–11969. doi: 10.1021/bi00037a038. [DOI] [PubMed] [Google Scholar]
- Proske D, Gilch S, Wopfner F, Schatzl HM, Winnacker EL, Famulok M. Prion-protein-specific aptamer reduces PrPSc formation. ChemBioChem. 2002;3:717–725. doi: 10.1002/1439-7633(20020802)3:8<717::AID-CBIC717>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Rosenbaum DM, Liu DR. Efficient and sequence-specific DNA-templated polymerization of peptide nucleic acid aldehydes. J. Am. Chem. Soc. 2003;125:13924–13925. doi: 10.1021/ja038058b. [DOI] [PubMed] [Google Scholar]
- Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- Ryadnov MG. Peptide α-helices for synthetic nanostructures. Biochem. Soc. Trans. 2007;35:487–491. doi: 10.1042/BST0350487. [DOI] [PubMed] [Google Scholar]
- Schmidt JG, Nielsen PE, Orgel LE. Information transfer from peptide nucleic acids to RNA by template-directed syntheses. Nucleic Acids Res. 1997a;25:4797–4802. doi: 10.1093/nar/25.23.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JG, Nielsen PE, Orgel LE. Enantiomeric cross-inhibition in the synthesis of oligonucleotides on a nonchiral template. J. Am. Chem. Soc. 1997b;119:1494–1495. doi: 10.1021/ja963563c. [DOI] [PubMed] [Google Scholar]
- Schmidt JG, Christensen L, Nielsen PE, Orgel LE. Information transfer from DNA to peptide nucleic acids by template-directed syntheses. Nucleic Acids Res. 1997c;25:4792–4796. doi: 10.1093/nar/25.23.4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Bernloehr H, Lohrmann R, Sulston J, Orgel LE, Miles HT. Specificity of template-directed synthesis with adenine nucleosides. J. Mol. Biol. 1970;47:257–260. doi: 10.1016/0022-2836(70)90344-x. [DOI] [PubMed] [Google Scholar]
- Sen S, Venkata Dasu V, Mandal B. Developments in directed evolution for improving enzyme functions. Appl. Biochem. Biotechnol. 2007;143:212–223. doi: 10.1007/s12010-007-8003-4. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- Shoyele SA. Controlling the release of proteins/peptides via the pulmonary route. Methods Mol. Biol. 2008;437:141–148. doi: 10.1007/978-1-59745-210-6_6. [DOI] [PubMed] [Google Scholar]
- Spangler CM, Spangler C, Schaerling M. Luminescent lanthanide complexes as probes for the determination of enzyme activities. Ann. N Y Acad. Sci. 2008;1130:138–148. doi: 10.1196/annals.1430.008. [DOI] [PubMed] [Google Scholar]
- Stormo GD. An overview of RNA structure prediction and applications to RNA gene prediction and RNAi design. Curr. Protoc. Bioinformatics. 2006;Chapter 12 doi: 10.1002/0471250953.bi1201s13. Unit 12.1. [DOI] [PubMed] [Google Scholar]
- Subtelny AO, Hartman MC, Szostak JW. Ribosomal synthesis of N-methyl peptides. J. Am. Chem. Soc. 2008;130:6131–6136. doi: 10.1021/ja710016v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J, Lohrmann R, Orgel LE, Schneider-Bernloehr H, Weimann BJ, Miles HT. Non-enzymic oligonucleotide synthesis on a polycytidylate template. J. Mol. Biol. 1969;40:227–234. doi: 10.1016/0022-2836(69)90471-9. [DOI] [PubMed] [Google Scholar]
- Summerer D, Marx A. 4′C-modified nucleotides as chemical tools for investigation and modulation of DNA polymerase function. Synlett. 2004:217–224. [Google Scholar]
- Summerer D, Marx A. 4′ C-ethynyl-thymidine acts as a chain terminator during DNA-synthesis catalyzed by HIV-1 reverse transcriptase. Bioorg. Med. Chem. Lett. 2005;15:869–871. doi: 10.1016/j.bmcl.2004.12.072. [DOI] [PubMed] [Google Scholar]
- Summers JS, Shaw BR. Boranophosphates as mimics of natural phosphodiesters in DNA. Curr. Med. Chem. 2001;8:1147–1155. doi: 10.2174/0929867013372409. [DOI] [PubMed] [Google Scholar]
- Tan Z, Forster AC, Blacklow SC, Cornish VW. Amino acid backbone specificity of the Escherichia coli translation machinery. J. Am. Chem. Soc. 2004;126:12752–12753. doi: 10.1021/ja0472174. [DOI] [PubMed] [Google Scholar]
- Tomasz J, Shaw BR, Porter K, Spielvogel BF, Sood A. Boron-containing nucleic-acids. 3. 5′-P-borane-substituted thymidine monophosphate and triphosphate. Angew. Chem. Int. Ed. Engl. 1992;31:1373–1375. [Google Scholar]
- Tuchscherer G, Scheibler L, Dumy P, Mutter M. Protein design: on the threshold of functional properties. Biopolymers. 1998;47:63–73. doi: 10.1002/(SICI)1097-0282(1998)47:1<63::AID-BIP7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Vasir JK, Tambwekar K, Garg S. Bioadhesive microspheres as a controlled drug delivery system. Int. J. Pharm. 2003;255:13–32. doi: 10.1016/s0378-5173(03)00087-5. [DOI] [PubMed] [Google Scholar]
- Vastmans K, Pochet S, Peys A, Kerremans L, Van Aerschot A, Hendrix C, Marliere P, Herdewijn P. Enzymatic incorporation in DNA of 1,5-anhydrohexitol nucleotides. Biochemistry. 2000;39:12757–12765. doi: 10.1021/bi001297g. [DOI] [PubMed] [Google Scholar]
- Vastmans K, Froeyen M, Kerremans L, Pochet S, Herdewijn P. Reverse transcriptase incorporation of 1,5-anhydrohexitol nucleotides. Nucleic Acids Res. 2001;29:3154–3163. doi: 10.1093/nar/29.15.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastmans K, Rozenski J, Van Aerschot A, Herdewijn P. Recognition of HNA and 1,5-anhydrohexitol nucleotides by DNA metabolizing enzymes. Biochim. Biophys. Acta. 2002;1597:115–122. doi: 10.1016/s0167-4838(02)00267-4. [DOI] [PubMed] [Google Scholar]
- Veedu RN, Vester B, Wengel J. In vitro incorporation of LNA nucleotides. Nucleosides Nucleotides Nucleic Acids. 2007a;26:1207–1210. doi: 10.1080/15257770701527844. [DOI] [PubMed] [Google Scholar]
- Veedu RN, Vester B, Wengel J. Enzymatic incorporation of LNA nucleotides into DNA strands. ChemBioChem. 2007b;8:490–492. doi: 10.1002/cbic.200600501. [DOI] [PubMed] [Google Scholar]
- Veedu RN, Vester B, Wengel J. Polymerase chain reaction and transcriptionusing locked nucleic acid nucleotide triphosphates. J. Am. Chem. Soc. 2008;130:8124–8125. doi: 10.1021/ja801389n. [DOI] [PubMed] [Google Scholar]
- Wang J, Verbeure B, Luyten I, Froeyen M, Hendrix C, Rosemeyer H, Seela F, Van Aerschot A, Herdewijn P. Cyclohexene nucleic acids (CeNA) form stable duplexes with RNA and induce RNase H activity. Nucleosides Nucleotides Nucleic Acids. 2001;20:785–788. doi: 10.1081/NCN-100002430. [DOI] [PubMed] [Google Scholar]
- Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- Wang K, Neumann H, Peak-Chew SY, Chin JW. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 2007;25:770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- White SR, Sottos NR, Geubelle PH, Moore JS, Kessler MR, Sriram SR, Brown EN, Viswanathan S. Autonomic healing of polymer composites. Nature. 2001;409:794–797. doi: 10.1038/35057232. [DOI] [PubMed] [Google Scholar]
- White RR, Roy JA, Viles KD, Sullenger BA, Kontos CD. A nuclease-resistant RNA aptamer specifically inhibits angiopoietin-1-mediated Tie2 activation and function. Angiogenesis. 2008;11:395–401. doi: 10.1007/s10456-008-9122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilds CJ, Pattanayek R, Pan C, Wawrzak Z, Egli M. Selenium-assisted nucleic acid crystallography: use of phosphoroselenoates for MAD phasing of a DNA structure. J. Am. Chem. Soc. 2002;124:14910–14916. doi: 10.1021/ja021058b. [DOI] [PubMed] [Google Scholar]
- Wolfe JL, Kawate T, Belenky A, Stanton V., Jr Synthesis and polymerase incorporation of 5′-amino-2′,5′-dideoxy-5′-N-triphosphate nucleotides. Nucleic Acids Res. 2002;30:3739–3747. doi: 10.1093/nar/gkf502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe JL, Wang BH, Kawate T, Stanton VP., Jr Sequence-specific dinucleotide cleavage promoted by synergistic interactions between neighboring modified nucleotides in DNA. J. Am. Chem. Soc. 2003;125:10500–10501. doi: 10.1021/ja035646g. [DOI] [PubMed] [Google Scholar]
- Woodley J. Bioadhesion: new possibilities for drug administration? Clin. Pharmacokinet. 2001;40:77–84. doi: 10.2165/00003088-200140020-00001. [DOI] [PubMed] [Google Scholar]
- Xie J, Schultz PG. An expanding genetic code. Methods. 2005;36:227–238. doi: 10.1016/j.ymeth.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Xie J, Schultz PG. A chemical toolkit for proteins—an expanded genetic code. Nat. Rev. Mol. Cell Biol. 2006;7:775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- Yliperttula M, Chung BG, Navaladi A, Manbachi A, Urtti A. High-throughput screening of cell responses to biomaterials. Eur. J. Pharm. Sci. 2008;35:151–160. doi: 10.1016/j.ejps.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Zhan ZYJ, Lynn DG. Chemical amplification through template-directed synthesis. J. Am. Chem. Soc. 1997;119:12420–12421. [Google Scholar]
- Zhang L, Peritz A, Meggers E. A simple glycol nucleic acid. J. Am. Chem. Soc. 2005;127:4174–4175. doi: 10.1021/ja042564z. [DOI] [PubMed] [Google Scholar]
- Zhang B, Tan Z, Dickson LG, Nalam MN, Cornish VW, Forster AC. Specificity of translation for N-alkyl amino acids. J. Am. Chem. Soc. 2007;129:11316–11317. doi: 10.1021/ja0734871. [DOI] [PMC free article] [PubMed] [Google Scholar]