Summary
Cyclin-dependent kinases (CDKs) are subunits of transcription factor (TF) IIH and positive transcription elongation factor b (P-TEFb). To define their functions, we mutated the TFIIH-associated kinase Mcs6 and P-TEFb homologs Cdk9 and Lsk1 of fission yeast, making them sensitive to bulky purine analogs. Selective inhibition of Mcs6 or Cdk9 blocks cell division, alters RNA polymerase (Pol) II carboxyl-terminal domain (CTD) phosphorylation and represses specific, overlapping subsets of transcripts. At a common target gene, both CDKs must be active for normal Pol II occupancy, and Spt5—a CDK substrate and regulator of elongation—accumulates disproportionately to Pol II when either kinase is inhibited. In contrast, Mcs6 activity is sufficient, and necessary, to recruit the Cdk9/Pcm1 (mRNA cap methyltransferase) complex. In vitro, phosphorylation of the CTD by Mcs6 stimulates subsequent phosphorylation by Cdk9. We propose that TFIIH primes the CTD and promotes recruitment of P-TEFb/Pcm1, serving to couple elongation and capping of select pre-mRNAs.
Introduction
Protein phosphorylation by cyclin-dependent kinases (CDKs) plays essential roles in eukaryotic gene expression. Three conserved components of the RNA polymerase (Pol) II transcription machinery have associated CDKs: transcription factor (TF) IIH, which includes the Cdk7/cyclin H/Mat1 subcomplex; Mediator, which contains the Cdk8/cyclin C pair; and positive transcription elongation factor b (P-TEFb), which comprises Cdk9 and cyclin T [reviewed by (Fisher, 2005; Peterlin and Price, 2006; Zhou and Yik, 2006)].
A common target of those CDKs is the carboxyl-terminal domain (CTD) of the Pol II large subunit Rpb1, which consists of repeats of the heptapeptide YSPTSPS. Phosphorylation of serine residues in positions 2 and 5 (Ser2 and Ser5) creates binding sites for accessory factors including mRNA-processing and chromatin-remodeling enzymes [reviewed by (Phatnani and Greenleaf, 2006)]. Cdk7, -8 and -9 phosphorylate the CTD in vitro with distinct but overlapping specificities (Ramanathan et al., 2001), possibly to recruit different proteins. However, the specific consequences of phosphorylations by individual kinases remain largely undefined.
TFIIH is a component of the preinitiation complex (PIC) assembled at the promoter (Lee and Young, 2000). In vitro, catalytic activity of Cdk7 is required after initiation for transcription of some but not all templates (Tirode et al., 1999). In vivo, impairment of TFIIH-associated kinase function has varying impact on gene expression, depending on the organism and type of mutant analyzed. In the budding yeast Saccharomyces cerevisiae, inactivation of the Cdk7 ortholog Kin28 by temperature-sensitive (ts) mutation shut down most transcription by Pol II (Holstege et al., 1998), apparently by destabilizing the PIC (Kanin et al., 2007). In contrast, chemical inhibition of Kin28 had no major effects on transcript abundance, but reduced 5′-end capping of poly(A)+ RNA (Kanin et al., 2007), suggesting that an active kinase is needed not in transcription per se but rather in mRNA maturation. In the fission yeast Schizosaccharomyces pombe, thermal inactivation of the Mcs6/Mcs2/Pmh1 complex (orthologous to Cdk7/cyclin H/Mat1) caused specific rather than global defects in transcription (Lee et al., 2005), possibly indicating a role in differential gene expression.
P-TEFb was identified as a factor that stimulates elongation by Pol II in vitro, where it antagonizes pausing enforced by the DRB-sensitivity inducing factor (DSIF, a heterodimer of Spt4 and Spt5 apparently conserved in all eukaryotes) and negative elongation factor (NELF, absent in yeast) (Peterlin and Price, 2006). Promoter-proximal pausing could facilitate recruitment of pre-mRNA processing machinery (Core and Lis, 2008; Glover-Cutter et al., 2008). Thus might P-TEFb and DSIF—with or without NELF depending on the organism—execute a quality control function that links elongation to processing, analogous to cell-cycle checkpoints that ensure the fidelity of chromosome duplication and segregation (Orphanides and Reinberg, 2002). Studies in S. cerevisiae implicitly challenged this idea, however, by showing a lack of coupling between the synthesis of full-length transcripts and their maturation into translatable mRNAs (Muratani et al., 2005; Kanin et al., 2007).
In budding yeast, the functions of P-TEFb appear to be split between the essential Bur1/Bur2 dimer and the non-essential Ctk1/Ctk2/Ctk3 trimer (Wood and Shilatifard, 2006); there may be a similar division of labor in fission yeast between the essential Cdk9/Pch1 and non-essential Lsk1/Lsc1 complexes (Pei et al., 2006; Karagiannis and Balasubramanian, 2007). Two-hybrid assays uncovered a potential interaction network comprising S. pombe Cdk9, its substrates—Rpb1 and Spt5—and the RNA triphosphatase Pct1 of the mRNA capping machinery (Pei and Shuman, 2002; Pei et al., 2003; Pei and Shuman, 2003). We previously isolated a Cdk9 complex from fission yeast cells, which contained apparently equimolar amounts of the cyclin Pch1 and the cap methyltransferase Pcm1 (Pei et al., 2006). Pcm1 catalyzes the last step of capping; its physical association and co-recruitment to chromatin with a P-TEFb homolog seemed to strengthen the case for an elongation checkpoint (Pei et al., 2006; Guiguen et al., 2007).
To dissect contributions of TFIIH and P-TEFb to the regulation of gene expression in S. pombe, we introduced analog-sensitive (AS) kinases into cells in place of the wild-type versions. Selective inhibition of Mcs6 or Cdk9 impaired late events in cell division. In each case only a subset of genes was repressed, and even the combined inhibition of Mcs6 and Cdk9 did not shut off most transcription. There was extensive overlap between the transcripts affected by inhibition of Mcs6 or Cdk9. At an Mcs6- and Cdk9-dependent gene, active Mcs6 promoted recruitment of the Cdk9/Pcm1 complex, and inhibition of either kinase increased the Spt5:Pol II crosslinking ratio. In vitro, phosphorylation by Mcs6 “primed” a CTD substrate, enhancing its subsequent phosphorylation by Cdk9. Together, these results suggest an explanation for the overlapping expression profiles: TFIIH and P-TEFb act sequentially and coordinately at specific genes, to link relief of Spt5-dependent pausing with methylation of the mRNA 5′-cap structure.
Results
Analog-Sensitive mcs6 and cdk9 Mutants
To achieve selective kinase inhibition in S. pombe, we introduced mutations of “gatekeeper” residues Mcs6-Leu87 and Cdk9-Thr120 to Gly, enlarging the ATP-binding pockets of the respective enzymes to accommodate bulky purine analogs that bind poorly to wild-type kinases (Knight and Shokat, 2007). Neither mutant strain was sensitive to 1-NM-PP1—a selective inhibitor of human Cdk7as containing the analogous mutation (Larochelle et al., 2006). We therefore screened related compounds (Figure S1) and identified 3-MB-PP1, a more potent inhibitor of Cdk7as (Merrick et al., 2008), as an inhibitor of growth in both mcs6L87G and cdk9T120G strains (hereafter mcs6as and cdk9as, respectively). We also changed gatekeeper residue Lsk1-Phe353 to Gly; 3-MB-PP1 did not selectively affect growth of the lsk1F353G strain (data not shown), indicating either that Lsk1 activity is not essential, or that Lsk1F353G is not AS (but see below).
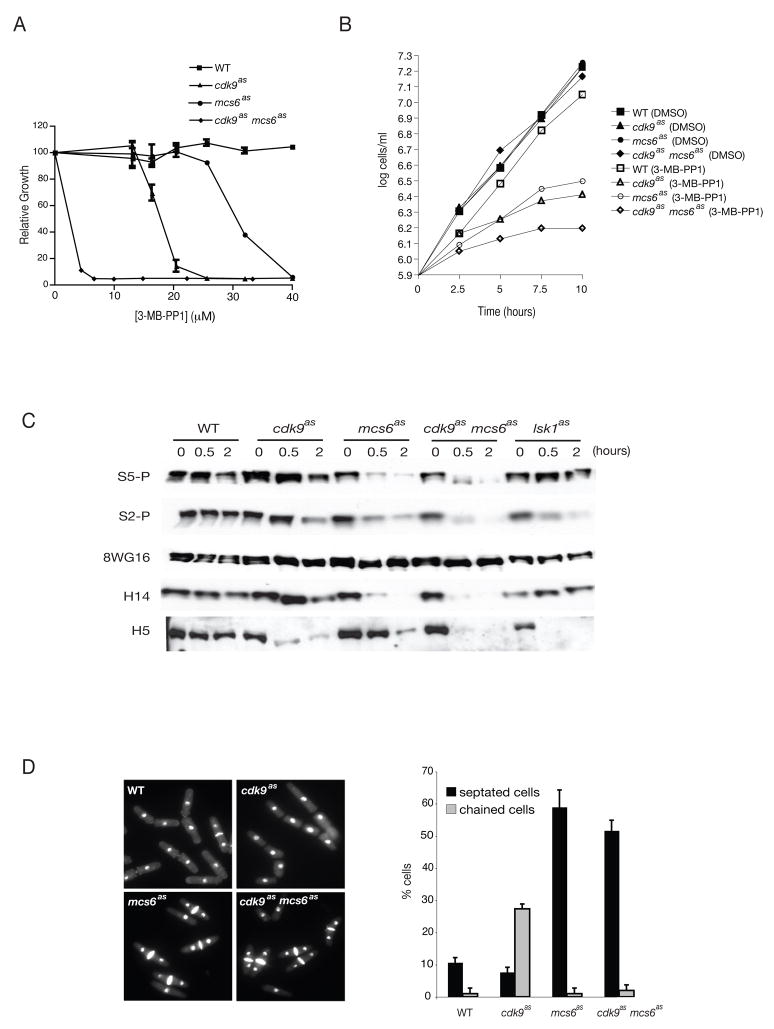
Treatment with 3-MB-PP1 inhibited division of mcs6as and cdk9as cells with IC50 values of ~30 and ~15 μM, respectively (Figure 1A). In an mcs6as cdk9as strain obtained by crossing the two single mutants, the IC50 was <5 μM—a synthetic effect reminiscent of interactions previously detected between the two genes (Pei et al., 2006). Addition of 40 μM 3-MB-PP1 to exponentially growing cultures arrested proliferation in all three mutant strains without affecting growth of wild-type cells (Figure 1B). We conclude that Mcs6 and Cdk9 activities are both required for viability, and perform both unique and overlapping functions.
Figure 1. Phenotypes of mcs6as and cdk9as Cells.
(A) Dose response of wild-type (JS78), cdk9as (LV7), mcs6as-3HA (SG2) and cdk9as mcs6as-3HA (LV42) strains to 3-MB-PP1. Cells were grown in 96-well microtitre dishes with indicated concentrations of inhibitor. Absorbance was measured and plotted relative to that of DMSO-treated cells of the same genotype (defined as 100%). Error bars denote standard error of mean (S.E.M) in triplicate measurements.
(B) Exponential growth curves of strains in (A), in rich medium to which 40 μM 3-MB-PP1 or DMSO, as indicated, was added at time 0.
(C) Time course of changes in Pol II phosphorylation after CDK inhibition. Exponentially growing cells of strains in (A) plus lsk1as (CS118) were collected before adding inhibitor (time 0), and after 0.5- and 1-hr treatments with 40 μM 3-MB-PP1. Extracts were immunoblotted with indicated antibodies: P-Ser5, P-Ser2, 8WG16, H14 and H5.
(D) Micrographs of indicated strains fixed and stained with 4′,6-diamidino-2-phenylindole and calcofluor after 10 hr in 40 μM 3-MB-PP1. Percentages of cells with septa, or attached in chains of three or more, were measured in triplicate with S.E.M denoted by error bars.
Chemical Genetic Dissection of Pol II Phosphorylation In Vivo
In vitro, Mcs6 generated Rpb1 CTD isoforms recognized by a monoclonal antibody specific for modification at Ser5 (H14) but not by one that reacts most strongly with repeats doubly phosphorylated at Ser2 and Ser5 (H5), whereas Cdk9 increased both H14- and H5-reactivity (Pei et al., 2006). In a previous chemical-genetic analysis, treatment of an S. pombe cdk9T120G strain with 1-NM-PP1 reduced H5 signals (Guiguen et al., 2007). In another study, however, disruption of either lsk1+ or lsc1+ abolished H5-reactivity, and phenocopied cytokinesis defects due to an rpb1 mutation eliminating all Ser2 residues, indicating that Lsk1/Lsc1 is the major Ser2 kinase, and that Cdk9 must have an essential function besides phosphorylation of Ser2, which is itself non-essential (Karagiannis and Balasubramanian, 2007).
To assess the contributions of Mcs6, Cdk9 and Lsk1 in vivo, we attempted to inhibit each CDK with 3-MB-PP1, and monitored Rpb1 phosphorylation both with H14 and H5 and with polyclonal antibodies raised against peptides phosphorylated at Ser5 (Ser5-P) or Ser2 (Ser2-P) (Figures 1C and S2). Inhibition of Mcs6 caused dose- and time-dependent decreases in Ser5 phosphorylation in vivo, measured with either Ser5-P or H14. In lsk1F353G cells treated with 3-MB-PP1, Ser2-P and H5 reactivity diminished, indicating that: 1) the mutant kinase is indeed AS; and 2) Lsk1 activity is required for Ser2 phosphorylation but not for survival, consistent with results in lsk1 and lsc1 deletion strains (Karagiannis and Balasubramanian, 2007). In contrast, inhibition of Cdk9 or Mcs6 each caused only partial diminution of Ser2-P signal. H5-reactivity also diminished to similar extents in mcs6as and cdk9as strains, albeit more rapidly in the latter (Figure 1C). In an mcs6as cdk9as double mutant, 3-MB-PP1 caused more severe reductions in H5 and Ser2-P signals, which were similar to those observed in lsk1as extracts. Therefore, Ser2 phosphorylation in vivo depends on the activities of Lsk1 and either Mcs6 or Cdk9.
Inhibition of Cdk9 alone did not abolish phosphorylation of Ser5 or Ser2, but caused reproducible loss of the slowest-migrating Rpb1 phospho-isoform (Figures 1C and S2), which did not occur in either the mcs6as or lsk1as single mutant. Taken together, the data suggest that Cdk9 activity is dispensable for phosphorylation of Ser2 or Ser5, but might be required for efficient, multisite phosphorylation of the CTD.
Inhibition of Mcs6 or Cdk9 Impairs Cell Division
We examined the morphologies of cells arrested by 3-MB-PP1 treatment (Figure 1D). The mcs6as cells accumulated with two nuclei and a division septum; after 10 hr the septation index reached ~60%. The cdk9as mutant did not become hyperseptated, but accumulated chains of three or more incompletely separated cells with a frequency of ~25%—lower than the septation index of mcs6as, but a greater fold-enrichment of this class relative to the controls. Both mcs6as and cdk9as strains therefore had analog-sensitive defects in cell separation. The mcs6as phenotype resembled those of mcs6 and pmh1 ts mutants at restrictive temperature (Saiz and Fisher, 2002; Lee et al., 2005), suggesting that chemical inhibition and thermal denaturation of the Mcs6 complex had similar effects on cell division. Moreover, the mcs6as cdk9as cell morphology mimicked that of mcs6as, indicating that Mcs6 and Cdk9 act in the same pathway.
Mcs6 and Cdk9 Activities Are Required to Express a Common Set of Genes
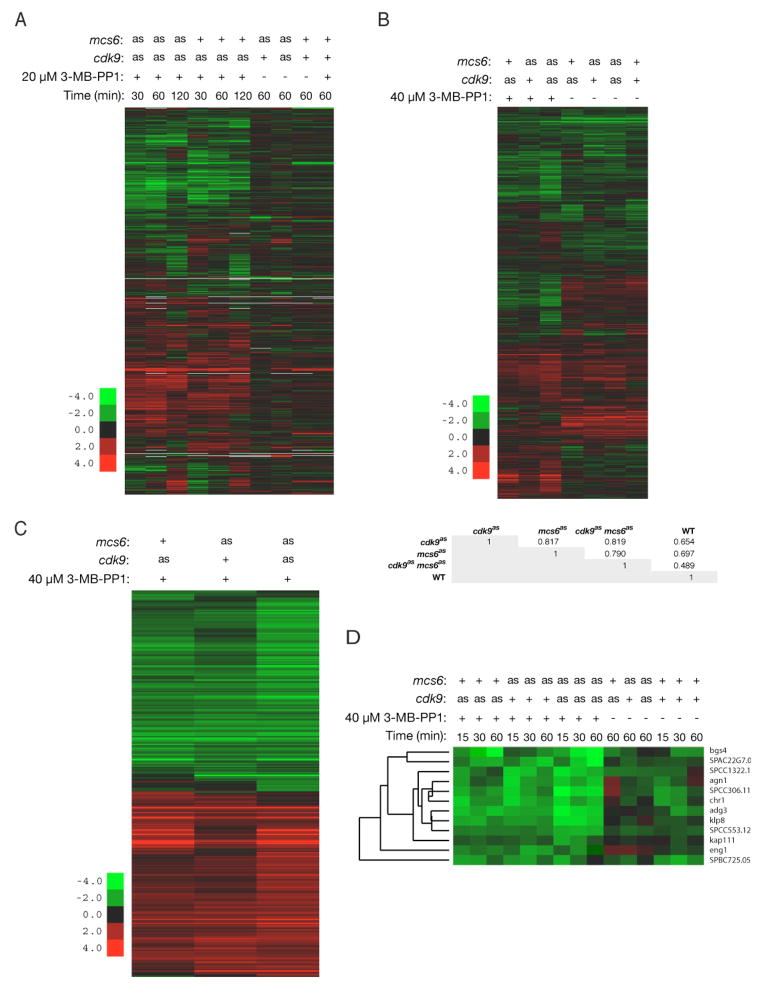
We next investigated requirements for Mcs6 and Cdk9 in gene expression, by microarray hybridization. In cdk9as and mcs6as cdk9as mutants treated with 20 μM 3-MB-PP1 for various times, 10–20% of transcripts increased or decreased by ≥2-fold in at least one condition, relative to mock-treated controls. The patterns of repression and induction were similar in cdk9as and mcs6as cdk9as cells (Figure 2A).
Figure 2. Mcs6 and Cdk9 are Required to Express a Common Subset of Genes.
(A) Gene expression changes of ≥2-fold in cdk9as (LV7) and cdk9as mcs6as-3HA (LV42) mutants, after treatment with 20 μM 3-MB-PP1 for indicated time, versus mock treatment of same strains (first six lanes). Last four lanes show as mutants in absence, and wild-type (JS78) cells in presence, of 3-MB-PP1, compared to mock-treated wild type.
(B) Genes differentially expressed in at least one of the mutants (as in [A] plus mcs6as-3HA [SG2]), treated for 15 min with 40 μM 3-MB-PP1 or DMSO, compared to 3-MB-PP1-treated wild type. Table below cluster diagram shows Pearson correlations for the ratios of all genes in 3-MB-PP1-treated samples.
(C) Transcripts from panel B differentially expressed in mutants relative to wild type (P < 0.001).
(D) Effects of Mcs6 and/or Cdk9 inhibition on Ace2-dependent transcripts.
Next, we compared the three mutant strains (mcs6as, cdk9as and mcs6as cdk9as) to the wild type, all treated with 40 μM 3-MB-PP1 (Figure 2B and data not shown). To filter out effects that were independent of the mutations at the higher dose, we selected genes differentially expressed in the mutants relative to the wild type at the 15-min time point. A statistical analysis of the data revealed highly significant overlap between genes differentially expressed in mcs6as and cdk9as strains (Figure 2C); the cdk9as mutant clustered only slightly more closely with the double mutant (Pearson correlation of differentially expressed genes: ~0.86) than with mcs6as (correlation: ~0.85). Correlation between mcs6as and cdk9as expression patterns was also >0.8 when all genes on the arrays were compared (Figure 2B). Dual kinase inhibition, in mcs6as cdk9as cells, increased the amplitude of changes but did not greatly expand the set of affected genes.
Most genes induced by Mcs6 or Cdk9 inhibition were involved in stress responses—a phenomenon also observed in mcs6 and pmh1 ts mutants at restrictive temperature (Lee et al., 2005). The ability to mount a transcriptional response to stress was also preserved in the absence of Mcs6 and Cdk9 activity (Figure 2C).
Thermal inactivation of Mcs6 repressed genes important for cytokinesis and cell separation (Lee et al., 2005), which are periodically expressed under control of transcription factors Sep1 and Ace2 (Rustici et al., 2004). Similarly, most Ace2-dependent genes were repressed by 3-MB-PP1 in mcs6as, cdk9as or mcs6as cdk9as strains (Figure 2D), while the ace2+ transcript itself was not affected (Supplemental data online). Taken together, the microarray results indicate that Mcs6 and Cdk9 are required in vivo to express largely similar subsets of genes, perhaps distinguished by their dependence on specific transcription factors.
CDK Inhibition Decreases Pol II Occupancy at Target Genes
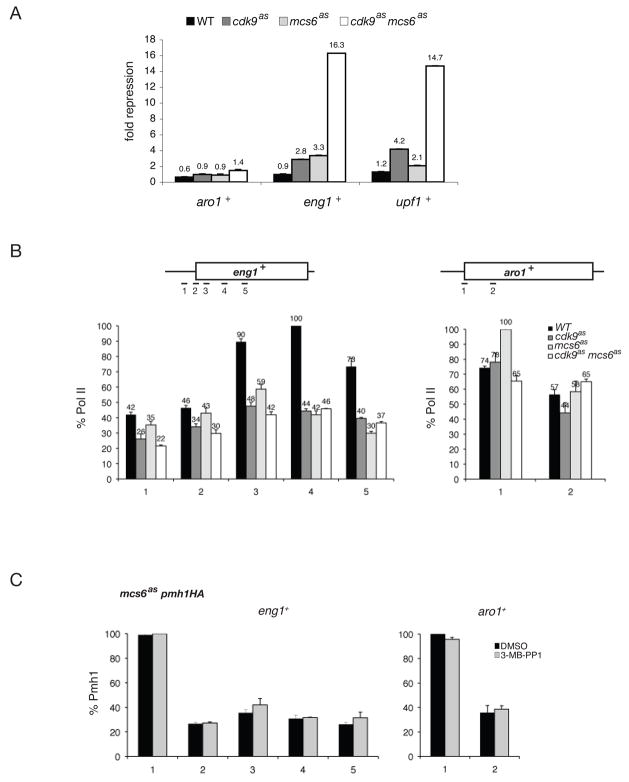
To validate the microarray data and select representative genes for further analysis, we quantified mRNA levels by real time PCR (qPCR). Based on results obtained with 20 μM 3-MB-PP1, we chose eng1+ and upf1+ as candidate genes repressed by CDK inhibition and aro1+ as a control, analog-unresponsive gene. In all three mutants, transcript levels of eng1+ and upf1+, but not aro1+, were reduced ≥2-fold by 3-MB-PP1, whereas none of the genes was similarly affected in wild-type cells. The effects of inhibiting Mcs6 and Cdk9 were approximately additive; for example, the eng1+ transcript was repressed ~3-fold in each single mutant but >10-fold in the double mutant (Figure 3A).
Figure 3. CDK Inhibition Decreases Pol II Occupancy at Target Genes.
(A) Levels of aro1+, eng1+ and upf1+ transcripts by qRT-PCR when Cdk9 and/or Mcs6 are inhibited by 20 μM 3-MB-PP1 treatment for 1 hr. Fold-repression is defined as the negative mean expression ratio of analog- and DMSO-treated samples (same strains as in Figure 1B).
(B) Pol II occupancy measured by ChIP at aro1+ and eng1+ genes after 1-hr treatment with 20 μM 3-MB-PP1. Primers used to amplify specific regions are shown schematically at top. Average enrichment is plotted relative to maximal signal in each series, defined as 100%.
(C) Occupancy of Pmh1 at eng1+ and aro1+ in mcs6as pmh1-HA (CS142) cells treated 1 hr with 20 μM 3-MB-PP1 or DMSO, as indicated.
Error bars in all panels denote S.E.M in triplicate measurements.
To investigate the mechanism of repression, we performed chromatin immunoprecipitation (ChIP) with anti-Rpb1 antibodies on wild-type, mcs6as, cdk9as and mcs6as cdk9as cells treated with 20 μM 3-MB-PP1 for 1 hr, and amplified sequences from eng1+ or aro1+ (Figure 3B). Pol II occupancy did not change consistently in either promoter-proximal or downstream regions of aro1+, or in the eng1+ promoter region (detected with primer pairs 1 and 2), in response to Mcs6 or Cdk9 inhibition. In contrast, crosslinking of Rpb1 was reduced at three consecutive positions in the downstream region of eng1+ (detected with primer pairs 3–5) in all three mutants relative to the wild type. PIC assembly occurred normally at the eng1+ promoter even when Mcs6as was inhibited; crosslinking of the TFIIH subunit Pmh1 was unaffected by 3-MB-PP1 treatment of mcs6as pmh1-HA cells (Figure 3C), and Mcs6 activity was not required for its own recruitment (Figure S3). This, together with depletion of Pol II from the eng1+ coding region and corresponding reduction in mRNA levels, suggests a specific defect in transcript elongation in the absence of Mcs6 or Cdk9 activity.
TFIIH Promotes Chromatin Recruitment of a Constitutive P-TEFb/Pcm1 Complex
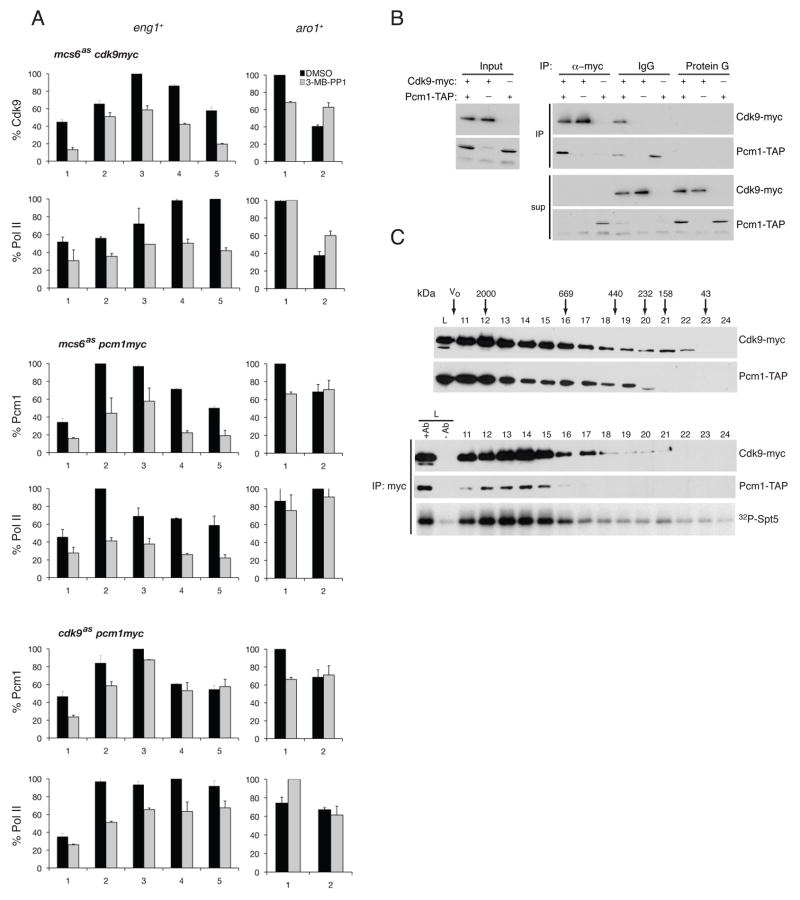
Because inhibition of either Mcs6 or Cdk9 caused similar perturbations of gene expression, we asked if Mcs6 activity was required for recruitment of Cdk9 and its partner Pcm1 to a target gene. In mcs6+ or mock-treated mcs6as cells, crosslinking of Cdk9-Myc and Pcm1-Myc peaked ~200 bp downstream of the transcription start site of eng1+. Treatment of mcs6as cells with 3-MB-PP1—40 μM for 15 min (Figure 4A) or 20 μM for 1 hr (Figure S4)—reduced Cdk9/Pcm1 occupancy along the entire eng1+ gene. Neither treatment caused consistent effects on binding to aro1+ in mcs6as cells (Figures 4A and S4) or to either gene in mcs6+ cells (Figure S5).
Figure 4. Mcs6 Promotes Recruitment of Cdk9/Pcm1 to Elongation Complexes.
(A) ChIP of Cdk9, Pcm1 and Pol II at eng1+ and aro1+ in mcs6as cdk9-myc (CS103), mcs6as pcm1-myc (LV81) and cdk9as pcm1-myc (CS165) cells treated 15 min with 40 μM 3-MB-PP1 or DMSO, as indicated. Error bars denote S.E.M in triplicate measurements.
(B) Co-precipitation of Cdk9-myc and Pcm1-TAP from extracts of cdk9-myc pcm1-TAP (CS54) cells. Protein G beads (no antibody) is negative control. Inputs, immunoprecipitates, and supernatants were immunoblotted to detect Cdk9-myc and Pcm1-TAP.
(C) Sizing column chromatography of cdk9-myc pcm1-TAP cell lysate. Top panels: Cdk9-myc and Pcm1-TAP detected as in (B). Bottom panels: immunoprecipitated Cdk9-myc from fractions tested for co-precipitating Pcm1-TAP and Spt5 kinase activity.
Decreased chromatin association of the Cdk9/Pcm1 complex could be due to a specific defect in recruitment, or an indirect effect of decreased Pol II density. To distinguish between these possibilities, we tested whether inhibition of Cdk9 also impeded Pcm1 recruitment. Treatment of cdk9as pcm1-myc cells with 20 or 40 μM 3-MB-PP1 reduced Pol II occupancy on eng1+ after 1 hr or 15 min, respectively, but had only small effects on Pcm1-Myc crosslinking to the promoter, and none on binding to the coding region (Figures 4A and S4). The results suggest that Mcs6 activity is specifically required to bring Cdk9/Pcm1 to the elongation complex.
A previous study showed that crosslinking of Cdk9 to chromatin was sensitive to levels of the Pcm1 protein (Guiguen et al., 2007). To address whether the two enzymes are recruited sequentially or as a preformed complex, we asked if any Pcm1 existed free of the Cdk9 complex. Immunodepletion of Cdk9 cleared Pcm1 from cdk9–13myc pcm1-TAP cell extracts, but extracts from which Pcm1 had been removed contained residual Cdk9 (Figure 4B). Therefore, there was little or no Cdk9-free Pcm1, but we could not rule out the presence of free Cdk9 by this method. The two proteins co-migrated in sizing columns, however, and Cdk9 activity co-purified with peaks of both Cdk9 and Pcm1 (Figure 4C). The amount of Pcm1 bound to Cdk9 was constant throughout the cell cycle, in proliferating versus stationary cells and in rich or minimal medium (data not shown). Inhibition of Mcs6 did not perturb the interaction (Figure S6). Cdk9/Pcm1 is therefore a constitutive complex, likely to be recruited to chromatin en bloc—an event that depends on Mcs6 activity at a subset of transcribed genes.
Priming Phosphorylation by Mcs6 Stimulates Cdk9 Activity on the Pol II CTD
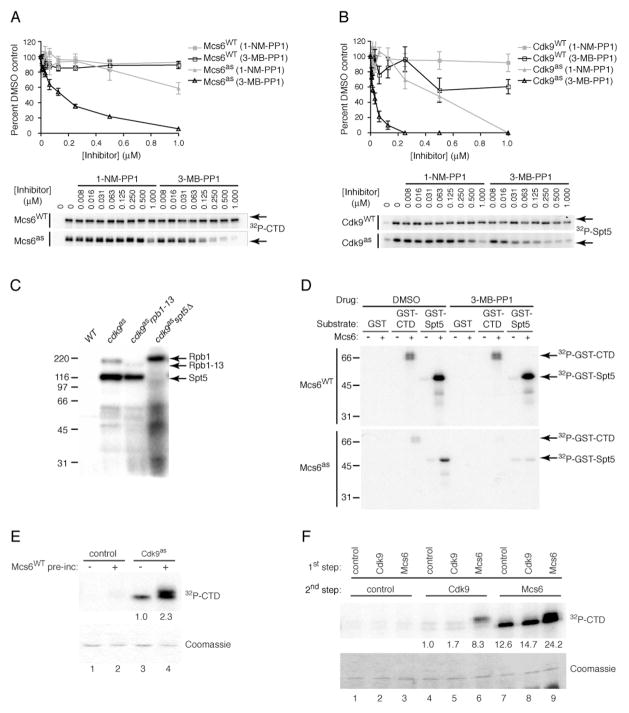
Results of ChIP, showing that Mcs6 activity is required for efficient Cdk9 recruitment at eng1+ (Figures 4A and S4), appeared to place Mcs6 function upstream of Cdk9, and suggest sequential action by the two CDKs in promoting transcript elongation. To explore this idea biochemically, we first affinity-purified Mcs6 and Cdk9 complexes from yeast after tagging Pmh1 or Pcm1, respectively. In kinase assays, 3-MB-PP1 inhibited Mcs6as with an IC50 of ~125 nM, whereas 1-NM-PP1 was a poor inhibitor, and neither compound inhibited wild-type Mcs6 at concentrations up to 1 μM (Figure 5A). Cdk9as was also more sensitive to 3-MB-PP1 (IC50 ~25 nM) than to 1-NM-PP1 (IC50 ~500 nM), whereas wild-type Cdk9 was resistant to both drugs (Figure 5B).
Figure 5. Mcs6 Primes the Pol II CTD for Phosphorylation by Cdk9.
(A) Mcs6WT and Mcs6as partially purified from pmh1-TAP strains (CS80 and CS141, respectively) were incubated with indicated amounts of 1-NM-PP1 or 3-MB-PP1 and tested for kinase activity towards GST -CTD relative to signal in absence of inhibitor. Error bars denote S.E.M in triplicate measurements.
(B) Cdk9WT and Cdk9as partially purified from pcm1-TAP strains (SUS3 and CS63, respectively) were incubated with indicated amounts of each inhibitor and tested for kinase activity towards GST-Spt5(801–990), quantified as in (A). Error bars denote S.E.M in triplicate measurements.
(C) Chemical-genetic identification of Rpb1 and Spt5 as Cdk9 substrates. Whole-cell extracts of wild-type (JS78), cdk9as (LV7), cdk9as rpb1–13 (LV40) and cdk9as spt5ΔC (LV101) strains were incubated with 1 mM ATP, an ATP-regenerating system, and [γ-32P]N6-(benzyl)-ATP.
(D) Mcs6 phosphorylates Rpb1 and Spt5 in vitro. Partially purified Mcs6WT or Mcs6as was tested for kinase activity towards GST, GST-CTD or GST-Spt5 in the absence or presence of 1 μM 3-MB-PP1, as indicated.
(E) Mcs6 promotes CTD hyper-phosphorylation by Cdk9. After a 1-hr incubation of GST-CTD with Mcs6WT (where indicated) and ATP, Cdk9as (where indicated) and [γ-32P]N6-(benzyl)-ATP were added and reactions continued for 15 min. Phosphorylation was quantified by phosphorimager, a background signal (in corresponding control reactions containing no AS kinase in second step) was subtracted and fold-stimulation (indicated by numbers below lanes of autoradiogram) was calculated relative to signal produced by Cdk9 on unprimed substrate (defined as 1.0). Coomassie-staining reveals no shift of bulk CTD substrate, indicating that phosphorylation sites were not saturated.
(F) Mcs6 primes the CTD for Cdk9. GST-CTD was incubated with unlabeled ATP and indicated kinase from 200 μg of lysate, immobilized on IgG beads (or control beads incubated in untagged extracts); the supernatant was removed and incubated with [γ-32P]-ATP and beads containing Cdk9, Mcs6 or no kinase, as indicated. Coomassie-staining shows CTD was not saturated. Signals were quantified as in (E).
To identify Cdk9 substrates, we incubated whole-cell extracts of wild-type and cdk9as cells with the substrate analog [γ-32P]N6-(benzyl)-ATP, as previously described for labeling of mammalian proteins by Cdk7as (Larochelle et al., 2006). Under these conditions, no proteins were labeled above background in a wild-type extract, whereas two polypeptides of ~220 and ~120 kDa were strongly labeled in the cdk9as extract (Figure 5C). Because these mobilities matched the predicted sizes of Rpb1 and Spt5, respectively, we crossed the cdk9as mutant to strains harboring non-lethal truncations of each putative Cdk9 substrate (S.S. and B.S., unpublished observations): 1) an rpb1–13 strain, in which only 13 of the 29 CTD repeats remain; and 2) an spt5ΔC strain, in which a carboxyl-terminal region of Spt5, containing 18 nonapeptide repeats phosphorylated by Cdk9 in vitro (Pei and Shuman, 2003), is removed. When we repeated the labeling in cdk9as rpb1–13 cell extracts, the ~220 kDa band was absent—replaced by a smaller, less heavily labeled species likely to be the truncated form of Rpb1. Likewise, the ~120 kDa polypeptide failed to become labeled in cdk9as spt5ΔC extracts, but no new signal appeared at a size expected for the truncated Spt5 protein, suggesting that it could not be phosphorylated by Cdk9. We conclude that Cdk9as selectively phosphorylated both Rpb1 and Spt5 in the presence of all soluble S. pombe proteins.
We were unable to establish conditions for analog-selective labeling in extracts from mcs6as cells, so to identify Mcs6 substrates we instead performed assays with partially purified kinases and natural ATP. Wild-type Mcs6 phosphorylated both Rpb1-and Spt5-derived substrates under these conditions, and was not inhibited by 3-MB-PP1. To determine whether phosphorylation was catalyzed by Mcs6 itself or by a co-purifying but distinct kinase, we repeated the assays with Mcs6as complexes; phosphorylation of both Rpb1 and Spt5 was inhibited by 1 μM 3-MB-PP1 (Figure 5D). Therefore, both Mcs6 and Cdk9 can phosphorylate both Rpb1 and Spt5, as appears to be the case for the human proteins (Larochelle et al., 2006).
We next asked if Mcs6 and Cdk9 acted synergistically on a common substrate, by first incubating the Rpb1 CTD with wild-type Mcs6 and unlabeled ATP, then adding Cdk9as and [γ-32P]N6-(benzyl)-ATP. Under these conditions, in which both CDKs were present in the final reaction but radiolabeling was due solely to the activity of the AS kinase, pre-incubation with Mcs6 caused a 2- to 3-fold increase in incorporation by Cdk9as. The increase occurred predominantly in a slow-migrating electrophoretic isoform, suggestive of multisite phosphorylation (Figure 5E). Phosphorylation by Mcs6 might thus “prime” the Rpb1 CTD for subsequent phosphorylation by Cdk9, despite the overlap in site-specificity—and potential competition for sites—between the two CDKs (Pei et al., 2006).
To distinguish a priming mechanism from one in which the two CDKs act cooperatively, we performed sequential incubations with wild-type enzymes and natural ATP, in which the substrate was exposed to only one CDK at a time. The CTD was first incubated with unlabeled ATP, without or with an immobilized active kinase (Mcs6 or Cdk9), then recovered and incubated with a second immobilized kinase in the presence of [γ-32P]-ATP. Prior phosphorylation by Mcs6 stimulated Cdk9 activity ~8–10-fold, and the labeled CTD was quantitatively shifted to a slower mobility, possibly indicating processive phosphorylation (Figure 5F). We also observed enhancement when a GST-CTD fusion protein was incubated sequentially with soluble Mcs6 and Cdk9 (data not shown). Paradoxically, preincubation with Mcs6 also enhanced subsequent phosphorylation by Mcs6, suggesting a “self-priming” ability. In this case, the stimulation was only ~2-fold, and both shifted and unshifted species were labeled. Under no conditions we tested, however, did prior phosphorylation by Cdk9 have a stimulatory effect on Mcs6 (Figure 5F and data not shown). CTD priming might therefore be a unique function of the TFIIH-associated kinase, consistent with a role upstream of P-TEFb in transcription.
Inhibition of Mcs6 or Cdk9 Promotes Crosslinking of Spt5 to Chromatin
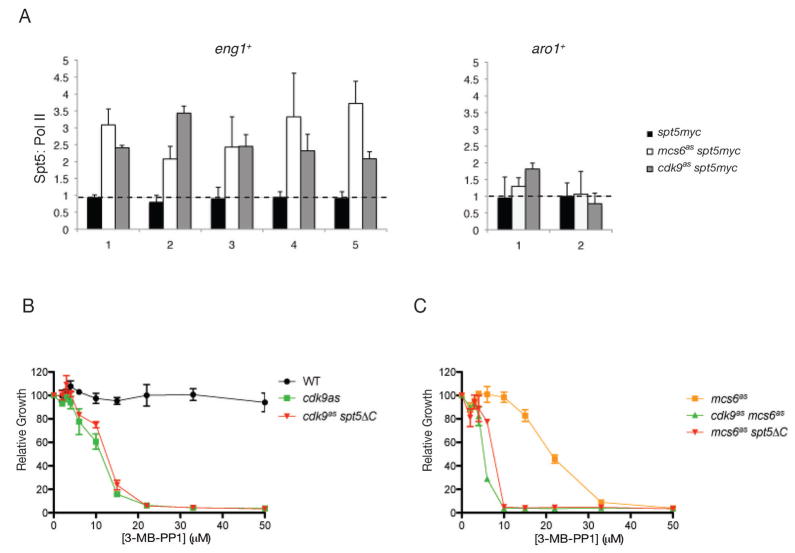
Spt5 interacts with the capping apparatus and is thought to regulate elongation (Pei and Shuman, 2002; Pei and Shuman, 2003). In other organisms, Spt5 and Pol II associate in extracts and track together on actively transcribed genes (Krogan et al., 2002; Glover-Cutter et al., 2008). Indeed, in mock-treated S. pombe cells or an spt5–13myc strain that lacked an AS kinase, Rpb1 and Spt5 distributions matched closely along the lengths of both eng1+ and aro1+ (Figure S7). When Mcs6 or Cdk9 was inhibited, however, Spt5 occupancy increased slightly but consistently in both promoter and coding regions of eng1+, even at downstream positions where Pol II was depleted. Therefore, both an initiation factor (TFIIH) and an elongation factor (Spt4/Spt5) continued to associate with eng1+ when Mcs6 activity was impaired—further evidence that depletion of P-TEFb was a specific, direct consequence of kinase inhibition. A plot of Spt5:Rpb1 ratios (Figure 6A) revealed disproportionate crosslinking of Spt5 and Pol II, which was restricted to eng1+ and might therefore contribute to the specific elongation defect.
Figure 6. Inhibition of Mcs6 or Cdk9 Enhances Chromatin Association of Spt5.
(A) The ratios of the signal intensity ratios (40 μM 3-MB-PP1, 15 min, versus DMSO) for Spt5 and Pol II at eng1+ and aro1+ in mcs6as spt5-myc (LV81), cdk9as spt5-myc (CS112) and spt5-myc (no as mutation, CS111) cells.
(B) Spt5 truncation is epistatic to cdk9as. Growth of indicated strains (JS78, LV7 and LV10) with increasing concentrations of 3-MB-PP1, measured as in Figure 1A.
(C) Spt5 truncation phenocopies Cdk9 inactivation in mcs6as strain. Growth of indicated strains (SG2, LV42 and LV105) with increasing concentrations of 3-MB-PP1.
Error bars in all panels denote S.E.M in triplicate measurements.
In mammalian cells, conversion of DSIF (Spt4/Spt5) from elongation-inhibitory to -stimulatory form depends on phosphorylation of Spt5 by P-TEFb (Yamada et al., 2006). Spt5 is a major Cdk9 substrate in fission yeast extracts (Figure 5C). To test genetically if that phosphorylation might neutralize its ability to block elongation, we crossed spt5ΔC to either mcs6as or cdk9as and measured sensitivities of the double mutants to growth inhibition by 3-MB-PP1. Truncation of Spt5 had no effect on IC50 in a cdk9as strain (Figure 6B), but was synergistic with inhibition of Mcs6; an mcs6as spt5ΔC strain was as sensitive to 3-MB-PP1 as a cdk9as mcs6as double mutant (Figure 6C). The epistasis between cdk9as and spt5ΔC, and similar synthetic interactions with mcs6as, suggest that Cdk9 can compensate for diminished Mcs6 activity by phosphorylating Spt5, perhaps to overcome blocks to elongation. Unlike the Spt5 phosphorylation motif, however, Cdk9 activity is essential, possibly because it is needed to phosphorylate Pol II. Taken together, the results suggest collaboration between TFIIH and P-TEFb both in phosphorylating Pol II and Spt5 and in coupling elongation with capping of specific pre-mRNAs.
Discussion
CDKs Act in Concert to Drive Pol II Transcription
The functions of protein phosphorylation in regulating gene expression are still being worked out, two decades after the first discovery of a Pol II CTD kinase (Lee and Greenleaf, 1989). For example, the initial assignment to Kin28 of a general, catalytic role in transcription was almost certainly incorrect. Instead, the global depletion of mRNAs in a kin28 ts mutant (Holstege et al., 1998) was probably due to destabilization of the PIC; small-molecule inhibition in a kin28as mutant had minimal effects on transcription (Kanin et al., 2007), suggesting that kinase activity was dispensable for most Pol II-dependent RNA synthesis in vivo.
In S. pombe, classical and chemical genetic studies of the Mcs6 complex are in closer agreement; thermal inactivation of multiple ts mutants (Lee et al., 2005) and chemical inhibition of Mcs6as (this work) both repressed transcription of specific genes. Most transcripts were produced at near-normal levels, however, in either setting. Therefore the catalytic activity of Mcs6 is needed for the efficient production of certain mRNAs in vivo, but that requirement is probably imposed by regulatory factors and not intrinsic to transcription per se.
The sets of transcripts dependent on Mcs6 or Cdk9 activity overlap extensively, suggesting that TFIIH and P-TEFb work in concert to coordinate fission yeast gene expression. Consistent with this model, the phenotype of an mcs6as cdk9as double mutant resembled that of an mcs6as single mutant. The different morphological effects of inhibiting either CDK alone could be due to the additional role of Mcs6, one of two CDK-activating kinases (CAKs), in cell cycle control (Saiz and Fisher, 2002). They could also indicate leakiness of cdk9as and a more pervasive effect on mRNA capping and cap-dependent translation when the pathway is blocked further upstream by Mcs6 inhibition. Perhaps consistent with the latter explanation, inhibition of protein synthesis in S. pombecauses cell separation defects resembling the mcs6as phenotype (Polanshek, 1977).
Inhibiting Mcs6 and Cdk9 together did not greatly increase the number of genes affected, but enhanced their repression (or induction), compared to the effects when either kinase was inhibited alone. Therefore, most transcripts that require either CDK for proper regulation depend on both. However, there were exceptions—a gene cluster that required only Mcs6 activity for full expression and genes repressed only in the double mutant—consistent with CDK-specific or combinatorial regulation, respectively.
CDKs Couple Transcription and Capping Enzyme Recruitment
Here we have shown that Cdk9 recruitment to a target gene depends on the catalytic activity of Mcs6. We can envision two mechanisms that might explain this dependency. First, priming phosphorylation by Mcs6 makes the CTD a better substrate for Cdk9 in vitro, and might contribute to P-TEFb recruitment in vivo. Second, TFIIH activity might facilitate recruitment of Pcm1 to transcription complexes, either directly, or by promoting the first two steps in capping to provide a substrate for methylation. Both the RNA triphosphatase Pct1 and guanylyltransferase Pce1 interact with Rpb1 and Spt5 (Pei et al., 2001; Pei and Shuman, 2002; Takagi et al., 2002). In budding yeast, Kin28 is implicated in capping enzyme recruitment (Komarnitsky et al., 2000; Rodriguez et al., 2000; Schroeder et al., 2000) and its inhibition decreases capping efficiency (Kanin et al., 2007). In vitro, CTD peptides phosphorylated on Ser5 bind and activate the mammalian cap guanylyltransferase (Ho and Shuman, 1999). Therefore, TFIIH and its substrates play conserved roles in mRNA 5′-end formation, which might suffice to couple TFIIH and P-TEFb functions in S. pombe, because Cdk9 is in a constitutive complex with the capping enzyme Pcm1.
Cdk9/Pcm1 crosslinked to aro1+ efficiently even when Mcs6as was inhibited, implying a relaxed requirement for phosphorylation or an alternative mode of recruitment to some genes (perhaps the majority). Threshold requirements for CDK activity must therefore be set locally at specific genes, rather than globally throughout the genome. Selective requirements for Pol II phosphorylation and for specific CTD kinases at different promoters have been detected in budding yeast (Lee and Lis, 1998; McNeil et al., 1998) and in mammalian cells (Gomes et al., 2006; Donner et al., 2007), suggesting a conserved role for CDKs in regulating gene expression, not simply in supporting global transcription.
What makes genes differentially dependent on CDK activity for full expression? An elongation factor such as Spt5, which accumulated out of proportion to Pol II on eng1+ but not aro1+, might play a role in defining CDK target genes, but what then influences Spt5 recruitment or retention? One possibility is the local chromatin environment; in S. cerevisiae, Bur1 regulates histone ubiquitinylation and methylation, possibly to permit elongation (Laribee et al., 2005; Wood et al., 2005)—a mechanism that could vary in importance at different genetic loci.
Unraveling Pol II Phosphorylation in Fission Yeast
Based on chemical-genetic analysis of the two P-TEFb homologs, Lsk1 appears to be uniquely required for phosphorylation of CTD Ser2, in agreement with a previous report (Karagiannis and Balasubramanian, 2007), but Ser2 phosphorylation additionally depends on either Mcs6 or Cdk9 being active. In budding yeast, Ctk1 seems to be primarily responsible for Ser2 phosphorylation (Cho et al., 2001). Our data therefore support the functional analogy between Lsk1 and Ctk1, neither of which is essential.
Fission yeast Cdk9 and budding yeast Bur1 are likewise analogous in function. Inactivation of either kinase decreased Pol II crosslinking in coding regions [(Keogh et al., 2003; Guiguen et al., 2007) and this work], and heterologous co-expression of Cdk9 and Pch1 rescued lethality due to deletion of BUR1 (Pei et al., 2003). Inactivation of Bur1 had modest effects on Ser2 and Ser5 phosphorylation in vivo, prompting the suggestion that it acts principally through another target (Keogh et al., 2003). In S. pombe, truncation of Spt5 to remove the region phosphorylated by Cdk9 mimicked inhibition of Cdk9as in an mcs6as background. It did not cause lethality or suppress cdk9as, however, indicating that Cdk9 acts on at least one other substrate to provide an essential function. Although we suspect that target is Pol II, the requirement for Cdk9 cannot simply be due to its ability to phosphorylate Ser2. Compared to Lsk1 inactivation, inhibiting Cdk9 modestly reduced Ser2 phosphorylation, but more effectively increased electrophoretic mobility of phosphorylated Rpb1. In vitro, Cdk9 generated hyperphosphorylated CTD isoforms—an effect potentiated by Mcs6-mediated priming phosphorylation. We posit that Cdk9 is required for multi-site Pol II phosphorylation, which in turn might promote processive elongation on CDK-dependent genes.
A Checkpoint in Transcription, Reconsidered
Studies in budding yeast seemed to argue against the existence of a general checkpoint that prevents elongation of improperly processed transcripts (Muratani et al., 2005; Kanin et al., 2007). Here we have shown that inactivation of TFIIH-associated kinase in fission yeast prevents recruitment of P-TEFb/Pcm1 and impedes elongation of select transcripts, perhaps through the unopposed action of Spt5 (Figure 7). The results suggest that a checkpoint-like control does operate at a subset of genes in S. pombe. They do not reveal how that subset is specified, but the mere fact of specificity could help reconcile observations in different organisms, by indicating that the checkpoint is a control mechanism conferring an additional layer of regulation on specific genes rather than a general feature of transcription by Pol II.
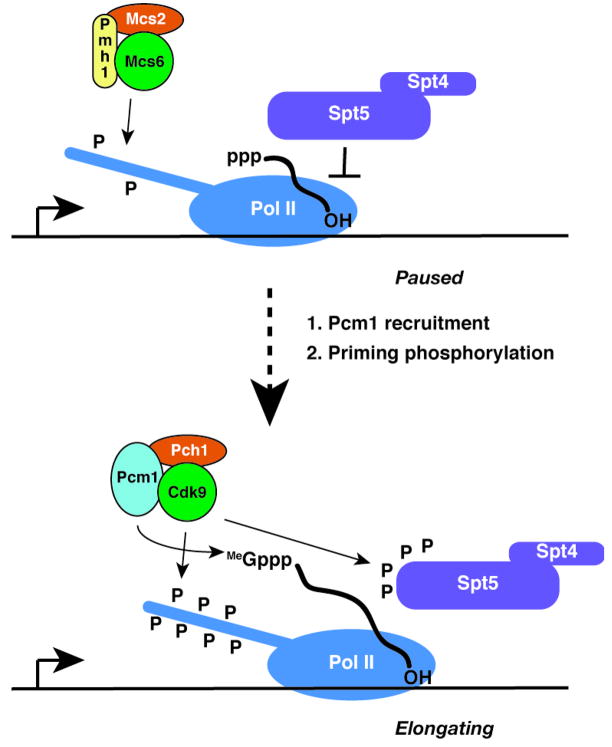
Figure 7. A Mechanism to Couple Capping and Elongation at Select Genes.
The Mcs6 complex phosphorylates and primes the Pol II CTD (top), which facilitates recruitment of the Cdk9/Pcm1 complex to phosphorylate the CTD and Spt5, complete the 5′-cap structure and relieve the Spt5-induced pause to elongation (bottom).
Experimental Procedures
Creation of as Mutant Strains
To introduce AS mutations in mcs6, cdk9 and lsk1, we transformed haploids with PCR products containing mutant sequences in tandem with a KanMX marker and flanking 3′ sequence. We also replaced mcs6+ with a cDNA containing an L87G mutation and a carboxyl-terminal hemagglutinin tag (mcs6L87G-3HA), as described for mcs6 ts strain creation (Saiz and Fisher, 2002). Correct integration was confirmed by PCR and sequencing. See Table S1 for strains used in this study.
Kinase Assays
Mcs6 and Cdk9 complexes were isolated from pmh1-TAP and pcm1-TAP extracts, respectively, by adsorption to IgG-agarose (Sigma) and elution with TEV protease. Eluates were incubated 15 min at 22°C with varying amounts of 1-NM-PP1 or 3-MB-PP1 in DMSO (at 0.5% [v/v] final concentration), followed by addition of assay mixture (10 mM Hepes [pH 7.4], 1 mM DTT, 100 μM ATP, 5 μCi [γ-32P]-ATP, plus 2.5 mM MnCl2, 1 μg GST-Rpb1-CTD for Mcs6; or 10 mM MgCl2, 150 mM NaCl, 1 μg GST-Spt5[801–990] for Cdk9). Reactions were stopped after 15 (Cdk9) or 60 min (Mcs6) at 22°C by addition of SDS to 2% (w/v) final concentration. Labeling with [γ-32P]N6-(benzyl)-ATP was performed in extracts supplemented with 1 mM unlabeled ATP and a regenerating system, or with purified proteins and 100 μM ATP, as described (Larochelle et al., 2006). Phosphorylation was detected by autoradiography and quantified by phosphorimager.
Immunological Methods
Immunoprecipitations were performed with antibody 9E10 and Protein G-Sepharose (GE Healthcare) for Cdk9-Myc, or IgG-Sepharose (GE Healthcare) for Pcm1-TAP, and washed with 10 mM Hepes (pH 7.4), 150 mM NaCl, 0.1% (v/v) Triton X-100. For immunodepletion, two consecutive immunoprecipitations were performed. Inputs (2.5%), supernatants (2.5%) and first immunoprecipitates (50%) were analyzed by immunoblot.
Microarray and qRT-PCR Analysis
Array data were collected in two groups. First, oligo(dT)-primed, fluorescently labeled cDNA from cdk9as or cdk9as mcs6as cells treated with 20 μM 3-MB-PP1 for 30, 60 or 120 min was competitively hybridized with cDNA from the same strain treated with DMSO. Second, cDNA from cdk9as, mcs6as or cdk9as mcs6as cells treated with 40 μM 3-MB-PP1 for 15, 30 or 60 min was hybridized against a mock-treated wild-type reference cDNA. Both sets included controls in which: 1) cDNA from each mutant and the wild type in the absence of 3-MB-PP1 was competitively hybridized, to determine effects of the mutations per se; and 2) wild-type cells treated with 3-MB-PP1 or DMSO were compared, to measure nonspecific drug effects. To determine differential expression between cdk9 or mcs6 strains and wild type and to reduce dye-bias effects, statistical contrasts were generated against 40 μM 3-MB-PP1-treated wild-type cells, as described in Supplemental Experimental Procedures. In addition, technical replication was performed for each experiment. The complete data set including raw and processed data is available through ArrayExpress under accession number E-MEXP-1746.
RNA extraction for qPCR analysis was performed with Qiagen RNeasy columns. First strand cDNA was synthesized from 1 μg total RNA, using SuperScript First-Strand Synthesis kit (Invitrogen), and quantified with a Stratagene MX3000P instrument with SYBRGreen Master Mix (Applied Biosytems).
Chromatin Immunoprecipitation
Cells (~5×108/100 ml) were treated 15 min or 1 hr with 40 or 20 μM 3-MB-PP1 (or DMSO), respectively, fixed with 1% (v/v) formaldehyde 15 min at room temperature, harvested and flash-frozen. 50 μl of protein extract was incubated with 1 μl 9E10 or 8WG16 (Covance) or anti-HA sc-7392 (Santa Cruz Biotechnology) antibody. Immune complexes were recovered with 20 μl Protein A- or Protein G-Sepharose. Regions of eng1+ and aro1+ were amplified by qRT-PCR as previously described (Tanny et al., 2007). Background subtraction was based on negative immunoprecipitation controls (no antibody for 8WG16, untagged strain for 9E10 and anti-HA). See Table S2 for sequences and locations of primers.
Gel Exclusion Chromatography
Extracts of mid-log cdk9–13myc pcm1–2XTAP cells were prepared as described (Pei et al., 2006) and chromatographed in Superose 6 columns (GE Healthcare). Each fraction (0.15 of 0.75 ml) was incubated with 100 ng Csk1 (Pei et al., 2006) in 10 mM Hepes (pH 7.4), 10 mM MgCl2, 1 mM DTT, 1 mM ATP to activate Cdk9, which was precipitated with 9E10 and tested for activity as described above.
Supplementary Material
Acknowledgments
We thank S. Larochelle for guidance in performing analog-selective labeling and for thoughtful comments on the manuscript, J. Tanny and D. Allis for help with quantitative PCR, S. Garrett for strain construction, S. Shuman for helpful discussions and T. Malcolm for critical review of the manuscript. This work was supported by U.S. National Institutes of Health grants GM076021 (R.P.F.), GM076272 (J.K.L.), EB001987 (K.M.S.) and GM052470 (B.S.), and by the Facility for Fission Yeast Microarrays (J.K.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RP. Secrets of a double agent: CDK7 in cell-cycle control and transcription. J Cell Sci. 2005;118:5171–5180. doi: 10.1242/jcs.02718. [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiguen A, Soutourina J, Dewez M, Tafforeau L, Dieu M, Raes M, Vandenhaute J, Werner M, Hermand D. Recruitment of P-TEFb (Cdk9-Pch1) to chromatin by the cap-methyl transferase Pcm1 in fission yeast. Embo J. 2007;26:1552–1559. doi: 10.1038/sj.emboj.7601627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Kanin EI, Kipp RT, Kung C, Slattery M, Viale A, Hahn S, Shokat KM, Ansari AZ. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc Natl Acad Sci U S A. 2007;104:5812–5817. doi: 10.1073/pnas.0611505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis J, Balasubramanian MK. A Cyclin-Dependent Kinase that Promotes Cytokinesis through Modulating Phosphorylation of the Carboxy Terminal Domain of the RNA Pol II Rpb1p Sub-Unit. PLoS ONE. 2007;2:e433. doi: 10.1371/journal.pone.0000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Podolny V, Buratowski S. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol Cell Biol. 2003;23:7005–7018. doi: 10.1128/MCB.23.19.7005-7018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–430. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr Biol. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Larochelle S, Batliner J, Gamble MJ, Barboza NM, Kraybill BC, Blethrow JD, Shokat KM, Fisher RP. Dichotomous but stringent substrate selection by the dual-function Cdk7 complex revealed by chemical genetics. Nat Struct Mol Biol. 2006;13:55–62. doi: 10.1038/nsmb1028. [DOI] [PubMed] [Google Scholar]
- Lee D, Lis JT. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature. 1998;393:389–392. doi: 10.1038/30770. [DOI] [PubMed] [Google Scholar]
- Lee JM, Greenleaf AL. A protein kinase that phosphorylates the C-terminal repeat domain of the largest subunit of RNA polymerase II. Proc Natl Acad Sci USA. 1989;86:3624–3628. doi: 10.1073/pnas.86.10.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Miklos I, Du H, Watt S, Szilagyi Z, Saiz JE, Madabhushi R, Penkett CJ, Sipiczki M, Bahler J, Fisher RP. Impairment of the TFIIH-associated CDK-activating Kinase Selectively Affects Cell Cycle-regulated Gene Expression in Fission Yeast. Mol Biol Cell. 2005;16:2734–2745. doi: 10.1091/mbc.E04-11-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- McNeil JB, Agah H, Bentley D. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 1998;12:2510–2521. doi: 10.1101/gad.12.16.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick KA, Larochelle S, Zhang C, Allen JJ, Shokat KM, Fisher RP. Distinct activation pathways confer cyclin binding selectivity on Cdk1 and Cdk2 in human cells. Mol Cell. 2008;32:662–672. doi: 10.1016/j.molcel.2008.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M, Kung C, Shokat KM, Tansey WP. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell. 2005;120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Pei Y, Du H, Singer J, St Amour C, Granitto S, Shuman S, Fisher RP. Cyclin-dependent kinase 9 (Cdk9) of fission yeast is activated by the CDK-activating kinase Csk1, overlaps functionally with the TFIIH-associated kinase Mcs6, and associates with the mRNA cap methyltransferase Pcm1 in vivo. Mol Cell Biol. 2006;26:777–788. doi: 10.1128/MCB.26.3.777-788.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, Hausmann S, Ho CK, Schwer B, Shuman S. The length, phosphorylation state, and primary structure of the RNA polymerase II carboxyl-terminal domain dictate interactions with mRNA capping enzymes. J Biol Chem. 2001;276:28075–28082. doi: 10.1074/jbc.M102170200. [DOI] [PubMed] [Google Scholar]
- Pei Y, Schwer B, Shuman S. Interactions between fission yeast Cdk9, its cyclin partner Pch1, and mRNA capping enzyme Pct1 suggest an elongation checkpoint for mRNA quality control. J Biol Chem. 2003;278:7180–7188. doi: 10.1074/jbc.M211713200. [DOI] [PubMed] [Google Scholar]
- Pei Y, Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J Biol Chem. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- Pei Y, Shuman S. Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation, and mutational analysis. J Biol Chem. 2003;278:43346–43356. doi: 10.1074/jbc.M307319200. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Polanshek MM. Effects of heat shock and cycloheximide on growth and division of the fission yeast, Schizosaccharomyces pombe. J Cell Sci. 1977;23:1–23. doi: 10.1242/jcs.23.1.1. [DOI] [PubMed] [Google Scholar]
- Ramanathan Y, Rajpara SM, Reza SM, Lees E, Shuman S, Mathews MB, Pe’ery T. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J Biol Chem. 2001;276:10913–10920. doi: 10.1074/jbc.M010975200. [DOI] [PubMed] [Google Scholar]
- Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol. 2000;20:104–112. doi: 10.1128/mcb.20.1.104-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bähler J. Periodic gene expression program of the fission yeast cell cycle. Nat Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- Saiz JE, Fisher RP. A CDK-activating kinase network is required in cell cycle control and transcription in fission yeast. Curr Biol. 2002;12:1100–1105. doi: 10.1016/s0960-9822(02)00903-x. [DOI] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi T, Cho EJ, Janoo RT, Polodny V, Takase Y, Keogh MC, Woo SA, Fresco-Cohen LD, Hoffman CS, Buratowski S. Divergent subunit interactions among fungal mRNA 5′-capping machineries. Eukaryot Cell. 2002;1:448–457. doi: 10.1128/EC.1.3.448-457.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 2007;21:835–847. doi: 10.1101/gad.1516207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirode F, Busso D, Coin F, Egly JM. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol Cell. 2005;20:589–599. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Wood A, Shilatifard A. Bur1/Bur2 and the Ctk complex in yeast: the split personality of mammalian P-TEFb. Cell Cycle. 2006;5:1066–1068. doi: 10.4161/cc.5.10.2769. [DOI] [PubMed] [Google Scholar]
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.