Abstract
Polycomb Group (PcG) proteins represent a conserved family of developmental regulators that mediate heritable transcriptional silencing by modifying chromatin states. One PcG complex, the PRC2 complex, is composed of several proteins, including the histone H3 lysine 27 (H3K27) methyltransferase EZH2 and the WD-repeat protein EED. Histone H3K27 can be mono- (H3K27me1), di- (H3K27me2), or trimethylated (H3K27me3). However, it remains unclear what regulates the number of methyl groups added to H3K27 in a particular nucleosome. In mammalian cells, EED is present as four distinct isoforms, which are believed to be produced by utilizing four distinct, in-frame translation start sites in a common Eed mRNA. A mutation that disables all four EED isoforms produces defects in H3K27 methylation.1 To assess the roles of individual EED isoforms in H3K27 methylation, we first characterized three of the four EED isoform start sites and then demonstrated that individual isoforms are not necessary for H3K27me1, H3K27me2, or H3K27me3. Instead, we show that the core WD-40 motifs and the histone binding region of EED alone are sufficient for the generation of all three marks, demonstrating that EED isoforms do not control the number of methyl groups added to H3K27.
Keywords: histone, methylation, Polycomb, WD motif, translational initiation
Introduction
Polycomb Group (PcG) proteins are a conserved family of development regulators that modify chromatin states in order to mediate heritable transcriptional silencing. PcG-mediated repression is important in diverse biological processes including X-chromosome inactivation, genomic imprinting, and segmental patterning.2; 3; 4; 5; 6; 7; 8; 9; 10 One PcG complex, the PRC2 complex, is composed of several bona fide PcG proteins, including the histone H3 lysine 27 (H3K27) methyltransferase EZH2 and the WD-repeat protein EED.10; 11; 12; 13
Histone H3K27 can be mono- (H3K27me1), di- (HeK27me2), or trimethylated (H3K27me3).14 H3K27me3 is a repressive histone modification that localizes to confirmed targets of PcG silencing, including the inactive X-chromosome.2; 3; 15; 16; 17 This mark mediates its repressive effect by recruiting to chromatin or at least stabilizing the association of a second PcG complex, PRC1.18; 19; 20; 21 Conversely, the functions of H3K27me1 and H3K27me2 are not known, although H3K27me1 was recently shown to be present throughout euchromatic regions but absent in the vicinity of the start sites of active genes.22
While EZH2’s role as the catalytic subunit of PRC2 has been clearly demonstrated, the functions of noncatalytic PRC2 subunits remain poorly defined. Both EED and the Zn-finger protein SUZ12 are required to maintain the integrity of PRC2, and mutations in both genes cause EZH2 to become destabilized.1; 23 Additionally, both in vivo and in vitro, EED and SUZ12 are required for normal PRC2-mediated H3K27 methylation.1; 23; 24 However, precise mechanistic details of these functions remain unclear.
Four EED isoforms are found in mammals, and these isoforms are thought to be produced by utilizing four in-frame translation start sites in the Eed mRNA. EED isoform usage is regulated developmentally, leading to speculation that EED could also function as a PRC2 regulatory subunit.25 Additionally, EED-2, which has only been observed in undifferentiated stem cells and in tumors, has been proposed to be important in maintaining developmental plasticity.26 However, definitive biochemical functions of the various EED isoforms have not been demonstrated. Previous work using in vitro methyltransferase assays postulated that EED isoforms control the substrate specificity of the PRC2 complex. In those initial studies, the largest isoforms, EED-1 and EED-2 appeared to direct EZH2 methyltransferase activity towards histone H1K26, whereas EED-3 and EED-4 appeared to direct EZH2 methyltransferase activity towards H3K27.25 However, a second recent study that also utilized in vitro methyltransferase assays failed to detect any difference in the histone substrate preference of PRC2 complexes with or without EED-1 or EED-2.27
Given this discrepancy, in the present study, we aimed to define regions of EED required for each H3K27 methylation state in vivo using mouse embryonic stem (ES) cells that are wild-type or mutant for Eed. Because all four known EED isoforms associate with EZH2, we examined whether these isoforms might control the number of methyl groups added to H3K27 in a particular nucleosome.25; 26 To this end, we definitively characterize three of the four EED isoform start sites and demonstrate that individual isoforms are not necessary for H3K27me1, H3K27me2, or H3K27me3. These results indicate that EED isoforms do not control the enzymatic activity of the PRC2 complex. Instead, we show that EED’s core WD-40 motifs and histone binding region alone are sufficient for the generation of all three marks.
Results
Distinct localization of H3K27 methylation states
Unlike mouse stem cell lines, differentiated mouse cells, such as murine embryonic fibroblasts (MEFs), have a striking nuclear architecture, in which regions of the genome packaged as part of the pericentric heterochromatin are clearly visible by DAPI staining as DNA-rich foci. Using this characteristic DNA staining pattern to provide landmarks, we assessed the localization of H3K27me1, H3K27me2 and H3K27me3 in CD1 MEFs (Figure 1). For these experiments, we used antibodies specific for each H3K27 methylation state. The specificities of these antibodies have been demonstrated previously by competition assays.14 Additionally, positive staining with all three antibodies is lost in Eed mutant embryonic stem (ES) cells and trophoblast stem (TS) cells.1 Because methylation defects at other lysine residues have not been observed in Eed mutant ES and TS cells, these results strengthen the conclusion that the antibodies do not cross-react with methyl marks at other residues.
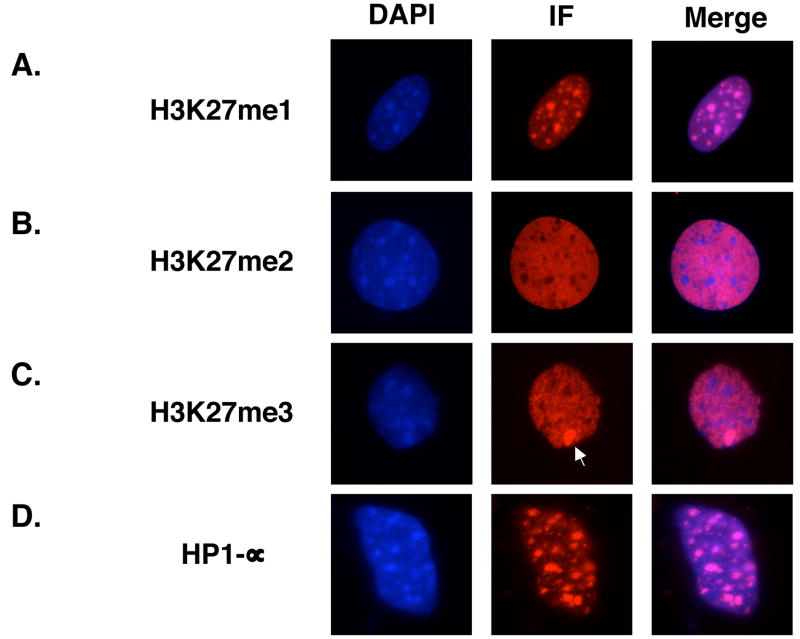
Figure 1. Localization of Histone H3K27 methylation marks.
Immunofluorescence analysis of (A) H3K27me1, (B) H3K27me2, (C) H3K27me3, and (D) HP1-α in CD1 murine embryonic fibroblasts. The inactive X-chromosome is indicated by an arrow in (C).
As previously reported, large foci of H3K27me1 colocalized with the DNA-rich pericentric heterochromatin (Figure 1A and 14). Conversely, H3K27me2 and H3K27me3 were specifically excluded from these regions, instead staining in a pattern reciprocal to that of H3K27me1 (Figures 1B and 1C, Supplementary Figure 1). HP1-α, an established marker of pericentric heterochromatin,28; 29; 30 was also enriched in the DNA-rich foci, confirming the identity of these regions (Figure 1D). Finally, H3K27me2 and H3K27me3, though largely colocalized (Supplementary Figure 2), were distinguished by the characteristic enrichment of H3K27me3 on the inactive X-chromosome in female MEFs (Figure 1B and 1C).
In addition to confirming and establishing global localization patterns of H3K27me1, -me2, and –me3, the distinct staining patterns confirm that the H3K27 antibodies are each specific for one H3K27 methylation state and do not cross-react with histones harboring other H3K27 methylation states.
Characterization of EED isoforms expressed in wild-type ES cells
The distinct localization patterns of the three H3K27 methylation states indicates that the number of methyl groups added to H3K27 in a particular nucleosome is a regulated process. Because EED has previously been posited to play a regulatory role in PRC2-mediated histone methylation,25 we aimed to define regions of EED required for H3K27me1, H3K27me2, and H3K27me3. As a first step, we attempted to discern which EED isoforms are required for H3K27 methylation by rescuing Eed mutant embryonic stem (ES) cells, which lack detectable levels of endogenous H3K27me1, H3K27me2, and H3K27me3,1 with constructs expressing individual EED isoforms. However, before a systematic series of rescue experiments was performed, we attempted to confirm the identities of the EED isoforms.
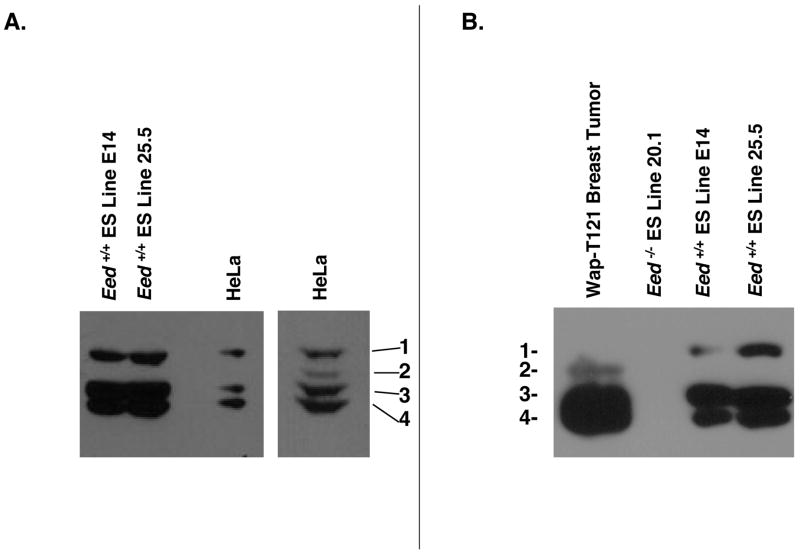
Previous work indicates that all four EED isoforms are present in wild-type, undifferentiated mouse ES cells.26 However, we were consistently only able to resolve three isoforms in multiple, independently-derived ES cell lines, as well as in TS cells (Figure 2 and data not shown). To assess whether our electrophoresis conditions would allow us to resolve all four EED isoforms and, if so, to confirm the identities of the three observed isoforms, we compared the isoforms present in our ES cells to the isoforms present in HeLa cells and in mouse Wap-T121 mammary tumors. Previous work has demonstrated that EED-1, EED-3, and EED-4 are expressed in HeLa cells and that EED-2 is upregulated in many mouse tumors.26 Consistent with those reports, we observed high levels of EED-1, EED-3 and EED-4 and much lower levels of EED-2 in HeLa cells (Figure 2A). Similarly, EED-2, along with EED-3 and EED-4, was observed in Wap-T121 breast tumors (Figure 2B). Comparison of the isoforms present in those sources with the isoforms present in our embryonic stem cells suggests that we observe isoforms 1, 3, and 4 but not 2 in our ES cells (Figure 2).
Figure 2. Confirmation of EED isoform identities.
Western blot analysis of EED comparing isoforms observed in embryonic stem cells to isoforms observed in (A) HeLa cells or (B) Mouse Wap-T121 breast tumors. On prolonged exposure, EED-2 becomes visible in HeLa lysates (Aright panel). EED isoforms 1–4 are indicated as 1, 2, 3, and 4.
Kuzmichev et al. demonstrated that EED-2 is rapidly downregulated when ES cells are stimulated to differentiate.25 Hence, the most parsimonious explanation for our failure to dectect four EED isoforms is that the ES cells employed were beginning to differentiate when they were harvested, causing a downregulation of EED-2. One possible explanation for such differentiation would be the removal of the cells from fibroblast feeders prior to harvesting in order to minimize feeder contamination in our immunoblotting assays. To avoid this complication, we also analyzed EED isoforms in E14 ES cells, which are feeder independent,31 but were still able to detect only three isoforms (Figure 2).
Embryonic stem cells, trophoblast stem cells, and primitive endoderm-derived XEN cells are the only known cell types that can be derived from mouse embryos prior to gastrulation, the embryonic stage when Eed mutant embryos arrest.32; 33; 34; 35; 36 Because only three EED isoforms were resolved in our ES and TS cells and because only trace levels of EED are detected in XEN cells (data not shown), we restricted our analysis to EED isoforms 1, 3, and 4.
Deletion mapping of EED isoform start sites
Although most proteins initiate translation at methionine-encoding AUG codons, an increasing number of proteins are recognized to initiate translation from non-AUG codons. Often, these alternative start sites generate upstream isoforms of proteins also translated from downstream, canonical AUG initiation codons, and typically, the alternative codons differ from the canonical AUG sequence at only one of the three nucleotide positions.37; 38 For instance, Fibroblast Growth Factor-2 (FGF2) is present as five isoforms in mammalian cells, with four CUG initiation codons upstream of a canonical AUG start site, and Vascular Endothelial Growth Factor (VEGF) is translated from both an upstream CUG and a downstream AUG.39; 40; 41; 42; 43 Consistent with both of these trends, the upstream EED-1 and EED-2 are postulated to initiate translation from non-canonical GUG codons at positions 169–171 and 274–276 in the Eed cDNA, respectively, and the putative start sites for EED-3 and EED-4 are canonical AUG sequences at positions 451–453 and 493–495 (Figure 3A and Supplementary Figure 3).25; 44
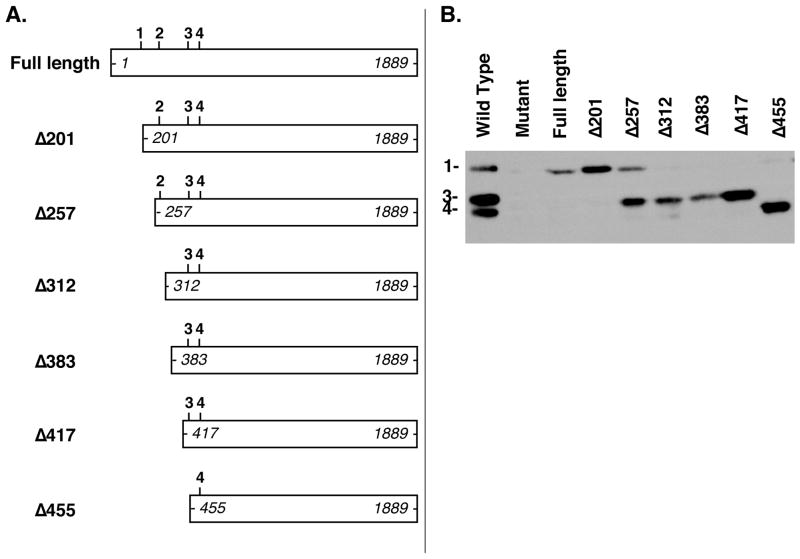
Figure 3. Deletion mapping of EED translational start sites.
(A) Constructs transfected into Eed mutant embryonic stem cell line 21 (below) are shown. Putative translation start sites for EED-1 (GUG 169–171), EED-2 (GUG 274–276), EED-3 (AUG 451–453), and EED-4 (AUG 493–495) are indicated in the schematic of the endogenous mRNA as 1,2,3, and 4 and correspond to codon locations in the mouse Eed transcript (Accession number: BC012966). The positions of the most 5′ and 3′ nucleotides of each construct are indicated in italics. Identifiers for each construct are shown to the left and indicate the number of nucleotides removed from the 5′ end of the Eed cDNA (e.g. Δ210 refers to a construct expressing an Eed cDNA lacking the 5′ 210 nucleotides of the full length message). (B) Whole-cell lysates from Eed+/+ ES cell line 25.5 (Wild-type), Eed−/− ES cell line 21 (Mutant), or Eed−/− ES cell line 21 transiently transfected with the Eed expression constructs shown in (A)were analyzed by Western blotting with an antibody detecting EED.
Overall, mouse and human EED are very similar proteins. In fact, the predicted EED-3 and EED-4 isoforms are 100% identical between the two species.45 However, the sequences between the putative EED-1 and EED-2 start sites, are much more variable.45 Given the abrupt boundary between highly variable and nearly identical sequences, we questioned whether the actual EED-1 start site might be further 3′ than previously reported.
To map EED isoform start sites, we generated a series of Eed expression constructs progressively truncated at the 5′ end of the Eed cDNA (Figure 3A). Individual constructs were then transiently transfected into Eed mutant ES cells, which lack detectable endogenous EED, and EED isoform expression was assessed by Western blotting. Because isoform expression could be lost either by deleting past an isoform start site or by deleting an upstream regulatory element required for translation from an intact start site, the absence of a band is uninformative in this assay. However, the continued presence of an isoform after its putative start site has been deleted is strong evidence that the actual start site must be further downstream. Notably, after transfection, the furthest 5′ intact translation start site was generally utilized preferentially to downstream translation start sites (Figure 3). This observation suggests that regulated usage of the various EED translation start sites is not simply a consequence of the interaction between trans-acting factors and sequences present in the message but instead may be influenced by upstream regulatory events.
Consistent with the relaxed sequence conservation between mouse and human Eed sequences beginning with and immediately downstream from GUG 169–171, EED-1 was expressed from constructs truncated 32 (Δ201) and even 88 (Δ257) nucleotides beyond the reported EED-1 start site, suggesting that EED-1 may not initiate translation at GUG 169–171 as previously proposed (Figure 3B). EED-1 expression was lost only after deleting the 5′ 312 nucleotides of the Eed cDNA, a deletion extending 143 nucleotides beyond the reported EED-1 start site and 38 nucleotides beyond the published EED-2 start site (Figure 3B). Consistent with the published identity of the EED-3 start site at AUG 451–453, this isoform was observed after deleting the 5′ 417 nucleotides but not after deleting the 5′ 455 nucleotides (Figure 3B). Finally, EED-4 was present even in the largest truncation, which deleted the 5′ 455 nucleotides, consistent with the published EED-4 start site residing at AUG 493–495 (Figure 3B). Notably, our ability to detect EED-1, EED-3, and EED-4 from this series of cDNA expression constructs confirms that alternative splicing of exons within the EED coding sequence is not responsible for the production of these three isoforms.
Confirmation of EED isoform start sites
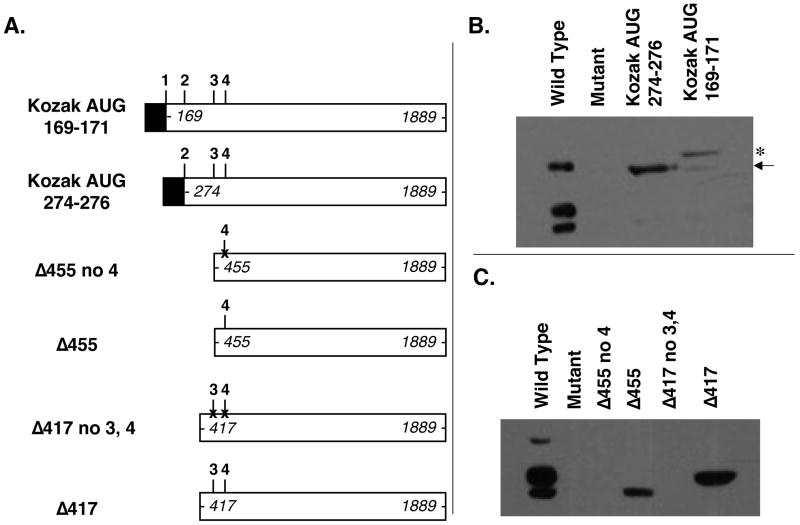
To verify that GUG 169–171 is upstream of the actual EED-1 start site, we forced expression from this codon by replacing the sequences encoding GUG 169–171 with a canonical translation start site consisting of a consensus Kozak sequence followed by an AUG initiator codon (Figure 4A). Consistent with the hypothesis that EED-1 translation actually initiates further downstream, the resulting product was substantially larger than EED-1 (Figure 4B, asterisk). A lower level of an EED-1 sized product, presumably initiating at the actual start site further downstream, was also observed after transfection with this construct (Figure 4B, arrow).
Figure 4. Verification of EED isoform start sites.
(A) Constructs transfected into Eed mutant embryonic stem cell line 21. The positions of the most 5′ and 3′ nucleotides of each construct are indicated in italics. 1,2,3, and 4 indicate reported EED translational start sites GUG 169–171, GUG 274–276, AUG 451–453, and AUG 493–495, respectively. Black boxes refer to strong Kozak-AUG sequences engineered into the expression construct in order to drive translation from the 169–171 codon (Kozak AUG 169–171) and from the 274–276 codon (Kozak AUG 274–276), respectively. “X” markings through the putative EED-3 and EED-4 start sites in Δ417 no3,4 and in Δ455 no4 indicate AUG→AUA mutations intended to disrupt translation intitation. (B and C) Whole-cell lysates from Eed+/+ ES cell line 25.5 (Wild-type), Eed−/− ES cell line 21 (Mutant), or Eed−/− ES cell line 21 transiently transfected with the Eed expression constructs shown in (A) analyzed by Western blotting with an antibody detecting EED.
Given that the deletion mapping data indicates that either the true EED-1 start site or some regulatory element required for translation from that true EED-1 start site must map between nucleotides 257 and 312, we hypothesized that EED-1 may be produced by translation initiating from the previously proposed EED-2 start site at GUG 274–276. To test that possibility, we forced expression of a protein initiating translation at that site by replacing GUG 274–276 with a consensus Kozak followed by an AUG initiator codon (Figure 4A). Consistent with EED-1 translation initiating at this location, the resulting product was the same size as EED-1 observed in mouse ES cells (Figure 4B).
Because EED-2 was not expressed in our wild-type ES cells or from any of our truncated expression constructs, we were unable to characterize this isoform. However, driving translation from a candidate start site at GUG 397–399 produced a protein that appears to be smaller than EED-2 (data now shown), suggesting that the EED-2 start site lies between GUG 274–276 and GUG 397–399 or that EED-2 is generated by alternative mechanisms, such as proteolytic processing.
Our deletion mapping data were consistent with presumptive EED-3 and EED-4 start sites residing at AUG 451–453 and AUG 493–495, respectively. To confirm that translation of those isoforms originates at those sites, we engineered AUG→AUA site directed point mutations into the AUG 451–453 and AUG 493–495 codons (Figure 4A). Consistent with those codons being the initiation codons for the two smaller isoforms, constructs harboring those mutations failed to express EED-3 and EED-4 (Figure 4C).
EED isoforms are dispensible for all H3K27 methylation states
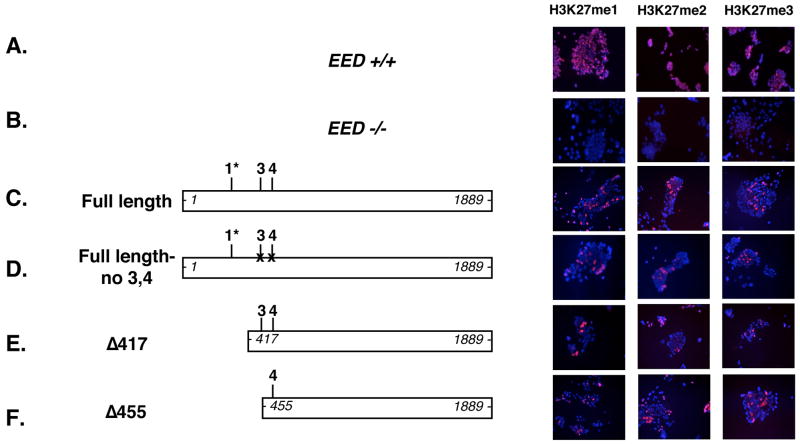
To determine whether all three EED isoforms present in our ES cells are required to mediate the three H3K27 methylation states, H3K27 methylation was monitored in Eed mutant ES cells transiently transfected with several of the Eed cDNA expression constructs. This rescue assay has previously been utilized to demonstrate that protein(s) expressed from a full-length Eed cDNA cassette can mediate all three H3K27 methylation states,1 although the role of individual isoforms in that rescue has not been directly assessed. This approach is advantageous relative to an isoform-specific, targeted loss-of-function approach, because it avoids the problematic interpretation of negative results in the event that individual isoforms are dispensible for H3K27 methylation.
Constructs harboring site-directed mutations to eliminate EED-3 and EED-4 expression or truncated to eliminate EED-1 expression were both able to rescue all three H3K27 methylation states in Eed mutant ES cells (Figure 5D and E). Additionally, a construct retaining only the EED-4 start site also rescued H3K27me1, H3K27me2, and H3K27me3 (Figure 5F). As previously reported, expression of the Eedl7Rn5–3354SB allele, which produces an unstable and nonfunctional protein harboring a L→P substitution, failed to rescue H3K27 methylation (data not shown and 1). Collectively, these results demonstrate that the three H3K27 methylation states are not dependent on individual EED isoforms.
Figure 5. Histone H3K27 methylation in cells lacking one or more EED isoforms.
Immunofluorescence analysis of H3K27me1, H3K27me2, and H3K27me3 in wild-type ES cell line 25.5 (Eed+/+) and in Eed mutant ES cell line 21 either mock transfected (Eed −/−) or transiently transfected with the indicated constructs. Eed expression constructs transfected into ES cell line 21 are shown on the left. The positions of the most 5′ and 3′ nucleotides of each construct are indicated in italics. Isoform start sites at GUG 274–276, AUG 451–453, and AUG 493–495 are shown as 1*, 3, 4, respectively. 1* discrimates the GUG 274–276 start site for EED-1 reported here from the GUG169–171 start site reported previously25; 44. “X” markings through the putative EED-3 and EED-4 starts sites in Full length no 3,4 and in Δ417 no3,4 represent AUG→AUA mutations intended to disrupt translation intitation. EED 1036–1038 L→P refers to Eedl7Rn5–3354SB, a point mutant protein previously demonstrated to lack H3K27 methyltransferase activity. DAPI-stained DNA is blue, and methylated histones are shown in red. In the transient transfection assay, approximately 10% of the ES cells are successfully transfected, and with constructs expressing functional EED, a similar percentage of cells are rescued.
Disruption of EED WD-40 motifs eliminates H3K27 methylation
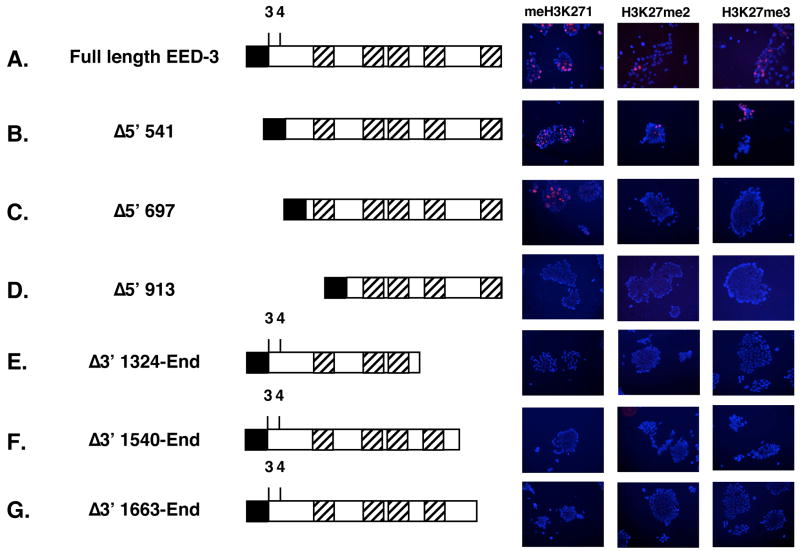
Having demonstrated that the sequences distinguishing EED isoforms are not required for H3K27 methylation, we attempted to assess which functional motifs common to all four EED isoforms might be essential for PRC2 activity. EED and its homologs in other organisms are WD-repeat proteins. However, there is disagreement about the number of WD-40 motifs present in EED, with estimates varying between five and seven.46; 47; 48 In functional studies assessing EED’s ability to bind EZH2 or its ability to mediate transcriptional repression when tethered to a GAL4 DNA binding domain, only five WD-40 motifs have appeared functionally necessary 49; 50. Those five motifs map to Eed cDNA sequences 721–808 (WD-40 motif 1), 1012–1105 (WD-40 motif 2), 1150–1240 (WD-40 motif 3), 1330–1444 (WD-40 motif 4), and 1672–1774 (WD-40 motif 5). To determine whether those same regions are required for EED’s ability to mediate H3K27 methylation, Eed mutant ES cells were transiently transfected with a series of Eed cDNA expression constructs progressively truncated from either the N- or C-terminus.
A protein lacking the N-terminal 16 amino acids of EED-4 (Δ5′ 541) was able to mediate all three H3K27 methylation states (Figure 6B), confirming that the N-terminal regions in EED, including those amino acids that distinguish individual isoforms, are not required for the catalytic activity of the PRC2 complex. Conversely, deletion of any of the five putative WD-40 motifs abolished EED’s ability to mediate H3K27 methylation (Figure 6D–G). An intermediate deletion, Δ5′ 697 also left the five putative WD-40 motifs intact. However, protein expressed from this construct retains only 8 amino acids upstream of WD-40 motif 1 and lacks a portion of EED recently shown to be necessary for histone binding.51 Although the expression of the Δ5′ 697 construct consistently rescued a low level of H3K27me1, little if any H3K27me2 and H3K27me3 were observed (Figure 6C).
Figure 6. Functional mapping of required WD-40 motifs in EED.
Immunofluorescence analysis of H3K27me3 in Eed mutant ES cell line 21 either mock transfected (Eed−/−) or transiently transfected with the indicated constructs. Below each schematic, nucleotides encoding for the N- and C-terminal amino acids of each protein are indicated in italics. Above each schematic, “3” and “4” refer to translation start sites for EED isoforms 3 and 4. Diagonally-lined boxes refer to putative WD-40 motifs encoded by cDNA sequences 721–808 (WD-40 motif 1), 1012–1105 (WD-40 motif 2), 1150–1240 (WD-40 motif 3), 1330–1444 (WD-40 motif 4), and 1672-1774 (WD-40 motif 5) 49. Black boxes refer to consensus Kozak + ATG initiator sequences engineered into the construct. DAPI-stained DNA is blue, and methylated histone are shown in red.
EED-EZH2 interaction assessed in living cells by YFP fragment complementation assays
The inability of several truncated Eed expression constructs to successfully rescue H3K27 methylation defects in Eed mutant cells could reflect an inability of these proteins to associate with EZH2, the catalytic subunit of the PRC2 complex. In order to assess EED-EZH2 interactions, we utilized a YFP fragment complementation assay.52 In this assay, test proteins are translationally fused to N- and C-terminal halves of Yellow Fluorescent Protein (YFP), called Venus 1 and Venus 2, respectively. If the test proteins are able to interact, the two YFP fragments will be close enough to reconstitute the functional fluorescent complex, which can be assessed by fluorescence microscopy or flow cytometry.
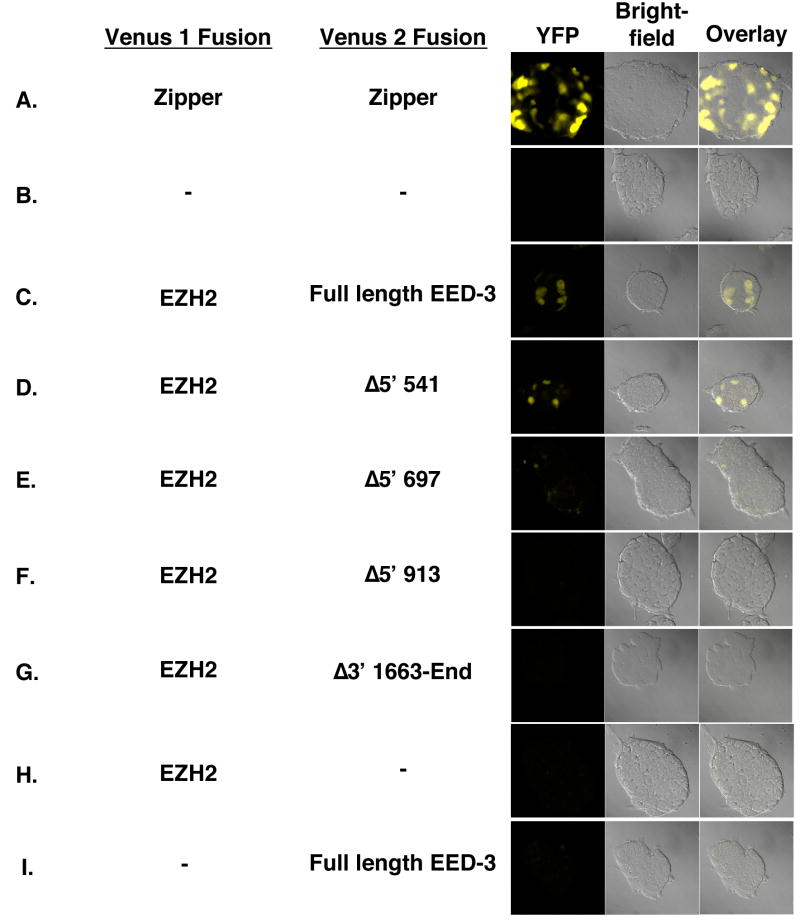
To demonstrate the validity of the YFP fragment complementation assay, control constructs expressing Venus 1 and Venus 2 fragments translationally fused to a homodimerizing leucine zipper protein, GCN4, were utilized. When co-expressed in E14 ES cells, robust YFP fluorescence was observed (Figure 7A). Similarly, Venus 1-EZH2 and EED-Venus 2 were able to interact and reconstitute functional YFP (Figure 7C, Table 1). This fluorescence was distributed throughout the cell and was specifically dependent on the presence of both constructs, as expression of individual constructs failed to produce YFP activity (Figure 7H and I).
Figure 7. EED-EZH2 interaction assessed by YFP fragment complentation assays.
YFP fragment complentation assays in E14 ES cells transiently transfected with constructs expressing the following Venus 1 and Venus 2 fusions: (A) Venus1-GCN4 and GCN4-Venus2, (B) no DNA, (C) Venus1-EZH2 and Full length EED3-Venus2, (D) Venus1-EZH2 and EED Δ5′ 541-Venus2, (E) Venus1-EZH2 and EED Δ5′ 697-Venus2, (F) Venus1- EZH2 and EED Δ5′ 913-Venus2, (G) Venus1-EZH2 and EED Δ3′ 1663-End-Venus2, (H) Venus1-EZH2 and no Venus 2, and (I) no Venus 1 and Full length EED3-Venus2.
Table 1.
Split YFP Complementation by EED-EZH2 Interaction
| Percentage of Positive Cells | |
|---|---|
| Full-length | 11.27 ± 0.33 |
| Δ5′ 541 | 6.21 ± 0.52 |
| Δ5′ 697 | 0.18 ± 0.04 |
| Δ5′ 913 | 0.12 ± 0.02 |
| Δ3′ 1663-End | 0.28 ± 0.05 |
| No transfect | 0 ± 0 |
Legend: E14 embryonic stem cells were cotransfected with Venus 1-EZH2 and EED-Venus 2 expression constructs. 24 hours post-transfection, triplicate samples were analyzed by FACS. The mean percentage of YFP-positive cells and corresponding standard deviation from a representative experiment are shown.
Next, the truncated Eed cDNAs, which were tested previously for their ability to rescue H3K27 methylation defects in Eed mutant cells, were fused to Venus 2. Consistent with previous reports,49; 50 deletion of even a single WD-40 motif from either the N- or C-terminus of EED appeared to abolish the EED-EZH2 interaction (Figure 7F and G, Table 1), just as they abolished the ability to rescue H3K27 methylation (Figure 6D and G).
The Eed Δ5′ 541 truncation fully rescues H3K27 methylation despite lacking the first 16 amino acids of EED-4 (Figure 6B). Consistent with that functional readout, in the YFP fragment complementation assay, this protein was able to robustly interact with Venus 1-EZH2, although at a lower level than the full length construct (Figure 7D, Table 1). Expression of the EedΔ5′ 697 alllele rescued H3K27me1 but not higher H3K27 methylation states. In the YFP fragment complentation assay, no fluorescence over background was observed upon coexpression of Venus 1-EZH2 and EED Δ5′ 697-Venus 2, suggesting that this more severe truncation severely limits EED’s ability to interact with EZH2.
Truncated EED protein levels
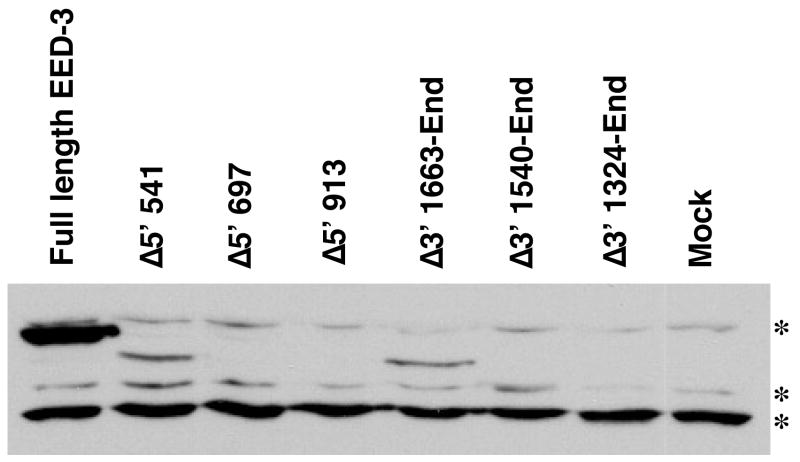
PRC2 subunits are frequently unstable outside of intact protein complexes.1; 23 As a result, the inability of several truncated proteins to interact with EZH2 raises the possibility that these truncated proteins are not stable. To address this possibility, levels of FLAG-tagged truncated EED proteins were assessed by Western blotting. Consistent with its ability to bind EZH2 and rescue H3K27 methylation, robust levels of EED Δ5′ 541 were detected in transfected cells (Figure 8). Conversely, EED Δ5′ 697, EED Δ5′ 913, EED Δ3′ 1540-End, and Δ3′ 1324-End, all of which fail to bind EZH2 in the Split-YFP assay, were undetectable (Figure 8). Consequently, there was a strong correlation between H3K27me2/me3 activity, EZH2 binding, and protein level (Figures 6–8). A striking exception to this trend was observed with the EedΔ3′ 1663-End allele. Despite failing to bind EZH2 robustly in the YFP complementation assay (Figure 7), this protein was stably detected in transfected cell populations (Figure 8).
Figure 8. Levels of truncated EED proteins.
The stability of truncated, tagged EED proteins was assessed by Western blotting lysates from transfected E14 embryonic stem cells with an anti-FLAG antibody. Three background bands observed with the FLAG antibody serve as loading controls and are indicated by asterisks.
Discussion
EED isoforms are not required for any H3K27 methylation state
Although the biological functions of H3K27me1 and H3K27me2 have not been defined, it is clear from localization studies that the three H3K27 methylation states are partitioned to nucleosomes associated with distinct regions of the genome (Figure 1 and 14; 22). Interestingly, whereas we and others have demonstrated by immunofluorescence that foci of H3K27me1 staining colocalize with the pericentric heterochromatin, a recent study utilizing chromatin immunoprecipitation demonstrated that H3K27me1 is found throughout chromatin (Figure 1 and 14; 22). Likely, the apparent pericentric enrichment of H3K27me1 observed by immunofluorescence simply reflects the local enrichment of histones in the compacted pericentric heterochromatin, as opposed to any specific enrichment of this mark as had previously been suggested.14
Nevertheless, H3K27me1, H3K27me2, and H3K27me3 do appear to localize to nucleosomes associated with distinct regions of the genome (Figure 1). This specificity, in turn, implies that the number of methyl groups added to a particular nucleosome is a regulated process. In the present study, we have defined those regions of EED required for H3K27 methylation. Initially, we hypothesized that EED isoforms could function as regulatory switches controlling H3K27 methylation states. However, none of the EED isoforms were specifically required in order to generate H3K27me1, H3K27me2, or H3K27me3 (Figure 5). These results demonstrate that the N-terminal extensions discriminating EED isoforms are not required for the enzymatic activity of the PRC2 complex.
EED WD-40 motifs and histone binding regions are required for H3K27 methylation
The ability of Eed expression constructs to rescue H3K27 methylation was dependent upon the presence of all five EED WD-40 motifs (Figure 6). Truncated proteins lacking the N- or C-terminal WD-40 motif were also unable to bind to EZH2 in a YFP fragment complentation assay (Figure 7E and F), confirming that EED-EZH2 association is likely necessary for normal H3K27 methylation. These observations are also consistent with earlier studies indicating that all five WD-40 motifs are required for EZH2 binding and EED-mediated transcriptional repression.49; 50
Interestingly, whereas the Δ5′ 541 construct was able to rescue all three H3K27 methylation states, the protein expressed from the Δ5′ 697 construct, which also retains all five WD-40 motifs, was able to mediate a low level of H3K27me1 but no H3K27me2 or H3K27me3 (Figure 6B and 6C). Recent work suggests that the N-termini of EED and of its fly homolog ESC bind to histone H3 and that the histone binding N-terminus of ESC is required for H3K27me3.51 The Δ5′ 697 construct deletes the region of EED required for histone H3 binding. Accordingly, the reduced levels of H3K27 methylation mediated by this construct suggest that, as in Drosophila, the histone H3-binding N-terminus of EED may be required for full activity of PRC2 complex. However, unlike the studies with ESC, binding between EED and EZH2 also appears to be compromised when the histone binding domain is deleted (Figure 7E). Furthermore, Δ5′ 697 protein levels were dramatically reduced, suggesting that the inability to bind EZH2 may destabilize this protein (Figure 8). Given that interpretation, it is unclear whether the reduced activity of the protein expressed from the Δ5′ 697 construct is a consequence of its inability to bind histones or its reduced ability to bind EZH2 and resultant instability. Finally, as a third explanation, it is possible that reduced levelsΔ5′ 697 protein could be due to reduced expression. While formally possible, this explanation is less likely, given that the Δ5′ 541 and Δ5′ 697 constructs have the same promoters, enhancers, 5′ and 3′ UTRs, and canonical Kozak sequences.
EED-1, EED-3, and EED-4 translation start sites map to GUG 274–276, AUG 451–453, and AUG 493–495
In mapping regions and motifs in EED required for H3K27 methylation, we have also defined in vivo the translation start sites for three of the four EED isoforms. Previous work has suggested that the four EED isoforms are generated by translation initiating at GUG 169–171, GUG 274–276, AUG 451–453, and AUG 493–495, respectively.25; 44 This interpretation followed from early studies by Denisenko and Bomsztyk, who assessed EED proteins translated in a cell free system.44 Subsequently, Kuzmichev et al. generated an isoform-restricted EED antibody, called αNT, which was raised to peptides encoded by Eed cDNA sequences from positions 258–453 (M26). Because αNT recognized EED-1 and EED-2 but not EED-3 and EED-4, those results demonstrated that EED-1 and EED-2 must include the αNT epitope and must initiate translation upstream of M451–453, as predicted by Denisenko and Bomsztyk.44
Here, we have directly assessed the identity of three EED translation start sites by a combination approach involving deletion mapping, forced translation from reported start sites, and site-directed mutagenesis of candidate initiation codons. These experiments definitively map EED-1, EED-3, and EED-4 start sites to GUG 274–276, AUG 451–453, and AUG 493–495, respectively. Importantly, because the informative αNT antibody utilized by Kuzmichev et al. recognizes amino acids that would be present not only in a hypothetical protein initiating at GUG 169–171 but also in a protein initiating at GUG 274–276, the EED-1 initiation site reported here, the immunoblotting data from Kuzmichev et al. are fully consistent with the results presented in the present work, even though our conclusions disagree with the conclusions made by those authors.25
On the other hand, these results do directly contradict the earlier work of Denisenko and Bomsztyk.44 Denisenko and Bomsztyk assessed EED proteins expressed from truncated and engineered Eed cDNAs comparable to those utilized here. However, whereas we assessed translation from those messages in living cells, Denisenko and Bomsztyk utilized a rabbit reticulocyte cell-free system. In their assay, a truncation that would have deleted the 5′ 222 nucleotides in our constructs eliminated EED-1 expression, even though a truncation extending 35 nucleotides further (Δ257) did not disrupt EED-1 expression in our assay (Figure 3B). Additionally, Denisenko and Bomsztyk observed apparent upregulation of EED-1 after mutating GUG 169–171 and flanking sequences to more closely resemble a canonical initiation sequence, whereas a similar experiment in our assays produced a protein larger than EED-1 (Figure 4B).
The differences in our respective methodologies may account for the discrepancy in our results. The major advantage to the transient transfection assay employed in this report is that it allows direct comparison of EED expressed from expression constructs to endogenous EED expressed in the same cell type. Moreover, a protein initiating at GUG 169–171 is predicted to be less than 4 kDa larger than a protein initiating at GUG 274–276. In our hands, resolving proteins initiating at those sites required large gels run at low voltage. Accordingly, differences in electrophoresis conditions could also explain discrepancies in our results.
Conclusions
The biological functions of the four reported EED isoforms remain unclear. Both in vivo and in vitro, all isoforms analyzed appear capable of facilitating PRC2-dependent histone methylation (Figure 5 and 27). Accordingly, it is uncertain whether the four isoforms are even functionally necessary in living cells. However, if isoform-specific functions do exist, the in vivo characterization of EED isoform start sites reported here is an important first step in defining those functions. One critical function common to the EED isoforms is the stabilization of EZH2 in PRC2 complexes. Here, we have demonstrated that the five WD40 motifs in EED are critical to this stabilization and to PRC2-mediated H3K27 methylation.
Materials and Methods
Cell lines and culture
CD1 murine embryonic fibroblasts (MEFs) were plated on gelatin-coated coverslips and grown to subconfluency. Wild-type embryonic stem cell (ES) line E14, wild-type embryonic stem (ES) cell line 25.5 and Eedl7Rn5–3354SB/l7Rn5–3354SB (herein referred to as Eed mutant or Eed−/−) ES cell line 21 were grown first on irradiated fibroblast feeders before being plated on coverslips without feeders and grown to subconfluency.31; 53 In transfection experiments, ES cells plated on coverslips were transfected using Lipofectamine 2000™ (Invitrogen) and harvested 24–48 hours later.
Immunofluorescence
Cells on coverslips were permeabilized with CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES [pH 6.8]) and fixed in 4% paraformaldehyde that had been diluted in phospho-buffered saline (PBS). To stain, cells were washed in PBS and incubated with blocking buffer (PBS, 5% goat serum, 0.2% Tween-20, 0.2% fish skin gelatin). After blocking, samples were incubated with primary antibody (anti-H3K27me1 [Upstate], anti-H3K27me214, anti-H3K27me314, or anti-HP1-∝ [Upstate]), which had been diluted 1:250 in blocking buffer. Subsequently, cells were washed in PBS/0.2% Tween-20 and then incubated with secondary antibody (Goat anti-Rabbit Alexa 594 [Molecular Probes]) diluted 1:250 in blocking buffer. Blocking and antibody incubations were always performed in a humid chamber at 37° C. Subsequently, cells were washed again in PBS/0.2% Tween-20 and mounted with Vectashield™ containing 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Vector Laboratories). Stained slides were visualized by fluorescence microscopy, and then black and white images were captured with a Spot CCD digital camera before being pseudo-colored and merged with Spot software V3.5.9 (Diagnostic Instruments Inc.).
Plasmid Construction
For mapping and rescue experiment, constructs expressing Eed cDNAs were cloned into the shuttle TA-cloning vector pGEM®-T Easy (Promega) and then subcloned into EcoRI-digested pTarget™ (Promega) by conventional molecular biology techniques. The orientation and identity of all inserted sequences were confirmed by fully sequencing the cDNA and the cloning junctions.
Eed cDNAs were truncated by PCR, utilizing forward primers that annealed within the Eed cDNA and reverse primers anchored in pTarget (primer sequences available upon request). Site-directed point mutations were generated by standard methods. Briefly, primers spanning EED-3 and EED-4 start sites but harboring ATG→ATA mutations were used as forward primers with a reverse primer anchored in pTarget. Finally, in order to engineer strong translation start sites, in frame consensus Kozak(GCCACC)-ATG 5′ extensions were included on forward primers.
For YFP fragment complementation assays, PRC-amplified Eed and Ezh2 fragments were cloned into split YFP expression vectors52. Briefly, Eed PCR products were TA-cloned into pGEM®-T Easy (Promega) and then subcloned into the NotI and ClaI sites at the 5′ end of Venus 2. Similarly, PCR-amplified Ezh2 fragments were TA-cloned before subcloning into the the BspEI and XbaI sites at the 3′ end of Venus 1.
Western blotting
Wild-type ES cell line 25.5, wild-type ES cell line E14, mock-transfected Eed mutant ES cell line 21, and Eed mutant ES cell line 21 transfected with various expression constructs were harvested in urea lysis buffer (7.5 M Urea, 0.01 M Tris [pH 8.0]), 0.1M NaH2PO4) 48 hours after transfection. Urea-lysates were also harvested from mouse Wap-T121 mammary tumor tissue, which is generated by tissue-specific expression of T121, a fragment of SV40 T antigen that interferes with the function of Retinoblastoma-family proteins 54. Proteins were separated on 10% SDS-PAGE gels in Tris-Glycine running buffer and transferred to Immun-Blot PVDF membranes (Bio-Rad) in Tris-Glycine Methanol transfer buffer. Membranes were blocked in 5% non-fat dried milk (NFDM [Food Lion])/TBST (50 mM Tris HCl [pH 7.4], 150 mM NaCl, 0.1% Tween-20) and then incubated overnight at 4° C with a mouse monoclonal anti-EED antibody55 diluted 1:400 in 3% NFDM/TBST or Sigma M2 anti-FLAG antibody diluted 1:1000 in 3% NFDM/TBST. Membranes were vigorously washed in TBST and then incubated overnight at 4°C with HRP-conjugated goat anti-rabbit antibody (Pierce) diluted 1:3000 in 5% NFDM/TBST. Membranes were then vigorously washed in TBST and TBS (50 mM Tris HCl [pH 7.4], 150 mM NaCl), before adding SuperSignal West Dura Extended Duration Substrate (Pierce) developing reagents. Finally, blots were exposed to film and developed.
YFP fragment complementation assays
E14 ES cells grown on coverslips were transiently transfected with YFP fragment complentation plasmids expressing GCN4, EZH2, or EED. 24 hours after transfection, the cells were washed in 1XPBS, fixed for 20 minutes in 4% paraformaldehyde, and then washed thoroughy in 1X PBS. Fixed cells were mounted in a polyvinyl alcohol/glycerol mounting medium with 2% N-propyl gallate and 0.25% 1,4-diazabicyclo[2,2,2]octane. Subsequently, YFP was visualized with a Carl Zeiss LSM5 fluorescence microscope.
For quantification of YFP fluorescence, cells were harvested 24 hours post-transfection, trypsinized, washed in 1X PBS, and then fixed in 1% formaldehyde/PBS. Relative levels of fluorescence were determined using a CyAn ADP flow cytometer (DakoCytomation, Inc.). For each sample, at least 200,000 total events were analyzed. Percent of YFP positive cells and YFP fluorescence intensity were determined using DakoCytomation Summit software version 4.3.
Supplementary Material
Immunofluorescence analysis of H3K27me1 (red, second panel) and H3K27me3 (green, third panel) HP1-∝ in CD1 murine embryonic fibroblasts.
Immunofluorescence analysis of H3K27me1 (red, second panel) and H3K27me3 (green, third panel) HP1-∝ in CD1 murine embryonic fibroblasts.
Schematic diagram indicating the positions of the four putative EED isoform translation start sites the five WD-40 motifs, and the TAA stop codon. Nucleotide positions for each are shown in parenthesis.
Acknowledgments
We thank A. Otte (EED antibody), T. Jenuwein (H3K27me2 and H3K27me3 antibodies), S. Bultman (Wap-T121 tumor tissue), and S. Michnick (YFP fragment complentation assay vectors) for generously sharing reagents. Additionally, we are grateful to B. Bagnell of the UNC Microscopy Services Laboratory, D. Ciavatta, and the UNC Flow Cytometry Core Facility for technical assistance. Finally, we thank D. Ciavatta and S. Chamberlain for helpful comments on the manuscript. This research was supported by a National Institutes of Health grant to T.M. and a Howard Hughes Medical Institute predoctoral fellowship to N.D.M.
Abbreviations
- EED
embryonic ectoderm development
- EZH2
enhancer of zeste homolog 2
- H3K27
histone H3 lysine 27
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–7. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 2.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–5. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 3.Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–95. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 4.Gebuhr TC, Bultman SJ, Magnuson T. Pc-G/trx-G and the SWI/SNF connection: developmental gene regulation through chromatin remodeling. Genesis. 2000;26:189–97. doi: 10.1002/(sici)1526-968x(200003)26:3<189::aid-gene4>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 5.Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36:1291–5. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- 6.Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36:1296–300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Mager J, Chen Y, Schneider E, Cross JC, Nagy A, Magnuson T. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet. 2001;28:371–5. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- 8.Mager J, Montgomery ND, de Villena FP, Magnuson T. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat Genet. 2003;33:502–7. doi: 10.1038/ng1125. [DOI] [PubMed] [Google Scholar]
- 9.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–43. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 10.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 11.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 12.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–96. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 14.Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 15.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 17.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–36. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 19.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–8. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–81. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–46. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–95. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasini D, Bracken AP, Jensen MR, Denchi EL, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. Embo J. 2004;23:4061–71. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 25.Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–93. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- 26.Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102:1859–64. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin C, Cao R, Zhang Y. Substrate preferences of the EZH2 histone methyltransferase complex. J Biol Chem. 2006;281:8365–70. doi: 10.1074/jbc.M513425200. [DOI] [PubMed] [Google Scholar]
- 28.James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–72. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.James TC, Eissenberg JC, Craig C, Dietrich V, Hobson A, Elgin SC. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol. 1989;50:170–80. [PubMed] [Google Scholar]
- 30.Jones DO, Cowell IG, Singh PB. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays. 2000;22:124–37. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 31.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–5. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 32.Faust C, Schumacher A, Holdener B, Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development. 1995;121:273–85. doi: 10.1242/dev.121.2.273. [DOI] [PubMed] [Google Scholar]
- 33.Faust C, Lawson KA, Schork NJ, Thiel B, Magnuson T. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development. 1998;125:4495–506. doi: 10.1242/dev.125.22.4495. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–5. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 35.Kunath T, Arnaud D, Uy GD, Okamoto I, Chureau C, Yamanaka Y, Heard E, Gardner RL, Avner P, Rossant J. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development. 2005;132:1649–61. doi: 10.1242/dev.01715. [DOI] [PubMed] [Google Scholar]
- 36.Rossant J. Stem cells and lineage development in the mammalian blastocyst. Reprod Fertil Dev. 2007;19:111–8. doi: 10.1071/rd06125. [DOI] [PubMed] [Google Scholar]
- 37.Touriol C, Bornes S, Bonnal S, Audigier S, Prats H, Prats AC, Vagner S. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell. 2003;95:169–78. doi: 10.1016/s0248-4900(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 38.Tikole S, Sankararamakrishnan R. A survey of mRNA sequences with a non-AUG start codon in RefSeq database. J Biomol Struct Dyn. 2006;24:33–42. doi: 10.1080/07391102.2006.10507096. [DOI] [PubMed] [Google Scholar]
- 39.Arnaud E, Touriol C, Boutonnet C, Gensac MC, Vagner S, Prats H, Prats AC. A new 34-kilodalton isoform of human fibroblast growth factor 2 is cap dependently synthesized by using a non-AUG start codon and behaves as a survival factor. Mol Cell Biol. 1999;19:505–14. doi: 10.1128/mcb.19.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huez I, Bornes S, Bresson D, Creancier L, Prats H. New vascular endothelial growth factor isoform generated by internal ribosome entry site-driven CUG translation initiation. Mol Endocrinol. 2001;15:2197–210. doi: 10.1210/mend.15.12.0738. [DOI] [PubMed] [Google Scholar]
- 41.Meiron M, Anunu R, Scheinman EJ, Hashmueli S, Levi BZ. New isoforms of VEGF are translated from alternative initiation CUG codons located in its 5′UTR. Biochem Biophys Res Commun. 2001;282:1053–60. doi: 10.1006/bbrc.2001.4684. [DOI] [PubMed] [Google Scholar]
- 42.Prats H, Kaghad M, Prats AC, Klagsbrun M, Lelias JM, Liauzun P, Chalon P, Tauber JP, Amalric F, Smith JA, et al. High molecular mass forms of basic fibroblast growth factor are initiated by alternative CUG codons. Proc Natl Acad Sci U S A. 1989;86:1836–40. doi: 10.1073/pnas.86.6.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Florkiewicz RZ, Sommer A. Human basic fibroblast growth factor gene encodes four polypeptides: three initiate translation from non-AUG codons. Proc Natl Acad Sci U S A. 1989;86:3978–81. doi: 10.1073/pnas.86.11.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denisenko ON, Bomsztyk K. The product of the murine homolog of the Drosophila extra sex combs gene displays transcriptional repressor activity. Mol Cell Biol. 1997;17:4707–17. doi: 10.1128/mcb.17.8.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schumacher A, Lichtarge O, Schwartz S, Magnuson T. The murine Polycomb-group gene eed and its human orthologue: functional implications of evolutionary conservation. Genomics. 1998;54:79–88. doi: 10.1006/geno.1998.5509. [DOI] [PubMed] [Google Scholar]
- 46.Schumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;384:648. doi: 10.1038/384648a0. [DOI] [PubMed] [Google Scholar]
- 47.Gutjahr T, Frei E, Spicer C, Baumgartner S, White RA, Noll M. The Polycomb-group gene, extra sex combs, encodes a nuclear member of the WD-40 repeat family. Embo J. 1995;14:4296–306. doi: 10.1002/j.1460-2075.1995.tb00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ng J, Li R, Morgan K, Simon J. Evolutionary conservation and predicted structure of the Drosophila extra sex combs repressor protein. Mol Cell Biol. 1997;17:6663–72. doi: 10.1128/mcb.17.11.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sewalt RG, van der Vlag J, Gunster MJ, Hamer KM, den Blaauwen JL, Satijn DP, Hendrix T, van Driel R, Otte AP. Characterization of interactions between the mammalian polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–95. doi: 10.1128/mcb.18.6.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–8. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 51.Tie F, Stratton CA, Kurzhals R, Harte PJ. The N-Terminus of Drosophila ESC Binds Directly to Histone H3 and is Required for E(Z)-Dependent Trimethylation of H3 Lysine 27. Mol Cell Biol. 2007 doi: 10.1128/MCB.01822-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nyfeler B, Michnick SW, Hauri HP. Capturing protein interactions in the secretory pathway of living cells. Proc Natl Acad Sci U S A. 2005;102:6350–5. doi: 10.1073/pnas.0501976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morin-Kensicki EM, Faust C, LaMantia C, Magnuson T. Cell and tissue requirements for the gene eed during mouse gastrulation and organogenesis. Genesis. 2001;31:142–6. doi: 10.1002/gene.10017. [DOI] [PubMed] [Google Scholar]
- 54.Simin K, Wu H, Lu L, Pinkel D, Albertson D, Cardiff RD, Van Dyke T. pRb inactivation in mammary cells reveals common mechanisms for tumor initiation and progression in divergent epithelia. PLoS Biol. 2004;2:E22. doi: 10.1371/journal.pbio.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamer KM, Sewalt RG, den Blaauwen JL, Hendrix T, Satijn DP, Otte AP. A panel of monoclonal antibodies against human polycomb group proteins. Hybrid Hybridomics. 2002;21:245–52. doi: 10.1089/153685902760213859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunofluorescence analysis of H3K27me1 (red, second panel) and H3K27me3 (green, third panel) HP1-∝ in CD1 murine embryonic fibroblasts.
Immunofluorescence analysis of H3K27me1 (red, second panel) and H3K27me3 (green, third panel) HP1-∝ in CD1 murine embryonic fibroblasts.
Schematic diagram indicating the positions of the four putative EED isoform translation start sites the five WD-40 motifs, and the TAA stop codon. Nucleotide positions for each are shown in parenthesis.