Abstract
Congenital infection with rubella virus (RUB) leads to persistent infection and congenital defects and we showed previously that primary human fetal fibroblasts did not undergo apoptosis when infected with RUB, which could promote fetal virus persistence (Adamo et al., 2004). To extend this observation, gene chip analysis was performed on a line of primary human fetal fibroblasts (ten weeks gestation) and a line of human adult lung fibroblasts (which underwent apoptosis in response to RUB infection) to compare gene expression in infected and uninfected cells. A total of 632 and 516 genes were upregulated or down regulated in the infected fetal and adult cells respectively in comparison to uninfected cells, however only 52 genes were regulated in both cell types. Although the regulated genes were different, across functional gene categories the patterns of gene regulation were similar. In general, regulation of pro- and apoptotic genes following infection appeared to favor apoptosis in the adult cells and lack of apoptosis in the fetal cells, however there was a greater relative expression of anti-apoptotic genes and reduced expression of pro-apoptotic genes in uninfected fetal cells versus uninfected adult cells and thus the lack of apoptosis in fetal cells following RUB infection was also due to the prevailing background of gene expression that is antagonistic to apoptosis. In support of this hypothesis, it was found that of a battery of five chemicals known to induce apoptosis, two induced apoptosis in the adult cells, but not fetal cells, and two induced apoptosis more rapidly in the adult cells than in fetal cells (the fifth did not induce apoptosis in either). A robust interferon-stimulated gene response was induced following infection of both fetal and adult cells and many of the genes upregulated in both cell types were those involved in establishment of an antiviral state; this is the first demonstration of an interferon response at this early stage of human embryonic development. In both fetal and adult cells, interferon controlled but did not eliminate virus spread and apoptosis was not induced in infected fetal cells in the absence of interferon. In addition to the interferon response, chemokines were induced in both infected fetal and adult cells. Thus, it is possible that fetal damage following congenital RUB infection, which involves cell proliferation and differentiation, could be due to induction of the innate immune response as well as frank virus infection.
Introduction
Rubella virus (RUB), a member of the Togaviridae family, is a positive-polarity, single-stranded RNA virus that causes a generally mild disease in children and adults, but is considered a pathogen of significant medical importance because of its potential to produce congenital defects, known collectively as congenital rubella syndrome (CRS), when the infection occurs in utero, especially during the first trimester of gestation (Banatvala and Brown, 2004; Lee and Bowden, 2000; Plotkin, 2001; Robertson et al, 2003; Wolinsky, 1996). The most pronounced CRS defects are associated with the eyes, ears, brain, heart, liver, spleen, and endocrine system. Affected organs exhibit reduced size and cell number and vascular damage can be widespread, often including necrotic lesions of adjoining tissue. Additionally, CRS infants are persistently infected at birth due in part to defects in cell-mediated immunity. Because of the effect of RUB infection in utero on cell number and organ development, studies on RUB-induced teratogenesis have focused on the effect of RUB replication on the infected cell. A number of these studies demonstrated that RUB interfered with cell division, RUB proteins bound to cell factors involved in cell division, or RUB altered cellular proliferative pathways (Atreya et al, 1995; Atreya et al, 1998; Atreya et al, 2004b; Beatch and Hobman, 2000; Bowden et al., 1987; Buzas et al, 2004; Cooray et al, 2005; Forng and Atreya, 1999; Lee and Bowden, 2000; Mohan et al, 2002; Sing et al, 1994; Yoneda et al., 1986). RUB also induces apoptosis in some cell culture lines including Vero (African green monkey kidney), RK13 (rabbit kidney), and rat oligodendrocytes, but not in others such as BHK-21 (hamster kidney), and it has been suggested that RUB-induced apoptosis may be associated with the development of CRS (Atreya et al, 2004a; Domegan and Atkins, 2002; Duncan et al, 1999; Hofmann et al, 1999; Lee and Bowden, 2000; Pugachev and Frey, 1998). However, in a previous study, we showed that while RUB induces apoptosis in nonproliferative primary cultures of cytotrophoblasts (CTB) and explants of chorionic villi (ECV) derived from human term placentas, it did not induce apoptosis in proliferative human fibroblasts derived from whole embryos of 10 weeks gestation (Adamo et al, 2004). Megyeri et al. (1999) also reported that RUB did not induce apoptosis in two lines of fetal lung fibroblasts. From these findings we hypothesized that the lack of apoptosis would promote virus persistence during congenital infection. Teratogenesis would then be due to disruption of cell growth and differentiation by the noncytocidal, persistent infection, rather than apoptosis, as proposed by Wolinsky (1996). To begin the study of how RUB infection alters the host cell, we used DNA microarrays to compare gene expression in uninfected and infected human fetal and adult cell cultures. Unexpectedly, we also found that comparison of gene expression between the uninfected and infected fetal and adult cultures was relevant to the differential induction of apoptosis by RUB infection in these two cell lines.
Results
RUB replication in embryonic and adult cell lines
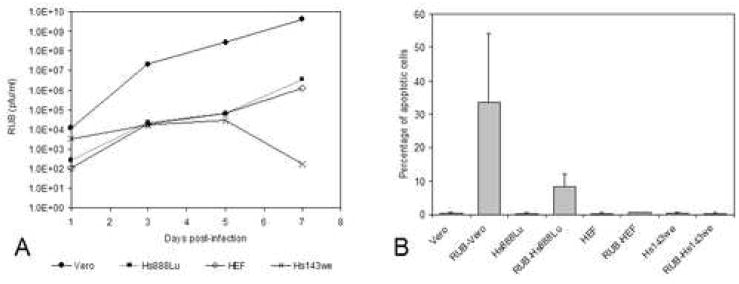
Two lines of human fetal cells (HEF and Hs143we) and one line of human adult cells (Hs888Lu) were infected with RUB at an MOI of 10 pfu/cell and the infection was followed through seven days post infection. Virus production was similar in one line of human fetal cells (HEF) and the line of human adult cells (Hs888Lu), reaching titers of ~104 pfu/ml by 3 days post-infection (dpi) and ~106 pfu/ml by 7 dpi, while in the second line of human fetal cells (Hs143we) titers never exceeded ~104 pfu/ml (Fig. 1A). By 3 dpi, roughly 35% of the cells in the adult Hs888Lu cultures and 50% of the cells in the fetal HEF cultures were infected, and these values rose to 45% and 70% by 5 dpi (Table 1). Such asynchronicity is characteristic of RUB infections, even in permissive cell lines, such as Vero cells, which were included as a control in many of these experiments. In contrast, less than 5% of fetal Hs143we cells were infected by 5 dpi. No cytopathic effect (CPE) was observed in the two fetal cell lines while moderate CPE was observed in the adult cell line. Some of the infected adult Hs888Lu cells exhibited morphological features characteristic of apoptosis (cell contraction, rounding, nuclear condensation and fragmentation) and using a TUNEL assay, roughly 10% of the cells in RUB-infected adult Hs888Lu cultures were found to be apoptotic while the percentage of apoptotic cells in infected fetal HEF or Hs143we cultures did not differ from the mock-infected controls (Fig. 1B), confirming our earlier results (Adamo et al., 2004).
FIG. 1.
Growth of RUB in human cell lines. Cultures of Hs888Lu, HEF, Hs143we, and Vero cells were infected with RUB (MOI = 10 PFU/cell) and titer of virus present in the culture medium was determined by plaque assay at 1, 3, 5, and 7 dpi (A) and the percentage of apoptotic cell was determined at 5 dpi (B). The data presented in Panel A are representative of 2 independent repetitions and the data presented in Panel B are the average of 3 independent repetitions.
Table 1.
Infected cell percentages
| Cell Line | % of infected cellsa | |
|---|---|---|
| 3 dpi | 5 dpi | |
| Vero | nd | 96.75 ± 3.3 |
| HEF | 50.7 ± 12.5 | 73.4 ± 9.1 |
| Hs888Lu | 34.3 ± 9.2 | 44.5 ± 8.1 |
| Hs143we | nd | 3.3 ± 2.3 |
Cultures of the indicated cell lines were infected with RUB (MOI = 10 pfu/cell) and the percentage of infected cells was determined by IFA at 3 and 5 days post-infection (dpi)
Gene chip analysis of regulation of gene expression in RUB-infected cells
Fetal HEF and adult Hs888Lu cells at 3 dpi were chosen for analysis of gene expression, a time at which one third to one half of the cells in the culture were infected. Of 33,000 genes interrogated, 632 and 516 genes were up or downregulated (using a two fold cutoff) in RUB-infected fetal HEF and adult Hs888Lu cells, respectively, in comparison to uninfected control cells (a list of the individual genes up- and down regulated in infected cells is given in Table 1 of the Supplemental Materials). The distribution of these regulated genes across standard functional gene categories was similar in the two cell types (Table 2 of the Supplemental Material). Strikingly, however, of 1096 genes regulated in either cell type, only 52 (4.7%) were regulated in both cell types (Table 2, individual genes regulated in both cell types are listed in Table 3 of the Supplemental Material).
Table 2.
Similarly and differentially regulated genes in RUB-infected HEF and Hs888Lu cells
| Subsets of genes | Number |
|---|---|
| Up regulated in both Hs888Lu and HEF | 38 |
| Up regulated in Hs888Lu but not changed in HEF | 271 |
| Up-regulated in Hs888Lu but down regulated in HEF | 1 |
| Not changed in Hs888Lu but up regulated in HEF | 298 |
| Not changed in Hs888Ly but down regulated in HEF | 282 |
| Down regulated in Hs888Lu but up regulated in HEF | 3 |
| Down regulated in Hs888Lu but not changed in HEF | 193 |
| Down regulated in both Hs888Lu and HEF | 10 |
Interferon response
Of the 38 genes upregulated in both cell types, sixteen were in the “Innate defense and immune regulation” category and it was among these genes that the greatest magnitudes of change in expression were observed. The majority of these were associated with the IFN response (Table 3). In spite of the robust IFN-stimulated response, none of the IFN-α genes were upregulated in either cell type and IFN-β gene induction was only detectable in fetal HEF at the 3 dpi time point (upregulated by 12-fold).
Table 3.
IFN-stimulated genes upregulated in both HEF and Hs888Lu by RUB infection
| Gene | Gene Symbol | Average fold change |
|
|---|---|---|---|
| HEF | Hs888Lu | ||
|
Cell defense/immune response | |||
| 2′,5′-oligoadenylate synthetase 1, 40/46kDa | OAS1 | 88.12 | 64.49 |
| 2′–5′-oligoadenylate synthetase 2, 69/71kDa | OAS2 | 24.21 | 16.95 |
| 2′–5′-oligoadenylate synthetase 3, 100kDa | OAS3 | 27.25 | 23.1 |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | DDX58 | 19.76 | 14.23 |
| Guanylate binding protein 5 | GBP5 | 13.28 | 23.31 |
| interferon, gamma-inducible protein 16 | IFI16 | 6.11 | 2.59 |
| Interferon-induced protein 44 | IFI44 | 9.49 | 12.39 |
| interferon-induced protein with tetratricopeptide repeats 1 | IFIT1 | 32.26 | 21.45 |
| interferon-induced protein with tetratricopeptide repeats 3 | IFIT3 | 32.98 | 20.89 |
| interferon-stimulated transcription factor 3, gamma 48kDa | ISGF3G | 2.54 | 2.97 |
| interleukin 24 | IL24 | 3.05 | 32.32 |
| major histocompatibility complex, class I, B | HLA-B | 2.5 | 5.16 |
| major histocompatibility complex, class I, F | HLA-F | 4.03 | 4.3 |
| myxovirus (influenza virus) resistance 1, interferon- inducible protein p78 (mouse) | MX1 | 105.34 | 161.43 |
| myxovirus (influenza virus) resistance 2 (mouse) | MX2 | 56.13 | 24.31 |
| tumor necrosis factor, alpha-induced protein 6 | TNFAIP6 | 28.79 | 3.64 |
|
| |||
|
Cell signaling/intracellular signaling cascades | |||
| CASP8 and FADD-like apoptosis regulator | CFLAR | 2.68 | 2.09 |
|
| |||
|
Regulation o transcription/transcription rate | |||
| E74-like factor 1 (ets domain transcription factor) | ELF1 | 2.9 | 2.54 |
| signal transducer and activator of transcription 2, 113kDa | STAT2 | 5.16 | 2.04 |
| SP110 nuclear body protein | SP110 | 10.2 | 3.82 |
| tripartite motif-containing 22 | TRIM22 | 2.41 | 2.72 |
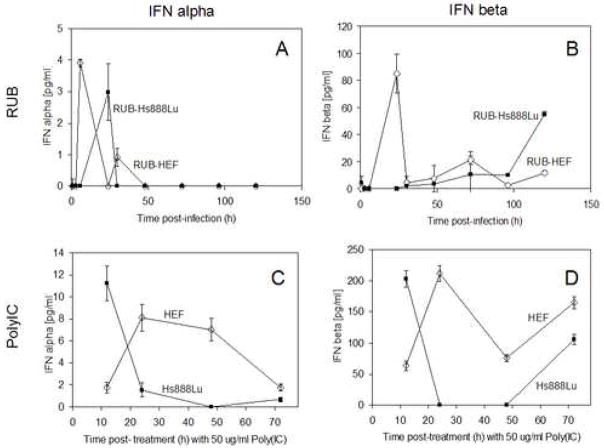
When IFN was assayed in the medium of infected cells at several time points post-infection (Fig. 2), both cell types showed a burst of IFN-αexpression in the first 24 hours after infection. The highest peak of IFNα in RUB-infected HEF was at 12 hpi followed by a maximal peak of IFN-β expression at 24 hpi. After 36 hpi, no IFN-α and a persisting low level of IFN-β were present throughout the experimental time course. In contrast, the peak of IFNα expression in RUB-infected Hs888Lu cells occurred at 24 hpi, no initial peak of IFNβ was detected, but similar to RUB-infected HEF, no IFNα and persisting low level of IFNβ were present throughout the remainder of the time course. This finding explained the lack of IFN-α gene upregulation at the 3 dpi time point in either cell line. In addition, in occasional specimens from RUB-infected Hs888Lu cells no IFN was detected at 3 dpi, likely explaining the lack of IFN-β gene upregulation in this cell line at this time point. When induced with a standard concentration of polyIC (50 μg/ml), both IFNα and -β were induced in both cell types; the peak amounts of both induced by polyIC were 2–4 times greater than the peak amounts induced by RUB infection and the kinetics of induction were different. Following polyIC induction of HEF, both IFNα and -β were expressed by 12 hours, peaked at 24 hours, and then persisted through 70 hours, the end of the experiment. In contrast, in Hs888Lu cells, peak expression was at 12 hours post induction after which expression fell off to low or undetectable levels at 24 and 48 hours post-induction.
FIG. 2.
Interferon induction in RUB-infected and poly IC treated cells. Cultures of Hs888Lu and HEF were infected with RUB (MOI = 10 PFU/cell) or treated with poly IC (50 μg/ml) and IFN’s in the culture medium were determined at the indicated time points. The data presented are the average of two (poly IC) or three (RUB infection) independent repetitions.
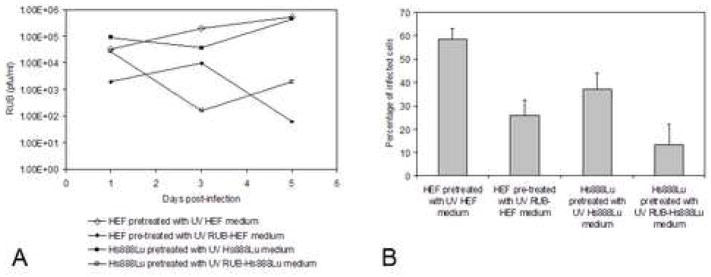
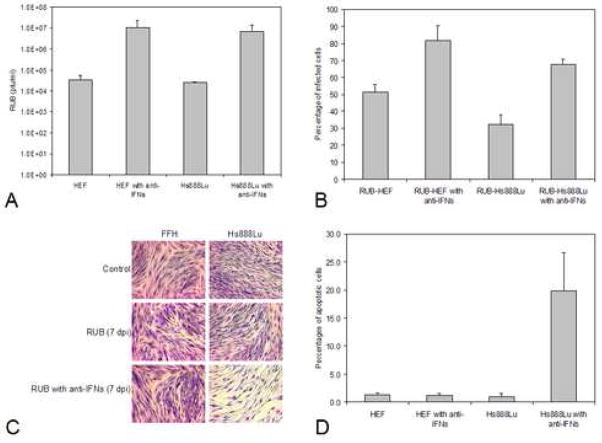
To determine the effect of the IFN present in the medium on RUB infection, two experiments were done. First, fresh cells were pre-incubated with medium from the same cell type infected with RUB [collected 3 dpi, at which time only IFN-β was detectable in the culture medium (Fig. 2)] that had been UV-irradiated to inactivate virus for 24 hours followed by RUB infection. As shown in Figs. 3A and 3B, virus titers were reduced by 2–3 logs (in comparison to cells pre-incubated with UV-irradiated mock-infected cell medium) and the percentage of infected cells was reduced by half (Figs. 3A and 3B). In the second, converse experiment, both cell types were infected in the presence of anti-IFN-α and -β antibodies and it was found that virus titers increased by over 2 logs and the percentage of infected cells at 3 dpi doubled in comparison to control cells infected in the absence of anti-IFN antibodies (Figs. 4A and 4B). Thus, the interferon response was effective reducing virus replication and the resulting rate of spread, although it did not eliminate the virus from the culture. Interestingly, in the presence of anti-IFN antibodies no CPE was observed in infected fetal HEF cells, however in infected adult Hs888Lu cells CPE was more profound than in the absence of antibodies (Fig. 4C) and the percentage of apoptotic cells doubled to ~20% (Fig. 4D). We also infected fetal Hs143we cells in the presence of anti-IFN antibodies with the result that virus titers rose by two logs and the percentage of infected cells increased by tenfold by 5 dpi. Thus, the IFN response was critical in the resistance of these cells to RUB infection.
FIG. 3.
Growth of RUB in cells pretreated with IFN-containing medium. Growth medium from mock-infected or RUB-infected Hs888Lu and HEF was harvested 3 dpi and virus was UV inactivated at 20,000 μJ/cm2 for 20 min on ice. Fresh Hs888Lu cells were then incubated with UV-inactivated medium from mock- or RUB-infected Hs888Lu cells for 24 hours followed by infection with RUB (10 PFU/cell) and likewise, fresh HEF were incubated with UV-inactivated medium from mock-or RUB infected HEF for 24 hours followed by infection with RUB. Titer of virus present in the culture medium was determined by plaque assay at 1, 3, and 5 dpi (A) and the percentage of infected cells was determined at 3 dpi (B). The data presented in Panel A are representative of 2 independent repetitions. The data presented in Panel B are the average of 3 independent repetitions.
FIG. 4.
Growth of RUB in cells pretreated with anti-IFN serum. Cultures of Hs888Lu cells and HEF were incubated with anti-Type I IFN antiserum for 24 hours and then infected with RUB (MOI = 10 PFU/cell) in the presence of the anti-interferon serum. At 3 dpi, titer of virus present in the culture medium was quantitated by plaque assay (A) and the percentage of infected cells was determined (B). In both cases, the data presented are the average of 2 independent repetitions. Cells were stained with Giemsa for microscopy at 7 dpi (C) and the percentages of apoptotic cells were determined by TUNEL assay at 5 dpi (D; average of 2 independent repetitions).
Genes associated with apoptosis
A number of pro- and anti-apoptotic genes were up-and down-regulated in RUB-infected fetal HEF and adult Hs888Lu (Table 4, Supplemental Material). With the exception of two genes, IL24 and CFLAR (CASP8 and FADD-like apoptosis regulator), and the interferon-inducible MX and OAS genes, the apoptotic response genes regulated in the two cell types were mutually exclusive and the magnitude of regulation was not as great as with the IFN-stimulated genes. We therefore corroborated our gene chip findings of up- or downregulated genes in this category using an RT-PCR array assay covering genes involved in apoptosis (only a subset of the up- or down-regulated genes listed in Table 4, Supplemental Materials, were included in this assay). As shown in Table 4, for the most part, the level of up- and down-regulation detected by the gene chips was confirmed by the RT-PCR assay.
Table 4.
Differential levels of gene expression as determined by DNA microarray and real time RT-PCR1
| Gene Symbol | Gene Title | DNA microarray | Real time RT-PCR | |
|---|---|---|---|---|
|
Gene expression in RUB-infected HEF compared to mock-infected HEF |
||||
| APAF1 | Apoptotic peptidase activating factor | −3.222 | −1.302* | |
| CASP1 | Caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | 15.37 | 6.63 | |
| CFLAR | CASP8 and FADD-like apoptosis regulator | 2.68 | 1.38 | |
| DAPK1 | Death-associated protein kinase 1 | −2.32 | −5.59 | |
| RIPK2 | Receptor-interacting serine-threonine kinase 2 | 2.13 | 1.76* | |
|
| ||||
|
Gene expression in RUB-infected Hs888Lu compared to mock-infected Hs888Lu | ||||
| CFLAR | CASP8 and FADD-like apoptosis regulator | 2.09 | 2.07 | |
|
| ||||
|
Gene expression in HEF compared to Hs888Lu | ||||
| BCL2L10 | BCL2-like 10 | 3.07 | 4.54 | |
| CASP1 | Caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | −9.87 | −4.72 | |
| CASP4 | Caspase 4, apoptosis-related cysteine peptidase | −2.93 | −2.17 | |
| CASP8 | Caspase 8, apoptosis-related cysteine peptidase | −2.74 | −6.44 | |
| LTBR | Lymphotoxin beta receptor (TNFR superfamily, member 3) | −4.3 | −1.62* | |
| TNFRSF11B | Tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) | −11.42 | −6.96 | |
The indicated pro- and apoptotic genes found to be up- or down-regulated by microarray (>2 fold) were also analyzed using a quantitative RT-PCR assay to confirm the level of up- or down-regulation.
Negative values indicate downregulation.
Regulation of < 2 fold that were statistically significant (p values < 0.05)
More pro-apoptotic genes were upregulated in RUB-infected adult Hs888Lu cells than in RUB-infected fetal HEF and, conversely, more anti-apoptotic genes were upregulated in RUB-infected fetal HEF than in RUB-infected adult Hs888Lu cells and concomitantly more anti-apoptotic genes were down regulated in RUB-infected adult Hs888Lu cells than in RUB-infected HEF (Table 5). Thus gene regulation in RUB-infected Hs888Lu cells would appear to favor apoptosis to a greater extent than in RUB-infected HEF. However, when we compared the basal level of expression of this gene set in uninfected fetal HEF vs adult Hs888Lu cells, we found that more than four times as many proapoptotic genes were down regulated in HEF as in Hs888Lu cells and roughly 50% more antiapoptotic genes were upregulated in HEF as in Hs888Lu cells (Table 5; individual genes are compiled in Table 4 of the Supplemental Materials).
Table 5.
Up- and down-regulated pro- and anti-apoptotic genes1
| RUB-HEF vs HEF | RUB-Hs888Lu vs Hs888Lu | HEF vs Hs888Lu | ||
|---|---|---|---|---|
| Pro-apoptotic genes | Up-regulated | 13 (2.06%)2 | 18 (3.49%)3 | 12 (1.42%)4 |
| Down-regulated | 7 (1.11%)2 | 6 (1.16%)3 | 45 (5.33%)4 | |
| Anti-apoptotic genes | Up-regulated | 12 (1.90%)2 | 7 (1.36%)3 | 45 (5.32%)4 |
| Down-regulated | 6 (0.95%)2 | 11 (2.13%)3 | 32 (3.79%)4 |
Individual up- and down-regulated genes are listed in Table 4 of the Supplementary Materials
Percentage is based on a total number of 632 genes which exhibited a ≥ 2 fold change in expression in this comparison.
Percentage is based on a total number of 516 genes which exhibited a ≥ 2 fold change in expression in this comparison.
Percentage is based on a total number of 845 genes which exhibited a ≥ 2 fold change in expression in this comparison.
Considering these results, we formulated the hypothesis that it was the prevailing background of expression, in addition to differential up-and-down regulation of pro- and anti-apoptotic genes following infection, that determined whether infection led to apoptosis. To test this hypothesis, we treated adult and fetal cell cultures with a battery of reagents (actinomycin D, camtothecin, cycloheximide, dexamethasone, and etoposide) known to induce apoptosis by interfering with different cellular processes in many cell types. By the time this experiment was done, the HEF line had expired and thus a new HEF line from a seven week embryo (HEF7) was employed. RUB titers produced following infection of this cell line and the percentage of infected cells were similar to HEF (data not shown). As shown in Table 6, HEF7 were less sensitive to the battery of apoptotic agents, exhibiting resistance to camptothecin and etoposide (Hs888Lu were sensitive to both) and a slower onset of morphological cell death following treatment with actinomycin D and cycoheximide compared with Hs888Lu. Neither cell line was sensitive to dexamethosone. These results were confirmed using a TUNEL assay to detect induction of apoptosis (data not shown)
Table 6.
CPE induced by apoptosis-inducing reagents in Vero, Hs888Lu, and HEF cells
| Cells | Treatment | Time of treatment (hours) | |||||
|---|---|---|---|---|---|---|---|
| 3 | 6 | 24 | 48 | 72 | 96 | ||
| Vero | DMSO | − | − | − | − | − | − |
| Actinomycin D | − | − | ++ | ++++ | +++++ | All dead | |
| Camptothecin | − | − | ++ | ++ | ++++ | All dead | |
| Cycloheximide | − | − | −/+ | ++ | +++ | ++++ | |
| Dexamethasone | − | − | − | − | −/+ | −/+ | |
| Etoposide | − | − | −/+ | ++ | ++ | +++ | |
| Cells | Treatment | Time of treatment (days) |
|||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | ||
| Hs888Lu | DMSO | − | − | − | − | − | − |
| Actinomycin D | −/+ | ++ | ++ | +++ | ++++ | +++++ | |
| Camptothecin | −/+ | + | ++ | +++ | ++++ | +++++ | |
| Cycloheximide | + | + | ++ | +++ | ++++ | +++++ | |
| Dexamethasone | − | − | −/+ | + | ++ | ++ | |
| Etoposide | −/+ | + | ++ | +++ | ++++ | +++++ | |
|
| |||||||
| HEF | DMSO | − | − | − | − | − | − |
| Actinomycin D | − | − | − | ++ | +++ | ++++ | |
| Camptothecin | − | − | − | − | + | + | |
| Cycloheximide | − | + | ++ | ++ | +++ | +++ | |
| Dexamethasone | − | − | − | − | − | − | |
| Etoposide | − | − | − | −/+ | + | + | |
CPE key: −/+. less than 5 % cell death; +. ~ 10 % cell death; ++. ~ 25 % cell death; +++. ~ 50 % cell death; ++++. ~ 75 % cell death; +++++. ~ 95 % cell death. The experiment was repeated twice. The data presented is from one experiment and is representative of both independent repetitions.
Discussion
In this and our previous study (Adamo et al., 2004), the cellular response to RUB infection was studied in human cells. Recent studies on induction of apoptosis, cell cycle perturbations, and alterations in survival and proliferation pathways by RUB were conducted in cells of simian, hamster, or rabbit origin selected because of lack of a functional interferon system leading to permissiveness for virus replication in the former two cases or susceptibility to RUB-induced cytopathic effect in the latter case. In our two studies, we have now studied RUB infection in explants of chorionic villi (ECV) taken from normal human term placentas (seven independent explants); primary cultures of cytotrophoblasts (CTB) isolated from normal human term placentas (three independent primary cultures); Hs888Lu, a diploid line of fibroblasts from human adult lung, Hs143we, a diploid line of human embryo fibroblasts derived at 6 weeks of gestation, and primary human embryo fibroblasts (HEF) derived at 7–10 weeks of gestation (ten independent primary cultures). In all but the Hs143we cell line, infection at a high multiplicity of infection led to an asynchronous infection, typical of RUB infection in permissive cell lines. In these cases the majority of cells were eventually infected and a moderate titer was produced. Thus, it is possible to conduct studies of RUB infection and the cell response in human cell lines.
The impetus for the present study was to follow up our previous observation that RUB induced apoptosis in nonproliferative, differentiated cells (ECV and CTB) but not in proliferative fetal cells and we confirmed that finding using cell lines independent from those used in the previous study [fetal Hs143we cells and HEF (a different isolation than used in the previous study) and adult Hs888Lu cells]. Using a standard two-fold cutoff, we found that ~500–600 genes out of 33,000 interrogated (<2%) were up- or downregulated following virus infection. Up- or downregulation of genes grouped in functional categories was roughly comparable between infected fetal and adult cells, but over 90% of the genes up or downregulated were cell type-specific and only 52 genes were regulated in both cell types. This is undoubtedly due to developmental differences in gene activity between fetal and adult cells, however it is also possible that tissue specific differences are also at play since the adult cells were derived from lung while the fetal cells derived from whole embryo. It will be important to generate similar data from additional independent cell lines following RUB infection to ascertain which genes are reproducibly up- or downregulated.
The one set of genes regulated in common by infection of both fetal and adult cells were interferon-stimulated genes (ISG’s). In both cell types, an initial spike of IFNα induction followed by a protracted induction of IFNβ was observed. Among the ISG’s detected, the OAS genes, MX genes, and IFIT genes were the most dramatically upregulated (>20 fold in both cells types); this is the first documentation of induction of specific ISG’s in response to RUB infection of human cells. It has been reported that the antiviral IFN response is active in cultures of human trophoblasts at the first trimester of gestation (Uchide et al., 2005). Our results document a potent IFN response functioning as an antiviral mechanism in fibroblasts derived from a human embryo of 10 weeks gestation; we are unaware of other reports in the literature of the presence of an IFN response this early in development. Despite the IFN-stimulated antiviral response, a majority of cells in both cell types were eventually infected. We found however that pretreatment of cells with medium from RUB-infected cells, containing a low level of IFN-β, reduced both the spread and titer of virus while inclusion of anti-IFN antiserum in the medium of the infected culture lead to increased titers and more rapid virus spread. Thus, the IFN response counteracts, but does not eliminate, virus in the infected culture. Similar conclusions were drawn in previous studies of RUB infection in several IFN-competent and -deficient cell lines (Mifune et al., 1970; Stanwick and Hallum, 1974; Wong et al., 1967).
This project was initiated to study the differential induction of apoptosis among cell types. In adult Hs888Lu cells, inclusion of anti-IFN anti-serum in the infected culture medium increased apoptosis, indicating that apoptosis could be induced by an IFN-independent mechanism. However, of the ISG’s upregulated in RUB-infected adult Hs888Lu were several known to be associated with induction of apoptosis, such as the MX genes, the OAS genes, and PKR. In addition, several of the other pro-apoptotic genes up-regulated in RUB-infected adult Hs888Lu cells have been associated with the interferon response, including PML, XIAP-associated factor-1 (BIRC4BP), FOXO3A, and IL24 (Chawla-Sarkar et al., 2003; Kim et al., 2005; Chada et al., 2004). Therefore, in RUB-infected adult Hs888Lu cells apoptosis is likely triggered by both IFN-dependent and -independent pathways.
In RUB-infected fetal HEF, apoptosis was not observed. Nevertheless, upregulation of pro-apoptotic genes such as MX, OAS, and IL24, similar to RUB-infected adult Hs888Lu cells, was detected. Apoptosis was likewise not observed in the absence of IFN, indicating a circumvention of both IFN-dependent and -independent pathways. In comparison with RUB-infected adult Hs888Lu cells, fewer pro-apoptotic cells were upregulated in RUB-infected fetal HEF and more anti-apoptotic genes were upregulated and fewer downregulated. Thus, differential pro- and anti-apoptotic gene regulation would appear to favor apoptosis in the adult cells and lack of apoptosis in the fetal cells following RUB infection. However, in addition to analyzing gene expression following RUB infection, we compared the relative level of expression of pro-and anti-apoptotic genes in the uninfected fetal and adult cell lines, with the finding that anti-apoptotic genes were expressed at a higher level in HEF than in Hs888Lu cells and pro-apoptotic genes were expressed at a lower level. This led us to hypothesize that RUB infection does not induce apoptosis in fetal cells due to the prevailing background of gene expression that is antagonistic to apoptosis. To test this hypothesis, we treated Hs888Lu and HEF with a standard battery of five apoptotic reagents and found an increased level of resistance of HEF to apoptosis induction by four of the five reagents (the fifth did not induce apoptosis in either cell line), congruent with our hypothesis.
As we hypothesized in our previous study, the failure to induce apoptosis in RUB-infected fetal cells would promote virus persistence. A vigorous humoral response against the virus is generated in infected fetuses; however it has been shown that virus persistence in cell culture can be maintained in the presence of neutralizing antibodies (Abernathy et al., 1990; Rawls and Melnick, 1966; Williams et al., 1981). CRS patients also have defects in RUB-specific cell-mediated immunity as measured by lymphocyte proliferative assays (as these studies were done >25 years ago, it would be of interest to study the cell mediated response in CRS patients with more recent reagents to gain understanding into the specific nature of the defect). While such defects explain the inability of CRS patients to clear virus persistence by the time of birth, at the critical time in gestation during which congenital defects are induced, i.e. the first twelve weeks, the immune system has yet to mature underscoring the importance of apoptosis as a potential mechanism of virus clearance particularly considering that the interferon system only appears to slow virus replication.
Fetal defects in CRS are associated with reduced tissue or organ size due to reduced cell number, leading to the hypothesis that the virus infection interferes with cell survival, proliferation and/or differentiation at early stages of organogenesis. There has been speculation that cell survival was compromised by induction of apoptosis (Pugachev and Frey, 1998; Duncan et al., 1999; Hofmann et al., 1999; Megyeri et al., 1999; Atreya et al., 2004b), although our findings that infected fetal cells do not undergo apoptosis would appear to rule this possibility out as a general mechanism, however RUB infection of specific fetal cell types may lead to induction of apoptosis. In terms of cell survival and proliferation, RUB infection both upregulated and downregulated a number of genes involved in cell growth and proliferation as well as signaling (Supplemental Material). It was recently shown that RUB infection of RK13 cells increased phosphorylation of components of the Ras-Raf-MEK-ERK pathway (Cooray et al., 2005). Fibroblast growth factor 2, associated with the induction of this pathway, is induced in Hs888Lu following RUB infection (Supplemental Material) and we are in the process of testing the phosphorylation state of these components in RUB-infected human cells to see if this cell survival pathway is altered or compromised. It was of interest that genes involved in development were also up-and down-regulated in RUB infected HEF (eg. three homeobox genes; Supplemental Material). Thus, RUB-infection could have a major effect on growth and differentiation of embryonic cells.
However, inter-cellular factors, namely interferons, were also upregulated in RUB-infected HEF and in the presence of a persistent infection, as during congenital RUB infection, interferon induction would be continuous. In addition, cytokines were upregulated as well; in particular, CCL5 or RANTES was upregulated 90-fold in RUB-infected HEF (Supplemental Material). The action of these factors could also profoundly disrupt growth and proliferation pathways in developing and differentiating cells and thus contribute to congenital defects. In this regard, chemokines have been suggested to play important roles in development, particularly neurodevelopment (Bakhiet et al., 2001; Rezaie et al., 2002). This scenario fits with the finding that very few cells (1/1000 or less) in a RUB infected fetus actually harbor virus (Rawls et al., 1968).
Materials and Methods
Cell cultures and virus
The Cordoba strain, an isolate from Cordoba, Argentina, of RUB was propagated and titered in Vero cells as previously described (Forng and Frey, 1995; Hemphill et al., 1988; Wang et al., 1994). Four human cell lines were employed: HEF and HEF-7, primary cultures of human embryo fibroblasts from 10 and 7 week human spontaneous abortuses, respectively, established as previously described (Adamo et al., 2004)(by morphological examination following H&E staining, 95.47% +/−2.78% of the cells in the HEF culture and 97.67% +/− 1.83% of the cells in the HEF7 culture were fibroblastic; Hs888Lu, a diploid line of human adult lung fibroblasts (ATCC# CCL-211); and Hs143we, a diploid fibroblast line derived from whole human embryo (ATCC# CRL-7092). The method for generating primary cultures of human embryo fibroblasts was approved by the Bioethics Committee at Facultad de Ciencias Medicas, Universidad Nacional de Cordoba, Argentina, and the utilization of these cells was approved by the IRB of Georgia State University. These cell lines were grown in RPMI 1640 (Gibco) supplemented with 10% FBS at 37°C under 5% CO2. The cell lines were generally maintained for 15 passages before senescence was observed, at which time the culture was regenerated from a vial of low passage number cells stored in LN2. ~90–100% confluent cultures were infected with RUB at an MOI of 10 pfu/cell in RPMI 1640 containing 5% FBS at 37°C. Mock-infected control cultures were processed in parallel without virus.
Microscopy
All microscopy was done using a Zeiss Axioplan 2 microscope with epifluorescence capability and images were captured with an Axio Cam HRc Zeiss camera and Axioplan 2 Imaging software.
Giemsa staining
Cell monolayers grown in 4-well chamber slides were rinsed gently with PBS and fixed with methanol for 10 min at −20°C. Cultures were stained (10% Gurrs improved RGG Giemsa) for 1 h, washed with distilled water, and mounted using Clarion (Santa Cruz).
Immunofluorescence
Cultures in 8-well chamber slides were rinsed with PBS and fixed with 4% paraformaldehyde. The cells were permeabilized with 1% NP40, blocked with 1% bovine serum albumin, and incubated with mouse monoclonal antibodies against RUB E1 and E2 (Viral Antigens) followed by incubation with secondary biotin-conjugated goat anti-mouse antibodies (Sigma) and Texas Red-conjugated Streptavidin (Gibco). Hoechst 33342 was used as a counter stain. To determine the percentage of infected cells, nuclei and cells stained for virus antigens were counted in at least three randomly selected fields at 20X magnification.
TUNEL assay
To detect apoptotic cells, an in situ cell death detection kit (Roche) was used following the manufacturer’s protocol. The percentage of apoptotic cells was determined by counting cells under white light and fluorescent apoptotic nuclei in at least three randomly selected fields at 20X magnification.
Experiments with interferon (IFN) and anti-IFN antibodies
Human IFNα-and human IFNβ-ELISA Kits (PBL Biomedical Laboratories) were used to detect IFN-α and -β in the medium of infected cultures and in cultures treated with 50 μg/ml Poly(I:C) (Amersham). To biologically test for IFN activity, medium was collected from RUB-infected or mock-infected Hs888Lu and HEF at 3 dpi and RUB was inactivated by exposure to UV light (20, 000 μJ/cm2, 20 min on ice). Fresh cultures of Hs888Lu and HEF were incubated with UV-inactivated medium from the corresponding mock- or RUB-infected culture for 24 hours prior to infection. Monoclonal anti-IFNα and anti-IFNβ antibodies were purchased from PBL. Twenty-four hours prior to infection, cultures were treated with 0.5 μg/ml of each antibody and during infection both antibodies were maintained in the culture medium at a concentration of 0.5μg/ml/day.
RNA extraction and target preparation
RNA was extracted from cells in 60 mm plates at 3 days post-infection with TRI Reagent (MRC) using the manufacturer’s protocol. RNA quality was assessed by electrophoresis in 1% agarose gels containing 1 μg/ml ethidium bromide and by absorbance at 260 nm and 280 nm. RNA concentration was quantified by absorbance at 260 nm. Aliquots containing 5 μg of total RNA were used for biotinylated copy RNA synthesis (biotin-cRNA) using the One Cycle Target Labeling and Control Reagents Kit (Affymetrix), strictly adhering to the instructions in the Affymetrix Expression Analysis Technical Manual (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). High quality biotin-cRNA was generated as assessed by agarose gel electrophoresis and by the housekeeping control signal output 3′/5′ ratios of internal control genes (provided by Affymetrix). Biotin-cRNA samples were fragmented by incubating the biotin-cRNA in the buffer provided by Affymetrix at 94° C, 35 min, as indicated in the GeneChip Expression Analysis Technical Manual, before hybridization.
Microarrays and hybridizations
The high-density oligonucleotide human genome HG U-133 Set (A+B) chips from Affymetrix were used in this study with which the expression of approximately 33,000 human genes could be interrogated. 15 μg of each biotin-cRNA sample was hybridized independently to the A and B chips of the set and eukaryotic hybridization controls (Affymetrix) were included in each hybridization cocktail. Following hybridization, the arrays were washed, stained with Streptavidin Phycoerythrin, and scanned using an Agilent GeneArray Scanner (Agilent Technologies). Images were analyzed with Affymetrix Microarray Suite v. 5.0 (MAS v.5.0) software. Two independent replicate hybridization assays were done: the biotin-cRNA’s used in these replicates were produced from two total RNA samples extracted from two independent infections/mock-infections of HEF and Hs888Lu cells. Each hybridization assay met the quality control standards of the manufacturer.
Gene expression analysis
Data from the hybridization assays were analyzed with Affymetrix MAS v.5.0 and Data Mining Tool 3.0 (DMT v.3.0) software. Comparisons between RUB-infected HEF (experiment) versus mock-infected HEF (baseline), RUB-infected Hs888Lu (experiment) versus mock-infected Hs888Lu (baseline), and mock-infected HEF (experiment) versus mock-infected Hs888Lu (baseline) were performed. Average signal, standard deviation, fold-changes and p values using the T-test were calculated. Changes in gene expression with p<0.05 were considered significant. The Gene Ontology tool obtained from the Affymetrix web site (https://www.affymetrix.com/analysis/netaffx/index.affx) was used to define gene groups according to function. Additionally, pro- and anti-apoptotic genes were ascertained through PubMed.
Real time RT-PCR
Total RNA extracted from three independent infections/mock-infections of HEF and Hs888Lu cells (3 days post-infection)(one of these RNA samples was also analyzed by gene chip while the other two were not) were treated with TURBO DNA-free (Ambion) to remove DNA and purified using an RNeasy Mini Kit (Qiagen). The quality and quantity of RNA samples were assessed by UV spectrophotometry and electrophoresis: RNA concentration and purity were evaluated by A260 and A260:A280, and A260:A230 ratios and integrity of rRNA bands following agarose gel electrophoresis and ethidium bromide staining. 0.5 μg aliquots of each RNA sample with demonstrated quality were subsequently retro-transcribed with RT2 PCR Array First Strand Kit form SuperArray, which synthesizes cDNA using both random hexamers and oligo-dT primers and utilizes a built-in external RNA control to test for inhibitors of reverse transcription. The level of RNA quality and transcription efficiency was further assessed by characterizing the cDNA products with the RT2 RNA QC PCR Array and RT2 Real-Time™ SYBR Green/ROX PCR Master Mix (SuperArray). All of the samples met the quality parameters specified by the manufacturer. Real time PCR was performed with RT2 Profiler™ PCR Array Human Apoptosis and RT2 Real-Time™ SYBR Green/ROX PCR Master Mix (SuperArray) using an Applied Biosystems 7500 Real-Time PCR System. Dissociation curves for each gene amplified showed only one peak. GADPH and HPRT1 threshold cycles were used for normalization of the threshold cycles of the other genes.
Treatment with chemical apoptosis inducers
Cultures were treated with 5 μM actinomycin D, 1 μM camptothecin, 50 μM cycloheximide, 5 μM dexamethasone, and 50 μM etoposide from BioVision. Control cultures were treated with DMSO, with which stock solutions of the apoptotic inducers were formulated. Cytopahic effect (cell detachment) was followed microscopically through 4 to 6 days post-treatment and a TUNEL assay was performed two days post-treament.
Supplementary Material
Acknowledgments
This research was supported by a grant from the National Institutes of Health (AI 21389). M.P.A. was supported by a Ph.D. fellowship from SeCyT, Universidad Nacional de Cordoba and an International Student Travel Award from the American Society for Microbiology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abernathy ES, Wang CY, Frey TK. Effect of antiviral antibody on maintenance of long-term rubella virus persistent infection in Vero cells. J Virol. 1990;64:5183–5187. doi: 10.1128/jvi.64.10.5183-5187.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo P, Asís L, Silveyra P, Cuffini C, Pedranti M, Zapata M. Rubella virus does not induce apoptosis in primary human embryo fibroblasts cultures: a possible way of viral persistence in congenital infection. Viral Immunol. 2004;17:87–100. doi: 10.1089/088282404322875485. [DOI] [PubMed] [Google Scholar]
- Atreya CD, Lee NS, Forng RY, Hofmann J, Washington G, Marti G, Nakhasi HL. The rubella virus putative replicase interacts with the retinoblastoma tumor suppressor protein. Virus Genes. 1998;16:177–183. doi: 10.1023/a:1007998023047. [DOI] [PubMed] [Google Scholar]
- Atreya CD, Mohan KVK, Kulkarni S. Rubella virus and birth defects: molecular insights into the viral teratogenesis at the cellular level. Birht Defects Res. 2004a;70:431–437. doi: 10.1002/bdra.20045. [DOI] [PubMed] [Google Scholar]
- Atreya CD, Singh NK, Nakhasi HL. The rubella virus RNA binding activity of human calreticulin is localized to the N-terminal domain. J Virol. 1995;69:3848–3851. doi: 10.1128/jvi.69.6.3848-3851.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya CD, Kulkarni S, Mohan KV. Rubella virus P90 associates with the cytokinesis regulatory protein Citron-K kinase and the viral infection and constitutive expression of P90 protein both induce cell cycle arrest following S phase in cell culture. Arch Virol. 2004b;149:779–789. doi: 10.1007/s00705-003-0267-6. [DOI] [PubMed] [Google Scholar]
- Bakhiet M, Mousaa A, Seiger A, Andersson J. Constitutive and inflammatory induction of and chemokines in human first trimester forebrain astrocytes and neurons. Molecular Immunology. 2001;38:921–929. doi: 10.1016/s0161-5890(02)00019-6. [DOI] [PubMed] [Google Scholar]
- Banatvala JE, Brown DW. Rubella. Lancet. 2004;363:1127–1137. doi: 10.1016/S0140-6736(04)15897-2. [DOI] [PubMed] [Google Scholar]
- Beatch MD, Hobman TC. Rubella virus capsid associates with host cell protein p32 and localizes to mitochondria. J Virol. 2000;74:5569–5576. doi: 10.1128/jvi.74.12.5569-5576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzás K, Miczák A, Degré M, Megyeri K. Rubella virus infection dysregulates the pattern of p63 expression. APMIS. 2004;112:656–662. doi: 10.1111/j.1600-0463.2004.apm1121004.x. [DOI] [PubMed] [Google Scholar]
- Bowen JM, Chamley L, Mitchell MD, Keelan JA. Cytokines of the placenta and extra-placental membranes: biosynthesis, secretion and roles in establishment of pregnancy in women. Placenta. 2002;23:239–256. doi: 10.1053/plac.2001.0781. [DOI] [PubMed] [Google Scholar]
- Bowden DS, Pedersen JS, Toh BH, Westaway EG. Distribution by immunofluorescence of viral products and actin-containing cytoskeletal filaments in rubella virus-infected cells. Arch Virol. 1987;92:211–219. doi: 10.1007/BF01317478. [DOI] [PubMed] [Google Scholar]
- Chada S, Mhashilkar AM, Ramesh R, Mumm JB, Sutton RB, Bocangel D, Zheng M, Grimm EA, Ekmekcioglu S. Bystander activity of Ad-mda7: human MDA-7 protein kills melanoma cells via an IL-20 receptor-dependent but STAT3-independent mechanism. Mol Ther. 2004;10:1085–1095. doi: 10.1016/j.ymthe.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Chawla-Sarkar M, Lindner DJ, Liu YF, Williams BR, Sen GC, Silverman RH, Borden EC. Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis. 2003;8:237–249. doi: 10.1023/a:1023668705040. [DOI] [PubMed] [Google Scholar]
- Chiou SK, Jones MK, Tarnawski AS. Survivin - an anti-apoptosis protein: its biological roles and implications for cancer and beyond. Med Sci Monit. 2003;9:PI25–PI29. [PubMed] [Google Scholar]
- Cooray S, Jin L, Best JM. The involvement of survival signaling pathways in rubella virus induced apoptosis. Virol J. 2005;2 doi: 10.1186/1743-422X-2-1. [Online.] http://www.virologyj.com/content/2/1/1. [DOI] [PMC free article] [PubMed]
- Degli-Esposti MA, Dougall WC, Smolak PJ, Waugh JY, Smith CA, Goodwin RG. The novel receptor TRAIL-R4 induces NF-kappaB and protects against TRAIL-mediated apoptosis, yet retains an incomplete death domain. Immunity. 1997;7:813–20. doi: 10.1016/s1074-7613(00)80399-4. [DOI] [PubMed] [Google Scholar]
- Domegan LM, Atkins GJ. Apoptosis induction by the Therien and vaccine RA27/3 strains of rubella virus causes depletion of olygodendrocytes from rat neural cell cultures. J Gen Virol. 2002;83:2135–2143. doi: 10.1099/0022-1317-83-9-2135. [DOI] [PubMed] [Google Scholar]
- Duncan R, Muller J, Lee N, Esmaili A, Nakhasi HL. Rubella virus-induced apoptosis varies among cell lines and is modulated by Bcl-XL and caspase inhibitors. Virology. 1999;255:117–128. doi: 10.1006/viro.1998.9562. [DOI] [PubMed] [Google Scholar]
- Forng RY, Atreya CD. Mutations in the retinoblastoma protein-binding LXCXE motif of rubella virus putative replicase affect virus replication. J Gen Virol. 1999;80:327–332. doi: 10.1099/0022-1317-80-2-327. [DOI] [PubMed] [Google Scholar]
- Forng RY, Frey TK. Identification of the rubella virus non-structural proteins. Virology. 1995;206:843–853. doi: 10.1006/viro.1995.1007. [DOI] [PubMed] [Google Scholar]
- Gurney AL, Marsters SA, Huang RM, Pitti RM, Mark DT, Baldwin DT, Gray AM, Dowd AD, Brush AD, Heldens AD, Schow AD, Goddard AD, Wood WI, Baker KP, Godowski PJ, Ashkenazi A. Identification of a new member of the tumor necrosis factor family and its receptor, a human ortholog of mouse GITR. Curr Biol. 1999;9:215–218. doi: 10.1016/s0960-9822(99)80093-1. [DOI] [PubMed] [Google Scholar]
- Hemphill ML, Forng RY, Abernathy ES, Frey TK. Time-course of virus-specific macromolecular synthesis in rubella virus infected Vero cells. Virology. 1988;162:65–75. doi: 10.1016/0042-6822(88)90395-9. [DOI] [PubMed] [Google Scholar]
- Hofmann J, Pletz MW, Liebert UG. Rubella virus-induced cytopathic effect in vitro is caused by apoptosis. J Gen Virol. 1999;80:1657–1664. doi: 10.1099/0022-1317-80-7-1657. [DOI] [PubMed] [Google Scholar]
- Khabar KS, Al-Haj L, Al-Zoghaibi F, Marie M, Dhalla M, Polyak SJ, Williams BR. Expressed gene clusters associated with cellular sensitivity and resistance towards anti-viral and anti-proliferative actions of interferon. J Mol Biol. 2004;342:833–846. doi: 10.1016/j.jmb.2004.07.065. [DOI] [PubMed] [Google Scholar]
- Kim HS, Skurk C, Maatz H, Shiojima I, Ivashchenko Y, Yoon SW, Park YB, Walsh K. Akt/FOXO3a signaling modulates the endothelial stress response through regulation of heat shock protein 70 expression. FASEB J. 2005;19:1042–4. doi: 10.1096/fj.04-2841fje. [DOI] [PubMed] [Google Scholar]
- Lee JY, Bowden DS. Rubella virus replication and links to teratogenicity. Clin Microbiol Rev. 2000;13:571–587. doi: 10.1128/cmr.13.4.571-587.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megyeri K, Berencsi K, Halazonetis TD, Prendergast GC, Gri G, Plotkin SA, Rovera G, Gönczöl E. Involvement of a p53-dependent pathway in rubella virus-induced apoptosis. Virology. 1999;259:74–84. doi: 10.1006/viro.1999.9757. [DOI] [PubMed] [Google Scholar]
- Mifune K, Desmyter J, Rawls WE. Effect of Exogenous Interferon on Rubella Virus Production in Carrier Cultures of Cells Defective in Interferon Production. Infect Immun. 1970;2:132–138. doi: 10.1128/iai.2.2.132-138.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan KVK, Ghebrehiwet B, Atreya CD. The N-terminal conserved domain of rubella virus capsid interacts with the C-terminal region of cellular p32 and over expression of p32 enhances the vira infectivity. Virus Res. 2002;85:151–161. doi: 10.1016/s0168-1702(02)00030-8. [DOI] [PubMed] [Google Scholar]
- Plotkin SA. Rubella erradication. Vaccine. 2001;19:3311–3319. doi: 10.1016/s0264-410x(01)00073-1. [DOI] [PubMed] [Google Scholar]
- Pugachev KV, Frey TK. Rubella virus induces apoptosis in culture cells. Virology. 1998;250:359–370. doi: 10.1006/viro.1998.9395. [DOI] [PubMed] [Google Scholar]
- Robertson SE, Featherstone DA, Gacic-Dobo M, Hersh BS. Rubella and congenital rubella syndrome: global update. Rev Panam Salud Publica. 2003;14:306–315. doi: 10.1590/s1020-49892003001000005. [DOI] [PubMed] [Google Scholar]
- Rawls WE, Melnick JL. Rubella virus carrier cultures derived from congenitally infected infants. J Exp Med. 1966;123:795–816. doi: 10.1084/jem.123.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls WE, Desmyter J, Melnick JL. Virus carrier cells and virus-free cells in fetal rubella. Proc Soc Exp Biol Med. 1968;129:477–483. doi: 10.3181/00379727-129-33348. [DOI] [PubMed] [Google Scholar]
- Rezaie P, Trillo-Pazos G, Everall IP, Male DK. Expression of b-Chemokines and Chemokine Receptors in Human Fetal Astrocyte and Microglial Co-cultures: Potential Role of Chemokines in the Developing CNS. Glia. 2002;37:64–75. doi: 10.1002/glia.1128. [DOI] [PubMed] [Google Scholar]
- Singh NK, Atreya CD, Nakhasi HL. Identification of calreticulin as a rubella virus RNA binding protein. Proc Natl Acad Sci USA. 1994;91:12770–12774. doi: 10.1073/pnas.91.26.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanwick TL, Hallum JV. Role of interferon in six cell lines persistently infected with rubella virus. Infect Immun. 1974;10:810–815. doi: 10.1128/iai.10.4.810-815.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchide N, Ohyama K, Bessho T, Toyoda H. Induction of pro-inflammatory cytokine gene expression and apoptosis in human chorion cells of fetal membranes by influenza virus infection: possible implications for maintenance and interruption of pregnancy during infection. Med Sci Monit. 2005;11:RA7–RA16. [PubMed] [Google Scholar]
- Wang CY, Dominguez G, Frey TK. Construction of rubella virus genome-length cDNA clones and synthesis of infectious RNA transcripts. J Virol. 1994;68:3550–3557. doi: 10.1128/jvi.68.6.3550-3557.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MP, Brawner TA, Riggs HG, Jr, Roehrig JT. Characteristics of a persistent rubella infection in a human cell line. J Gen Virol. 1981;52:321–328. doi: 10.1099/0022-1317-52-2-321. [DOI] [PubMed] [Google Scholar]
- Wolinsky JS. Rubella. In: Fields BN, Knipe DM, Howley PM, et al., editors. Fields Virology. 3. Lippincott - Raven; Philadelphia: 1996. pp. 899–929. [Google Scholar]
- Wong KT, Baron S, Ward TG. Rubella virus: role of interferon during infection of African green monkey kidney tissue cultures. J Immunol. 1967;99:1140–1149. [PubMed] [Google Scholar]
- Yoneda T, Urade M, Sakuda M, Miyazaki T. Altered growth, differentiation, and responsiveness to epidermal growth factor of human embryonic mesenchymal cells of palate by persistent rubella virus infection. J Clin Invest. 1986;77:1613–1621. doi: 10.1172/JCI112477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.