Abstract
Ultrasensitive responses are crucial for cellular regulation. Protein sequestration, where an active protein is bound in an inactive complex by an inhibitor, can potentially generate ultrasensitivity. Here, in a synthetic genetic circuit in budding yeast, we show that sequestration of a basic leucine zipper transcription factor by a dominant-negative inhibitor converts a graded transcriptional response into a sharply ultrasensitive response, with apparent Hill coefficients up to 12. A simple quantitative model for this genetic network shows that both the threshold and the degree of ultrasensitivity depend upon the abundance of the inhibitor, exactly as we observed experimentally. The abundance of the inhibitor can be altered by simple mutation; thus, ultrasensitive responses mediated by protein sequestration are easily tuneable. Gene duplication of regulatory homodimers and loss-of-function mutations can create dominant negatives that sequester and inactivate the original regulator. The generation of flexible ultrasensitive responses is an unappreciated adaptive advantage that could explain the frequent evolutionary emergence of dominant negatives.
Keywords: all-or-none, inhibitor, threshold, titration, transcription
Introduction
Protein sequestration is a widespread mechanism in regulatory networks. Many transcription factors (e.g. sigma factors or basic leucine zippers (bzip), basic helix–loop–helix (bHLH) proteins) are sequestered into an inactive complex either by anti-sigma or by naturally occurring dominant-negative inhibitors (Benezra et al, 1990; Van Doren et al, 1991; Hughes and Mathee, 1998). In signaling pathways, kinases or morphogenic proteins are often sequestered by stoichiometric inhibitors (Morgan, 2007) or protein antagonists into inactive complexes (Yanagita, 2005). For high-affinity inhibitors, protein sequestration is an effective mechanism of repression because it strongly buffers against fluctuations in the accumulation of active regulatory molecules. Moreover, protein sequestration can provide a rapid response to biological signals by the regulated release of active regulatory molecule from a pre-established pool of inhibited complex. As a molecular tool, regulated expression of dominant-negative inhibitors has been a successful strategy for conditionally repressing the activity of regulatory proteins (Herskowitz, 1987; Krylov et al, 1997).
As the level of abundance of regulatory protein rises to a level just sufficient to deplete the inhibitor (the ‘equivalence point' in titration theory (Buchler and Louis, 2008)), an ultrasensitive or ‘all-or-none' response can theoretically occur (McCarrey and Riggs, 1986; Ferrell, 1996); see our threshold model in Figure 1. The hallmarks of an ‘all-or-none' response are strong buffering below a concentration threshold and amplification of a small fold change in input into a larger fold change in output at the threshold (i.e. ultrasensitivity). Such buffering and ultrasensitivity is expected to occur for any regulatory response in which free, active regulatory molecule is stoichiometrically bound by an inhibitor into an inactive complex (Buchler and Louis, 2008). Indeed, both theoretical and experimental studies on diverse regulatory molecules have shown ultrasensitive regulation of mRNA levels by small RNAs (Lenz et al, 2004; Levine et al, 2007; Shimoni et al, 2007; Legewie et al, 2008; Mehta et al, 2008), and ultrasensitive regulation of enzyme activity through binding competition between enzymes and substrates (LaPorte et al, 1984; Ferrell, 1996; Legewie et al, 2005b; Kim and Ferrell, 2007).
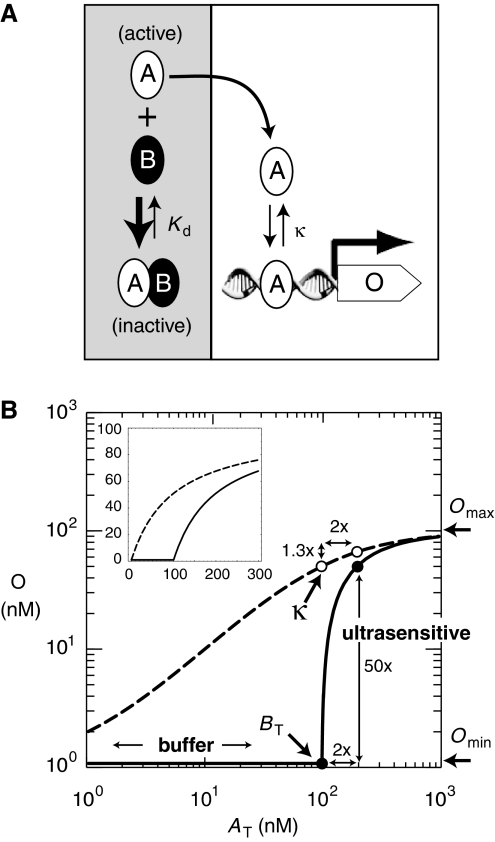
Figure 1.
A threshold model: How protein sequestration generates an ultrasensitive transcriptional response. (A) Active transcription factor A is sequestered by inhibitor B into an inactive AB complex that cannot bind DNA. The concentration of free, active A depends on the dimer dissociation constant (Kd) and the total concentration of transcription factor (AT=A+AB) and inhibitor (BT=B+AB) in a cell. For stoichiometric-binding conditions (BT/Kd≫1), the inhibitor serves as a finite sink that buffers and titrates free, activator A such that A≈AT/(BT/Kd) when AT<BT and A≈AT−BT when AT>BT (Buchler and Louis, 2008). At steady state, transcription of a target gene (output, O) by free activator A is Omin+Omax·A/(A+κ) and κ is the DNA-binding dissociation constant. The transcriptional response is non-cooperative (i.e. Hill coefficient nH=1) because the promoter contains a single DNA-binding site for the activator. (B) Logarithmic plot of the input–output response as a function of input (AT) for parameters κ=100 nM, Omax=100 nM, and Omin=1 nM (nanomolar). In bacteria or yeast, 1 nM≈1–25 molecules per cell, respectively. Inset: Linear plot of the input–output response. For a range of parameters, the transcriptional output of gene O in the presence of protein sequestration is well approximated by a threshold model where O≈Omin when AT<BT and O≈Omin+Omax·(AT−BT)/((AT−BT)+κ) when AT>BT (see Supplementary information). Consider the transcriptional response to a two-fold change in input (AT(low)=100 nM, AT(high)=200 nM, drawn as circles). In the absence of inhibitor (BT=0 nM, dashed curve and open circles), the transcriptional response will always be less than or equal to linear (e.g. two-fold change in input will be attenuated to a two-fold change or less in output). In the presence of inhibitor (BT=100 nM, solid curve and closed circles), the original input–output response is simply shifted to the right by BT (see inset) and the transcriptional response is now ultrasensitive at the equivalence point (AT≈BT). A process is ultrasensitive when a small fold change in input (2 × ) is amplified to a larger fold change (50 × ) in output, or when the response coefficient or logarithmic sensitivity is greater than one (Savageau, 1976; Goldbeter and Koshland, 1984). Both buffering and ultrasensitivity are a simple consequence of resetting the ‘zero point' of the original transcriptional response to the equivalence point, such that O≈Omin when AT<BT.
At a systems level, both buffering and ultrasensitivity are crucial for robust bistability, an important feature for resilient epigenetic switches or the oscillation of circadian clocks and the cell cycle. Indeed, protein sequestration is a common regulatory mechanism in many bistable switches and oscillatory genetic networks (Young and Kay, 2001; Dubnau and Losick, 2006; Buchler and Louis, 2008). However, despite the prevalence of protein sequestration in natural regulatory networks, it has not been experimentally tested whether protein sequestration generates an ultrasensitive response in a genetic network. Here, we construct a synthetic genetic network controlled by sequestration, and explicitly show that protein sequestration of a transcriptional activator by a dominant-negative inhibitor can generate a flexible ultrasensitive response in gene expression.
Results
We quantified the ability of protein sequestration to generate ultrasensitivity in a simple genetic network in yeast (Figure 2A). Our synthetic gene circuit consisted of three components: (i) transactivator, (ii) dominant-negative inhibitor, and (iii) reporter. Both the transactivator and dominant-negative inhibitor are part of the basic leucine zipper (bZIP) protein family, a large class of dimeric transcription factors in eukaryotes. We used a heterologous mammalian bZIP (CEBPα) as our transactivator to minimize unwanted endogenous interactions in budding yeast. Our dominant-negative inhibitor (‘3HF') was designed earlier to bind CEBPα with high affinity (Kd≈0.04 nM) into an inactive heterodimer (Krylov et al, 1995). We produced a range of transactivator and dominant-negative concentrations in yeast by expressing them under the control of different yeast promoters (Figure 2A). The CEBPα transactivator was fused to a red fluorescent protein (mCherry), so that we could measure the transactivator (CEBPα–RFP) in individual cells. The downstream reporter was yellow fluorescent protein (yEVenus) gene under the control of a promoter containing a single binding site for CEBPα–RFP transactivator. We intentionally used a single DNA-binding site to minimize cooperative binding of the transactivator (Wolf and Eeckman, 1998; Bintu et al, 2005), such that any ultrasensitivity in YFP transcription should be a consequence of protein sequestration of the transactivator by a dominant-negative inhibitor.
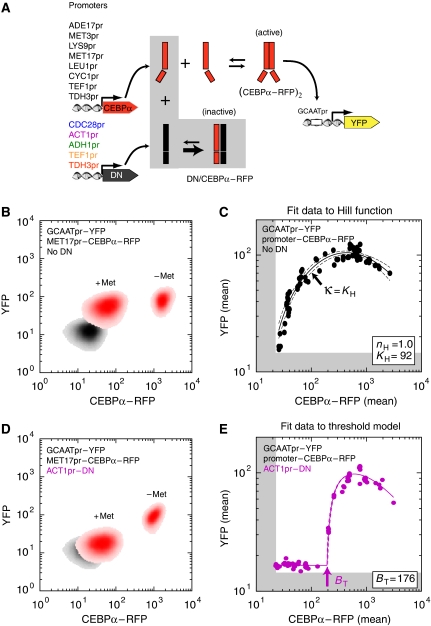
Figure 2.
Protein sequestration generates an ultrasensitive transcriptional response: an experimental approach. (A) Scheme of our synthetic circuit in budding yeast. Our transactivator (CEBPα–RFP) consisted of an N-terminal fusion of a compact VP16 activation sequence (F2) and a C-terminal fusion of red fluorescent protein (mCherry) to a minimal basic leucine zipper (mouse CEBPα); see Supplementary information. All yeast strains contained yellow fluorescent protein (yEVenus) reporter, whose transcription was controlled by a ‘zero-background' MEL1 promoter with a single ‘GCAAT' binding site for the CEBPα–RFP dimer. Different strains had a high-affinity ‘3HF' dominant-negative inhibitor (DN), which sequesters CEBPα into an inactive complex (Krylov et al, 1995), expressed under constitutive promoters of increasing strength (from blue to red). Both reporter and dominant-negative plasmids were chromosomally integrated in a single copy at ADE2, and URA3 loci, respectively. For each reporter-only (No DN) or dominant-negative+reporter strain, we generated a range of steady-state transactivator concentrations by integrating different promoter–CEBPα–RFP plasmids with variable copy number in the LEU2 locus. (B) A density plot of fluorescence concentration (CEBPα–RFP, YFP) for a strain in which MET17pr–CEBPα–RFP plasmid was integrated into a reporter-only strain (No DN). MET17 is an amino-acid repressed promoter and we measured the steady-state response in two growth conditions (+Met, −Met). The strain with no reporter and transactivator (control for autofluorescence background) is shown in gray. (C) The full input–output response in reporter-only strain (No DN). Each data point is the mean CEBPα–RFP (input) and mean YFP (output) of a single transformant. All the data were least-squares fit with a Hill function Omin+OmaxċATnH/(ATnH+KHnH) modified to include squelching and subtraction of autofluorescence background determined from cells lacking reporter and transactivator (indicated by the edge of the gray shading); see Supplementary information. The solid line corresponds to the best fit and dashed lines are the 95% confidence intervals. The best-fit Hill coefficient (nH) and half-maximum concentration (KH) are shown in the box. The measured transcriptional response is graded and non-cooperative, thus the DNA-binding dissociation constant κ=KH (D) A density plot of fluorescence concentration (CEBPα–RFP, YFP) for a strain in which MET17pr–CEBPα–RFP plasmid was integrated into ACT1pr–DN+reporter strain. (E) The full input–output response in ACT1pr–DN+reporter strain. All data were least-squares fit with the threshold model (presented in Figure 1) modified to include squelching and subtraction of autofluorescence background (indicated by the edge of the gray shading); see Supplementary information. The best-fit threshold (indicated by arrow) is shown in the box.
To establish a baseline for the input–output relationship between transactivator (CEBPα–RFP, input) and the transcriptional response (YFP, output), in the absence of protein sequestration, we measured the correlation between CEBPα–RFP and YFP fluorescence in individual cells by flow cytometry. All RFP and YFP fluorescence measurements were normalized by the forward scatter (related to cell size) to create a metric for fluorescence concentration (see Materials and methods). A density plot of fluorescence concentration for an MET17pr–CEBPα–RFP strain (Figure 2B), with or without the induction of CEBPα–RFP (obtained by growth in medium without or with methionine), shows that the response of YFP to a 40-fold change in activator concentration is only 1.5-fold after subtraction of autofluorescent background levels (control strain lacking reporter and transactivator; gray area in Figure 2B). In contrast, the full dynamic range of YFP response to CEBPα–RFP determined in Figure 2C is ∼16-fold. The low responsiveness of the Met-regulated system reflects the fact that the repressed MET17 promoter (+Met) still produces enough CEBPα–RFP transactivator to significantly induce the YFP reporter. Such promoter ‘leakiness' is a typical feature of most regulated promoters and often precludes their use to conditionally express target genes, an important issue to which we will return.
To measure the full input–output response of YFP to CEBPα–RFP, we needed to generate a range of CEBPα–RFP concentrations at steady state. For this purpose, we developed a simple approach to generate a broad range of CEBPα–RFP concentrations, using (i) different promoters with variable expression strength to drive the production of CEBPα–RFP (Figure 2A) and (ii) the variable copy number of plasmid constructs integrated into the same genomic locus by ends-in homologous recombination (Orr-Weaver et al, 1981). This approach generated a nearly continuous range of transactivator concentrations (Figure 2C). We could not readily use standard GAL1 or amino-acid-repressed promoters to reliably generate a broad, continuous range of transactivator concentrations at steady state. This was a consequence of cryptic carbon-source response elements in many yeast shuttle plasmids (Pauwels et al, 1999) and the fact that intermediate levels of metabolites (e.g. methionine, adenine, leucine, and lysine) were rapidly consumed by yeast, thus causing varying degrees of induction during culture growth (data not shown).
We quantified the full input–output response for CEBPα–RFP by fitting a modified Hill function to the data in Figure 2C. The Hill function was modified to include a ‘squelching' term to reflect the observed decrease in YFP for large transactivator concentrations (see Supplementary information). Squelching is thought to arise when the abundant transactivator begins to titrate components of the transcriptional machinery into unproductive complexes (Gill and Ptashne, 1988). Despite observing mild squelching in the YFP response, we did not detect toxic phenotypes such as slow growth rates or abnormal cell morphology, which were observed with stronger VP16-based activators (data not shown). The best-fit Hill coefficient (nH) is 1.0, which indicates that the transcriptional response was graded and non-cooperative, as might be expected for a promoter with a single transactivator-binding site that is driven by a strong dimerizing CEBPα, Kd≈10 nM (Krylov et al, 1995).
Addition of the 3HF dominant-negative inhibitor (‘DN') under the control of a strong constitutive promoter markedly changed the YFP transcriptional response. First, YFP transcription was suppressed to a basal level (approximately the level expressed in cells lacking any transactivator) at transactivator concentrations that earlier yielded nearly full induction in the absence of dominant-negative inhibitor (compare +Met in Figure 2B and D). Second, an ultrasensitive transcriptional response occurred at a threshold concentration, which we predict to be the equivalence point where CEBPα–RFP approximately equals DN (Figure 2E). Amounts of transactivator greater than the equivalence point yielded full YFP induction. Thus, by tuning the expression of the dominant-negative inhibitor to an appropriate level in our synthetic gene circuit, we have obtained a simple method for transforming any leaky, conditional promoter (e.g. MET17, LEU1, ADE17, LYS9) into a digital ‘on/off' transcriptional response of a downstream target gene (e.g. YFP).
We developed a simple quantitative model (threshold model in Figure 1) to predict the transcriptional response of YFP in the presence of protein sequestration of CEBPα–RFP by DN. We subsequently determined the equivalence point (BT) by least-squares fitting of this threshold model to the full input–output data of Figure 2E. Importantly, fitting the threshold model is highly constrained by the parameters determined for the original input–output response in the absence of a dominant-negative inhibitor (Figure 2C): the only free parameter to be fit is BT, the total effective dominant-negative inhibitor concentration in units of CEBPα–RFP equivalents.
The threshold model makes the strong prediction that the location of the threshold (amount of CEBPα–RFP at the transition from low to high YFP expression) should increase linearly with increasing dominant-negative inhibitor concentration. To test this, we measured the full input–output response for CEBPα–RFP in the presence of increasing levels of dominant–negative inhibitor DN, obtained by varying the strength of the constitutive promoter driving DN expression (Figure 3A, color coding as in Figure 2A). The dominant-negative inhibitor concentration (BT) of each input–output response was determined by a least-squares fit of the threshold model, as in Figure 2E, for each promoter driving DN expression. We also quantified the abundance of HA-tagged DN using quantitative western blotting (see Materials and methods). There is excellent agreement between the measured concentration of DN and the best-fit dominant-negative inhibitor concentration (BT) of the full input–output response, after standardizing both to the data for the strongest promoter (TDH3) (Figure 3B).
Figure 3.
Input–output threshold and ultrasensitivity are controlled by the concentration of dominant-negative inhibitor. (A) Full input–output response of the different dominant-negative inhibitor+reporter strains (data are color-coded similarly to Figure 2A). Best fit of the data to the modified threshold model is shown by solid lines (BT=0, 9, 176, 411, 1163, 1631; threshold indicated by arrows). (B) The abundance of DN was measured by quantitative western blotting and compared with the measured threshold (BT) of the input–output response. (C) Full input–output response of the different dominant-negative inhibitor+reporter strains. Best fit of data to the modified Hill function is shown by solid lines (nH=1.0, 1.3, 2.7, 5.7, 11.8, 11.4). (D) Plot of the fitted Hill coefficient from (C) as a function of fitted input–output threshold BT from (A) normalized by the fitted DNA-binding dissociation constant κ (from Figure 2C). All these quantities are dimensionless. Theoretical expectation (solid curve) was obtained by a least-squares fit of the Hill function to an ideal threshold model (infinite data points; no measurement error); see Supplementary information. The error bars for western blot data and fitted parameters are the s.e.m. and s.d. (at 68% confidence intervals), respectively.
The threshold model also makes quantitative predictions for the sharpening of the response (increased ultrasensitivity) with increasing BT. In this study, we used the apparent Hill coefficient (nH) as a coarse, global measure of ultrasensitivity in the transcriptional response; see (Gunawardena, 2005; Legewie et al, 2005a). The nH was determined by a least-squares fit of the modified Hill function, as in Figure 2C, for each promoter driving DN expression (Figure 3C). The nH increased as a function of BT, as predicted by the threshold model (Figure 3D). In our experimental series, nH varies from 2 to 12 depending on promoter strength. For comparison, the synergistic or cooperative interaction of transactivators bound at multiple DNA-binding sites typically yields nH values from 2–5 (Burz et al, 1998; Rossi et al, 2000; Hooshangi et al, 2005; Pedraza and van Oudenaarden, 2005; Rosenfeld et al, 2005; Ajo-Franklin et al, 2007; Gregor et al, 2007; Kim and O'Shea, 2008). In principle, larger Hill coefficients for cooperativity could be obtained through multi-gene cascades or bifurcations via positive feedback loops (Goldbeter and Koshland, 1982; Ferrell, 1997; Kholodenko et al, 1997). However, such enhanced ultrasensitivity requires careful matching of concentrations and biophysical parameters (Yokobayashi et al, 2002; Hooshangi et al, 2005; Pedraza and van Oudenaarden, 2005). In contrast, our synthetic two-gene circuit only required the addition of a dominant-negative inhibitor to achieve an equivalent, sharp threshold response.
Discussion
Our observations imply that protein sequestration is a highly flexible regulatory mechanism for generating ultrasensitive responses in genetic networks. We have shown that both the threshold and the degree of ultrasensitivity can be tuned by varying the expression of the dominant-negative inhibitor. In addition, the ultrasensitivity can be tuned independently of the threshold simply by changing the DNA-binding dissociation constant of the transactivator (κ); see Figure 3D. We note that mutations that alter the expression levels of the dominant-negative inhibitor or modify the transactivator's DNA-binding affinity are likely to be frequent mutational events. This is in contrast with molecular cooperativity, in which the threshold and degree of ultrasensitivity is a complicated function of the number of DNA-binding sites, the DNA-binding affinity, and protein–protein affinity between transactivators (Wolf and Eeckman, 1998; Bintu et al, 2005). However, the advantage of a concentration-dependent threshold might also pose a challenge because inhibitor concentrations in vivo can show significant fluctuations. The extent to which natural genetic circuits favor cooperativity, protein sequestration, or other regulatory mechanisms to generate ultrasensitivity is an important question that remains to be empirically addressed.
There are hundreds of different bZIPs or bHLH transcription factors with distinct DNA-binding sites for which high-affinity dominant-negative inhibitors (Kd≈0.01–0.1 nM) can be designed (Krylov et al, 1995, 1997). Thus, our approach could yield a large number of independently regulated promoters with ultrasensitive responses by using different naturally regulated promoters (not initially ultrasensitive) coupled to different bZIP/bHLH/dominant-negative pairs. This would provide a highly expanded toolkit of ‘on/off' promoters for synthetic biology, as the basal expression of our reporter in the repressed state is very low (Figure 2D). Furthermore, by coupling protein sequestration to regulatory feedback, such a toolkit lays a foundation for building robust bistable and oscillatory genetic circuits with tuneable properties (Francois and Hakim, 2005).
For the first time, we show experimentally that protein sequestration can provide strong buffering and tuneable ultrasensitivity in genetic networks. By varying the abundance of the inhibitor, evolutionarily selectable variation in ultrasensitivity could occur much more easily by this mechanism than by the protein and/or promoter re-design required to alter molecular cooperativity. In regulatory networks, buffering and ultrasensitivity are important for promoting robust bistable or oscillatory regulatory networks. We note that many bistable and oscillatory genetic networks show sequestration as a regulatory mechanism (Young and Kay, 2001; Dubnau and Losick, 2006; Buchler and Louis, 2008). Our results suggest that sequestration might play an unsuspected role in facilitating the evolution of bistable or oscillatory circuits in natural regulatory systems (Francois and Hakim, 2004).
Materials and methods
Strain construction
All yeast strains are derivatives of W303 (Thomas and Rothstein, 1989). Yeast shuttle plasmids of reporter, transactivator, and dominant–negative gene constructs were linearized and integrated into chromosomal ADE2, LEU2, and URA3 loci, respectively. We followed standard protocols of high-efficiency yeast transformation (Burke et al, 2000). A full description of how yeast shuttle plasmids and strains were constructed can be found in Supplementary information.
Flow cytometry
All strains were grown overnight in selective SCD medium at 30°C on a roller drum. In the morning, these strains were subsequently diluted 100-fold into non-selective SCD medium and allowed to grow at 30°C on a roller drum for ∼5–7 h until they reached mid-log phase. Cultures were put on ice and sonicated. Fluorescence was measured using a Becton–Dickinson LSR II flow cytometer equipped with 488 and 561 nm lasers that maximally excite YFP and CEBPα–RFP, respectively. Side-scatter fluorescence from 488 and 561 nm lasers was filtered and collected between wavelengths of 515–545 nm (488-H) and 605–615 nm (561-H), respectively. As yEVenus and mCherry excitation and emission spectra are well separated, no color compensation was necessary between the raw 488-H and 561-H signals. All flow cytometry data were gated on FSC-A/SSC-A to exclude debris and on FSC-A/FSC-W to exclude yeast doublets. There was a positive correlation between 488-H, 561-H (proportional to total abundance of fluorescent protein) and FSC-H (proportional to cell size). This correlation arises mostly because larger cells have more total fluorescent protein than smaller cells (Volfson et al, 2006). We divided 488-H and 561-H by FSC-H to create pseudo fluorescence concentrations of YFP, CEBPα–RFP.
Quantitative western blotting
Strains were grown to mid-log phase and proteins were extracted by bead-breaking cells in SDS buffer with protease inhibitors. All protein concentrations were balanced, run on 4–20% Tris–Glycine SDS–PAGE gels (Invitrogen), and transferred to PVDF membrane through electro-blotting (25 V for 75 min). Western blot was probed with two primary antibodies: 1:10 000 dilution of mouse, monoclonal anti-HA antibody (anti-HA.11, Covance) and 1:10 000 dilution of mouse, monoclonal anti-PGK1 antibody (A6457, Invitrogen, Molecular Probes). After washing, western blot was probed with secondary antibody: 1:10 000 dilution of anti-mouse-HRP antibody (GE HealthCare). After washing, western blots were quantitated through chemiluminescence using LAS-3000 imaging system (FujiFilm). Blots were exposed until the first pixel started to saturate. All bands of interest were normalized by local background. Our measure of relative abundance was the total, normalized intensity of HA-3HF band (∼14 kDa) divided by the total, normalized intensity of internal control PGK1 band (∼45 kDa). The error bars in Figure 3B are the s.e.m. as determined for 6–7 replicates.
Supplementary Material
This file contains Supplementary Methods, Supplementary figures S1-3, Supplementary tables S1-4, Supplementary Notes (References)
Flow cytometry data for different base strains transformed with different promoters driving CEBPα-RFP.
Acknowledgments
We are grateful to C Vinson, R Weiss, G Charvin, S Di Talia, and F Tulin for plasmids. We thank L Bai, S Di Talia, P Francois, W Loomis, M Louis, Y Lu, C Oikonomou, J Robbins, D Rogulja, L Saez, J Skotheim, N Stavropoulos, S Syed, F Tulin, C Vinson, and J Widom for comments on the paper. NEB was supported by a Career Award at the Scientific Interface from Burroughs Wellcome Fund. FRC was supported by the National Institutes of Health.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EP, Landgraf D, Phillips I, Silver PA (2007) Rational design of memory in eukaryotic cells. Genes Dev 21: 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H (1990) The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61: 49–59 [DOI] [PubMed] [Google Scholar]
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Kuhlman T, Phillips R (2005) Transcriptional regulation by the numbers: applications. Curr Opin Genet Dev 15: 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchler NE, Louis M (2008) Molecular titration and ultrasensitivity in regulatory networks. J Mol Biol 384: 1106–1119 [DOI] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T (2000) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press [Google Scholar]
- Burz DS, Rivera-Pomar R, Jackle H, Hanes SD (1998) Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J 17: 5998–6009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D, Losick R (2006) Bistability in bacteria. Mol Microbiol 61: 564–572 [DOI] [PubMed] [Google Scholar]
- Ferrell JE Jr (1996) Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem Sci 21: 460–466 [DOI] [PubMed] [Google Scholar]
- Ferrell JE Jr (1997) How responses get more switch-like as you move down a protein kinase cascade. Trends Biochem Sci 22: 288–289 [DOI] [PubMed] [Google Scholar]
- Francois P, Hakim V (2004) Design of genetic networks with specified functions by evolution in silico. Proc Natl Acad Sci USA 101: 580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois P, Hakim V (2005) Core genetic module: the mixed feedback loop. Phys Rev E Stat Nonlin Soft Matter Phys 72: 031908. [DOI] [PubMed] [Google Scholar]
- Gill G, Ptashne M (1988) Negative effect of the transcriptional activator GAL4. Nature 334: 721–724 [DOI] [PubMed] [Google Scholar]
- Goldbeter A, Koshland DE Jr (1982) Sensitivity amplification in biochemical systems. Q Rev Biophys 15: 555–591 [DOI] [PubMed] [Google Scholar]
- Goldbeter A, Koshland DE Jr (1984) Ultrasensitivity in biochemical systems controlled by covalent modification. Interplay between zero-order and multistep effects. J Biol Chem 259: 14441–14447 [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF, Bialek W (2007) Probing the limits to positional information. Cell 130: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena J (2005) Multisite protein phosphorylation makes a good threshold but can be a poor switch. Proc Natl Acad Sci USA 102: 14617–14622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I (1987) Functional inactivation of genes by dominant negative mutations. Nature 329: 219–222 [DOI] [PubMed] [Google Scholar]
- Hooshangi S, Thiberge S, Weiss R (2005) Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proc Natl Acad Sci USA 102: 3581–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KT, Mathee K (1998) The anti-sigma factors. Annu Rev Microbiol 52: 231–286 [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Hoek JB, Westerhoff HV, Brown GC (1997) Quantification of information transfer via cellular signal transduction pathways. FEBS Lett 414: 430–434 [DOI] [PubMed] [Google Scholar]
- Kim HD, O'Shea EK (2008) A quantitative model of transcription factor-activated gene expression. Nat Struct Mol Biol 15: 1192–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Ferrell JE Jr (2007) Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell 128: 1133–1145 [DOI] [PubMed] [Google Scholar]
- Krylov D, Kasai K, Echlin DR, Taparowsky EJ, Arnheiter H, Vinson C (1997) A general method to design dominant negatives to B-HLHZip proteins that abolish DNA binding. Proc Natl Acad Sci USA 94: 12274–12279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov D, Olive M, Vinson C (1995) Extending dimerization interfaces: the bZIP basic region can form a coiled coil. EMBO J 14: 5329–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPorte DC, Walsh K, Koshland DE Jr (1984) The branch point effect. Ultrasensitivity and subsensitivity to metabolic control. J Biol Chem 259: 14068–14075 [PubMed] [Google Scholar]
- Legewie S, Bluthgen N, Herzel H (2005a) Quantitative analysis of ultrasensitive responses. FEBS J 272: 4071–4079 [DOI] [PubMed] [Google Scholar]
- Legewie S, Bluthgen N, Schafer R, Herzel H (2005b) Ultrasensitization: switch-like regulation of cellular signaling by transcriptional induction. PLoS Comput Biol 1: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legewie S, Dienst D, Wilde A, Herzel H, Axmann IM (2008) Small RNAs establish delays and temporal thresholds in gene expression. Biophys J 95: 3232–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL (2004) The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118: 69–82 [DOI] [PubMed] [Google Scholar]
- Levine E, Zhang Z, Kuhlman T, Hwa T (2007) Quantitative characteristics of gene regulation by small RNA. PLoS Biol 5: e229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarrey JR, Riggs AD (1986) Determinator-inhibitor pairs as a mechanism for threshold setting in development: a possible function for pseudogenes. Proc Natl Acad Sci USA 83: 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, Goyal S, Wingreen NS (2008) A quantitative comparison of sRNA-based and protein-based gene regulation. Mol Syst Biol 4: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO (2007) The Cell Cycle: Principles of Control. Sunderland, MA: New Science Press [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ (1981) Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci USA 78: 6354–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels K, Abadjieva A, Hilven P, Crabeel M (1999) A strong carbon source effect is mediated by pUC plasmid sequences in a series of classical yeast vectors designed for promoter characterization. Yeast 15: 1269–1274 [DOI] [PubMed] [Google Scholar]
- Pedraza JM, van Oudenaarden A (2005) Noise propagation in gene networks. Science 307: 1965–1969 [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB (2005) Gene regulation at the single-cell level. Science 307: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Rossi FM, Kringstein AM, Spicher A, Guicherit OM, Blau HM (2000) Transcriptional control: rheostat converted to on/off switch. Mol Cell 6: 723–728 [DOI] [PubMed] [Google Scholar]
- Savageau MA (1976) Biochemical Systems Analysis: A Study of Function and Design in Molecular Biology. Reading, Mass.: Addison-Wesley Pub. Co., Advanced Book Program [Google Scholar]
- Shimoni Y, Friedlander G, Hetzroni G, Niv G, Altuvia S, Biham O, Margalit H (2007) Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol Syst Biol 3: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630 [DOI] [PubMed] [Google Scholar]
- Van Doren M, Ellis HM, Posakony JW (1991) The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development 113: 245–255 [DOI] [PubMed] [Google Scholar]
- Volfson D, Marciniak J, Blake WJ, Ostroff N, Tsimring LS, Hasty J (2006) Origins of extrinsic variability in eukaryotic gene expression. Nature 439: 861–864 [DOI] [PubMed] [Google Scholar]
- Wolf DM, Eeckman FH (1998) On the relationship between genomic regulatory element organization and gene regulatory dynamics. J Theor Biol 195: 167–186 [DOI] [PubMed] [Google Scholar]
- Yanagita M (2005) BMP antagonists: their roles in development and involvement in pathophysiology. Cytokine Growth Factor Rev 16: 309–317 [DOI] [PubMed] [Google Scholar]
- Yokobayashi Y, Weiss R, Arnold FH (2002) Directed evolution of a genetic circuit. Proc Natl Acad Sci USA 99: 16587–16591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MW, Kay SA (2001) Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2: 702–715 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Methods, Supplementary figures S1-3, Supplementary tables S1-4, Supplementary Notes (References)
Flow cytometry data for different base strains transformed with different promoters driving CEBPα-RFP.