Abstract
The NMR structures of three single-amino acid variants of the C-terminal domain of the human prion protein, hPrP(121–230), are presented. In hPrP(M166V) and hPrP(R220K) the substitution is with the corresponding residue in murine PrP, and in hPrP(S170N) it is with the corresponding Syrian hamster residue. All three substitutions are in the surface region of the structure of the cellular form of PrP (PrPC) that is formed by the C-terminal part of helix 3, with residues 218–230, and a loop of residues 166–172. This molecular region shows high species variability and has been implicated in specific interactions with a so far not further characterized “protein X,” and it is related to the species barrier for transmission of prion diseases. As expected, the three variant hPrP(121–230) structures have the same global architecture as the previously determined wild-type bovine, human, murine, and Syrian hamster prion proteins, but with the present study two localized “conformational markers” could be related with single amino acid exchanges. These are the length and quality of definition of helix 3, and the NMR-observability of the residues in the loop 166–172. Poor definition of the C-terminal part of helix 3 is characteristic for murine PrP and has now been observed also for hPrP(R220K), and NMR observation of the complete loop 166–172 has so far been unique for Syrian hamster PrP and is now also documented for hPrP(S170N).
The three-dimensional structures of the recombinant prion proteins (PrP) from mouse (mPrP; refs. 1–3), Syrian hamster (shPrP; refs. 4 and 5), humans (hPrP; ref. 6), and cattle (bPrP; ref. 7) share a common architecture, consisting of a globular domain with residues 125–228 and an N-terminal, flexibly disordered “tail” of about 100 residues from position 23 to position 124. The globular domains in the four PrP structures differ only by strictly localized conformational variations, which include differences in atom positions, variable surface charge distribution, and locally different dynamic properties. To further investigate the origin of these species variations on the three-dimensional structure level, we exchanged individual residues in the globular domain of hPrP with the corresponding amino acid types in mPrP and shPrP (8). On the basis of initial characterization by 1H NMR and circular dichroism spectroscopy, three of these hPrP variants were selected for complete structure determination of the fragment composed of residues 121–230. In these three variant proteins the residues Met-166 and Arg-220 in hPrP were replaced with Val and Lys, respectively, which are the corresponding residues in mPrP, and Ser-170 in hPrP was replaced with Asn, which is the corresponding amino acid in shPrP. The present paper reports the NMR structures of hPrP(M166V), hPrP(S170N), and hPrP(R220K).
The “species barrier” for transmissible spongiform encephalopathies (TSEs) describes the fact that although transmission between different individuals of the same mammalian species may occur efficiently, the process of infection between different species may be either inefficient or completely ineffective (9, 10). If one accepts the protein-only hypothesis (9–13) so as to attribute a key role to PrP in the TSE-infection process, either as the infective agent or in a supporting role, one will quite necessarily infer that the stringency of the species barrier is related to species variations in the PrP structure. By correlating species variations in the amino acid sequence (8) with variations in the three-dimensional structure of the cellular, monomeric form of PrP (PrPC), the present project attempts to gain new insight into the structural basis of variable stringency of the species barrier between different combinations of mammalian species.
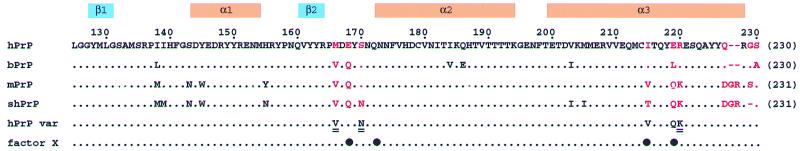
The selection of the variant hPrP proteins for this study was based on the data in Fig. 1: The polypeptide segments 166–170 and 215–230 have an outstandingly high incidence of nonconservative species variations, which are located in close proximity in a contiguous surface area of the three-dimensional structure (14). Evidence has also been presented, from experiments with transgenic laboratory animals, that this surface area of the molecule is recognized by a species-specific “protein X,” which has been suggested to be a mediator of the transformation of PrPC into the TSE-related “scrapie form,” PrPSc (15). Intriguingly, among the presently available human, bovine, murine, and Syrian hamster PrPC structures, significant conformational variations are observed in this very region of the molecule, where we can actually identify two “conformational markers” for the globular domain of mammalian PrPs (Table 1). These are the regular nature of the C-terminal part of helix 3, and the presence in the [15N,1H]-COSY spectra of all of the backbone resonances in a loop formed by residues 166–172. In hPrP, shPrP, and bPrP a regular, straight helix 3 extends from residue 200 to approximately residue 226, while it is well defined only up to residue 219 in mPrP. In hPrP (6), mPrP (1–3), and bPrP (7) the backbone amide resonances of three amino acids in the loop 166–172 and of Phe-175 were not observed, probably because of line broadening, whereas in shPrP all backbone amide resonances were observed and assigned (5). The presently described variant proteins were selected with the purpose of changing the aforementioned markers from hPrP-type to either mPrP-type for helix 3, or to shPrP-type for the loop 166–172.
Figure 1.
Amino acid sequence alignment of the fragment 125–230 (numeration of hPrP following ref. 8; for each protein the number in parentheses on the right indicates the sequence position attributed to the C-terminal residue) of the human, bovine, murine, and Syrian hamster PrPs. At the top the locations of the regular secondary structure elements of hPrP(121–230) are indicated. The red letters identify residue positions with amino acid exchanges or insertions in the region of the three-dimensional structure that are discussed in this paper. The row “hPrP var” lists the amino acid exchanges in the single-residue variants of hPrP studied in this paper, where the doubly underlined residues indicate those variants for which a complete structure determination is presented. In the row “factor X” the black circles identify the sequence positions 168, 172, 215, and 219, which have been suggested to form an epitope or part of an epitope for interactions with a species-specific “factor X” or “protein X” (15, 33).
Table 1.
Conformational markers in mammalian PrPs
| Protein | TM,* K | 166–172, 175† | Helix 3‡ |
|---|---|---|---|
| hPrP(121–230) | 334 | + (0.49/—) | |
| mPrP(121–231) | 335 | (0.68/1.59) | |
| bPrP(121–230) | 336 | + (0.38/0.71) | |
| shPrP(90–231) | + | + (0.33/0.65) | |
| hPrP(M166V) | 336 | + (0.49/0.91) | |
| hPrP(S170N) | 332 | + | + (0.49/0.44) |
| hPrP(R220K) | 333 | (0.62/1.12) |
TM, denaturation temperature.
A + sign indicates that [15N,1H]-correlated spectroscopy (COSY) cross peaks were observed and assigned for the entire loop 166–172 and for residue 175.
A + sign indicates that for the residues 200–226 the root-mean-square deviation (rmsd) to the mean coordinates for N, Cα, and C′ among the 20 conformers used to describe the NMR structure is <0.5 Å, and that the rmsd of the mean structure relative to the mean hPrP(121–230) structure is <1.0 Å (the two rmsd values are given in parentheses).
Materials and Methods
Sample Preparation and Characterization.
For the cloning, expression, and purification of hPrP(121–230) variants in unlabeled form and with uniform 13C,15N-labeling we closely followed the strategy used for the preparation of wild-type hPrP (6, 16). Single-point mutants of hPrP were constructed by following the Quickchange site-directed mutagenesis protocol (Stratagene). Melting temperatures, TM, were evaluated from the temperature dependence of the circular dichroism spectrum at 222 nm. Concentrated protein solutions for NMR spectroscopy were obtained by using Ultrafree-15 Centrifugal Filter Biomax Devices (Millipore), resulting in 1 mM protein solutions either in 90% H2O/10% 2H2O (D2O) or in 99.9% D2O, and containing 10 mM sodium acetate and 0.05% sodium azide at a pH meter reading of 4.5 and 20°C.
NMR Measurements and Structure Calculations.
NMR measurements were performed on Bruker DRX600 and DRX750 spectrometers. The programs Prosa (17) and Xeasy (18) were used for data processing and spectral analysis, respectively. Sequence-specific resonance assignments for the variant proteins were obtained by standard triple-resonance NMR experiments (19), starting from the chemical shift lists of wild-type hPrP(121–230) (6).
For each mutant protein, distance constraints for the structure calculation were obtained from three nuclear Overhauser enhancement spectroscopy (NOESY) experiments recorded at a proton frequency of 750 MHz with a mixing time of 40 ms: a three-dimensional (3D) 13C-resolved [1H,1H]-NOESY spectrum for NOEs with aliphatic CH moieties, a 3D 15N-resolved [1H,1H]-NOESY spectrum, and a 3D 13C-resolved [1H,1H]-NOESY spectrum for NOEs with aromatic CH moieties. NOE cross peak assignments were initially generated by using an automatic NOE assignment module (T. Herrmann, P.G., and K.W., unpublished work) implemented in the program Dyana (20), and then confirmed by visual inspection of the spectra. The program Dyana was also used to convert NOE intensities into upper distance bounds according to an inverse sixth power peak volume-to-distance relationship, to remove meaningless constraints, to derive constraints for the backbone torsion angles φ and ψ from Cα chemical shift values (21), and to perform a systematic grid search analysis of the local conformation along the polypeptide backbone with the subroutine Found (22). Final structure calculations using the torsion angle dynamics protocol of Dyana with 8,000 steps (20) were started from 100 randomized conformers. The 20 conformers with the lowest final Dyana target function value were energy-minimized in a water shell with the program Opalp (23, 24), using the Amber force field (25). The program Molmol (26) was used to analyze the results of the structure calculations (Table 2) and to prepare the drawings of the structures.
Table 2.
Input for the structure calculation and characterization of the energy-minimized NMR structures of three single-amino acid variants of hPrP(121–230)
| Quantity | Value
|
||
|---|---|---|---|
| hPrP (M166V) | hPrP (S170N) | hPrP (R220K) | |
| NOE upper distance limits | 1,592 | 1,797 | 1,670 |
| Dihedral angle constraints | 418 | 488 | 414 |
| Residual target function, Å2 | 0.19 ± 0.05 | 0.84 ± 0.19 | 0.18 ± 0.05 |
| Residual NOE violations | |||
| Number >0.1 Å | 9 ± 3 | 20 ± 3 | 16 ± 2 |
| Maximum, Å | 0.19 ± 0.09 | 0.16 ± 0.07 | 0.12 ± 0.03 |
| Residual angle violations | |||
| Number >2.0° | 0.2 ± 0.4 | 0.4 ± 0.6 | 0.1 ± 0.2 |
| Maximum, ° | 2.5 ± 0.7 | 2.0 ± 0.5 | 1.3 ± 0.6 |
| Amber energies, kcal/mol | |||
| Total | −4,669 ± 78 | −4,689 ± 84 | −4,615 ± 85 |
| van der Waals | −335 ± 16 | −324 ± 15 | −354 ± 13 |
| Electrostatic | −5,234 ± 73 | −5,304 ± 94 | −5,157 ± 78 |
| rmsd to the mean coordinates, Å | |||
| N, Cα, C′ (125–228) | 0.84 | 0.82 | 0.96 |
| All heavy atoms (125–228) | 1.33 | 1.40 | 1.23 |
Except for the top two entries the average for the 20 conformers with the lowest residual Dyana target function values and the standard deviation among them are given.
Besides the addition of the aforementioned new routine for automatic NOE assignments, the presently used, so far unreleased new version of Dyana varies from the original program (20) by a modified implementation of the torsion angle dynamics algorithm (27) and by a different treatment of distance constraints to groups of chemical shift-equivalent protons and pairs of diastereotopic substituents that have not been individually assigned (T. Herrmann, P.G., and K.W., unpublished work). Since the main interest in the structures presented in this paper is focused on quite subtle, local conformational variations relative to the bPrP structures in the preceding paper (7) and the previously published structures of mPrP (2), hPrP (6), and shPrP (5), we reevaluated the input data and repeated the calculations of the previously published PrP structures from our laboratory (hPrP, mPrP) with the new version of Dyana. In this way we ensure that the structure comparisons in this and the preceding paper (7) are not influenced by systematic differences that might arise from the use of somewhat different protocols for data analysis and structure calculation.
Results
In a group of five single-residue variants of hPrP(121–230), which represent amino acid substitutions in the loop 166–172 and the helix 3 between the human, murine, and Syrian hamster PrPs (see Fig. 1 and the Introduction), we concluded from inspection of one- and two-dimensional 1H NMR spectra that the proteins hPrP(E219Q) and hPrP(I215V) showed at most minimal conformational differences relative to the wild-type protein. The remaining three proteins, hPrP(M166V), hPrP(S170N), and hPrP(R220K), were uniformly 13C,15N-labeled for a complete structure determination. Solutions containing 1 mM protein and 10 mM sodium acetate at pH 4.5 and 20°C were used for the structure determinations.
Resonance Assignment and Structure Determination.
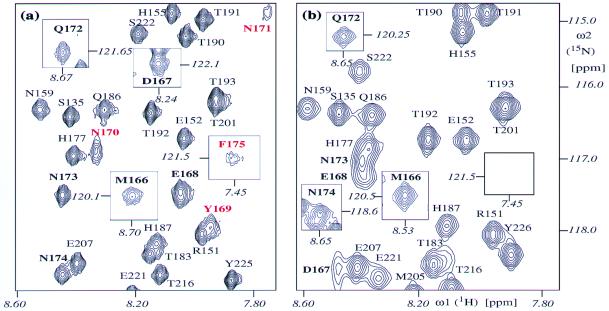
Sequence-specific NMR backbone assignments of the three aforementioned variant hPrP domains were obtained based on intraresidual and sequential connectivities of 15N, 1HN, and 13C resonances, using standard triple-resonance experiments (19). For hPrP(R220K) and hPrP(M166V) the polypeptide backbone assignments are nearly complete, the exceptions being the amide protons and amide nitrogens of Asp-167, Tyr-169, Ser-170, Asn-171, and Phe-175, for which no resonances were detected in any of the spectra recorded. For hPrP(S170N) all polypeptide backbone resonances were observed and assigned, including the amide nitrogens and amide protons of all of the residues in the loop 166–172 and of Phe-175 (Fig. 2a).
Figure 2.
Two-dimensional [15N,1H]-COSY spectra. (a) hPrP(S170N). (b) hPrP(121–230). A spectral region is shown that contains most of the resonances of the loop 166–172 in hPrP(S170N). The following notation is used for the peak assignments: Cross peaks detected in both proteins are black. Cross peaks present only in hPrP(S170N) are red. Cross peaks belonging to residues 166–175 are bold. The rectangular frames display cross peaks outside of the region shown here, with the chemical shifts of the peaks indicated in italics. The spectra were recorded at 600 MHz with 1 mM protein solutions at pH 4.5 and 20°C.
For each variant protein the same protocol has been used for the collection of the structural constraints and the determination of a bundle of 20 conformers that represent the NMR structure (for details see Materials and Methods). Table 2 gives a survey of the input of conformational constraints and the results of the structure calculations with the programs Dyana (20) and Opalp (23, 24). The small residual constraint violations show that the structures are consistent with the experimental constraints, and the spread among the bundles of 20 conformers described by the global rmsd values is representative of a high-quality structure determination.
Comparison of the hPrP(121–230) Variants with Wild-Type hPrP(121–230).
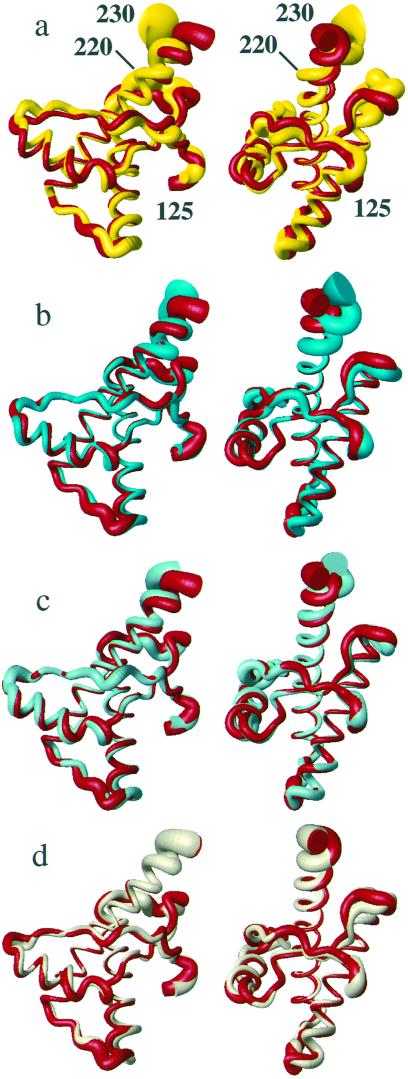
The three variant proteins have the same global fold as wild-type hPrP, which is in turn very similar to mPrP (2), shPrP (5), and bPrP (7). hPrP(121–230) and its presently used variants contain a short two-stranded antiparallel β-sheet with residues 128–131 and 161–164, α-helix 1 with residues 144–154, α-helix 2 with residues 173–194, and α-helix 3, which is somewhat variable in length and degree of definition within the segment 200–228. The N terminus comprising residues 121–125, the residues 166–172 that form a loop connecting the β-strand 2 with helix 2, the residues 190–200 that form the last turn of helix 2 and the loop connecting the helices 2 and 3, and the C terminus of helix 3 are noticeably less well defined than the remainder of the molecules (Fig. 3).
Figure 3.
Superposition of the polypeptide backbone from residues 125–230 in different PrP NMR structures for best fit of the backbone heavy atoms of residues 125–220. The radius of the cylindrical rods representing the polypeptide chains is proportional to the mean global backbone displacement per residue (35) among the 20 energy-minimized conformers used to represent the NMR structures. The views on the right were obtained from those on the left by a −90° rotation about a vertical axis. (a) hPrP(121–230) (red) and mPrP(121–231) (yellow). (b) hPrP(121–230) (red) and hPrP(R220K) (cyan). (c) hPrP(121–230) (red) and hPrP(M166V) (turquoise). (d) hPrP(121–230) (red) and hPrP(S170N) (amber). For these comparisons we recalculated the NMR structures of mPrP(121–231) (2) and hPrP(121–230) (6), using the same protocol as for the hPrP(121–230) variants (see Materials and Methods).
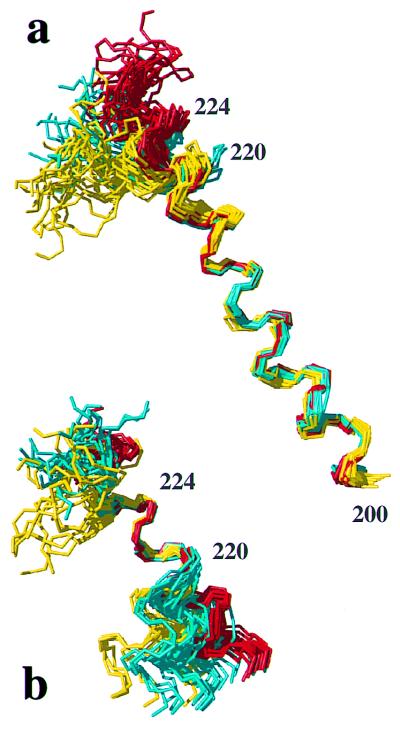
Within the framework of the preserved global structural scaffold, the three proteins show variations in the conformational markers of Table 1. Helix 3 is as well defined in hPrP(S170N) as in hPrP (Fig. 3d), and the differences between these two proteins fall within the conformation space spanned by the 20 conformers (Table 1). In hPrP(M166V) the helix 3 is well defined up to residue 226 but shows a slight deviation from the straight helix axis after residue 220 (Fig. 3c), which is manifested in an increase of the local rmsd value for the residues 200–226 in hPrP(M166V) when compared with hPrP (Table 1). In hPrP(R220K) the helix 3 is well characterized up to residue 219, but then it is less well ordered and shows a more pronounced deviation from the straight helix axis than hPrP(M166V), which is again clearly manifested in an increase of the rmsd value for residues 200–226 (Table 1). These observations in the calculated structures correlate with different experimental distance constraints within the segment of residues 217–224. In hPrP(R220K) some of the medium-range NOE distance constraints expected for a regular helix (28) are either missing or extremely weak, whereas essentially complete sets of constraints are present in hPrP, hPrP(M166V), and hPrP(S170N) (Table 3). Further insight into the conformational variability of helix 3 was obtained from comparison of the different proteins after superposition for best fit of different polypeptide segments (Fig. 4). It is readily apparent that the residues 221–225 form a quite regular α-helix structure in both hPrP and hPrP(R220K), but in hPrP(R220K) the helix 3 is less regular and slightly kinked at residues 218–220.
Table 3.
Medium-range NOE constraints in the C-terminal part of helix 3 in five PrP structures
| NOE connectivity | hPrP(121–230) | hPrP(M166V) | hPrP(S170N) | hPrP(R220K) | mPrP(121–231) |
|---|---|---|---|---|---|
| Hα217–HN/Hβ220 | w/m | w/m | m/m | w/m | −/w |
| Hα218–HN/Hβ221 | −/− | −/− | −/w | −/− | −/− |
| Hα219–HN/Hβ222 | w/m | w/m | m/s | −/− | −/− |
| Hα220–HN/Hβ223 | m/s | w/m | m/m | m/m | w/w |
| Hα221–HN/Hβ224 | w/m | m/m | m/m | −/m | −/w |
s, m, and w represent strong, medium, and weak NOEs. Absence of peak intensity at the (known) position for the NOE cross peak is indicated by a minus sign. The first entry refers to the dαN(i, i + 3) NOEs, the second one to the more intense of the dαβ(i, i + 3) NOEs (28).
Figure 4.
Comparison of helix 3 in hPrP(121–230) (red), mPrP(121–231) (yellow), and hPrP(R220K) (cyan) with the NMR structures represented by bundles of 20 conformers (28). (a) Polypeptide backbone of residues 200–229 superimposed for best fit of the residues 200–218. (b) Residues 213–229 superimposed for best fit of the residues 221–225.
For the second conformational marker there is a clear-cut qualitative difference among the three proteins. In hPrP(R220K) and hPrP(M166V) the [15N,1H]-COSY cross peaks for the residues 167, 169, 170, 171, and 175 were not observed, which is similar to previous observations in hPrP (6), whereas in hPrP(S170N) all of the cross peaks in the polypeptide segment 166–175 have been detected (Fig. 2).
Discussion
In the group of the four prion proteins from humans, cattle, mice, and Syrian hamsters the large number of species variations in the sequence locations 166–170 and 215–230 (Fig. 1) results in variability of the two conformational markers characterized in Table 1. Furthermore, as was previously pointed out by Billeter et al. (14), the propensity for intermolecular hydrogen bond formation in this surface area changes from species to species as a result of nonconservative exchange of polar and charged amino acids (Fig. 1). If this area of the PrP molecule is involved in intermolecular contacts that influence disease-related conformational transitions, as has been postulated from observations with transgenic laboratory animals (15), these structure variations could possibly provide a basis for rationalizing differences in the stringency of the species barrier for transmission of prion diseases between different combinations of mammalian species.
In Results it was shown that the three variants of hPrP differ in qualitative aspects from the wild-type protein. If we now consider the other species in Table 1, it is apparent that the variants of hPrP show similarities with either mPrP or shPrP. The amino acid substitutions M166V and R220K both result in significant changes in helix 3, with M166V affecting the straightness of the helix axis, and R220K affecting both the helix axis and the precision of the structure determination for helix 3 (Fig. 3 and Table 1). In the variants of hPrP this conformational marker thus changes from the structure type seen in hPrP, bPrP, and shPrP toward the mPrP structure. Although the individual amino acid exchanges in hPrP result in structures intermediate between hPrP and mPrP (Fig. 3, Table 1), the combined effects from M166V and R220K can account for the difference between hPrP and mPrP. The effect of the replacement S170N is particularly clear-cut, with the polypeptide segment 166–172 changing from the structure type seen for hPrP, bPrP, and mPrP to that reported for shPrP (Table 1). Both conformational markers do not correlate with the net charge of the peptide segments 166–172 and 215–230, which is −3 in hPrP and −1 in the other three species.
Overall, the present experiments demonstrate that the conformational markers in Table 1 can be related to specific amino acid exchanges between the different species. Since the molecular region considered here is not well defined in the three-dimensional structure (Fig. 3), we refrain from a detailed discussion of the nature of the nonbonding interactions that cause the observed local conformational changes, except to state that the residues 166 and 220 are clearly involved in anchoring the helix 3 against the loop 166–172 and the residues following the first strand of the β-sheet, respectively. It has previously been argued that the observation of rather narrow NMR signals of the residues 166–172 in shPrP is due to rapid exchange between different conformations (29). In this context it is intriguing that there are large variations of the chemical shifts between hPrP and hPrP(S170N) for those residues of this loop that can be observed in both proteins. This would be compatible with a large change, between the wild-type protein and the variant, of the relative populations in a dynamic equilibrium of two or multiple conformers. This same exchange process would presumably lead to broadening of some resonances in wild-type hPrP (6).
Considering the high incidence of species variations in the amino acid sequence of the presently considered molecular region (Fig. 1), it is not surprising that evidence has accumulated for its involvement in species-specific intermolecular interactions, including intermolecular recognition related to species barriers for TSEs. For example, the species barrier for transmissible mink encephalopathy between ferret and mink must relate to the only two residue variations between the two species, which are in the positions 175 and 220 (30), and the species barrier between human and rabbit has been related to the replacement of Asn-174 in hPrP by Ser (31). One of three polymorphisms in sheep PrP that have been associated with increased susceptibility to scrapie is located in the position corresponding to residue 168 in hPrP (32). A discontinuous epitope of residues 168, 172, 215, and 219 (numeration for hPrP) has been suggested to support binding to a conversion factor, “protein X,” which would mediate the transformation of PrPC to PrPSc (15, 33). Finally, the fragment 225–231 represents an epitope for a monoclonal antibody (R2) that strongly reacts with mPrP and shPrP, but not with hPrP and bPrP (34).
In conclusion, the presently described effects of single amino acid variations on static and dynamic aspects of the hPrP conformation (Table 1, Figs. 3 and 4) demonstrate that the local conformational differences between hPrP, bPrP, mPrP, and shPrP in the surface area of helix 3 and the loop 166–172 (Table 1) are significant, although they are located in a poorly ordered molecular region (Fig. 3). The data of Table 1 provide a frame of reference for future investigations on the structural basis of species-specific intermolecular recognition involving the PrPC isoform, and in particular for processes that relate to species barriers for transmission of prion diseases.
Acknowledgments
We thank Mrs. M. Geier for careful processing of the manuscript. Financial support was obtained from the Schweizerischer Nationalfonds (projects 31.49047.96 and 438+050287). Use of the computing facilities of the Competence Center for Computational Chemistry of the Eidgenössische Technische Hochschule Zürich is gratefully acknowledged.
Abbreviations
- PrP
prion protein
- PrPC
cellular form of PrP
- PrPSc
scrapie form of PrP
- hPrP(121–230)
fragment of human PrP comprising residues 121–230
- hPrP(M166V)
variant of hPrP(121–230) with Met-166 replaced by Val
- hPrP(S170N)
variant of hPrP(121–230) with Ser-170 replaced by Asn
- hPrP(R220K)
variant of hPrP(121–230) with Arg-220 replaced by Lys
- bPrP
bovine PrP
- shPrP
Syrian hamster PrP
- mPrP
mouse PrP, TSE, transmissible spongiform encephalopathy
- COSY
correlated spectroscopy
- rmsd
root-mean-square deviation
- NOE
nuclear Overhauser enhancement
- NOESY
NOE spectroscopy
Footnotes
Data deposition: Atomic coordinates for hPrP(M166V), hPrP(S170N) and hPrP(R220K) have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1E1G, 1E1J, 1E1P, 1E1S, 1E1U, and 1E1W).
References
- 1.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. Nature (London) 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 2.Riek R, Wider G, Billeter M, Hornemann S, Glockshuber R, Wüthrich K. Proc Natl Acad Sci USA. 1998;95:11667–11672. doi: 10.1073/pnas.95.20.11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K. FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 4.Donne D G, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E, Dyson H J. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Farr-Jones S, Ulyanov N B, Llinas M, Marqusee S, Groth D, Cohen F E, Prusiner S B, James T L. Biochemistry. 1999;38:5362–5377. doi: 10.1021/bi982878x. [DOI] [PubMed] [Google Scholar]
- 6.Zahn R, Liu A, Lührs T, Riek R, von Schroetter C, López-García F, Billeter M, Calzolai L, Wider G, Wüthrich K. Proc Natl Acad Sci USA. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.López García F, Zahn R, Riek R, Wüthrich K. Proc Natl Acad Sci USA. 2000;97:8334–8339. doi: 10.1073/pnas.97.15.8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schätzl H M, Da Costa M, Taylor L, Cohen F E, Prusiner S B. J Mol Biol. 1995;245:362–374. doi: 10.1006/jmbi.1994.0030. [DOI] [PubMed] [Google Scholar]
- 9.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weissmann C. FEBS Lett. 1996;389:3–11. doi: 10.1016/0014-5793(96)00610-2. [DOI] [PubMed] [Google Scholar]
- 11.Alper T, Cramp W A, Haig D A, Clarke M C. Nature (London) 1967;214:764–766. doi: 10.1038/214764a0. [DOI] [PubMed] [Google Scholar]
- 12.Griffith J S. Nature (London) 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 13.Prusiner S B. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 14.Billeter M, Riek R, Wider G, Hornemann S, Glockshuber R, Wüthrich K. Proc Natl Acad Sci USA. 1997;94:7281–7285. doi: 10.1073/pnas.94.14.7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 16.Zahn R, von Schroetter C, Wüthrich K. FEBS Lett. 1997;417:400–404. doi: 10.1016/s0014-5793(97)01330-6. [DOI] [PubMed] [Google Scholar]
- 17.Güntert P, Dötsch V, Wider G, Wüthrich K. J Biomol NMR. 1992;2:619–629. [Google Scholar]
- 18.Bartels C, Xia T, Billeter M, Güntert P, Wüthrich K. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 19.Bax A, Grzesiek S. Acc Chem Res. 1993;26:131–138. [Google Scholar]
- 20.Güntert P, Mumenthaler C, Wüthrich K. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 21.Luginbühl P, Szyperski T, Wüthrich K. J Magn Reson B. 1995;109:229–233. [Google Scholar]
- 22.Güntert P, Billeter M, Ohlenschläger O, Brown L R, Wüthrich K. J Biomol NMR. 1998;12:543–548. doi: 10.1023/A:1008391403193. [DOI] [PubMed] [Google Scholar]
- 23.Luginbühl P, Güntert P, Billeter M, Wüthrich K. J Biomol NMR. 1996;8:136–146. doi: 10.1007/BF00211160. [DOI] [PubMed] [Google Scholar]
- 24.Koradi R, Billeter M, Güntert P. Comput Phys Commun. 2000;124:139–147. [Google Scholar]
- 25.Cornell W D, Cieplak P, Bayly C I, Gould I R, Merz K M, Jr, Ferguson D M, Spellmeyer D C, Fox T, Caldwell J W, Kollman P A. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 26.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 27.Jain A, Vaidehi N, Rodriguez G. J Comp Phys. 1993;106:258–268. [Google Scholar]
- 28.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 29.James L J, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartz J C, McKenzie D I, Bessen R A, Marsh R F, Aiken J M. J Gen Virol. 1994;75:2947–2953. doi: 10.1099/0022-1317-75-11-2947. [DOI] [PubMed] [Google Scholar]
- 31.Loftus B, Rogers M. Gene. 1997;184:215–219. doi: 10.1016/s0378-1119(96)00598-7. [DOI] [PubMed] [Google Scholar]
- 32.Belt P B, Muileman I H, Schreuder B E, Bos-de Ruijter J, Gielkens A L, Smits M A. J Gen Virol. 1995;76:509–517. doi: 10.1099/0022-1317-76-3-509. [DOI] [PubMed] [Google Scholar]
- 33.Kaneko K, Zulianello L, Scott M, Cooper C M, Wallace A C, James T L, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williamson R A, Peretz D, Pinilla C, Ball H, Bastidas R B, Rozenshteyn R, Houghten R A, Prusiner S B, Burton D R. J Virol. 1998;72:9413–9418. doi: 10.1128/jvi.72.11.9413-9418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Billeter M, Kline A D, Braun W, Huber R, Wüthrich K. J Mol Biol. 1989;206:677–687. doi: 10.1016/0022-2836(89)90575-5. [DOI] [PubMed] [Google Scholar]