Abstract
Mycobacterium abscessus is an emerging rapidly growing mycobacterium (RGM) causing a pseudotuberculous lung disease to which patients with cystic fibrosis (CF) are particularly susceptible. We report here its complete genome sequence. The genome of M. abscessus (CIP 104536T) consists of a 5,067,172-bp circular chromosome including 4920 predicted coding sequences (CDS), an 81-kb full-length prophage and 5 IS elements, and a 23-kb mercury resistance plasmid almost identical to pMM23 from Mycobacterium marinum. The chromosome encodes many virulence proteins and virulence protein families absent or present in only small numbers in the model RGM species Mycobacterium smegmatis. Many of these proteins are encoded by genes belonging to a “mycobacterial” gene pool (e.g. PE and PPE proteins, MCE and YrbE proteins, lipoprotein LpqH precursors). However, many others (e.g. phospholipase C, MgtC, MsrA, ABC Fe(3+) transporter) appear to have been horizontally acquired from distantly related environmental bacteria with a high G+C content, mostly actinobacteria (e.g. Rhodococcus sp., Streptomyces sp.) and pseudomonads. We also identified several metabolic regions acquired from actinobacteria and pseudomonads (relating to phenazine biosynthesis, homogentisate catabolism, phenylacetic acid degradation, DNA degradation) not present in the M. smegmatis genome. Many of the “non mycobacterial” factors detected in M. abscessus are also present in two of the pathogens most frequently isolated from CF patients, Pseudomonas aeruginosa and Burkholderia cepacia. This study elucidates the genetic basis of the unique pathogenicity of M. abscessus among RGM, and raises the question of similar mechanisms of pathogenicity shared by unrelated organisms in CF patients.
Introduction
Mycobacteria form a group of over one hundred species, ranging from harmless saprophytic organisms to major human pathogens. The well known pathogenic species, such as Mycobacterium tuberculosis, Mycobacterium leprae and Mycobacterium ulcerans, belong to the subgroup of slowly growing mycobacteria (SGM). By contrast, rapidly growing mycobacteria (RGM) —almost 60 species of which have been identified—usually live in the soil or water and only rarely cause human infections [1]. Mycobacterium abscessus is one of the few RGM able to infect humans and is undoubtedly the most frequently isolated and the most difficult to combat [2].
M. abscessus was first described by Moore and Frerichs in 1953 [3]. These authors reported the isolation of a previously unknown mycobacterium from a human knee infection with subcutaneous abscess-like lesions (type strain M. abscessus ATCC 19977T), hence the name “abscessus”. With the recognition of Mycobacterium chelonei (now M. chelonae) in 1972, these two RGM organisms were classified as two subspecies of the same species. Over two decades, they were collectively designated “M. chelonae”, or even grouped with the RGM Mycobacterium fortuitum under the designation “M. fortuitum complex” [4]. It was only in 1992 that M. abscessus was separated from M. chelonae [5], and this separation soon resulted in the recognition that M. abscessus has a particular pathogenicity in humans [6]. Very recently, M. abscessus itself (now M. abscessus sensu lato) was shown to consist of three species: M. abscessus sensu stricto, M. massiliense and M. bolletii [7], [8]. These species are very closely related and cause a similar spectrum of human infections [9], [10]. Thus, hereafter, unless otherwise stated, they will be collectively referred to as “M. abscessus”.
Following its recognition as a distinct entity, and the development of molecular methods of identification for mycobacteria, M. abscessus has emerged as an important human pathogen over the last 10 years, causing many more cases of infection than M. chelonae and M. fortuitum—historically the most important pathogenic RGM [2], [11]. M. abscessus is responsible for more than 80% of all pulmonary infections due to RGM in the United States and is associated with a much higher fatality rate than any other RGM [6]. M. abscessus lung infection usually, but not exclusively, develops in subjects with underlying lung disorders (e.g. bronchiectasis, cystic fibrosis [CF]) [6]. The infection of CF patients is becoming a major issue: M. abscessus is being recovered with increasing frequency from CF patients, including young children. It causes a serious, life-threatening lung disease and is responsible for disseminated, often fatal infections following lung transplantation [12]–[18]. M. abscessus is also a leading cause of sporadic and epidemic cases of skin and soft-tissue RGM infections, following the use of contaminated syringes or needles, and after plastic or cardiac surgery [19], [20]. M. abscessus is not only pathogenic, it is also one of the most antibiotic-resistant RGM species [1]. It is resistant to most disinfectants and biocides and thrives in the most hostile environments—a feature associated with its propensity to cause outbreaks of healthcare-associated disease [1].
The pathogenicity of M. abscessus has been investigated in recent studies in various cell and mouse models. M. abscessus is an intracellular bacterium able to grow in macrophages and free-living amebas [8], [21]. M. abscessus infection in mice is associated with granulomatous lesions spontaneously evolving toward caseous lesions [22]. Interferon gamma (IFN-γ) and tumor necrosis factor (TNF) are the key cytokines of the murine host response, and are absolutely required to control infection [22]. Studies have also identified major differences in pathogenic profile between the two forms in which M. abscessus is isolated from humans: the S (smooth) form and the R (rough) form [21]. The R form lacks a surface polyketide compound, glycopeptidolipid (GPL) [23], [24], and causes more severe infections in mice, strongly inducing TNF secretion by macrophages [23].
Over the last decade, genomic studies have shown how the ecological and pathogenic characteristics of certain SGM have changed through evolution. For example, M. leprae, the causal agent of leprosy, represents a model case of adaptation through massive genome reduction [25]. Gene deletion and decay have resulted in the elimination from M. leprae of many of the major metabolic activities present in the closely related species, M. tuberculosis, the tubercle bacillus. This process of gene deletion is associated with the divergent evolution of M. leprae towards an obligate intracellular lifestyle. Other mycobacteria have acquired plasmid-borne virulence factors. The presence of a giant plasmid involved in the synthesis of a potent macrolide toxin forms the basis, for example, of the unique pathogenic properties of M. ulcerans, the causal agent of Buruli ulcer [26]. Genomic studies have also revealed how the deletion of large chromosomal regions led to the attenuation of Mycobacterium bovis bacillus Calmette-Guérin, the only vaccine against tuberculosis currently available [27], [28].
Very few genomic studies have been performed in the RGM group, and none has dealt with a “pathogenic” RGM. The first RGM to be sequenced—M. smegmatis—is a model mycobacterium widely used in research laboratories as a surrogate host for the expression of heterologous mycobacterial genes. The other RGM organisms sequenced (e.g., M. vanbaalenii) have been studied because they are able to degrade polycyclic aromatic hydrocarbons and are therefore of potential interest for use in environmental bioremediation [29]. We report here the complete genome sequence of M. abscessus (sensu stricto) and the insights it has provided into the genetic basis of its the pathogenicity of this bacterium, which is highly unusual among RGM. Whole-genome analysis not only revealed the presence of many “mycobacterial” virulence genes, but also showed that M. abscessus had a large series of specific genes in common with two pathogens most frequently isolated from CF patients—Pseudomonas aeruginosa and Burkholderia cepacia. These genes were presumably acquired from distantly related environmental bacteria via horizontal gene transfer (HGT).
Results
The M. abscessus genome
General features and comparison with other Mycobacterium species
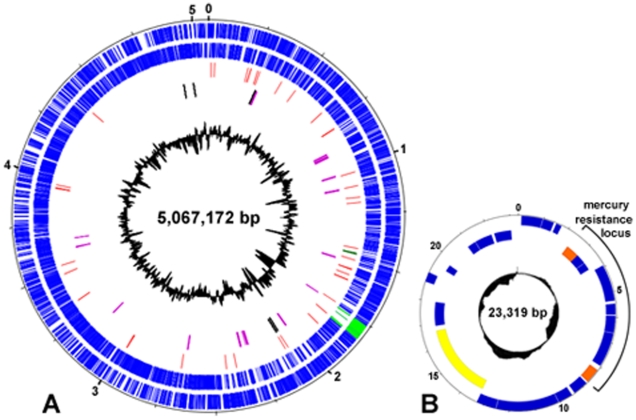
The M. abscessus (sensu stricto) genome consists of a circular chromosome of 5,067,172 base pairs (bp) including 4,920 predicted coding sequences (CDS) with a coding capacity of 93%, and a G+C content of 64% other than in the prophage region (59.5%) (Table 1 and Fig. 1A). A circular 23 kb mercury resistance plasmid (23,319 bp; G+C content, 68%) was also detected (Fig. 1B) and shown to have a nucleotide sequence 99.9% identical to that of the 23 kb pMM23 mercury resistance plasmid from Mycobacterium marinum (strain ATCC BAA-535) [30]. Like pMM23, the 23 kb M. abscessus plasmid carries a mercury resistance operon flanked by two genes encoding site-specific recombinases; it also encodes a relaxase/helicase that may function in conjugation or mobilization (Fig. 1B).
Table 1. General features of the M. abscessus genome and comparison with other Mycobacterium species.
| Features | RGM | SGM | ||||||
| Mabs | Msmeg(a) | M. gilvum (a) | Mvanba(b) | Mtb(c) | M. avium (a) | M. marinum (d) | M. ulcerans (e) | |
| Genome size, bp | 5,067,172 | 6,988,209 | 5,619,607 | 6,491,865 | 4,411,532 | 5,475,491 | 6,636,827 | 5,631,606 |
| G+C content, % | 64,1 | 67 | 67 | 67 | 65,6 | 68 | 65 | 65 |
| Protein coding, % | 93 | 90 | 92 | 91 | 90,8 | 88 | 89 | 72 |
| Proteins | 4920 | 6716 | 5241 | 5979 | 3959 | 5120 | 5424 | 4160 |
| tRNAs | 47 | 47 | 47 | 50 | 45 | 45 | 46 | 45 |
| rRNA operons | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 |
| Prophage elements, no. | 4(f) | NR | NR | NR | 2 | NR | 10 | 2 |
| IS, total no. of copies | 5 | 112 | NR | NR | 54 | NR | 7 | 302 |
[29].
[31].
[30].
[87].
Including the 81-kb full-length prophage and three prophage-like elements detailed in Table S1.
Abbreviations: RGM, rapidly growing mycobacteria ; SGM, slowly growing mycobacteria ; Mabs, M. abscessus ; Msmeg, M. smegmatis; Mvanba, M. vanbaalenii; Mtb, M. tuberculosis; nt, nucleotide; IS, insertion sequence; NR, not reported.
Figure 1. The M. abscessus CIP 104536T genome.
(A) Circular representation of the chromosome. The initiation codon for the dnaA gene was chosen as the starting point for numbering. The scale is in Mb. Moving inward, the first two circles show forward and reverse genes (blue lines); light-green lines indicate phage genes. The third circle shows tRNA genes (red) and rRNA operon (dark-green). The fourth circle shows genes presumably acquired “en bloc” from non mycobacterial organisms by HGT (purple) and IS elements (black). The inner black histogram represents the local G+C content (scale: 50% to 69%). (B) Circular representation of the 23-kb mercury resistance plasmid. The scale is in kb. Forward and reverse genes and the local G+C content are indicated with the same code as for the chromosome map. The plasmid carries a mercury resistance operon flanked by two genes encoding site-specific recombinases (MAB_p04c and MAB_p10, orange); it also encodes a relaxase/helicase that may function in conjugation or mobilization (MAB_p15c, yellow).
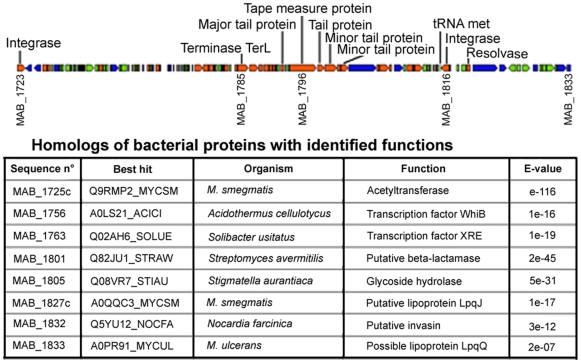
The M. abscessus chromosome is about 1.92 Mb smaller than the M. smegmatis genome; these two genomes are collinear, with no evidence of extensive rearrangements (Fig. S1). The M. abscessus chromosome includes 47 tRNA genes, but has a single ribosomal RNA operon, a feature of SGM genomes [31]. It contains a full-length prophage (81 kb) resembling the members of a recently characterized group of dsDNA tailed mycobacteriophages [32]. This prophage is integrated into a Met tRNA and contains 112 CDS, 8 (7.1%) of which are similar to bacterial proteins with identified functions (Fig. 2). There are also three prophage-like elements (Table S1). Unlike other sequenced mycobacteria, M. abscessus has very few insertion sequences (IS) in its genome: there are only five IS, each present as a single copy (Table 1). These elements include the composite element ISMab1 [33], which is probably part of an integrated plasmid (gene encoding a putative plasmid replication initiator protein in its vicinity [MAB_2100]).
Figure 2. The M. abscessus full-length prophage.
Each arrow represents a predicted protein-coding gene (length approximately to scale). Orange, similar to other phage proteins; blue, similar to other bacterial proteins; green, hypothetical protein. The table shows homologs of bacterial proteins with identified functions (Uniprot Blast search).
Functional information
It was possible to assign a biological function to 60.5% of the CDS on the M. abscessus chromosome; 27.5% were found to be conserved hypothetical proteins and 12% were unique. The distributions of M. abscessus and M. smegmatis proteins, according to the Kegg classification, were different, with a lower proportion of M. abscessus proteins involved in xenobiotic biodegradation and metabolism (p<10−4, chi-squared test), and in biosynthesis of secondary metabolites (p = 0.02) (Table S2). Consistent with the smaller size of the M. abscessus genome, most paralogous families were found to be underrepresented in M. abscessus with respect to M. smegmatis (Table S3). This was particularly true for paralogs involved in the adaptation of microorganisms to diverse environments (e.g. ABC transporters, two-component sensor histidine kinases). Most of the small number of protein families found to be overrepresented in M. abscessus are known to be associated with mycobacterial pathogenicity (e.g. PE and PPE proteins, MCE and YrbE proteins, lipoprotein LpqH precursors, lipases/esterases/monooxygenases). Others, such as the members of the ArsC family, salicylate hydroxylases and cysteine desulfurases, are hallmarks of organisms living in soil or water.
Transfers of blocks of genes from non mycobacterial environmental organisms
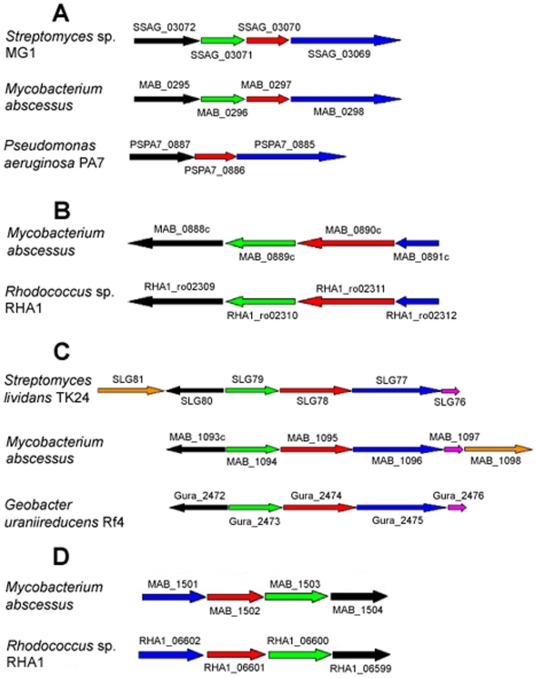
We detected 17 gene clusters syntenic with regions from non mycobacterial organisms, and which are absent from other sequenced mycobacterial species (Tables 2 and S4), suggesting multiple en bloc HGTs. The factors encoded by these gene clusters are much more similar to proteins from syntenic organisms than to proteins from other mycobacteria, or have no significant mycobacterial homologs (Table S4). The hypothesis of multiple HGT of blocks of genes is also supported by significant differences in codon usage for proline and arginine between this subset of genes and other M. abscessus genes (p<10−4, chi-squared test). The organisms with the best conserved syntenies are all environmental bacteria with a high G+C content, mostly actinobacteria (Rhodococcus sp., Streptomyces sp., Nocardia sp.), but also Pseudomonas sp. and Burkholderia sp. (Tables 2 and S4). There are more horizontally acquired gene clusters in the 5′ half of the genome than in the 3′ half (Fig. 1), with most concentrated into two “hot-spots” (MAB_0888c-1098, MAB_2027-2286). Examples of such gene clusters are shown in Fig. 3.
Table 2. Gene clusters syntenic with non mycobacterial organisms and not observed in other mycobacteria(a).
| Cluster no. | Mabs CDS. | Main synteny(b) | Putative function |
| 1 | MAB_0295-0298(c) | Streptomyces sp. MG1 (B4V5P4_9ACTO)(d) | Phenazine biosynthesis |
| 2 | MAB_0300c-0303 | Nocardia farcinica (Q5YP17_NOCFA) | Resistance to salicylic acid-mediated defense mechanisms |
| 3 | MAB_0888c-0891c | Rhodococcus sp. (Q0SEC1_RHOSR)(e) | Homogentisate catabolic pathway |
| 4 | MAB_0899c-0911(f) | Nocardia farcinica (Q5YXU1_NOCFA)(g,h) | Phenylacetic acid degradation |
| 5 | MAB_1014c-1019c | Rhodococcus sp. (Q0S4M6_RHOSR) | Unknown |
| 6 | MAB_1093c-1098 | Streptomyces lividans (Q460H9_STRLI)(i) | DNA degradation (dnd locus) |
| 7 | MAB_1501-1504 | Rhodococcus sp. (strain RHA1) (Q0S261_RHOSR)(j) | Iron uptake |
| 8 | MAB_1720-1722 | Rhodococcus sp. (Q0S6K3_RHOSR) | Two-component system |
| 9 | MAB_2027-2032 | Pseudomonas putida F1 (A5W4P6_PSEP1) | Biosynthesis of phytotoxic peptides and antibiotics |
| 10 | MAB_2251-2253 | Burkholderia cepacia complex (Q0B890_BURCM) | Unknown |
| 11 | MAB_2255-2257(k). | Myxococcus xanthus (Q1D6A2_MYXXD) | Polyketide biosynthesis |
| 12 | MAB_2257-2258(l,m) | Streptomyces ambofaciens (A3KI34_STRAM) | Polyketide biosynthesis |
| 13 | MAB_2278-2286(n). | Streptomyces coelicolor (Q9K3F5_STRCO) | Unknown |
| 14 | MAB_2610-2613 | Bacillus pumilus (B4AMJ6_BACPU) | Carbohydrate transport |
| 15 | MAB_3112-3115 | Nocardia farcinica (Q5YN04_NOCFA) | Unknown |
| 16 | MAB_3569c-3574c(o ,p) | Streptomyces antibioticus (Q0R4L5_STRAT) | Biosynthesis of secondary metabolites |
| 17 | MAB_3621c-3623 | Rhodococcus sp. (Q0SA51_RHOSR)(q) | Taurine metabolism |
Clusters comprising ≥3 syntenic genes.
The organism and the homolog of the 5′ M. abscessus gene product (in brackets, entry name in Swiss-Prot/TrEMBL database) are indicated.
Upstream, MAB_0292c is homologous to an ISX08 transposase from Saccharopolyspora erythraea.
Also A6UZN8_PSEA7-A6UZN6_PSEA7 from Pseudomonas aeruginosa (strain PA7) (see also Fig. 3).
Partial synteny (MAB_0888c-0890c) in B. cepacia.
Downstream, MAB_0920 is homologous to a phenylacetic acid-responsive trancriptional repressor gene from Kineococcus radiotolerans.
Also Q0SCR6_RHOSR–Q0SCS7_RHOSR from Rhodococcus sp. (strain RHA1).
Partial synteny (MAB_0906-0910) in B. cepacia.
Also A5G4D2_GEOUR- A5G4D6_GEOUR from Geobacter uraniireducens (strain Rf4) (see also Fig. 3)
Also syntenic regions in B. cepacia and in the pathogens Salmonella Paratyphi A, Salmonella Typhimurium, Salmonella Typhi, and Burkholderia mallei.
Just upstream, MAB_2254c is homologous to a PPE protein from Mycobacterium vanbaalenii.
This cluster has only two genes but is part of a composite region encoding various polyketide synthases (see upstream, MAB_2255-MAB_2257).
Just downstream, MAB_2259 is homologous to a putative O-methyltransferase gene from Myxococcus xanthus.
MAB_2284 (homologous to Q9K3G5) is also homologous to the protein PrnC gene from Burkholderia cepacia.
Except MAB_3570c, which is homologous to a 4′-phosphopantetheinyl transferase gene from the Actinomycetales (A3R4S1_9ACTO).
MAB_3574c is also homologous to a 3-oxoacyl-[acyl-carrier-protein] synthase III from Frankia alni.
Partial synteny (MAB_3621c-MAB_3622c) in B. cepacia.
Abbreviations: Mabs, M. abscessus; CDS, coding sequence.
Figure 3. Examples of gene blocks presumably inherited from non mycobacterial organisms.
(A) MAB_0295-0298 (phenazine biosynthesis). (B) MAB_0888c-0891c (homogentisate catabolism). (C) MAB_1093c-1098 (DNA degradation [dnd locus]). (D) MAB_1501-1504 (iron uptake). Genes are drawn approximately to scale and are indicated according to their names in the Embl-Ebi database.
Virulence factors involved in intracellular parasitism
Many factors known to be involved in M. tuberculosis virulence have orthologs in the M. abscessus genome and best hits with proteins from other mycobacteria (Table S5). In addition to these “mycobacterial” factors, the M. abscessus genome encodes a number of “non mycobacterial” factors known to play a major role in microbial pathogenesis.
“Mycobacterial” factors
The PE- PPE and ESAT-6 families
The PE and PPE proteins, with their characteristic proline-glutamate (PE), or proline-proline-glutamate (PPE) N-terminal motifs, are often found associated with ESX gene clusters, which encode ATP-dependent specific secretion systems and are named after the 6 kDa early secretory antigenic target ESAT-6 [31], [34], [35]. There are three PE and six PPE genes in M. abscessus, and three ESX loci, all similar to the essential and highly immunogenic ESX-3 gene cluster of M. tuberculosis.
MCE and yrbE proteins
MCE (mammalian cell entry) proteins allow mycobacteria to invade host cells [36]. There are seven mce operons in M. abscessus and only four in M. smegmatis, one of which is interrupted by an IS element in M. smegmatis (not shown). It has recently been suggested that the number of mce operons may be related to pathogenicity in actinomycetes: there are six mce operons in Nocardia farcinica, one of the agents causing nocardiosis, whereas Streptomyces avermitilis and Streptomyces coelicolor, both nonpathogenic soil bacteria, each have only one copy of the mce operon [37].
LpqH-like proteins
LpqH, also known as the 19 kDa protein, is an immunodominant antigen recognized by T cells and sera from patients with tuberculosis [38]. M. abscessus possesses four genes encoding LpqH-like proteins, scattered throughout the genome, suggesting that these molecules may be involved in the pathogenicity of M. abscessus, possibly through modification of the host response.
Regulators of virulence factors
M. abscessus has homologs of a large number of regulators known to control virulence factors in M. tuberculosis. Most appear to have counterparts in M. smegmatis, but there are a few exceptions, consistent with the specialization of M. abscessus towards pathogenicity. For example, M. abscessus possesses homologs of the five sigma factors shown to be involved in M. tuberculosis virulence (SigA, SigC, SigD, SigE, SigH), whereas M. smegmatis has homologs of only four of these factors (SigA, SigD, SigE, SigH). M. abscessus also has a protein homologous to the VirS virulence transcription factor of M. tuberculosis, whereas M. smegmatis does not.
“Non mycobacterial” factors
Phospholipase C
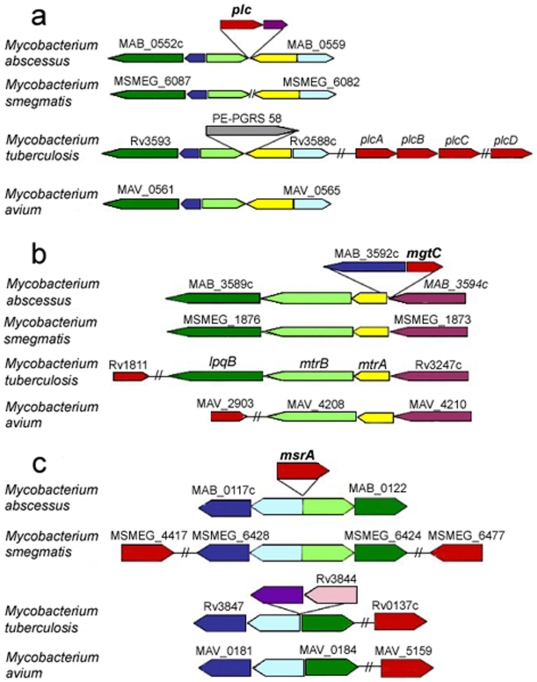
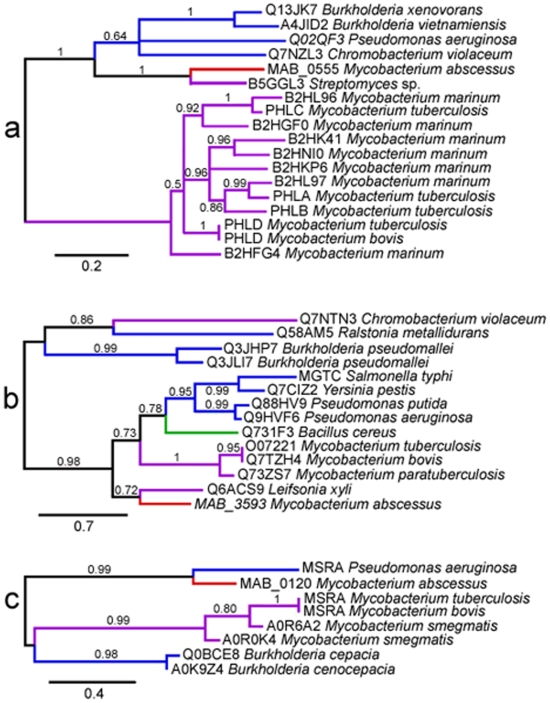
Bacterial phospholipases C are key virulence factors allowing intracellular pathogens to escape phagosomal vacuoles by disrupting eukaryotic membranes [39]. M. tuberculosis has four phospholipase C-encoding genes: plcABC and plcD [31]. Triple (plcABC) and quadruple (plcABCD) mutants have negligible enzyme activity and attenuated virulence in mouse models, suggesting that phospholipase C activity is required for the growth of mycobacteria in vivo [40]. Phospholipase C activity may be particularly critical for mycobacteria infecting human hosts, as suggested by the presence of the region encompassing plcABC in clinical Mycobacterium microti isolates, but not in attenuated isolates from voles [41]. The M. abscessus phospholipase C closely resembles proteins from Streptomyces sp., Chromobacterium violaceum and P. aeruginosa. The locus containing the corresponding gene differs from those of the phospholipase C genes in M. tuberculosis (Fig. 4), and phylogenetic analysis is also consistent with horizontal acquisition from non mycobacterial organisms (Fig. 5).
Figure 4. plc, mgtC and msrA loci in M. abscessus: comparison with other mycobacteria.
a. plc: note that MAB_0557 (transcriptional regulatory protein AraC) has no homolog at the counterpart of the M. abscessus plc locus in the other mycobacterial species, suggesting an insertion; also note the substitution for a PE-PGRS gene in the corresponding M. tuberculosis region. b. mgtC: note that the gene encoding MgtC is located at other genomic sites in M. tuberculosis and M. avium; also note that MAB_3592c (probable chain fatty acid-CoA ligase, blue) has no homolog at the counterpart of the M. abscessus mgtC locus in the other mycobacterial species, also suggesting an insertion. c. msrA: note that MsrA-encoding genes are located at other genomic sites in other mycobacteria; and the presence of sodA (light blue) upstream of M. abscessus msrA; there is a substitution for a transposase gene (Rv3844) in the corresponding M. tuberculosis region. Out of scale.
Figure 5. Phylogenetic trees (maximum likelihood) of PlC (a), MgtC (b) and MsrA (c) proteins.
Branch supports values are indicated at the nodes. Branch colors indicate proteins from M. abscessus (red), Actinobacteria (purple), Proteobacteria (blue) and Firmicutes (green). Labels at the leaves show the Uniprot identifier of the proteins and the species they belong to.
MgtC
Intracellular pathogens make use of MgtC proteins to increase intracellular Mg2+ concentration, facilitating their survival within cells. As first demonstrated in Salmonella, these proteins are essential for bacterial growth within professional phagocytes, such as macrophages [42]. M. tuberculosis has an mgtC gene, the disruption of which strongly attenuates virulence in both macrophages and mice [43]. The acquisition of mgtC genes by HGT is common among microbes and has been associated with pathogenicity [44]. There is an mgtC gene in M. abscessus, but not in M. smegmatis, providing further evidence that this gene is important for the intracellular lifestyle developed by M. abscessus. The M. abscessus mgtC gene seems to have been acquired by HGT, probably from actinobacteria (Fig. 4 and 5) [44].
MsrA
The peptide methionine sulfoxide reductase (MsrA) of M. tuberculosis is thought to protect this organism against the oxidative damage caused by the reactive nitrogen intermediates produced by macrophages [45]. The M. abscessus msrA gene is located at another genomic site, in the immediate vicinity of sodA (Fig. 4). The MsrA protein of M. abscessus closely resembles a protein from Rhodococcus sp. and does not cluster with mycobacterial proteins in the phylogenetic tree for MsrA (Fig. 5), providing further evidence for horizontal acquisition from a non mycobacterial gene pool.
Iron uptake
Bacterial pathogens have developed a number of systems for acquiring iron, which is present in limiting concentrations in living hosts. M. abscessus has a four-gene cluster very similar to a locus encoding an ABC Fe(3+) transporter in Rhodococcus sp. This cluster also has closely related homologs in a number of major bacterial pathogens, including Salmonella sp. and Burkholderia sp. (Tables 2 and S4; Fig. 3D).
“Non mycobacterial” factors relevant to the infection of CF patients (Tables 2 and S4; Fig. 3)
Phenazine biosynthesis
Phenazines are secondary metabolites with broad-spectrum antibiotic activity against bacteria, fungi and parasites, produced by Pseudomonas sp. and Streptomyces sp. Pyocyanin, the blue phenazine synthesized by P. aeruginosa, is a key virulence determinant in vivo, and is thought to be involved in the persistence of this pathogen in CF patients [46].
Homogentisate catabolism
The homogentisate catabolic pathway results in homogentisate being broken down into two compounds of central metabolism: fumarate and acetoacetate [47]. However, homogentisate is also a precursor for the biosynthesis of pyomelanin, a brown pigment generated through the extracellular accumulation and polymerization of homogentisate. Pyomelanin production appears to be a key step in the process by which P. aeruginosa adapts to the respiratory tract of CF patients, possibly facilitating iron acquisition [48].
Phenylacetic acid degradation
The region involved in phenylacetic acid degradation—the largest region of the M. abscessus genome acquired by HGT—is located downstream from the “homogentisate catabolism” region. The proteins encoded by this region constitute a complex functional unit, the phenylacetyl-CoA catabolon, which transforms various aromatic compounds (e.g., styrene, 2-phenylethylamine) into phenylacetyl-CoA, which is subsequently catabolized into TCA intermediates [49]. The five-gene cluster putatively encoding the enzymes of the ring-hydroxylating complex (MAB_0906-0910) is also present in B. cepacia and the homolog of MAB_0910 is essential for B. cepacia survival in a rat model of chronic lung infection [50].
Cysteine desulfurases and the “Dnd phenotype”
The M. abscessus genome encodes a large number of cysteine desulfurases (Table S3). One is part of a locus closely resembling the S. lividans dndA-E locus (Tables 2 and S4; Fig. 3), which is involved in the Dnd (DNA degradation) phenotype observed in vitro during DNA extraction [51]. A recent pulsed-field gel electrophoresis study showed that M. abscessus strains with a Dnd phenotype belonged to a small number of closely related clones playing a major role in human disease [52]. The sputum of CF patients is extremely rich in DNA, which may constitute an important source of nutrients for dnd-positive strains.
Resistance to antimicrobial compounds
M. abscessus has achieved notoriety as one of the most drug-resistant mycobacterial species [1]. Much of this multidrug resistance may result from weak permeability of the cell wall, but analysis of the M. abscessus genome has also revealed the presence of many potential drug resistance determinants. The hydrolytic or drug-modifying enzymes present in this species include an Ambler class A beta-lactamase, a rifampin ADP-ribosyl transferase, an aminoglycoside 2′-N-acetyltransferase and at least 12 homologs of aminoglycoside phosphotransferases. We also found four homologs of monooxygenases potentially involved in resistance to rifampin and tetracyclines, two FolP homologs conferring resistance to cotrimoxazole, one homolog of UDP-N-acetylglucosamine 1-carboxyvinyltransferase MurA conferring resistance to fosfomycin, and two homologs of 23S rRNA methylases conferring resistance to macrolides, including the erm gene product (MAB_2297) recently shown to be involved in inducible macrolide resistance in M. abscessus [53]. Most of these drug resistance determinants are mycobacterial, with the notable exception of the Ambler class A beta-lactamase (MAB_2875), which closely resembles beta-lactamases from the gram-negative bacteria Pseudomonas luteola and Serratia fonticola (not shown). The genome of M. abscessus also encodes many proteins potentially involved in drug-efflux systems, including members of the major facilitator family, ABC transporters and MmpL proteins. Finally, the presence of a single rRNA operon favors the occurrence of dominant mutations conferring resistance to aminoglycosides and macrolides [54].
We identified three putative ars operons scattered over the chromosome of M. abscessus [55]. M. abscessus is therefore likely to be resistant to high concentrations of arsenic [56]. Finally, due to the presence of merB within the mer operon, the 23 kb plasmid probably confers resistance to a wide range of organomercury compounds [57].
Discussion
Deciphering the ecology and biology of M. abscessus
The genetic information contained in the genome of M. abscessus tells us a great deal about the lifestyle of this microorganism in natural conditions. The presence of a large number of genes and operons involved in resistance to arsenic or encoding cysteine desulferases is clearly a hallmark of an environmental organism living in soil or aquatic environments. However, M. abscessus also contains a whole series of genes known to be involved in intracellular survival (e.g., mgtC, msrA, plc), and is well-equipped to obtain energy from the degradation of eukaryotic host-derived lipids (numerous lipase-encoding genes), as observed for mycobacteria adapted to an intracellular lifestyle [58]. The low level of metabolic versatility (e.g., far fewer ABC transporters or two-component sensor histidine kinases than M. smegmatis) suggests that this bacterium tends to specialize in intracellular parasitism.
The most plausible hypothesis is that M. abscessus has evolved to escape predators, such as free-living amebas [8] sharing the same ecosystem. Soil-dwelling amebas are known to be most abundant at plant-soil interfaces, because these interfaces support the growth of various plant parasites, including bacteria, on which amebas feed [59]. Consistent with this hypothesis, the genome of M. abscessus encodes a particularly large number of salicylate hydroxylases, enabling this bacterium to resist the salicylic acid-mediated defense mechanisms of plants [60]. This suggests that M. abscsessus lives in close contact with plants and therefore has to deal with amebas. This hypothesis may explain an extraordinary paradox in the epidemiology of M. abscessus: despite all the evidence to suggest that M. abscessus lives in soil and water—our own genomic data and the large number of epidemics linked to the direct or indirect use of non sterile water—this bacterium is detected much less frequently in such environments than other closely related RGM, such as M. chelonae [61].
We analyzed an S phenotype strain. A major challenge for the future will be to determine the role of S↔R switches in the natural lifecycle of M. abscessus (controlling whether this bacterium grows in the form of a biofilm) and its interaction with its hosts, including humans (modulation of the host response). GPL may be required for biofilm establishment or for escape from amebas in aquatic environments [62], but seems to hinder the development of infection, probably by acting as a target of the specific immune response of the host [22]. We recently reported the in vivo isolation of an R variant from the type strain CIP 104536T [22]. Transcriptomic studies are currently underway to determine the mechanisms responsible for the loss of GPL production in this R variant and the associated events potentially accounting for its “hypervirulence” in mice. The data obtained should make it possible to identify the external signals involving in triggering the switching process.
Evolutionary mechanisms
This study highlights the major role of horizontal gene transfers in the evolution of RGM. It is hardly surprising that this evolutionary mechanism, which has also been described in SGM [63], [64], is particularly important in RGM, and that it involves a reservoir of genes from different bacteria with a high G+C content widely present in soil or water, such as Streptomyces sp., Rhodococcus sp. and pseudomonads. RGM come into contact with many other bacteria in the environment—often as part of a biofilm [65] —and they may exchange genetic material with these other bacteria [66]. Mycobacteriophages—or other bacteriophages with a wide host spectrum—may play a key role in such transfers, as they display extensive mosaicism, combining viral and bacterial genes in a vast gene pool [32]. Such a role in gene transfer is consistent with the presence of a full-length prophage sequence containing non mycobacterial genes in the M. abscessus genome. However, the presence of this prophage sequence does not exclude a role for other genetic vectors, such as plasmids, which are frequently found free or integrated into the genome within RGM [67].
The demonstration that pathogenicity genes of non mycobacterial origin are present in M. abscessus raises questions about the timing of their acquisition. The fact that the closely related species M. chelonae is also pathogenic in humans—an exceptional feature among RGM—strongly suggests that many of these genes were acquired before the separation of these two species. We are currently carrying out a comparative genomics study of M. abscessus and M. chelonae (http://www.genoscope.cns.fr/spip/Mycobacterium-chelonae-and.html), which should make it possible to confirm or to infirm this hypothesis. We have also recently made use of the genome sequence of M. abscessus to develop a multilocus sequence typing (MLST) approach. Our preliminary analyses on more than a hundred M. abscessus (sensu lato) strains suggest that there are three highly homogeneous groups, corresponding to the three previously described species (M. abscessus sensu stricto, M. massiliense, M. bolletii), with less than 1% divergence within groups and around 2% divergence between groups. The species of M. abscessus sensu lato therefore seem to have emerged relatively recently. However, it should be stressed that most of the strains of M. abscessus available from collections were isolated recently and mostly in a clinical context. Indeed, as stated above, M. abscessus is only very rarely isolated from the environment. There may therefore be a bias in the results, because we cannot rule out the possibility that strains capable of infecting humans constitute an unusual subpopulation.
One of the findings of this study was entirely unexpected: the presence of a mercury resistance plasmid almost identical to the pMM23 from the M. marinum strain recently sequenced by the team of Stinear (strain ATCC BAA-535) [30]. The pMM23 plasmid discovered in this strain, isolated from a patient in 1992 (Moffett Hospital, San Francisco), is exceptional in M. marinum, as none of the more than 40 other isolates of this species studied by the team of Stinear has been found to carry this plasmid [30]. The presence of this plasmid in M. abscessus is, thus, particularly interesting, as it demonstrates that exchanges may occur between M. marinum, a SGM, and M. abscessus, a RGM, either directly or via another organism, probably a mycobacterium. It also suggests that M. abscessus and M. marinum may live in the same ecosystems and may be transmitted to humans by similar mechanisms. Future work should determine the prevalence of this plasmid in M. abscessus and should assess whether this plasmid constitutes a useful marker (e.g., for epidemicity).
We were also surprised by the very low frequency of IS in the genome of the strain of M. abscessus that we sequenced, much lower than usually found in mycobacteria. Confirmation of this result is required, with a representative panel of isolates. If confirmed, this characteristic would have a major impact on the plasticity of the genome of M. abscessus. As elegantly demonstrated in Escherichia coli, reducing the number of IS elements renders bacterial genomes more stable, with a greater capacity for acquiring foreign DNA [68].
Key factors shared with other major CF pathogens
This study provides new insight into the emergence of M. abscessus as a pathogen in CF patients. We were surprised to discover that the largest tranferred regions detected in M. abscessus contained genes involved in the metabolism of aromatic compounds. Such systems are characteristic of pseudomonads in general, and of two major CF pathogens, P. aeruginosa and B. cepacia, in particular [49]. This implies that M. abscessus is able to live in the same ecosystems as P. aeruginosa and B. cepacia, with patients becoming infected from the same microbial reservoir. Another, not necessarily exclusive possibility is that these metabolic characteristics provide a selective advantage in CF patients, due either to their illness or the treatments increasingly used over recent years, such as aerosolized drug administration [69]. According to this hypothesis, M. abscessus may benefit from factors promoting its extracellular development and its implantation in the bronchial tract, before going on to cause deeper infection of the pulmonary parenchyma and ganglions.
Conversely, several M. abscessus factors typical of intracellular parasites are also present in P. aeruginosa and B. cepacia, the most notable examples being phospholipase C and the MgtC protein [70], [71]. Both P. aeruginosa and B. cepacia produce two phospholipases C and two MgtC proteins [70], [71]. An MgtC-like protein is also found in Aspergillus fumigatus—the main pathogenic fungus in CF patients—but not in closely related nonpathogenic species such as Aspergillus nidulans. Pseudomonads and other related organisms infecting CF patients have previously been considered to be exclusively “extracellular” pathogens. Our data raise questions about the interaction of these organisms with macrophages or other monocyte-derived cells in CF patients. This is consistent with the finding that the production of MgtC is required for the survival of Burkholderia cenocepacia–the main B. cepacia complex pathogen infecting CF patients–within macrophages [72], [73].
A recent analysis of the genomes of various CF and non-CF P. aeruginosa isolates revealed mosaic structures, consisting of a conserved core component interrupted by strain-specific genomic islands acquired by HGT, which seem to provide CF isolates with specific metabolic pathways involved in infection [74]. The identification of multiple episodes of HGT in M. abscessus strongly suggests that a similar evolutionary trend occurs within RGM. Along the same lines as the studies carried out in P. aeruginosa by the team of Lowry [74], comparative genomic studies of CF and non-CF M. abscessus isolates could prove particularly fruitful for elucidating the tropism of certain organisms for the respiratory tract of CF patients, opening up promising new possibilities for the control of microbial infections in CF patients.
Materials and Methods
We sequenced M. abscessus (sensu stricto) CIP 104536T ( = ATCC 19977T), using a whole-genome shotgun strategy (EMBL accession numbers: CU458896, chromosome; CU458745, plasmid). This strain is of the S phenotype, and can switch in vivo to an R phenotype [22]. Mycobacteria were grown in Middlebrook 7H9 broth supplemented with Tween 80. M. abscessus DNA, prepared using standard methods, was manipulated in the presence of 50 µM thiourea (DNA in solution) or by replacing Tris buffer by HEPES at the same molarity (DNA in plugs), to prevent Tris-dependent DNA degradation [39]. We constructed three genomic libraries (inserts of 3–4, 8–10 and ∼20 kb, respectively) and generated ∼80,000 sequences (50,000, 20,000 and 10,000 sequences, respectively, giving 11-fold coverage). Putative protein-coding sequences were predicted by SHOW (http://migale.jouy.inra.fr/outils/select_mig_outils_zpt), tRNA genes by tRNAscan, and rRNA genes by RNAmmer [75], [76]. Sequences were analyzed with the BIOFACET package and the BLAST software suite [77], [78]. General features, such as G+C content (%), were assessed with ARTEMIS software [79]. The origin of replication was identified with ORILOC [80]. The circular representations of chromosome and plasmid were generated with DNAPlotter (http://www.sanger.ac.uk/Software/Artemis/circular). The M. abscessus full-length prophage was drawn with BugView (http://www.gla.ac.uk/~dpl1n/BugView/index.html). Whole genome dotplot comparison of M. abscessus versus M. smegmatis was drawn with Gepard (http://mips.gsf.de/services/analysis/gepard). CLUSTER-C was used to cluster genes into paralogous families [81]. Alien Hunter was used to screen the genome for regions with “atypical” sequence content [82]. Transfers of blocks of genes from non mycobacterial organisms were identified as follows. We first identified CDS more similar to proteins from non mycobacterial organisms than to mycobacterial proteins (no mycobacterial protein among the 50 best hits). We then used GeneTeam, with a delta value of 3 and visual inspection to search for areas of synteny with relevant non mycobacterial organisms [83]. Only clusters with at least 3 syntenic genes not found in other sequenced mycobacteria were retained. Phylogenetic analyses were carried out with the “Phylogeny.fr” web server (http://www.phylogeny.fr), using Muscle for multiple alignment and GBlocks for alignment curation, and constructing the phylogenetic trees with PhyML [84], [85]. Branch supports were calculated with the approximate likelihood ratio test [86]. Distributions of M. abscessus and M. smegmatis proteins, according to the Kegg classification, were compared using chi-squared tests with continuity correction. To account for multiple testing, p-values were corrected according to Hochberg's method. Differences were considered as statistically significant if corrected p-values were <0.05.
Supporting Information
Whole genome dotplot comparison of M. abscessus (horizontal axis) versus M. smegmatis.
(0.08 MB TIF)
M. abscessus prophage-like elements
(0.03 MB DOC)
M. abscessus and M. smegmatis proteins involved in metabolism, according to the Kegg classification
(0.04 MB DOC)
A selection of paralogous families
(0.06 MB DOC)
Proteins encoded in the 17 horizontally acquired gene clusters, and their syntenic non mycobacterial homologs
(0.06 MB DOC)
(0.06 MB DOC)
Acknowledgments
We thank Veronique Vincent and Cristina Gutierrez (Institut Pasteur, Paris, France) for kindly providing us with strain CIP 104536T, and Raphaël Porcher, Alain Billault and Isabelle Sénégas for assistance. We also thank Stewart Cole for helpful comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work received financial support from the association “Vaincre la Mucoviscidose”. The funders (Vaincre la Mucoviscidose) had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brown-Elliott BA, Wallace RJ., Jr Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin Microbiol Rev. 2002;15:716–746. doi: 10.1128/CMR.15.4.716-746.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 3.Moore M, Frerichs JB. An unusual acid-fast infection of the knee with subcutaneous, abscess-like lesions of the gluteal region; report of a case with a study of the organism, Mycobacterium abscessus, n. sp. J Invest Dermatol. 1953;20:133–169. doi: 10.1038/jid.1953.18. [DOI] [PubMed] [Google Scholar]
- 4.Kubica GP, Baess I, Gordon RE, Jenkins PA, Kwapinski JB, et al. A co-operative numerical analysis of rapidly growing mycobacteria. J Gen Microbiol. 1972;73:55–70. doi: 10.1099/00221287-73-1-55. [DOI] [PubMed] [Google Scholar]
- 5.Kusunoki S, Ezaki T. Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov. Int J Syst Bacteriol. 1992;42:240–245. doi: 10.1099/00207713-42-2-240. [DOI] [PubMed] [Google Scholar]
- 6.Griffith DE, Girard WM, Wallace RJ., Jr Clinical features of pulmonary disease caused by rapidly growing mycobacteria. An analysis of 154 patients. Am Rev Respir Dis. 1993;147:1271–1278. doi: 10.1164/ajrccm/147.5.1271. [DOI] [PubMed] [Google Scholar]
- 7.Adekambi T, Berger P, Raoult D, Drancourt M. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int J Syst Evol Microbiol. 2006;56:133–143. doi: 10.1099/ijs.0.63969-0. [DOI] [PubMed] [Google Scholar]
- 8.Adekambi T, Reynaud-Gaubert M, Greub G, Gevaudan MJ, La Scola B, et al. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J Clin Microbiol. 2004;42:5493–5501. doi: 10.1128/JCM.42.12.5493-5501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HY, Yun YJ, Park CG, Lee DH, Cho YK, et al. Outbreak of Mycobacterium massiliense infection associated with intramuscular injections. J Clin Microbiol. 2007;45:3127–3130. doi: 10.1128/JCM.00608-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viana-Niero C, Lima KV, Lopes ML, Rabello MC, Marsola LR, et al. Molecular characterization of Mycobacterium massiliense and Mycobacterium bolletii in isolates collected from outbreaks of infections after laparoscopic surgeries and cosmetic procedures. J Clin Microbiol. 2008;46:850–855. doi: 10.1128/JCM.02052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffith DE. Emergence of nontuberculous mycobacteria as pathogens in cystic fibrosis. Am J Respir Crit Care Med. 2003;167:810–812. doi: 10.1164/rccm.2301001. [DOI] [PubMed] [Google Scholar]
- 12.Chalermskulrat W, Sood N, Neuringer IP, Hecker TM, Chang L, et al. Non-tuberculous mycobacteria in end stage cystic fibrosis: implications for lung transplantation. Thorax. 2006;61:507–513. doi: 10.1136/thx.2005.049247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jönsson BE, Gilljam M, Lindblad A, Ridell M, Wold AE, et al. Molecular epidemiology of Mycobacterium abscessus, with focus on cystic fibrosis. J Clin Microbiol. 2007;45:1497–1504. doi: 10.1128/JCM.02592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olivier KN, Weber DJ, Wallace RJ, Jr, Faiz AR, Lee JH, et al. Nontuberculous mycobacteria. I: multicenter prevalence study in cystic fibrosis. Am J Respir Crit Care Med. 2003;167:828–834. doi: 10.1164/rccm.200207-678OC. [DOI] [PubMed] [Google Scholar]
- 15.Pierre-Audigier C, Ferroni A, Sermet-Gaudelus I, Le Bourgeois M, Offredo C, et al. Age-related prevalence and distribution of nontuberculous mycobacterial species among patients with cystic fibrosis. J Clin Microbiol. 2005;43:3467–3470. doi: 10.1128/JCM.43.7.3467-3470.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanguinetti M, Ardito F, Fiscarelli E, La Sorda M, D'Argenio P, et al. Fatal pulmonary infection due to multidrug-resistant Mycobacterium abscessus in a patient with cystic fibrosis. J Clin Microbiol. 2001;39:816–819. doi: 10.1128/JCM.39.2.816-819.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sermet-Gaudelus I, Le Bourgeois M, Pierre-Audigier C, Offredo C, Guillemot D, et al. Mycobacterium abscessus and children with cystic fibrosis. Emerg Infect Dis. 2003;9:1587–1591. doi: 10.3201/eid0912.020774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomashefski JF, Jr, Stern RC, Demko CA, Doershuk CF. Nontuberculous mycobacteria in cystic fibrosis. An autopsy study. Am J Respir Crit Care Med. 1996;154:523–528. doi: 10.1164/ajrccm.154.2.8756832. [DOI] [PubMed] [Google Scholar]
- 19.Galil K, Miller LA, Yakrus MA, Wallace RJ, Jr, Mosley DG, et al. Abscesses due to Mycobacterium abscessus linked to injection of unapproved alternative medication. Emerg Infect Dis. 1999;5:681–687. doi: 10.3201/eid0505.990509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villanueva A, Calderon RV, Vargas BA, Ruiz F, Aguero S, et al. Report on an outbreak of postinjection abscesses due to Mycobacterium abscessus, including management with surgery and clarithromycin therapy and comparison of strains by random amplified polymorphic DNA polymerase chain reaction. Clin Infect Dis. 1997;24:1147–1153. doi: 10.1086/513656. [DOI] [PubMed] [Google Scholar]
- 21.Byrd TF, Lyons CR. Preliminary characterization of a Mycobacterium abscessus mutant in human and murine models of infection. Infect Immun. 1999;67:4700–4707. doi: 10.1128/iai.67.9.4700-4707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rottman M, Catherinot E, Hochedez P, Emile JF, Casanova JL, et al. Importance of T cells, gamma interferon, and tumor necrosis factor in immune control of the rapid grower Mycobacterium abscessus in C57BL/6 mice. Infect Immun. 2007;75:5898–5907. doi: 10.1128/IAI.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Catherinot E, Clarissou J, Etienne G, Ripoll F, Emile JF, et al. Hypervirulence of a rough variant of the Mycobacterium abscessus type strain. Infect Immun. 2007;75:1055–1058. doi: 10.1128/IAI.00835-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howard ST, Rhoades E, Recht J, Pang X, Alsup A, et al. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology. 2006;152:1581–1590. doi: 10.1099/mic.0.28625-0. [DOI] [PubMed] [Google Scholar]
- 25.Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 26.Stinear TP, Mve-Obiang A, Small PL, Frigui W, Pryor MJ, et al. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci U S A. 2004;101:1345–1349. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, et al. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 28.Gordon SV, Brosch R, Billault A, Garnier T, Eiglmeier K, et al. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol Microbiol. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 29.Kim SJ, Kweon O, Jones RC, Edmondson RD, Cerniglia CE. Genomic analysis of polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Biodegradation. 2008;19:859–881. doi: 10.1007/s10532-008-9189-z. [DOI] [PubMed] [Google Scholar]
- 30.Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008;18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 32.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, et al. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 33.Howard ST, Byrd TF, Lyons CR. A polymorphic region in Mycobacterium abscessus contains a novel insertion sequence element. Microbiology. 2002;148:2987–2996. doi: 10.1099/00221287-148-10-2987. [DOI] [PubMed] [Google Scholar]
- 34.Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, et al. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol. 2006;6:95. doi: 10.1186/1471-2148-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorensen AL, Nagai S, Houen G, Andersen P, Andersen AB. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect Immun. 1995;63:1710–1717. doi: 10.1128/iai.63.5.1710-1717.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley LW. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science. 1993;261:1454–1457. doi: 10.1126/science.8367727. [DOI] [PubMed] [Google Scholar]
- 37.Ishikawa J, Yamashita A, Mikami Y, Hoshino Y, Kurita H, et al. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc Natl Acad Sci U S A. 2004;101:14925–14930. doi: 10.1073/pnas.0406410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thoma-Uszynski S, Stenger S, Takeuchi O, Ochoa MT, Engele M, et al. Induction of direct antimicrobial activity through mammalian toll-like receptors. Science. 2001;291:1544–1547. doi: 10.1126/science.291.5508.1544. [DOI] [PubMed] [Google Scholar]
- 39.Titball RW. Bacterial phospholipases C. Microbiol Rev. 1993;57:347–366. doi: 10.1128/mr.57.2.347-366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raynaud C, Guilhot C, Rauzier J, Bordat Y, Pelicic V, et al. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol Microbiol. 2002;45:203–217. doi: 10.1046/j.1365-2958.2002.03009.x. [DOI] [PubMed] [Google Scholar]
- 41.Brodin P, Eiglmeier K, Marmiesse M, Billault A, Garnier T, et al. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect Immun. 2002;70:5568–5578. doi: 10.1128/IAI.70.10.5568-5578.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moncrief MB, Maguire ME. Magnesium and the role of MgtC in growth of Salmonella typhimurium. Infect Immun. 1998;66:3802–3809. doi: 10.1128/iai.66.8.3802-3809.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchmeier N, Blanc-Potard A, Ehrt S, Piddington D, Riley L, et al. A parallel intraphagosomal survival strategy shared by Mycobacterium tuberculosis and Salmonella enterica. Mol Microbiol. 2000;35:1375–1382. doi: 10.1046/j.1365-2958.2000.01797.x. [DOI] [PubMed] [Google Scholar]
- 44.Blanc-Potard AB, Lafay B. MgtC as a horizontally-acquired virulence factor of intracellular bacterial pathogens: evidence from molecular phylogeny and comparative genomics. J Mol Evol. 2003;57:479–486. doi: 10.1007/s00239-003-2496-4. [DOI] [PubMed] [Google Scholar]
- 45.St John G, Brot N, Ruan J, Erdjument-Bromage H, Tempst P, et al. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc Natl Acad Sci U S A. 2001;98:9901–9906. doi: 10.1073/pnas.161295398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lau GW, Hassett DJ, Ran H, Kong F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med. 2004;10:599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Arias-Barrau E, Olivera ER, Luengo JM, Fernandez C, Galan B, et al. The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J Bacteriol. 2004;186:5062–5077. doi: 10.1128/JB.186.15.5062-5077.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ernst RK, D'Argenio DA, Ichikawa JK, Bangera MG, Selgrade S, et al. Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis. Environ Microbiol. 2003;5:1341–1349. doi: 10.1111/j.1462-2920.2003.00518.x. [DOI] [PubMed] [Google Scholar]
- 49.Luengo JM, Garcia JL, Olivera ER. The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol Microbiol. 2001;39:1434–1442. doi: 10.1046/j.1365-2958.2001.02344.x. [DOI] [PubMed] [Google Scholar]
- 50.Hunt TA, Kooi C, Sokol PA, Valvano MA. Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infect Immun. 2004;72:4010–4022. doi: 10.1128/IAI.72.7.4010-4022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X, He X, Liang J, Li A, Xu T, et al. A novel DNA modification by sulphur. Mol Microbiol. 2005;57:1428–1438. doi: 10.1111/j.1365-2958.2005.04764.x. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Yakrus MA, Graviss EA, Williams-Bouyer N, Turenne C, et al. Pulsed-field gel electrophoresis study of Mycobacterium abscessus isolates previously affected by DNA degradation. J Clin Microbiol. 2004;42:5582–5587. doi: 10.1128/JCM.42.12.5582-5587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nash KA, Brown-Elliott BA, Wallace RJ., Jr A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob Agents Chemother. 2009;53:1367–1376. doi: 10.1128/AAC.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prammananan T, Sander P, Brown BA, Frischkorn K, Onyi GO, et al. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J Infect Dis. 1998;177:1573–1581. doi: 10.1086/515328. [DOI] [PubMed] [Google Scholar]
- 55.Silver S, Phung LT. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 56.Ordonez E, Letek M, Valbuena N, Gil JA, Mateos LM. Analysis of genes involved in arsenic resistance in Corynebacterium glutamicum ATCC 13032. Appl Environ Microbiol. 2005;71:6206–6215. doi: 10.1128/AEM.71.10.6206-6215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barkay T, Miller SM, Summers AO. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev. 2003;27:355–384. doi: 10.1016/S0168-6445(03)00046-9. [DOI] [PubMed] [Google Scholar]
- 58.Wheeler PR, Ratledge C. Bloom BR, editor. Tuberculosis: Pathogenesis, Protection and Control. American Society for Microbiology. 1994. pp. 353–385.
- 59.Rodriguez-Zaragoza S. Ecology of free-living amoebae. Crit Rev Microbiol. 1994;20:225–241. doi: 10.3109/10408419409114556. [DOI] [PubMed] [Google Scholar]
- 60.Prithiviraj B, Bais HP, Weir T, Suresh B, Najarro EH, et al. Down regulation of virulence factors of Pseudomonas aeruginosa by salicylic acid attenuates its virulence on Arabidopsis thaliana and Caenorhabditis elegans. Infect Immun. 2005;73:5319–5328. doi: 10.1128/IAI.73.9.5319-5328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le Dantec C, Duguet JP, Montiel A, Dumoutier N, Dubrou S, et al. Occurrence of mycobacteria in water treatment lines and in water distribution systems. Appl Environ Microbiol. 2002;68:5318–5325. doi: 10.1128/AEM.68.11.5318-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Danelishvili L, Wu M, Stang B, Harriff M, Cirillo SL, et al. Identification of Mycobacterium avium pathogenicity island important for macrophage and amoeba infection. Proc Natl Acad Sci U S A. 2007;104:11038–11043. doi: 10.1073/pnas.0610746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Becq J, Gutierrez MC, Rosas-Magallanes V, Rauzier J, Gicquel B, et al. Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol Biol Evol. 2007;24:1861–1871. doi: 10.1093/molbev/msm111. [DOI] [PubMed] [Google Scholar]
- 64.Rosas-Magallanes V, Deschavanne P, Quintana-Murci L, Brosch R, Gicquel B, et al. Horizontal transfer of a virulence operon to the ancestor of Mycobacterium tuberculosis. Mol Biol Evol. 2006;23:1129–1135. doi: 10.1093/molbev/msj120. [DOI] [PubMed] [Google Scholar]
- 65.Hall-Stoodley L, Lappin-Scott H. Biofilm formation by the rapidly growing mycobacterial species Mycobacterium fortuitum. FEMS Microbiol Lett. 1998;168:77–84. doi: 10.1111/j.1574-6968.1998.tb13258.x. [DOI] [PubMed] [Google Scholar]
- 66.Davey ME, O'Toole G A. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jang J, Becq J, Gicquel B, Deschavanne P, et al. Horizontally acquired genomic islands in the tubercle bacilli. Trends Microbiol. 2008;16:303–308. doi: 10.1016/j.tim.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 68.Posfai G, Plunkett G, 3rd, Feher T, Frisch D, Keil GM, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 69.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 71.Weingart CL, Hooke AM. A nonhemolytic phospholipase C from Burkholderia cepacia. Curr Microbiol. 1999;38:233–238. doi: 10.1007/pl00006793. [DOI] [PubMed] [Google Scholar]
- 72.Guina T, Purvine SO, Yi EC, Eng J, Goodlett DR, et al. Quantitative proteomic analysis indicates increased synthesis of a quinolone by Pseudomonas aeruginosa isolates from cystic fibrosis airways. Proc Natl Acad Sci U S A. 2003;100:2771–2776. doi: 10.1073/pnas.0435846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maloney KE, Valvano MA. The mgtC gene of Burkholderia cenocepacia is required for growth under magnesium limitation conditions and intracellular survival in macrophages. Infect Immun. 2006;74:5477–5486. doi: 10.1128/IAI.00798-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A. 2008;105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Glemet E, Codani JJ. LASSAP, a LArge Scale Sequence compArison Package. Comput Appl Biosci. 1997;13:137–143. doi: 10.1093/bioinformatics/13.2.137. [DOI] [PubMed] [Google Scholar]
- 79.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 80.Frank AC, Lobry JR. Oriloc: prediction of replication boundaries in unannotated bacterial chromosomes. Bioinformatics. 2000;16:560–561. doi: 10.1093/bioinformatics/16.6.560. [DOI] [PubMed] [Google Scholar]
- 81.Mohseni-Zadeh S, Brezellec P, Risler JL. Cluster-C, an algorithm for the large-scale clustering of protein sequences based on the extraction of maximal cliques. Comput Biol Chem. 2004;28:211–218. doi: 10.1016/j.compbiolchem.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 82.Vernikos GS, Parkhill J. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics. 2006;22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- 83.Luc N, Risler JL, Bergeron A, Raffinot M. Gene teams: a new formalization of gene clusters for comparative genomics. Comput Biol Chem. 2003;27:59–67. doi: 10.1016/s1476-9271(02)00097-x. [DOI] [PubMed] [Google Scholar]
- 84.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 86.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 87.Stinear TP, Seemann T, Pidot S, Frigui W, Reysset G, et al. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 2007;17:192–200. doi: 10.1101/gr.5942807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole genome dotplot comparison of M. abscessus (horizontal axis) versus M. smegmatis.
(0.08 MB TIF)
M. abscessus prophage-like elements
(0.03 MB DOC)
M. abscessus and M. smegmatis proteins involved in metabolism, according to the Kegg classification
(0.04 MB DOC)
A selection of paralogous families
(0.06 MB DOC)
Proteins encoded in the 17 horizontally acquired gene clusters, and their syntenic non mycobacterial homologs
(0.06 MB DOC)
(0.06 MB DOC)