Abstract
The transforming growth factor-β (TGFβ) and Wnt/wingless pathways play pivotal roles in tissue specification during development. Activation of Smads, the effectors of TGFβ superfamily signals, results in Smad translocation from the cytoplasm into the nucleus where they act as transcriptional comodulators to regulate target gene expression. Wnt/wingless signals are mediated by the DNA-binding HMG box transcription factors lymphoid enhancer binding factor 1/T cell-specific factor (LEF1/TCF) and their coactivator β-catenin. Herein, we show that Smad3 physically interacts with the HMG box domain of LEF1 and that TGFβ and Wnt pathways synergize to activate transcription of the Xenopus homeobox gene twin (Xtwn). Disruption of specific Smad and LEF1/TCF DNA-binding sites in the promoter abrogates synergistic activation of the promoter. Consistent with this observation, introduction of Smad sites into a TGFβ-insensitive LEF1/TCF target gene confers cooperative TGFβ and Wnt responsiveness to the promoter. Furthermore, we demonstrate that TGFβ-dependent activation of LEF1/TCF target genes can occur in the absence of β-catenin binding to LEF1/TCF and requires both Smad and LEF1/TCF DNA-binding sites in the Xtwn promoter. Thus, our results show that TGFβ and Wnt signaling pathways can independently or cooperatively regulate LEF1/TCF target genes and suggest a model for how these pathways can synergistically activate target genes.
The transforming growth factor-β (TGFβ) and Wnt/wingless pathways function during numerous developmental stages to regulate cell fate determination (1, 2). These unrelated extracellular factors mediate their effects through two distinct signaling cascades. For TGFβ, initiation of signaling occurs when ligand binds to a heteromeric receptor complex composed of type I and type II serine/threonine kinase receptors, which then propagate the signal to the Smad family of proteins (3–7). Activation of the type I receptor kinase leads to association and phosphorylation of a specific receptor-regulated Smad. This phosphorylation causes dissociation of the receptor-regulated Smad from the receptor, stimulates the assembly of a heteromeric complex between the phosphorylated receptor-regulated Smad and the co-Smad, Smad4, and induces the nuclear accumulation of this complex. In the nucleus, these heteromeric complexes function to regulate gene expression by directly interacting with resident DNA-binding proteins and by recruiting coactivators or corepressors to the promoter (4, 5, 7).
The Wnt/wingless pathway is distinct from that of TGFβ and is mediated by β-catenin and members of the lymphoid enhancer binding factor 1/T cell-specific factor (LEF1/TCF) transcription factor family (8–10). In the absence of Wnt signaling, the adenomatous polyposis coli protein binds to β-catenin in a complex that also contains glycogen-synthase kinase-3β (GSK-3β) and axins. Phosphorylation of β-catenin by glycogen-synthase kinase-3β induces the degradation of β-catenin by the ubiquitin-proteasome pathway. Activation of Wnt signaling by binding of Wnt to the Frizzled family of receptors inhibits glycogen-synthase kinase-dependent phosphorylation of β-catenin and results in an increase in β-catenin protein levels. The accumulation of β-catenin promotes its nuclear translocation where it associates with LEF1/TCF transcription factors and activates Wnt target genes.
Several studies have shown that cooperation between TGFβ and Wnt/wingless signaling pathways plays a significant role in controlling certain developmental events. In Xenopus, activin and Wnt both cooperate to restrict spatially early gene transcription to the Spemann organizer (11). In Drosophila, the vestigial gene, which is important for wing development, and Ultrabithorax, a homeotic gene that initiates endoderm induction, coordinately receive inputs from both dpp (a TGFβ superfamily member) and wingless (12, 13). However, the molecular mechanism for this cooperative regulation is not known.
Methods
Xtwn Promoter and Reporter Constructs.
The 322-bp Xtwn promoter fragment (−359 to −38; ref. 14) was amplified by PCR from Xenopus laevis genomic DNA with the primers 5′-TATAACTGGTTTATAGTTGCA-3′ and 5′-AACAGAAAGAAGTGGGAGTG-3′ and then subcloned into SacI/BglII sites of a modified pGL2-promoter vector (Promega) as described (15). Mutation of the second Smad-binding element (SBE) in Xtwn 609 (−160 to −38) was done by PCR with the primer 5′-GGATTACTGAAATTTGAGTAAGATCA-3′. The LEF1/TCF-binding sites were mutated in 605 mutLEF1 (−263 to −38) and 606 mutLEF1 (−128 to −38) as described (14). The improved Topflash reporter in pGL3 (pOT) was obtained from B. Vogelstein (Johns Hopkins University, Baltimore). Twntop was constructed by subcloning a portion of Xtwn (−263 to −116) upstream of the three LEF1/TCF sites of Topflash by using SacI/SmaI.
Mammalian, Glutathione S-Transferase (GST) Fusion, and in Vitro Expression Constructs.
Smad and receptor mammalian expression constructs have been described (15). Hemagglutinin (HA) and myc epitope-tagged LEF1 constructs were prepared in pCMV5B by PCR with pCG-mLEF1-HA or pCG-mLEF1-myc as template. LEF1-GST fusion constructs were prepared by subcloning from pCMV5B into pGEX4T1. The pBS-KS-LEF1-myc construct was prepared by subcloning from pCG-mLEF1-myc. The pCMV5B-myc-β-catenin was obtained by subcloning from pBAT-myc-β-catenin.
Transcriptional Response Assays.
For luciferase assays, HepG2 cells were transiently transfected by using the calcium phosphate DNA precipitation method as described (15). Unless otherwise indicated, transfections typically contained 0.15 μg of Xtwn–Lux or 0.07 μg of Topflash or Twntop reporter, 0.025 μg of each Smad, 0.007 μg of LEF1, 0.23 μg of β-catenin, 0.025 μg of constitutively activated Alk6, 0.07 μg of pCMVβ-gal, and pCMV5 empty vector to a total of 0.75 μg per well.
Immunoprecipitation, GST Pull-Down, and Electrophoretic Mobility-Shift Assays (EMSAs).
For immunoprecipitations, DEAE-dextran-transfected COS-1 cells or calcium phosphate-transfected HEK 293T cells were lysed in lysis buffer [50 mM Tris⋅HCl/150 mM NaCl/1 mM EDTA/0.5% Triton X-100/1 mM DTT/10% (vol/vol) glycerol] containing phosphatase and protease inhibitors. Cell extracts were immunoprecipitated with anti-FLAG M2 monoclonal antibody (Sigma) or anti-human LEF1/TCF polyclonal antibody (Exalpha, Boston) and collected by using protein-G Sepharose. Immunoprecipitates were separated by SDS/PAGE and immunoblotted with anti-FLAG or anti-HA 12CA5 (Roche Diagnostics) monoclonal antibodies.
For GST pull-downs, GST fusion constructs of full-length (FL) Smads, Smad3-MH1 (amino acids 1–144), Smad3-NC (amino acids 145–234), and Smad3-MH2 (amino acids 199–440), as well as LEF1 FL and LEF1 Δ20 were purified by using glutathione-Sepharose 4B beads (Amersham Pharmacia). Cell lysates from transfected COS-1 cells were incubated with glutathione-Sepharose-bound fusion proteins on ice for 2 h. The beads were washed five times in wash buffer [50 mM Tris⋅HCl/150 mM NaCl/1 mM EDTA/0.1% Triton X-100/10% (vol/vol) glycerol], and associated proteins were detected by immunoblotting. In vitro transcription/translation GST pull-downs with in vitro transcribed and translated LEF1 (Promega) were conducted with pBS-KS-LEF1-myc as described (16).
EMSAs were conducted exactly as described (15). For supershifting, anti-FLAG (Sigma), anti-LEF1/TCF (Exalpha), or anti-Smad4 (17) antibodies were added with the probe.
Results
Identification of Xtwn as a Target of TGFβ and Wnt Signaling Pathways.
Activin and Wnt signaling pathways cooperate to control expression of the homeobox gene Siamois during Xenopus embryo development (11). To gain molecular insights into the mechanism of this cooperativity, we investigated whether twin (Xtwn), a gene that is closely related to Siamois and that shares a similar expression pattern (14), might also be coordinately regulated. Previous work in Xenopus demonstrated that Xtwn expression is induced by Wnt and requires LEF1/TCF-binding sites in the promoter (14). Thus, we subcloned a 322-bp fragment of the Xtwn promoter upstream of a luciferase reporter gene and tested for LEF1 responsiveness in HepG2 cells, which harbor an activated form of β-catenin (18). Consistent with previous work on other Wnt target genes, coexpression of LEF1 was sufficient to mediate induction of the Xtwn–lux reporter in these cells (Fig. 1A).
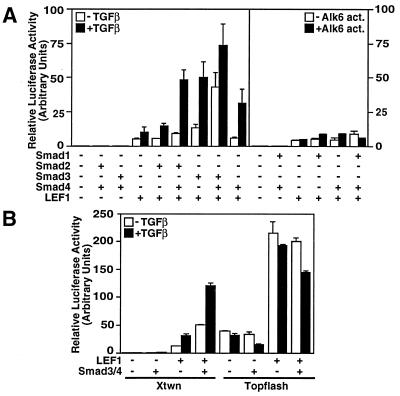
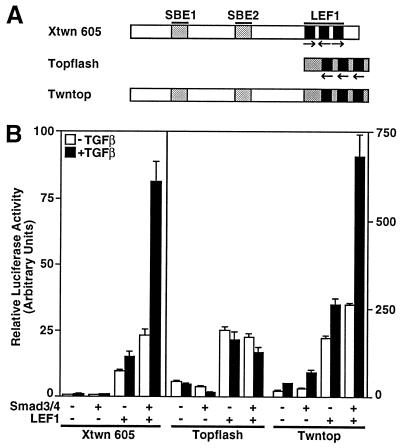
Figure 1.
TGFβ and Wnt signaling pathways activate Xtwn but not Topflash reporters. HepG2 cells were transfected with Xtwn-Lux (A and B) or Topflash (B) reporters alone or together with LEF1 and Smads. Cells were incubated overnight with or without 100 pM TGFβ (A Left and B) or were cotransfected with the constitutively active BMP type I receptor ALK6 (A Right). Luciferase activity was normalized to β-galactosidase activity and is expressed as the mean ± SD.
Activin and TGFβ both regulate Smad2 and Smad3 signaling through closely related receptors (3–7). Therefore, to examine the effect of the Smad2 and Smad3 pathways on the Xtwn promoter, we used TGFβ and HepG2 cells that contain endogenous TGFβ receptors and Smads (15, 19). In the absence of LEF1 expression, TGFβ had minimal effects on the Xtwn promoter, even when Smad2 or Smad3 was coexpressed with Smad4 (Fig. 1A). However, in the presence of LEF1, activation of the Xtwn promoter was stimulated by TGFβ treatment of the cells. Furthermore, coexpression of Smad2 and Smad4, Smad3 alone, or Smad3 and Smad4 resulted in strong enhancement of LEF1-dependent transcriptional activity (Fig. 1A). Activation of the promoter was specific for the TGFβ/activin pathway, because coexpression of a constitutively active bone morphogenetic protein (BMP) receptor or of Smad1, a mediator of BMP signals, did not enhance LEF1-dependent signaling (Fig. 1A). Of note, Smad4 alone had no effect on ligand-independent LEF-1 mediated activation of Xtwn.
We also examined the effect of TGFβ on Topflash, a Wnt-responsive reporter that contains three multimerized LEF1/TCF-binding sites (20) but no SBEs. In contrast to the synergistic activation of the Xtwn promoter by Smads and LEF1, we observed that coexpression of Smad3 and Smad4 had no effect on LEF1-dependent induction of luciferase activity from the Topflash reporter (Fig. 1B). Thus, although both Xtwn and Topflash can be activated by the β-catenin/Wnt signaling pathway, TGFβ-dependent enhancement of LEF1-dependent signaling is observed only on the Xtwn promoter.
LEF1/TCF Transcription Factors Associate with Smad2, Smad3, and Smad4.
To determine whether TGFβ signaling through LEF1 might reflect physical interactions between pathway components, we next examined whether Smads can associate with LEF1. Thus, COS-1 cells were transfected with HA-tagged LEF1, and cell lysates were incubated with bacterially expressed Smad fusion proteins. Anti-HA immunoblotting of bound proteins revealed that LEF1 interacted with the TGFβ/activin-regulated Smads, Smad2 and Smad3, and the common Smad, Smad4 (Fig. 2A). Although not evident here, a weak interaction of LEF1 with Smad1 was also detected in some experiments (data not shown). Association of bacterially expressed Smad3 and in vitro transcribed and translated LEF1 was also observed, indicating that the interaction is direct (Fig. 2C). To confirm that LEF1 also bound to Smad2 and Smad3 in mammalian cells, we expressed HA-tagged LEF1 in COS-1 cells together with Flag-tagged Smads in the presence of the constitutively active, activin type I receptor ActRIB. In anti-Flag immunoprecipitates of cells expressing Smad2 and Smad3, coprecipitation of LEF1-HA was observed (Fig. 2B). In similar experiments, association between Smad4 and LEF1 was also detected (data not shown).
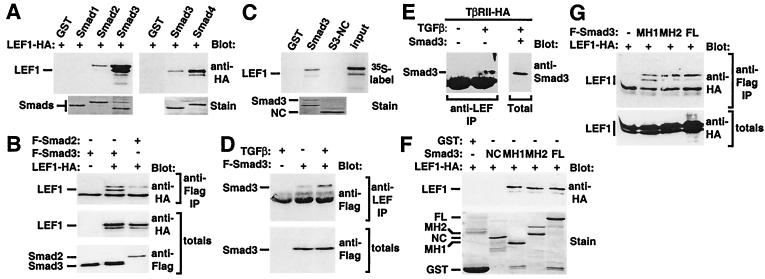
Figure 2.
LEF1/TCF transcription factors associate with Smads. (A) In vitro interaction of LEF1 with bacterially expressed Smads. Lysates from LEF1-transfected COS-1 cells (A) or in vitro transcribed and translated LEF1 (C) were incubated with bacterially expressed GST–Smad fusion proteins, and bound material was detected by immunoblotting. Levels of GST fusion proteins were confirmed by SDS/PAGE and Coomassie blue staining (stain). (B) Interaction of LEF1 with Smads in mammalian cells. COS-1 cells were transfected with constitutively activated ActRIB, Flag-tagged Smad2, or Smad3 and HA-tagged LEF1. Cell lysates were subjected to anti-Flag antibody immunoprecipitation and were analyzed by anti-HA immunoblotting. Protein levels were determined by anti-Flag or anti-HA immunoblotting of total cell lysates. (D) TGFβ-dependent enhancement of the interaction of Smad3 with endogenous LEF1/TCF. HEK 293T cells were transfected with Flag–Smad3, Smad4, and TβRII, and cells were incubated overnight in the presence or absence of TGFβ. Endogenous LEF1/TCF was collected by immunoprecipitation with anti-LEF1/TCF antibodies, and associated Smad3 was detected by anti-Flag immunoblotting. Smad3 protein levels were determined by anti-Flag immunoblotting of total cell lysates. (E) TGFβ-dependent association of endogenous Smad3 and LEF1/TCF. HEK 293T cells, transfected with TβRII, were incubated overnight with or without TGFβ. Endogenous LEF1/TCF was collected by anti-LEF1/TCF immunoprecipitation, and associated Smad3 was detected by immunoblotting with anti-Smad2/3 antibodies. In parallel, cells were transfected with untagged Smad3, and an aliquot of total cell extracts was subjected to anti-Smad2/3 immunoblotting to confirm comigration with endogenous Smad3. (F) In vitro interaction of LEF1 with bacterially expressed FL, MH1, MH2, or nonconserved domains of Smad3. Association of LEF1 with the various Smad3 domains was conducted as in A. (G) Interaction of LEF1 with FL, MH1, or MH2 domains of Smad3 in mammalian cells. Association of LEF1 with the various Smad3 domains was conducted as in B.
Smad4 is a common component of BMP and TGFβ pathways, whereas the receptor-regulated Smads are critical for mediating distinct signals (3–7). Because Xtwn is not regulated by the BMP pathway via Smad1/Smad4, we focused further studies on the association of the TGFβ-regulated Smad, Smad3, with LEF1. To examine whether the interaction of Smad3 with LEF1 depended on TGFβ signaling, we used 293T cells, which express endogenous LEF1/TCF family members. Because these cells have very low levels of the TGFβ type II receptor TβRII, this receptor was also coexpressed with Smad3 and Smad4. We observed that there was association between endogenous LEF1/TCFs and transfected Smad3 and that this association was enhanced on TGFβ treatment (Fig. 2D). Moreover, TGFβ-dependent association of endogenous Smad3 with endogenous LEF1/TCFs was also observed in these cells (Fig. 2E). Together, these data show that Smads and LEF1/TCFs can interact physically in a TGFβ-dependent manner.
Determination of the Domains in Smads and LEF1 That Mediate Their Association.
To determine the domains in Smad3 that mediate its association with LEF1, we expressed HA-tagged LEF1 in COS-1 cells and examined the ability of FL LEF1 to interact with the MH1, MH2, or nonconserved linker regions of Smad3 expressed as bacterial fusion proteins. We observed that LEF1 interacted with both the MH1 and MH2 domains but not the nonconserved linker region of Smad3 (Fig. 2F). Consistent with this observation, FL LEF1 was also shown to interact with the Smad3 MH1 and MH2 domains in transiently transfected COS-1 cells (Fig. 2G).
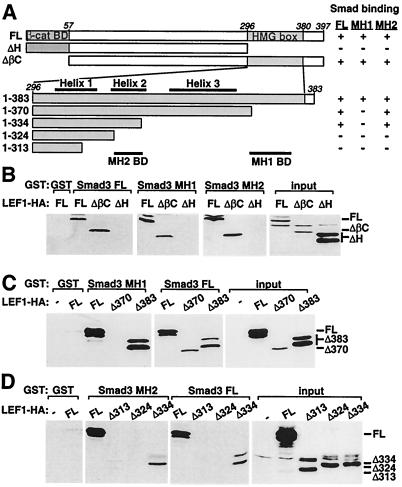
To identify the Smad-interacting domains in LEF1, we prepared various LEF1 deletions constructs (Fig. 3A). We observed that LEF1 ΔβC, which lacks the amino-terminal β-catenin-binding domain, interacted normally with Smad3, whereas LEF1 ΔH, which lacks the carboxyl-terminal HMG box, was unable to associate with the FL, MH1, or MH2 domains of Smad3 (Fig. 3B). To define the Smad-interaction domain further, we next created a series of LEF1 constructs containing varying deletions of the carboxyl terminus (Fig. 3A). We determined that amino acids 324–334 mediate binding to the Smad3 MH2 domain, whereas a lysine- and arginine-rich region between amino acids 370 and 383 is required for association with the MH1 domain (Fig. 3 A–D).
Figure 3.
Determination of the domains in LEF1 that mediate association with Smad3. (A) A schematic representation of mutant versions of LEF1 are shown. The β-catenin binding domain (β-cat BD) and the HMG box are marked. The location of the three helices within the HMG box (overline) and the MH1- and MH2-binding domains (BD; underline) are indicated. A summary of the interaction of LEF1 with FL, MH1, and MH2 domains of Smad3 is shown (right). (B–D) Interaction of wild-type or mutant LEF1 with bacterially expressed FL, MH1, or MH2 domains of Smad3. COS-1 cells were transfected with wild-type or mutant versions of LEF1 and cell lysates incubated with bacterially expressed GST fusion proteins. The associated LEF1 was visualized by anti-HA immunoblotting. Total LEF1 protein expression was confirmed by immunoblotting (input).
Smad and LEF1/TCF-Binding Elements Are Required for Synergistic Activation of Xtwn.
The Xtwn promoter has two clusters of putative SBEs at positions −218 to −203 and −160 to −155 located upstream of the three LEF1-binding sites identified previously (ref. 14; Fig. 4A). To determine the importance of these sites, we prepared a series of deletions and examined their effects on signaling. In parallel, the DNA-binding activity of LEF1 or Smad3 MH1 domain fusion proteins on these DNA fragments was determined by EMSA (Fig. 4B).
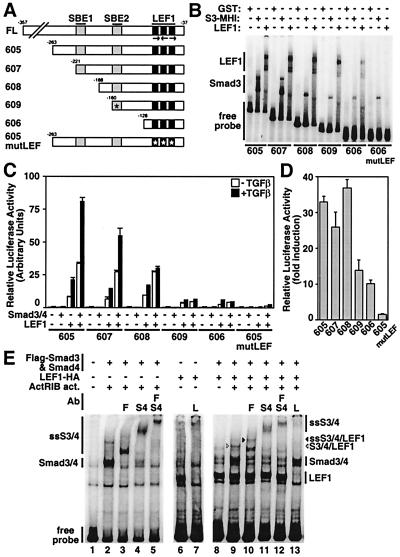
Figure 4.
The SBEs and the LEF1/TCF-binding sites are required for synergistic activation of Xtwn by TGFβ and Wnt signaling pathways. (A) A schematic representation of the Xtwn promoter and the 5′ end deletion constructs with the location of the SBEs, the triple LEF1/TCF-binding sites (LEF), and the presence of point mutations (asterisk) is shown. (B) EMSA analysis of Xtwn promoter deletions. Bacterially expressed GST–LEF1, GST–Smad3 MH1 domain, or GST control were subjected to gel shift assays with 32P-labeled Xtwn promoter probes corresponding to the indicated DNA fragments. (C) HepG2 cells were transfected with Xtwn–Lux reporter constructs containing various promoter fragments alone or together with combinations of LEF1, Smad3, and Smad4 as in Fig. 1. (D) LEF1-dependent activation of luciferase activity. Luciferase activity from C is replotted as fold induction for transfectants expressing LEF1 in the absence of TGFβ treatment compared with controls transfected with Xtwn reporters alone. (E) Smads and LEF1 expressed in mammalian cells bind to the Xtwn promoter. Extracts from COS-1 cells transfected with combinations of Smad3, Smad4, LEF1, and the constitutively active activin type I receptor ActRIB were tested for DNA-binding activity on the 322-bp Xtwn promoter as in B. For supershifting assays, anti-Flag (F), anti-Smad4 (S4), or anti-LEF1/TCF (L) antibodies (Ab) were added. The migration of DNA binding and supershifted (SS) complexes are indicated. The complex containing Smad3, Smad4, and LEF1 is marked (arrow).
Deletion of the first 136 nucleotides had little or no effect on TGFβ- and Smad3/4-dependent enhancement of activation of the Xtwn promoter in LEF1 expressing cells (Xtwn 605 and 607). However, removal of an additional 33 bp that included the first SBE reduced the Smad3/4-dependent enhancement of LEF1 signaling by approximately 50% (Fig. 4C). Further deletion of the second SBE (Xtwn 606) or introduction of point mutations in the second SBE (Xtwn 609) strongly reduced Smad3 MH1 DNA-binding activity (Fig. 4B) and blocked Smad-dependent activation of the Xtwn promoter (Fig. 4C). In contrast to the loss of TGFβ-dependent signaling, activation of Xtwn by LEF1 alone was affected only modestly by removal of the SBEs (Fig. 4 C and D). Introduction of point mutations into the LEF1-binding motifs prevented LEF1 DNA binding and abolished the activity of the Xtwn promoter (Fig. 4 B–D). Together, these results indicate that the two SBEs and the LEF1-binding motif are both required for TGFβ-dependent activation of the Xtwn promoter.
Smads and LEF1 Expressed in Mammalian Cells Can Bind to the Xtwn Promoter.
To determine whether Smads and LEF1 could bind simultaneously to the Xtwn promoter, we conducted EMSA analysis with extracts from COS-1 cells transiently expressing FL Smads and LEF1. Comparison of DNA-binding complexes from control extracts with those expressing Smad3 and Smad4 revealed the appearance of a DNA-binding complex (Fig. 4E, lane 2), which was confirmed to contain Smad3 and Smad4 by supershift analysis. LEF1 alone also bound to the DNA probe, and the complex was supershifted quantitatively in the presence of anti-LEF1 antibodies (Fig. 4E, lanes 6 and 7). In extracts from cells coexpressing Smad3, Smad4, and LEF1, two shifts in the DNA probe that comigrated with those of the LEF1 alone or the Smad3/4 DNA complex were detected (Fig. 4E, lane 9). In addition, a more slowly migrating complex that was supershifted or lost in the presence of antibodies directed toward LEF1, Smad3, and Smad4 was observed, demonstrating that Smad3, Smad4, and LEF1 can bind simultaneously to the Xtwn promoter. Of note, this complex was observed only in the presence of the activated receptor, indicating that simultaneous binding of Smads and LEF1 requires activation of TGFβ signaling. Although Smad3 and Smad4 are capable of binding DNA in the absence of LEF1, this binding is not sufficient to mediate activation of the Xtwn promoter (Fig. 1A). Thus, our observations suggest that Smad and LEF1 binding to specific DNA elements is required for TGFβ-dependent activation of the Xtwn promoter.
Introduction of SBE Converts Topflash into a TGFβ-Responsive Promoter.
To confirm that Smad-binding sites are essential for mediating TGFβ-dependent activation of LEF1 target genes, we introduced the SBEs from the Xtwn promoter upstream of the LEF1/TCF-binding sites in Topflash to generate Twntop. As described above, TGFβ treatment enhanced LEF1-dependent activation of Xtwn but had no effect on Topflash reporter activity (Fig. 5). However, in LEF1-transfected cells, activation of the Twntop promoter was stimulated by TGFβ treatment, and this stimulation was strongly enhanced on coexpression of Smad3 and Smad4 (Fig. 5). Thus, the presence of SBEs in the promoters of LEF1-dependent target genes can mediate TGFβ-dependent enhancement of transcriptional activity.
Figure 5.
Introduction of SBEs into Topflash confers TGFβ-responsiveness to the promoter. (A) A schematic representation of the Xtwn, Topflash, and Twntop promoters is shown. (B) HepG2 cells were transfected with Xtwn–Lux, Topflash, or Twntop reporters alone or together with various combinations of LEF1, Smad3, and Smad4 as in Fig. 1.
TGFβ-Dependent Activation of LEF1 Target Genes Occurs Independently of β-Catenin.
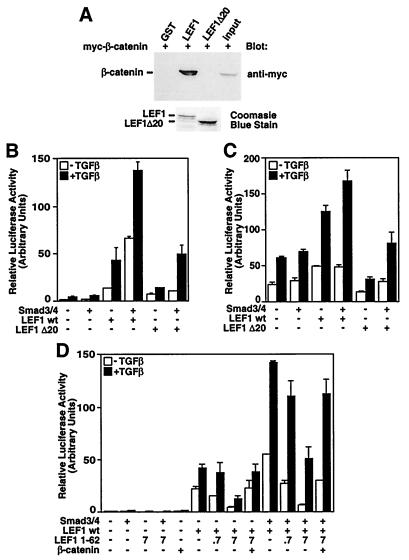
Activation of Wnt signaling leads to association of β-catenin with LEF1/TCF transcription factors, which then results in transcriptional activation of target genes (8–10). To investigate whether Smads require β-catenin function to activate the Xtwn promoter, we made a version of LEF1, LEF1 Δ20, which lacks a functional β-catenin-binding domain. GST pull-down assays confirm that this mutant does not interact with β-catenin (Fig. 6A), and as described for similar types of mutants (21, 22), LEF1 Δ20 is unable to mediate transcriptional activation of the LEF1 target, Topflash (data not shown). As described above, transient transfection of wild-type LEF1 into HepG2 cells, which express endogenous constitutively active β-catenin (18), yielded TGFβ-dependent activation of the Xtwn reporter gene that was increased by coexpression of Smad3 and Smad4. Importantly, Smad3/Smad4-transfected cells coexpressing the mutant LEF1 (LEF1 Δ20) also yielded TGFβ-dependent activation of Xtwn (Fig. 6B). Similar results were obtained by using the Twntop reporter (Fig. 6C). In cells expressing LEF1 Δ20, a reduction in both the basal activity of the Xtwn and Twntop reporters and in the level of TGFβ-dependent activation of these promoters was observed. The LEF1 mutant cannot bind β-catenin, indicating that this decrease reflects a loss of β-catenin-mediated activation of the two promoters.
Figure 6.
TGFβ-dependent activation of LEF1 target genes occurs independently of β-catenin. (A) In vitro interaction of bacterially expressed LEF1 and LEF1 Δ20 with β-catenin. Cell lysates from COS-1 cells transfected with β-catenin were incubated with bacterially expressed GST fusion proteins of LEF1, LEF1 Δ20, or control. Bound material was visualized by anti-myc immunoblotting. (B and C) Smad enhances LEF1-dependent signaling in the absence of β-catenin. HepG2 cells were transfected with Xtwn–Lux (B) or Twntop (C) reporter alone or with Smad3, Smad4, wild-type LEF1, or LEF1 Δ20. (D) Overexpression of the β-catenin-binding domain of LEF1 does not disrupt Smad-dependent enhancement of LEF1-dependent signaling. HepG2 cells were transfected with Xtwn–Lux reporter alone or with Smad3, Smad4, wild-type LEF1, LEF1 1–62, or β-catenin. For LEF1 1–62 the amount of DNA (ng per well) is indicated.
To obtain further evidence that Smad-dependent activation of LEF1-target genes can occur in the absence of β-catenin, we expressed the β-catenin-binding domain of LEF1, LEF1 1–62, and examined its effect on Smad-dependent induction of the Xtwn reporter. Increasing expression of LEF1 1–62 efficiently blocked LEF1-dependent activation of Xtwn (Fig. 6D), and this decrease was reversed by coexpression of FL β-catenin. Importantly, although the overall level of luciferase activity was reduced as a result of the loss of β-catenin-mediated signaling, TGFβ- and Smad-dependent induction of luciferase activity was still observed in the presence of LEF1 1–62. Although these results need to be confirmed by using endogenous proteins in vivo, our observations indicate that in the absence of β-catenin binding, Smads can stimulate transcriptional activation of the Xtwn and Twntop promoters in a LEF1-dependent manner. Moreover, the results show that activation of both TGFβ and Wnt pathways results in cooperative enhancement of Xtwn and Twntop promoter activation.
Discussion
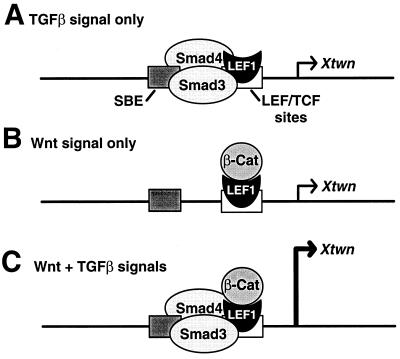
TGFβ and Wnt proteins are two groups of secreted proteins that can cooperate to regulate various developmental events (1, 2). Herein, we provide a molecular description of the mechanism for this cooperative effect. We demonstrate that synergistic activation of a specific Wnt and TGFβ target gene is mediated by a physical association between Smads and LEF1/TCFs. Furthermore, we show that both Smad and LEF1/TCF DNA-binding sites are required in the Xtwn promoter for this cooperative effect. These data are consistent with observations on other TGFβ target genes that show that Smads are recruited to specific regulatory elements through their association with distinct DNA-binding proteins and that, once recruited, Smad binding to DNA at adjacent sites stabilizes the transcriptional activation complex (3–7). Importantly, we observed that another LEF1-regulated element, Topflash, which does not contain Smad-binding sites, was insensitive to TGFβ signaling. Thus, we propose a model in which Smad regulation of LEF1 target elements depends not only on the physical association of the two proteins but also on the presence of a SBE adjacent to the LEF1 site (Fig. 7). This mechanism may provide the basis for promoter specificity in TGFβ-dependent regulation of LEF1 target genes, because LEF1-binding sites that are not adjacent to a SBE will not be regulated by the Smad pathway. Mapping of the domains on LEF1 that mediate protein–protein interactions showed that β-catenin and Smad3 bind to different regions on LEF1, namely, the amino terminus and the HMG box, respectively. Thus, distinct binding sites for β-catenin and Smads allows for simultaneous association of these proteins with LEF1 and provides a mechanism for the synergistic activation of specific target genes.
Figure 7.
A model for activation of specific target genes by TGFβ and Wnt pathways. Promoters with SBEs adjacent to the LEF1/TCF-binding sites can be activated by the TGFβ pathway in the absence of β-catenin (A), by Wnt signaling alone (B), or synergistically in the presence of both TGFβ and Wnt signals (C).
In addition to their cooperative activity, TGFβ and Wnt pathways can also independently regulate LEF1 target genes (Fig. 7). Wnt-dependent activation of LEF1 through stabilization of β-catenin is well established (8–10), and in this study, we show that TGFβ can stimulate activation of LEF1 target genes in the absence of β-catenin-binding to LEF1. β-catenin-independent activation of the TCRα enhancer has also been reported (21), suggesting that other transcriptional activators can regulate LEF1 through β-catenin-independent binding domains. In the future, it will be important to confirm whether β-catenin-independent activation of LEF1 target genes can occur with endogenous proteins in vivo.
Intriguingly, a recent report suggests that the common Smad, Smad4, plays an important role in Wnt-dependent activation of LEF1 target genes that occurs independently of TGFβ/activin signaling (16). It was shown that activation of the Wnt pathway alone enhanced Smad4/β-catenin interaction and, in Xenopus animal caps, resulted in a concomitant nuclear accumulation of both proteins. The use of Smad4 as a common mediator of both Wnt and TGFβ signaling adds an additional level of complexity to how these two distinct pathways might control multiple developmental events.
Our analysis of two LEF1-regulated reporter genes together with studies in Xenopus and Drosophila suggest that interaction between components of the TGFβ and Wnt signaling pathways is likely to play an important role during development. We propose that context-dependent dual regulation of distinct genes is important for the specification of diverse cell fates and that the cooperation of two pathways permits tight control of critical developmental processes.
Acknowledgments
We are grateful to R. Grosschedl for providing pCG-mLEF1-HA, P. Hamel and K. Boras for pCG-mLEF1-myc, W. Birchmeier for pBAT-myc-β-catenin, B. Vogelstein for the Topflash reporter, and J. Wrana for discussions. This work was supported by grants to L.A. from the Medical Research Council of Canada and the National Cancer Institute of Canada with funds from the Canadian Cancer Society. L.A. is a Medical Research Council scholar. E.L. is supported by an Medical Research Council studentship, and A.L. is supported by the Beca Postdoctoral del Gobierno Vasco.
Abbreviations
- TGFβ
transforming growth factor-β
- LEF1
lymphoid enhancer binding factor
- TCF
T cell-specific factor
- SBE
Smad-binding element
- GST
glutathione S-transferase
- HA
hemagglutinin
- EMSA
electrophoretic mobility-shift assay
- FL
full-length
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.150152697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.150152697
References
- 1.Cadigan K M, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 2.Whitman M. Genes Dev. 1998;12:2445–2462. doi: 10.1101/gad.12.16.2445. [DOI] [PubMed] [Google Scholar]
- 3.Massagué J. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 4.Wrana J L. Cell. 2000;100:189–192. doi: 10.1016/s0092-8674(00)81556-1. [DOI] [PubMed] [Google Scholar]
- 5.Derynck R, Zhang Y, Feng X-H. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 6.Heldin C-H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 7.Attisano L, Wrana J L. Curr Opin Cell Biol. 2000;12:235–243. doi: 10.1016/s0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- 8.Eastman Q, Grosschedl R. Curr Opin Cell Biol. 1999;11:233–240. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 9.Willert K, Nusse R. Curr Opin Genet Dev. 1998;8:95–102. doi: 10.1016/s0959-437x(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 10.Clevers H, van de Wetering M. Trends Genet. 1997;13:485–489. doi: 10.1016/s0168-9525(97)01305-x. [DOI] [PubMed] [Google Scholar]
- 11.Crease D J, Dyson S, Gurdon J B. Proc Natl Acad Sci USA. 1998;95:4398–4403. doi: 10.1073/pnas.95.8.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein T, Martinez Arias A. Development (Cambridge, UK) 1999;126:913–925. doi: 10.1242/dev.126.5.913. [DOI] [PubMed] [Google Scholar]
- 13.Riese J, Yu X, Munnerlyn A, Eresh S, Hsu S-C, Grosschedl R, Bienz M. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- 14.Laurent M N, Blitz I L, Hashimoto C, Rothbächer U, Cho K W-Y. Development (Cambridge, UK) 1997;124:4905–4916. doi: 10.1242/dev.124.23.4905. [DOI] [PubMed] [Google Scholar]
- 15.Labbé E, Silvestri C, Hoodless P A, Wrana J L, Attisano L. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 16.Nishita M, Hashimoto M K, Ogata S, Laurent M N, Ueno N, Shibuya H, Cho K W Y. Nature (London) 2000;403:781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- 17.Macìas-Silva M, Hoodless P A, Tang S J, Buchwald M, Wrana J L. J Biol Chem. 1998;273:25628–25636. doi: 10.1074/jbc.273.40.25628. [DOI] [PubMed] [Google Scholar]
- 18.de La Coste A, Romagnolo B, Billuart P, Renard C A, Buendia M A, Soubrane O, Fabre M, Chelly J, Beldjord C, Kahn A, et al. Proc Natl Acad Sci USA. 1998;95:8847–8851. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macías-Silva M, Abdollah S, Hoodless P A, Pirone R, Attisano L, Wrana J L. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 20.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 21.Hsu S-C, Galceran J, Grosschedl R. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roose J, Huls G, van Beest M, Moerer P, van der Horn K, Goldschmeding R, Logtenberg T, Clevers H. Science. 1999;825:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]