Abstract
Cyclic nucleotide phosphodiesterases (PDEs) hydrolyze the intracellular second messengers cAMP and cGMP to their corresponding monophosphates. PDEs play an important role in signal transduction by regulating the intracellular concentration of cyclic nucleotides. We have previously shown that the individual haplotype GAACC in the PDE11A gene was associated with major depressive disorder (MDD) based on block-by-block analysis. There are two PDE genes, PDE11A and PDE1A, located in chromosome 2q31–q32. In this study, we have further explored whether the whole region 2q31–q32 contribute to MDD or antidepressant response 278 depressed Mexican-American participants and 321 matched healthy controls. Although there is no significant interaction between the two genes, the remission rate of individual carrying the combination genotype at rs1880916 (AG/AA) and rs1549870 (GG) is significantly increased. We analyzed the global haplotype by examining 16 single-nucleotide polymorphisms (SNPs) in PDE11A and six SNPs in PDE1A. None of the haplotypes consisting of six SNPs in the PDE1A have a significant difference between depressed and control groups. Among haplotypes consisting of 16 SNPs across 440 kb in the PDE11A gene, 18 common haplotypes (with frequency higher than 0.8%) have been found in the studied population. Six haplotypes showed significantly different frequencies between the MDD group and the control group. The phylogenetic network result for the 16 SNPs showed that several historic recombination events have happened in the PDE11A gene. The frequency of one haplotype is significantly lower in the remitter group than in the nonremitter group for the depressed participants treated with either desipramine or fluoxetine. Thus, our data suggest that the PDE11A global haplotype is associated with both MDD and antidepressant drug response.
Keywords: PDE11A, major depression, antidepressant, haplotype, linkage disequilibrium, recombination
Introduction
The 3′, 5′-cyclic nucleotides cAMP and cGMP function as second messengers in a wide variety of signal transduction pathways. 3′, 5′-cyclic nucleotide phosphodiesterases (PDEs) catalyze the hydrolysis of cAMP and cGMP to the corresponding 5’-monophosphates and provide a mechanism to downregulate cAMP- and cGMP-signaling. Human PDEs are composed of 21 genes and are categorized into 11 families based on sequence homology, enzymatic properties, and sensitivity to inhibitors.1–3
PDE11A gene is the most recently identified and cloned member of the PDE protein superfamily.4 Four N-terminal transcript variants (PDE11A1, A2, A3, and A4) encoding different isoforms have been found for this gene.5,6 The PDE11A1, A2, and A3 have a catalytic domain and an N-terminus GAF domain, while the full-length form, PDE11A4, contains two GAF domains and a catalytic domain.4–7 The GAF domain in PDE11A is homologous to other signaling molecule as found in PDE2, PDE5, PDE6, and PDE10.1–3,8 The GAF domain is involved in high affinity allosteric cGMP and other small signaling molecules binding. Because of their unique ligand-binding topologies, the GAF domains of PDEs are likely to offer good new targets for rational drug design.9 PDE11A exhibits properties of dual-substrate PDE and hydrolyzes both cAMP and cGMP into AMP and GMP respectively with similar Km values.4–6 The Km for cAMP and cGMP were similar for all the four PDE11A variants.4–6
Haplotypes provide a record of evolution history more accurately than individual single-nucleotide polymorphisms (SNPs), they can also capture linkage disequilibrium (LD) and recombination hotspots information of a genomic region more completely. Individual haplotype results detected by haplotype blocks may vary considerably from different algorithms since the methods for block definition are still under devising and no single approach is appropriate for all datasets.10 If a recombination hotspot occurs within one gene, using this gene as candidate may not be appropriate. An alternative to individual haplotype association study is to perform multimarker association analysis with multiple adjacent markers,11 that is global haplotype. In unrelated individuals, PHASE program has been widely used to infer the most likely haplotype that an individual has inherited.11 Since we previously demonstrated a significant association between an individual haplotype in the PDE11A gene and major depressive disorder (MDD),12 and two PDE genes (PDE11A and PDE1A) are located in chromosome 2q31–q32, we now evaluate if the whole region 2q31–q32 is associated with MDD by analyzing the gene–gene interaction. Combinations of the individual haplotypes in multiple loci might determine the complexity of an individual’s susceptibility for MDD and outcome of antidepressant treatment. Therefore, we aimed to do the haplotype phasing before block partition using PHASE software by examining 16 SNPs across the PDE11A and six SNPs on the PDE1A gene in this study.
Materials and methods
Participants
This study was approved by the IRBs of the University of California Los Angeles and the University of Miami, and it has been registered in ClinicalTrials.gov (NCT00265292). Genomic DNA samples from 599 Mexican-Americans were used in this study: 278 depressed participants enrolled in a pharmacogenetic study of antidepressant treatment response to desipramine or fluoxetine, and 321 age- and sex-matched control participants recruited from the same Mexican-American community in Los Angeles.12,13 Controls were in general good health but were not screened for medical or psychiatric illness. Inclusion criteria included DSM-IV diagnosis of a current unipolar major depressive episode, with a 21-item Hamilton Depression Rating Scale (HAM-D21) score14 of ≥18 with item number 1 (depressed mood) rated ≥2. Detailed inclusion and exclusion criteria for both control and depressed participants were described previously.12,13,15 We used diagnostic and ratings instruments that have been fully validated in English and Spanish, and all assessments were conducted in the participants’ primary language. All Mexican-American participants aged 21–68 years had at least three grandparents born in Mexico. Written informed consent was obtained from each participant.
All patients had an initial comprehensive psychiatric and medical assessment and, if enrolled, had nine weeks of structured follow up assessments. The study consisted of two phases: a 1-week single-blind placebo lead-in phase to minimize the impact of placebo responders followed, if participants continue to meet the inclusion criteria after phase 1, by random assignment to one of the two treatment groups: fluoxetine 10–40 mg/day or desipramine 50–200 mg/day, administered in a double-blind manner for eight weeks, with blind dose escalation based on clinical outcomes. In the depressed group, 230 participants received treatment in our double-blind clinical trial. Of those, 143 patients completed the study, 61 attained remission, defined as having a final HAM-D21 score of <8, and the remainder (n = 82) were categorized as nonremitters.
Genotype procedure
The Illumina human SNP linkage analysis platform (Illumina, San Diego, CA) was used for genotyping.16,17 Briefly, all genomic DNAs were quantitated using PicoGreen and diluted into 100 ng/μl concentration. Then, the DNAs were transferred into seven bar-coded plastic 96-well conical microtiter plates. Genotyping of 27 SNPs on the PDE1A and PDE11A genes was done at the Wellcome Trust Sanger Institute (Hinxton, UK) using the Golden Gate assay (Illumina) and following the manufacturers’ instruction.16 All genotypes were evaluated using a quantitative quality score called GenCall score. A GenCall score ranges from 0 to 1 and reflects the proximity within a cluster plot of the intensities of that genotype to the centroid of the nearest cluster. Poorly performing samples were removed from the genotyping report files, and individual genotypes with Gen-Call scores below 0.25 were assigned a nocall. Data quality was assessed by duplicate DNAs (n = 26) across all plates. Genotypes from nonmatching duplicates were dropped; they were also dropped if they had one missing data point. Deviation from Hardy–Weinberg equilibrium (HWE) was tested separately for the control and depressed groups by using the ALLELE procedure in SAS/Genetics 9.1.3 (SAS Institute, Cary, NC). SNPs that were not in HWE in the control group (p < 0.05) and SNPs that were monoallelic in both groups were excluded. After the cleaning and filtering steps, 599 DNA samples genotyped for 22 SNPs on the PDE11A and PDE1A genes (Table 1) were used for the global haplotype analysis. Given the number of comparisons by a Bonferonni or similar adjustment would be too conservative, thus we have stated the nominal p-value so that the reader can assess the magnitude of significance. Chi-square test with Monte Carlo permutation test and Fisher’s exact test conducted to evaluate the association between the SNP and MDD as well as anti-depressant drug response. We concluded that the allele was associated with MDD or antidepressant drug response when all the methods had the concordant association results.
Table 1.
List of the 22 SNPs on chromosome 2q31–q32 that have been included for global haplotype analysis
| Order | SNPa | Allele | Chromosome position | Gene | Region | Distance between SNPs (BP) |
Minor allele frequency |
P valueb | |
|---|---|---|---|---|---|---|---|---|---|
| Depression | Control | ||||||||
| 1 | rs1997207 | C/G | 178331945 | PDE11A | Intron 19 | 0.06115 | 0.07886 | 0.2339 | |
| 2 | rs3770018 | A/C | 178366311 | PDE11A | Intron 16 | 34,367 | 0.05755 | 0.112 | 0.000863 |
| 3 | rs1465831 | G/T | 178394581 | PDE11A | Intron 13 | 28,271 | 0.02698 | 0.04416 | 0.113 |
| 4 | rs3770034 | G/A | 178426123 | PDE11A | Intron 10 | 31,543 | 0.1367 | 0.1577 | 0.3077 |
| 5 | rs1866212 | A/G | 178428793 | PDE11A | Intron 10 | 2671 | 0.1787 | 0.1577 | 0.3342 |
| 6 | rs1561320 | T/C | 178429529 | PDE11A | Intron 10 | 737 | 0.1241 | 0.1562 | 0.1131 |
| 7 | rs1866209 | G/A | 178432866 | PDE11A | Intron 10 | 3,338 | 0.1241 | 0.1562 | 0.1131 |
| 8 | rs1370656 | A/C | 178433505 | PDE11A | Intron 10 | 640 | 0.06835 | 0.09621 | 0.08242 |
| 9 | rs998059 | G/T | 178437777 | PDE11A | Intron 10 | 4273 | 0.1241 | 0.1577 | 0.09731 |
| 10 | rs1438048 | C/A | 178438353 | PDE11A | Intron 10 | 577 | 0.1241 | 0.1562 | 0.1131 |
| 11 | rs3770045 | C/G | 178454734 | PDE11A | Intron 10 | 16,382 | 0.2428 | 0.2855 | 0.09622 |
| 12 | rs1370661 | G/A | 178529090 | PDE11A | Intron 6 | 74,357 | 0.2104 | 0.235 | 0.3098 |
| 13 | rs2037757 | G/A | 178561572 | PDE11A | Intron 5 | 32,483 | 0.3201 | 0.3785 | 0.03523 |
| 14 | rs1405647 | A/C | 178584369 | PDE11A | Intron 4 | 22,798 | 0.2176 | 0.235 | 0.4748 |
| 15 | rs1880916 | G/A | 178620937 | PDE11A | Intron 2 | 36,569 | 0.1151 | 0.142 | 0.1686 |
| 16 | rs744397 | T/C | 178684625 | PDE11A | Intron 2 | 63,689 | 0.1619 | 0.1845 | 0.3033 |
| 17 | rs1432531 | A/G | 182833008 | PDE1A | Intron 14 | 4,148,384 | 0.2518 | 0.2508 | 0.968 |
| 18 | rs1991765 | A/G | 182864440 | PDE1A | Intron 14 | 31,433 | 0.2284 | 0.224 | 0.855 |
| 19 | rs1432534 | G/T | 182882952 | PDE1A | Intron 11 | 18,513 | 0.2176 | 0.2271 | 0.6942 |
| 20 | rs3769788 | A/G | 182884887 | PDE1A | Intron 11 | 1936 | 0.2302 | 0.2256 | 0.8482 |
| 21 | rs1432518 | A/G | 182923743 | PDE1A | Intron 5 | 38,857 | 0.3489 | 0.3281 | 0.4481 |
| 22 | rs1549870 | A/G | 182943110 | PDE1A | Intron 3 | 19,368 | 0.05576 | 0.05047 | 0.6847 |
Notes: aThree SNPs, rs1866209, rs998059, and rs1438048, were first reported here. Other SNPs have been analyzed in previous published paper;12
The p values correspond to the comparison of allele frequencies between the depression and control groups.
Abbreviations: BP, base pair; SNP, single-nucleotide polymorphisms; PDE, phosphodiesterase.
A confounding factor that may impact the internal validity of case-control studies is the presence of population stratification. We used two approaches to test for the hidden population stratification in our data.18 Firstly, 54 unlinked SNPs across 22 autosomal chromosomes were employed to analyze a combined sample with the genotype data download from three HapMap ethnic samples using STRUCTURE program. Three distinct clusters were identified with an average proportion of at least 92% of individuals correctly assigned to the given ethnic populations (CEU, CHB +JPT, YRI). We then used this panel of SNPs to test our sample and observed an almost equal proportion assigned to each clusters given K = 2, 3, 4 in both cases and controls. Secondly, genotype frequencies from each of the 54 unlinked SNPs were compared between cases and controls using the previous described method.18 In our study population, no significant difference was found based on an overall test statistic (χ2 = 100.50, df = 108, p = 0.68). Therefore, our data show that there is no significant evidence of stratification in the study population.
Haplotype analysis with PHASE software and statistic analysis
Maximum likelihood estimates of haplotype frequencies were calculated in control, depressed, remitter, and non-remitter groups using the program PHASE (v2.1.1).19,20 The haplotypes were estimated with PHASE in the whole group of participants, and then compared their distribution between groups. The phase reconstruction method regards the unknown haplotypes as unobserved random quantities and aims to evaluate their conditional distribution in light of the genotype data via using Markov chain–Monte Carlo algorithm. We analyzed the haplotype diversity including 16 SNPs on the PDE11A gene and six SNPs on the PDE1A gene. The haplotypes obtained by PHASE have been confirmed by other two haplotype estimation approaches, ie, PLINK v-1.01 and Haploview v-4.0. As shown in Tables 1 and 3, for identification and naming purposes, the order of SNPs for haplotyping started with the intron 19 of PDE11A and continued 3′ to 5′ up the PDE11A and PDE1A genes. Fisher’s exact test was conducted to evaluate the association between the haplotype/genotype, and MDD as well as anti-depressant drug response. Fisher’s exact test is a procedure that one can use for data in a two by two contingency table and allows one or two of the cell values less than five.
Table 3.
Estimated frequency for 16-locus (SNP1–16)a haplotype in depression (MDD), control groups (CT), remitter (R), and nonremitter groups (NR)
| Order | Sequenceb |
Diagnosis |
Antidepressant treatment |
Note | ||||
|---|---|---|---|---|---|---|---|---|
| MDD (%) N = 556 | CT (%) N = 642 | p value | R (%) N = 164 | NR (%) N = 122 | p value | |||
| H1 | CAGAGCACGCCAAAGC | 341 (61.3) | 374 (58.3) | 0.288 | 104 (63.4) | 73 (59.8) | 0.541 | Common |
| H2 | CAGGATGCTAGGGCGC | 27 (4.7) | 26 (4.1) | 0.574 | 7 (4.3) | 6 (4.9) | 0.783 | MDD |
| H3 | CAGAGCACGCGAGAAT | 26 (4.7) | 49 (7.6) | 0.042 | 10 (6.1) | 6 (4.9) | 0.797 | Common |
| H4 | CAGAACACGCCAAAGC | 18 (3.2) | 0 (0) | <0.001 | 6 (3.7) | 4 (3.3) | 1.000 | MDD |
| H5 | CAGAGCACGCCGGCGC | 14 (2.5) | 19 (3.0) | 0.598 | 4 (2.4) | 5 (4.1) | 0.503 | Common |
| H6 | CAGGATGATACGGCGC | 9 (1.6) | 8 (1.3) | 0.631 | 3 (1.8) | 1 (0.8) | 0.639 | Common |
| H7 | GAGAGCACGCCAAAGC | 8 (1.4) | 15 (2.3) | 0.205 | 0 (0) | 3 (2.5) | 0.077 | Normal |
| H8 | CAGAGCACGCGGGCGC | 8 (1.4) | 12 (1.9) | 0.654 | 0 (0) | 5 (4.1) | 0.013 | Common |
| H9 | GAGAGCACGCGAGAAT | 8 (1.4) | 10 (1.6) | 1.000 | 2 (1.2) | 0 (0) | 0.509 | Common |
| H10 | CAGAGCACGCGGGCGT | 6 (1.1) | 4 (0.6) | 0.528 | 1 (0.6) | 3 (2.5) | 0.316 | Common |
| H11 | CCGGATGATACGGCGC | 6 (1.1) | 17 (2.7) | 0.057 | 0 (0) | 2 (1.6) | 0.181 | Normal |
| H12 | CAGGATGATACAGAAT | 5 (0.9) | 0 (0) | 0.021 | 1 (0.6) | 0 (0) | 1.000 | MDD |
| H13 | CAGGATGATAGGGCGC | 5 (0.9) | 5 (0.8) | 0.759 | 1 (0.6) | 0 (0) | 1.000 | Common |
| H14 | CCGAGCACGCGAGAAT | 4 (0.7) | 9 (1.4) | 0.280 | 1 (0.6) | 0 (0) | 1.000 | Normal |
| H15 | CAGAGCACGCCAGAAT | 0 (0) | 6 (0.9) | 0.033 | 0 (0) | 0 (0) | 1.000 | Normal |
| H16 | CCGAGCACGCGGGCGC | 0 (0) | 8 (1.3) | 0.009 | 0 (0) | 0 (0) | 1.000 | Normal |
| H17 | CCGGATGCTAGGGCGT | 0 (0) | 5 (0.8) | 0.065 | 0 (0) | 0 (0) | 1.000 | Common |
| H18 | GAGAGCACGCCGGCGC | 0 (0) | 7 (1.1) | 0.017 | 0 (0) | 0 (0) | 1.000 | Normal |
| Total | 485 (87.2) | 574 (89.4) | 140 (85.4) | 109 (89.3) | ||||
Notes: aSee Table 1 for definitions of the SNP1–16, and the haplotype is from 3′ to 5′ in PDE11A;
The SNPs are bold and underlined in the haplotype that is different with most common haplotype.
Abbreviations: MDD, major depressive disorder; SNP, single-nucleotide polymorphisms; PDE, phosphodiesterase.
Additive gene–gene interaction was analyzed to test the combined effect from both PDE11A and PDE1A for antidepressant response. Briefly, Rothman’s synergy index (S)21 measure proposed by Rothman was used to assess the joint effect of the two SNPs, rs1880916 on the PDE11A gene and rs1549870 on the PDE1A gene. The S index is the ratio of the observed joint effect divided by the expected joint effect assuming additivity of the effects, defined as: S = (OR11−1)/(OR10 +OR01−2) in which subscript 0 denotes the absence of the risk genotype at the SNP and OR denotes the odds ratio. No interaction corresponds to S = 1, whereas S > 1 (S < 1) can be interpreted as a measure of relative increase (decrease) in the effect among those exposed to risk genotypes at both SNPs.
Phylogenetic network analysis in the PDE11A gene
To analyze the phylogenetic relationship of haplotypes consisting of 16 SNPs in the PDE11A gene among 278 depressed Mexican-Americans and 321 controls, a reduced-median-network was constructed using Network 4.2.0.1 program (http://www.fluxus-engineering.com/).22
Results
SNPs associated with MDD and antidepressant response
For the 22 SNPs crossing region 2q31–q32 (Table 1), three SNPs, including rs1866209, rs998059, and rs1438048, are first reported here to examine the PDE11A gene further density. Two SNPs on the PDE11A gene, rs3770018 and rs2037757, are associated with MDD; another two SNPs, rs1880916 on the PDE11A gene and rs1549870 on the PDE1A gene, are associated with attaining remitter and nonremitter status within the entire depressed group treated with either desipramine and fluoxetine, which is concordance with our previous findings.12 The remission rate of individuals carrying the minor allele A at rs1880916 in PDE11A or the homozygous for the major allele G at rs1549870 in PDE1A is about 50% (Table 2). Moreover, the remission rate is 81% in the combined genotype at rs1880916 (AG/AA) and rs1549870 (GG). The dramatically increased remission rate suggested that the combination genotype of these two SNPs could predict the antidepressant response, although there is no significant interaction between the two genes (S ≈ 1; Table 2).
Table 2.
Combined effect of PDE11A and PDE1A genes on the antidepressant response
| No. of risk allele | rs1880916 | rs1549870 | Remitter/nonremitter | Remission rate % | RR (95% CI) | pvalue |
|---|---|---|---|---|---|---|
| 0 | GG | AG/AA | 2/11 | 0.15 | 1.00 | – |
| 1 | GG | GG | 55/42 | 0.57 | 3.69 (1.02–13.34) | 0.047 |
| 2 | AG/AA | AG/AA | 3/3 | 0.50 | 3.25 (0.72–14.64) | 0.125 |
| 3 | AG/AA | GG | 22/5 | 0.81 | 5.30 (1.46–19.19) | 0.011 |
Note: Rothman synergy index = (5.30–1)/(3.25 +3.69 − 2) ≈1.
Abbreviations: CI, confidence interval; RR, relative risk.
Haplotype diversity in the PDE11A gene in depression and control groups
There is no significant frequency difference between the MDD and control groups when running haplotype analysis for six SNPs in the PDE1A gene. When including 16 SNPs across 440kb in the PDE11A gene, a total of 70 different haplotypes were identified. The haplotype H1 (3′-CAGAGCACGCCAAAGC-5′), with the highest frequency in the population, has frequencies of 61.3%, 58.3%, 63.8%, and 59.8% in the MDD, control, remitter, and nonremitter groups, respectively (Table 3). Eighteen common haplotypes (with frequency higher than 0.8%) account for frequencies of 87.2%, 89.4%, 85.4%, and 89.3% in MDD, control, remitter, and non-remitter groups, respectively (Table 3). The frequency of two haplotypes (H4: 3′-CAGAACACGCCAAAGC-5′ and H12: 3′-CAGGATGATACAGAAT-5′) is significantly higher in the MDD group than in the control group (p < 0.05). The haplotype H4 consists of the individual haplotype GAACC, which has been previously reported to be associated with MDD.12 Four other haplotypes (H3, H15, H16, and H18) have lower frequency in the MDD group than that in control group (p < 0.05). We also calculated the first ten highest frequency genotypes in the MDD group and compared between the MDD and control groups. The frequencies of genotypes H1/H4 and H2/H4 are significant higher in MDD than that of control group (Table 4). Therefore, combined with the genotype distribution between the MDD and control groups, the frequencies of the haplotypes H2, H4, and H12 are higher in the depressed group, and the frequencies of the haplotypes H7, H11, H14, H15, H16, and H18 are lower in this same group (Tables 3 and 4). The frequency of the haplotype H8: 3′-CAGAGCACGCGGGCGC-5′ is significantly lower in the remitter than that of nonremitter group for the depressed participants treated with either desipramine or fluoxetine (Table 3).
Table 4.
Estimated frequency for 16-locus (SNP1–16)a genotype in depression (MDD), control groups (CT), remitter (R), and nonremitter groups (NR)
| Genotype |
Diagnosis |
Antidepressant response |
||
|---|---|---|---|---|
| MDD (%) (N = 278) | CT (%) (N = 321) | R (%) (N = 82) | NR (%) (N = 61) | |
| H1/H1b | 115 (41.4) | 120 (37.4) | 34 (41.5) | 26 (42.6) |
| H1/H2 | 14 (5.0) | 14 (4.4) | 5 (6.1) | 3 (4.9) |
| H1/H3 | 18 (6.5) | 26 (8.1) | 8 (9.8) | 4 (6.6) |
| H1/H4 | 8 (2.9) | 0 (0)** | 3 (3.7) | 1 (1.6) |
| H1/H5 | 10 (3.6) | 13 (4.0) | 3 (3.7) | 4 (6.6) |
| H1/H6 | 8 (2.9) | 5 (1.6) | 2 (2.4) | 1 (1.6) |
| H1/H8 | 4 (1.4) | 7 (2.2) | 0 (0) | 2 (3.3) |
| H1/H9 | 5 (1.8) | 6 (1.9) | 2 (2.4) | 0 (0) |
| H2/H3 | 4 (1.4) | 4 (1.2) | 1 (1.2) | 2 (3.3) |
| H2/H4 | 4 (1.4) | 0 (0)* | 1 (1.2) | 1 (1.6) |
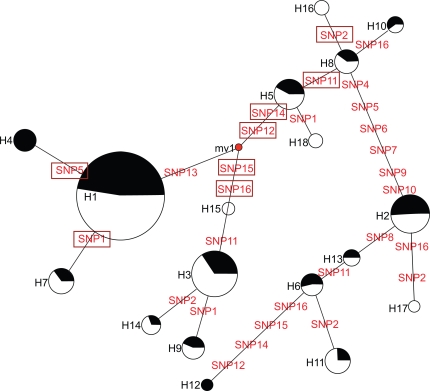
The results from both linkage disequilibrium analysis and phylogenetic network for the 16 SNPs showed that eight historic recombinations have happened in the PDE11A gene, including SNP1, 2, 5, 11, 12, 14, 15, and 16, showed in the boxes in Figure 1.
Figure 1.
Phylogenetic network for the 16 SNPs of the PDE11A haplotype. The reduced median network constructed using software Network 4.2.0.1. The circles represent the haplotypes shown in Table 2. The circle size was proportional to haplotype frequency. The black and white colors represent the depressed and control groups, respectively. The distances between haplotypes were indicated by SNPs (SNP1–16 in Table 1). The mv1 means median vector 1. A median vector is a hypothesized sequence that is required to connect existing sequences within the network with maximum parsimony principle. The boxed SNPs were probable recombination loci.
Discussion
The ultimate goal of our ongoing research is to find genetic variations that are associated with susceptibility to depression/antidepressant drug response and to use this information to identify genetic markers. Several lines of investigation support that hyperactivity of the hypothalamic-pituitary-adrenal (HPA) axis resulting from hyperactive hypothalamic corticotropin-releasing hormone (CRH) neurons and elevated cortisol levels are associated with major depression.23,24 It is well known that Cushing syndrome25 may be caused by increased levels of cortisol resulting from adrenocorticotropic hormone (ACTH)-producing pituitary tumors that are resistant to glucocorticoid negative feedback. Increased cAMP signaling has been linked to genetic forms of cortisol excess and leads to adrenocortical tumors and Cushing syndrome26 and cAMP levels are regulated by PDEs. Recently, a genetic form of Cushing has been linked to PDE dysfunction. Genome-wide SNP genotyping study of adrenocortical tumor tissues showed that they had 2q31–2q35 loss of heterozygosity (LOH), decreased protein expression and high cyclic nucleotide levels and cAMP-responsive element-binding protein (CREB) phosphorylation.27 Mutations in the PDE11A gene are a cause of Cushing disease and adrenocortical hyperplasia.27,28 In this study, we tested the association of the region 2q31–q32, which includes the PDE11A and PDE1A genes, and MDD. We found the remission rate of individual who had the minor allele A at rs1880916 in PDE11A and homozygous for the major allele G at rs1549870 in PDE1A is dramatically increased. This result is potentially useful for depression treatment as a clinical guide in the future. However, we did not find association between PDE1A and MDD or antidepressant response by studying six-SNP haplotype on the PDE1A gene.
PDE11A contains a single GAF domain at the N terminus similar to but distinct from the tandem GAF domains found in PDE2, PDE5, PDE6, and PDE10.1–3,8 GAF acronym comes from the names of the first three classes of proteins recognized to contain this domain: mammalian cGMP-binding PDEs, Anabaena adenylyl cyclases, and Escherichia coli FhlA.29 There are now 2,334 GAF domains in 1,996 proteins in the nonredundant SMART domain database (as of 10/08/2007), of which only 15 (0.66%) GAF domains in 8 (0.41%) proteins pertain to humans and nearly all of the GAF domains identified are in PDEs.30,31 GAF domains in PDEs provide multiple functional roles and therefore PDEs genes may associate with the diagnosis and treatment outcome of MDD through modulating intracellular levels of cGMP. The GAF domain makes PDEs particularly attractive therapeutic targets for manipulation of cAMP and cGMP signaling. The PDE11A and PDE1A genes have been included in the study because (1) they are located within 2q31–35, (2) as we indicated previously, the PDE11A individual haplotype is associated with MDD and three SNPs on PDE11A and PDE1A are associated with antidepressant drug response,12 and (3) the PDE11A gene has the GAF domain. PDE11A functions as a dual-substrate PDE that regulate both cGMP and cAMP under physiological conditions,4–6 and is a large gene with 440 kb. Eight recombination spots have been found in the 16 SNPs crossing the PDE11A gene. The linkage disequilibrium map of the PDE11A gene in the studied Mexican-Americans is similar to the haplotype blocks defined in other studies.32 Our data showed that the global haplotype on the PDE11A gene is associated with MDD as well as antidepressant response, which is in concordance with our previous results from individual SNPs and individual haplotype analysis.12 Our result is different from another study, which did not find association between the variants in PDE11A and citalopram response.33 The difference may result from different population studied or different antidepressant medication used in two studies. We have found that the genotypes H1/H4 and H2/H4 have significantly higher frequency in the MDD group than in the control group. Furthermore, the haplotype H4 has a high frequency in the MDD with 3.2% and is absent in the control group, which rises the probability of the haplotype contribution to MDD. However, the present haplotypes could only be estimated statistically and the frequencies under 5% may result in inconsistent associations. Therefore, the real haplotypes of the PDE11A gene must be determined experimentally. This would provide an understanding of the mechanisms underlying the genetic variation in MDD and antidepressant drug response among the population and would enable the prediction of the risk of MDD. It might be possible to perform the required experiments using a recently reported new method for long-range haplotype determination.34 Also, some of the SNPs on the PDE11A gene should be tested if they affect enzymatic function in vitro with variable changes in cAMP and/or cGMP levels in lymphocyte blood cells.
In conclusion, we identified three PDE11A global haplotypes that are associated with MDD and one with antidepressant drug response. These findings suggest that drugs targeted to affect PDE function, particularly GAF-related PDEs, could represent a new treatment strategy for major depression. A more focused study of functions related to the haplotypes in the PDE11A gene will improve our understanding of how genetic factors might contribute to individual susceptibility for major depression and antidepressant drug response.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Conti M. Phosphodiesterases and cyclic nucleotide signaling in endocrine cells. Mol Endocrinol. 2000;14:1317–1327. doi: 10.1210/mend.14.9.0534. [DOI] [PubMed] [Google Scholar]
- 2.Lugnier C. Cyclic nucleotide phosphodiesterase (PDE) superfamily: a new target for the development of specific therapeutic agents. Pharmacol Ther. 2006;109:366–398. doi: 10.1016/j.pharmthera.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Omori K, Kotera J. Overview of PDEs and their regulation. Circ Res. 2007;100:309–327. doi: 10.1161/01.RES.0000256354.95791.f1. [DOI] [PubMed] [Google Scholar]
- 4.Fawcett L, Baxendale R, Stacey P, et al. Molecular cloning and characterization of a distinct human phosphodiesterase gene family: PDE11A. Proc Natl Acad Sci U S A. 2000;97:3702–3707. doi: 10.1073/pnas.050585197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hetman JM, Robas N, Baxendale R, et al. Cloning and characterization of two splice variants of human phosphodiesterase 11A. Proc Natl Acad Sci U S A. 2000;97:12891–12895. doi: 10.1073/pnas.200355397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuasa K, Kotera J, Fujishige K, et al. Isolation and characterization of two novel phosphodiesterase PDE11A variants showing unique structure and tissue-specific expression. J Biol Chem. 2000;275:31469–31479. doi: 10.1074/jbc.M003041200. [DOI] [PubMed] [Google Scholar]
- 7.Weeks JL, 2nd, Zoraghi R, Francis SH, et al. N-Terminal domain of phosphodiesterase-11A4 (PDE11A4) decreases affinity of the catalytic site for substrates and tadalafil, and is involved in oligomerization. Biochemistry. 2007;46:10353–10364. doi: 10.1021/bi7009629. [DOI] [PubMed] [Google Scholar]
- 8.Yuasa K, Kanoh Y, Okumura K, et al. Genomic organization of the human phosphodiesterase PDE11A gene. Evolutionary relatedness with other PDEs containing GAF domains. Eur J Biochem. 2001;268:168–178. doi: 10.1046/j.1432-1327.2001.01866.x. [DOI] [PubMed] [Google Scholar]
- 9.Martinez SE, Beavo JA, Hol WG. GAF domains: Two-billion-year-old molecular switches that bind cyclic nucleotides. Mol Interv. 2002;2:317–323. doi: 10.1124/mi.2.5.317. [DOI] [PubMed] [Google Scholar]
- 10.Cardon LR, Abecasis GR. Using haplotype blocks to map human complex trait loci. Trends Genet. 2003;19:135–140. doi: 10.1016/S0168-9525(03)00022-2. [DOI] [PubMed] [Google Scholar]
- 11.Silverman EK. Haplotype thinking in lung disease. Proc Am Thorac Soc. 2007;4:4–8. doi: 10.1513/pats.200607-145JG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong ML, Whelan F, Deloukas P, et al. Phosphodiesterase genes are associated with susceptibility to major depression and antidepressant treatment response. Proc Natl Acad Sci U S A. 2006;103:15124–15129. doi: 10.1073/pnas.0602795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Licinio J, O’Kirwan F, Irizarry K, et al. Association of a corticotropin-releasing hormone receptor 1 haplotype and antidepressant treatment response in Mexican-Americans. Mol Psychiatry. 2004;9:1075–1082. doi: 10.1038/sj.mp.4001587. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong ML, Dong C, Maestre-Mesa J, et al. Polymorphisms in inflammation-related genes are associated with susceptibility to major depression and antidepressant response. Mol Psychiatry. 2008;13:800–812. doi: 10.1038/mp.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan JB, Oliphant A, Shen R, et al. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 17.Gunderson KL, Kruglyak S, Graige MS, et al. Decoding randomly ordered DNA arrays. Genome Res. 2004;14:870–877. doi: 10.1101/gr.2255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pritchard JK. Modern Epidemiology. Philadelphia, PA: Lippincott-Raven Publishers; 1998. [Google Scholar]
- 19.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalilani L, Atashili J. Measuring additive interaction using odds ratios. Epidemiol Perspect Innov. 2006;3:5. doi: 10.1186/1742-5573-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandelt HJ, Forster P, Sykes BC, et al. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gold PW, Goodwin FK, Chrousos GP. Clinical and biochemical manifestations of depression. Relation to the neurobiology of stress (2) N Engl J Med. 1988;319:413–420. doi: 10.1056/NEJM198808183190706. [DOI] [PubMed] [Google Scholar]
- 24.Nemeroff CB, Widerlov E, Bissette G, et al. Elevated concentrations of CSF corticotropin-releasing factor-like immunoreactivity in depressed patients. Science. 1984;226:1342–1344. doi: 10.1126/science.6334362. [DOI] [PubMed] [Google Scholar]
- 25.Bilodeau S, Vallette-Kasic S, Gauthier Y, et al. Role of Brg1 and HDAC2 in GR trans-repression of the pituitary POMC gene and misexpression in Cushing disease. Genes Dev. 2006;20:2871–2886. doi: 10.1101/gad.1444606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourdeau I, Stratakis CA. Cyclic AMP-dependent signaling aberrations in macronodular adrenal disease. Ann N Y Acad Sci. 2002;968:240–255. doi: 10.1111/j.1749-6632.2002.tb04339.x. [DOI] [PubMed] [Google Scholar]
- 27.Horvath A, Boikos S, Giatzakis C, et al. A genome-wide scan identifies mutations in the gene encoding phosphodiesterase 11A4 (PDE11A) in individuals with adrenocortical hyperplasia. Nat Genet. 2006;38:794–800. doi: 10.1038/ng1809. [DOI] [PubMed] [Google Scholar]
- 28.Horvath A, Giatzakis C, Robinson-White A, et al. Adrenal hyperplasia and adenomas are associated with inhibition of phosphodiesterase 11A in carriers of PDE11A sequence variants that are frequent in the population. Cancer Res. 2006;66:11571–11575. doi: 10.1158/0008-5472.CAN-06-2914. [DOI] [PubMed] [Google Scholar]
- 29.Aravind L, Ponting CP. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 30.Letunic I, Copley RR, Pils B, et al. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultz J, Milpetz F, Bork P, et al. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 33.Teranishi KS, Slager SL, Garriock H, et al. Variants in PDE11A and PDE1A are not associated with citalopram response. Mol Psychiatry. 2007;2:1061–1063. doi: 10.1038/sj.mp.4002074. [DOI] [PubMed] [Google Scholar]
- 34.Zhang K, Zhu J, Shendure J, et al. Long-range polony haplotyping of individual human chromosome molecules. Nat Genet. 2006;38:382–387. doi: 10.1038/ng1741. [DOI] [PubMed] [Google Scholar]