Abstract
Solavetivone, a potent antifungal phytoalexin, is derived from a vetispirane-type sesquiterpene, premnaspirodiene, by a putative regio- and stereo-specific hydroxylation, followed by a second oxidation to yield the α,β-unsaturated ketone. Mechanistically, these reactions could occur via a single, multifunctional cytochrome P450 or some combination of cytochrome P450s and a dehydrogenase. We report here the characterization of a single cytochrome P450 enzyme, Hyoscyamus muticus premna-spirodiene oxygenase (HPO), that catalyzes these successive reactions at carbon 2 (C-2) of the spirane substrate. HPO also catalyzes the equivalent regio-specific (C-2) hydroxylation of several eremophilane-type (decalin ring system) sesquiterpenes, such as with 5-epi-aristolochene. Moreover, HPO displays interesting comparisons to other sesquiterpene hydroxylases. 5-Epi-aristolochene di-hydroxylase (EAH) differs catalytically from HPO by introducing hydroxyl groups first at C-1, then C-3 of 5-epi-aristolochene. HPO and EAH also differ from one another by 91-amino acid differences, with four of these differences mapping to putative substrate recognition regions 5 and 6. These four positions were mutagenized alone and in various combinations in both HPO and EAH and the mutant enzymes were characterized for changes in substrate selectivity, reaction product specificity, and kinetic properties. These mutations did not alter the regio- or stereo-specificity of either HPO or EAH, but specific combinations of the mutations did improve the catalytic efficiencies 10–15-fold. Molecular models and comparisons between HPO and EAH provide insights into the catalytic properties of these enzymes of specialized metabolism in plants.
Plants possess a wide range of defense mechanisms to combat pathogens including the accumulation of phytoanticipins, phytochemicals synthesized constitutively as a pre-existing defense barrier, and by the inducible accumulation of phytoalexins, natural products possessing anti-microbial activities (1). Interestingly, these chemical defenses often encompass specific chemical classes for specific plant families. For instance, plants within the legume family accumulate phenylpropanoid derivatives in response to pathogen challenge (2), whereas plants in the mustard family such as cabbage and Arabidopsis accumulate indole-based compounds (3). The biosynthesis of these diverse phytochemicals have historical and contemporary significance because of the broad biological activity of these specialized compounds (4) and because of considerable interest in defining the mechanisms responsible for the biosynthesis of such structurally complex molecules (5).
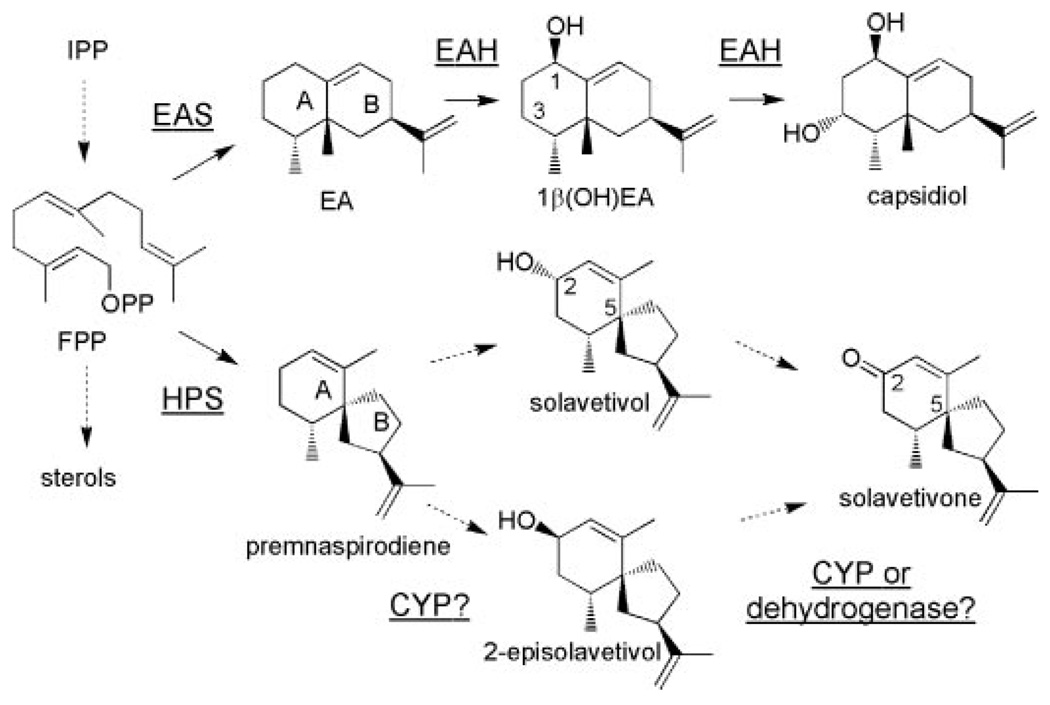
The biosynthesis of sesquiterpene phytoalexins by solanaceous plants, including potato, tobacco, tomato, and pepper, has also received considerable attention for several reasons. First, members of this family of compounds are known to inhibit mycelial growth of Phytophthora infestans (6) and Rhizoctonia solani (7), spore germination of Fusarium oxysporum (8), and the growth of the bacteria Staphylococcus aureus and Bacillus subtilis (9). Second, the induced biosynthesis of these phytoalexins coincides with a metabolic switch at a key branch point in the isoprenoid biosynthetic pathway. Under normal growth conditions, about 40% of the carbon flux through the mevalonate pathway is directed to C30 sterol biosynthesis. Upon pathogen or elicitor challenge, sterol biosynthesis is suppressed and ~20% of the carbon flux is diverted into a new branch pathway for C15 sesquiterpene biosynthesis (10, 11). Third, sesquiterpenes encompass over 300 structural classes including macro-, mono-, bi-, and tricyclic skeletons and a variety of rearranged structures (12). For instance, capsidiol, the dominant sesquiterpenoid accumulating in pepper and tobacco upon elicitation, has an eremophilane nucleus consisting of fused 6-membered rings, whereas potato and henbane (Hyoscyamus muticus) accumulate solavetivone, derived from a spirodecane structure (Scheme 1). Sesquiterpene synthases catalyze the biosynthesis of these hydrocarbon skeletons by ionizing the diphosphate substituent from the farnesyl diphosphate substrate leading to a series of carbocation intermediates en route to the final hydrocarbon reaction product.
SCHEME 1. Biosynthetic pathways for capsidiol and solavetivone.
FPP is diverted from the central mevalonate pathway via the action of sesquiterpene synthases (EAS and HPS)to generate EA and premnaspirodiene. Successive hydroxylations of EA at C-1 and C-3 are catalyzed by EAH to yield capsidiol. Successive hydroxylations of premnaspirodieneat C-2 with a preferred order, or hydroxylation followed by dehydration, for solavetivone biosynthesis has not been established. Relevant carbon atoms and rings are numbered.
Another reason for interest in sesquiterpene metabolism relates to the recent elucidations of the capsidiol biosynthetic pathway. Farnesyl diphosphate (FPP)2 is cyclized to 5-epi-aristolochene by the action of 5-epi-aristolochene synthase (EAS) (11, 13), and this hydrocarbon is then regio- and stereo-specifically hydroxylated at carbons 1 and 3 by epi-aristolochene dihydroxylase (EAH) (14). At a molecular level, EAH is a typical cytochrome P450 within the CYP71 family. However, unlike other CYP71 family members, EAH catalyzes an ordered, sequential dihydroxylation. The mechanistic details of the EAH-catalyzed reaction were previously reported using a variety of experimental approaches (15). Detailed kinetic analyses demonstrated an initial hydroxylation at C-1, followed by a more rapid hydroxylation at C-3. Kinetically, the second hydroxylation occurred under steady state conditions with a much lower Km and a higher turnover rate (kcat) for the C-1 monohydroxylated intermediate than for the C-3 monohydroxylated regioisomer. Site-specific mutations within substrate recognition domain (SRS) 6 (16), also abolished the second hydroxylation reaction without perturbing the catalytic efficiency (kcat/Km) of the first.
The complete biosynthetic pathway for solavetivone has yet to be defined (Scheme 1). Whereas the cyclization of FPP to the vetispirane hydrocarbon intermediate premnaspirodiene has been well documented (17), how the ketone carbonyl is introduced is unknown. One possibility is that a hydroxyl group is introduced at C-2 by a CYP-mediated mechanism followed by a subsequent oxidation to the ketone via the action of a dehydrogenase (Scheme 1). Alternatively, sequential hydroxylations at C-2 followed by elimination of H2O could yield solavetivone (Scheme 1). Precedent for both mechanisms can be found in the literature and include multiple hydroxylations at a single carbon position (18) or hydroxylation followed by a dehydrogenase-mediated oxidation (19). Nonetheless, both mechanisms are predicated on the initial regio-specific introduction of the hydroxyl group at C-2. Because early work demonstrated that the tobacco EAH was able to utilize premnaspirodiene (20) as a substrate, we reasoned that a similar hydroxylase might exist in henbane and that its gene could be identified based on sequence similarity to the tobacco EAH. Isolation of this gene would then allow for a more detailed and comparative dissection of evolutionarily related CYP-mediated oxidation reactions in specialized plant metabolism involving a chemically and biologically diverse group of sesquiterpenes.
EXPERIMENTAL PROCEDURES
Chemical Reagents
Standard laboratory reagents were purchased from Fisher, Sigma, and Aldrich. Authentic standards of EA, 4-epi-eremophilene (EE), 1β-hydroxy-EA (1β-(OH)EA), 3α-hydroxy-EA (3α-(OH)EA), and 3β-hydroxy-EA (3β-(OH)EA) were available from previous work (21). Premnaspirodiene was biosynthesized from FPP using premnaspirodiene synthase (17). Ten-ml reactions were set up with FPP (350 µg) and Hyoscyamus premnaspirodiene synthase (HPS) (1 mg) in 20-ml glass vials. After incubation at 30 °C for 20 min, premnaspirodiene was extracted with hexane (3 × 10 ml), and the combined extracts were applied to a 230–400-mesh silica gel column (0.5 × 4 cm). The hydrocarbon fraction was eluted with five column volumes of hexane. Combined hexane fractions were concentrated on ice under a N2 stream to limit loss of the volatile premnaspirodiene. The resultant highly pure oil, as determined by GC-MS profiling, was suspended in 500 µl of Me2SO for enzyme assays.
Amplification of the HPO cDNA from H. muticus mRNA
H. muticus cell suspension cultures, initially provided by Dr. Wayne Curtis (Pennsylvania State University), were maintained in B5 media with 0.2 mg/liter of 2,4-dichlorophenoxyacetic acid on a weekly subculturing regime. Cultures in rapid growth phase, 3 days after subculturing and doubling in mass every 24 h, were induced by the addition of a fungal elicitor preparation from Rhizoctonia solani at 100-µg glucose equivalents per ml of cell culture. The elicitor was prepared according to Ayers et al. (22) using fungal mycelia provided by Dr. Curtis. To increase the chances of isolating the full range of elicitor-inducible mRNAs, cells were collected at several time points (4, 6, and 8 h) and combined prior to RNA isolation.
Total RNA was extracted from the combined cells using the TRIzol reagent, followed by oligo(dT) cellulose column chromatography to isolate the poly(A)+ RNA fraction, both according to the manufacturer’s (Invitrogen) recommendations. First stand cDNA was subsequently generated by reverse transcription of isolated poly(A)+ RNA using oligo(dT) as the initial primer and Superscript II Reverse Transcriptase (Invitrogen), followed by PCR amplification using the primers shown in supplemental materials Fig. S1 according to Ralston et al. (14). The PCR primers were designed according to DNA sequence alignments of functionally characterized terpene hydroxylase genes 71D16 (GenBank accession number AF166332) and 71D20 (AF368376; EAH), and related CYP clones 71D4 (AJ296346), 71D6 (U48434), and 71D7 (U48435) that possess significant sequence similarity to EAH. The amplified cDNA fragments of appropriate size (>1.5 kb) were cloned into the pGEM-T Easy vector (Promega) for automated DNA sequencing using the Big-Dye Terminator Cycle sequencing protocol of PerkinElmer Life Sciences in combination with an ABI Prism 310 Genetic Analyzer (Applied Biosystems, Foster City, CA). The DNA and deduced amino acid sequences for the H. muticus clone, HPO or CYP71D55, based upon established nomenclature, are deposited in GenBank as accession number EF569601.
Yeast Expression and Microsome Preparations
Yeast expression vector (pYeDP60) harboring EAH or HPO was introduced into the WAT11 yeast strain, a strain previously engineered with an Arabidopsis NADPH-cytochrome P450 reductase gene (23) and kindly provided by Dr. P. Urban (Centre de Génétique Moléculaire, CNRS, Gif-sur-Yvette, France). Transformants were grown from single colonies, expression of the cDNAs was induced by galactose addition to the growth media, and microsomes were prepared as previously described (14).
CO Difference Spectra
CO difference spectra were measured to estimate the amount of properly folded P450s in microsomal preparations. The amount of functional P450 was calculated using the extinction coefficient of 91 mm−1 cm−1 at 450 nm (24). Wild type and mutant enzymes all exhibited an absorption maximum at 450 nm (supplemental materials Fig. S6).
CYP Assays
Standard assays were performed in 1-ml glass vials in a final reaction volume of 200 µl containing 4% (v/v) Me2SO, 2.4 mm NADPH, 100 mm Tris-HCl, pH 7.5, and micro-somes containing 90 pmol of HPO or EAH (measured by CO difference spectroscopy) per ml. Substrate concentrations varied from 6 to 60 µm for premnaspirodiene, 6 to 100 µm for valencene, 3 to 60 µm for EA, 5 to 50 µm for EE, and 5 to 60 µm for (−)-α-cedrene. After preincubation of the reaction mixtures at 30 °C for 5 min, reactions were initiated with the addition of 2.4 mm NADPH and allowed to proceed for 0.5 (premnaspirodiene to solavetivol), 5 (premnaspirodiene to solavetivone), 0.5 (valencene to nootkatol), 2 (valencene to nootkatol), 2 (EA), 2 (EE), 5 ((−)-α-cedrene), or 3 min (solavetivol). These time periods afforded 5–20% conversion of substrate to reaction products. Reactions were terminated by the rapid addition of 400 µl of ethyl acetate with rapid mixing by vortexing, followed by removal of the upper ethyl acetate layer. A second ethyl acetate extraction was combined with the first. The combined organic extracts were concentrated on ice under N2 gas, adjusted to 30 µl with ethyl acetate, and 10 ng/µl of (−)-α-cedrene or valencene added as an internal standard, culminating in GC-MS analyses. Assays were performed in triplicate for each substrate concentration, and kinetic constants were calculated by a nonlinear regression fit to the Michaelis-Menten equation using EnzymeKinetics version 1.5 software (Trinity Software). Rates were measured from reactions that were linear with respect to product formation and corresponding to a 5–20% conversion of substrate to product. Under these conditions satisfying the Michaelis-Menten assumptions, reactions were efficient enough to generate greater than 1 ng of reaction product that was readily quantified by GC/MS.
GC and GC-MS Analyses of CYP Reactions
Quantification of reaction products and isoprenoid standards were routinely referenced to (−)-α-cedrene (Fluka, Buchs, Switzerland) as an internal standard. Accuracy of the (−)-α-cedrene standard was periodically verified relative to valencene and nootkatone standards (Fluka, Buchs, Switzerland). Reaction products (1-µl aliquots) were quantified and identified using a Thermo Finnigan DSQ GC/MS system equipped with a Restec Rtx-5 capillary column (30 m × 0.32 mm, 0.25 µm phase thickness). Samples were injected in the splitless mode at 250 °C with an initial oven temperature of 70 °C for 1 min followed by an 8 °C per min gradient to 230 °C. Mass spectra were recorded at 70 eV, scanning from 35 to 300 atomic mass units and compared with authentic standards for verification. The regio- and stereo-specific hydroxylation products of HPO were analyzed by GC using a chiral capillary cyclosil B column (30 m × 0.25 mm, 0.25 µm phase thickness) and an HP5890-FID GC with the oven temperature maintained at 100 °C for 5 min and then increased to 230 °C at 8 °C per min.
Isolation and Purification of Products from HPO Incubations
Large scale reactions (20 × 5 ml) of premnaspirodiene, valencene, epi-aristolochene, and epi-eremophilene with HPO were performed under the standard assay conditions. After the incubations the dominant reaction products were isolated by three extractions with 20-ml portions of ethyl acetate, the combined extracts were carefully concentrated under a N2 stream, and the concentrates were re-dissolved/re-suspended in hexane such that the final ethyl acetate concentration was less than 2%. The solutions or suspensions were then applied to a 230–400-mesh silica gel column (2 × 15 cm). Remaining hydrocarbons were eluted with hexane and 1:24 (v/v) ethyl acetate:hexane, and the hydroxylated products were then eluted with 1:9 (v/v) ethyl acetate:hexane. The latter fractions were further purified using silica TLC (cyclohexane:ethyl acetate:triethylamine = 15:5:0.1). The relative mobilities (Rf values) for monohydroxylated sesquiterpenes are as follows: solavetivol, nootkatol, epiaristolochen-2β-ol, and 4-epieremophilen-2β-ol were 0.27, 0.26, 0.25, and 0.32, respectively. Additional purification of the alcohols was accomplished by C18 reverse phase TLC (MeOH:H2O = 85:15). Rf values of 0.35, 0.3, 0.3, and 0.3 were observed for solavetivol, nootkatol, epiaristolochen-2β-ol, and 4-epieremophilen-2β-ol.
Reference standards of nootkatol (β-nootkatol) and 2-epi-nootkatol (α-nootkatol) were prepared by LiAlH4 reduction of nootkatone in ether according to literature procedures (25, 26). Purification by flash chromatography on silica gel (5:1, hexane/ethyl acetate) provided pure samples of the nootkatol isomers in a 96.5:3.5 ratio (68% combined yield). The 500 MHz 1H NMR data and assignments in CDCl3 (see supplemental materials analytical and NMR information) are in good agreement with the available high field data in the literature, and the 2-R configuration (pseudo equatorial OH) for nootkatol follows from the large axial/axial H2–H3 and small H1–H2 couplings (δH 4.21, ddt, 1H, J = 9.8, 6.2, 2.1 Hz, CHOH) as noted in the literature (25–29).
Characterization and Identification of Purified Monohydroxylated Products
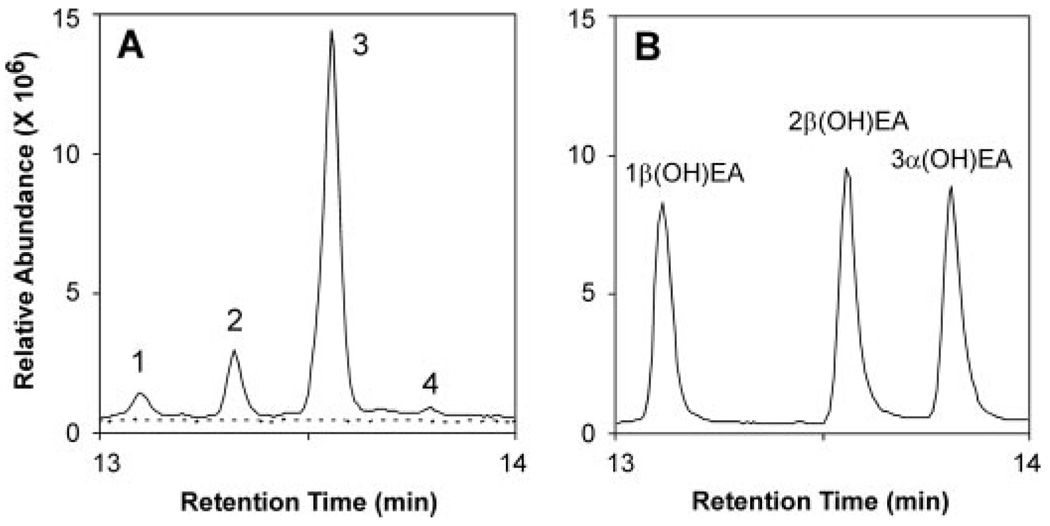
The hydroxylated sesquiterpenes solavetivol, epiaristolochen-2β-ol, and epieremophilen-2β-ol are all new compounds according to searches of the Chemical Abstracts data base through SciFinder. These fermentation products were extensively characterized by physical and spectral data, and the structures and stereochemistry were established by conformational considerations, proton NMR couplings, COSY plots, and NOE measurements. The systematic names given in parentheses are based on recommended skeletal names and positional numbering by Buckingham (30). The GC retention times, mass spectra, and chromatographic Rf values of epiaristolochen-2β-ol and epieremophilen-2β-ol were distinctly different from those of epiaristolochen-1β, 3α-, and 3β-ols, and epieremophilen-3α-ol (Fig. 4) (21).
FIGURE 4. Reaction product analysis of HPO incubations with EA.
Shown are total ion chromatograms for reaction products generated by HPO in the presence (solid line) and absence of NADPH (dotted line). Assays were incubated with 40 µm EA for 10 min before extracting the reaction products into ethyl acetate. Total ion chromatograms of the reaction products (A) in comparison to those for standards of 1β-(OH)EA and 3α-(OH)EA, and to 2β-(OH)EA isolated from large-scale incubations (B). Relevant mass spectra are shown in supplemental Fig. S4 and the time dependence of the reaction is shown in supplemental materials Fig. S7.
Solavetivol (Premnaspirodien-2α-ol or (2S,4R,5S,7R)-Spirovetiva-1(10),11(12)-dien-2-ol)
The peak at 4.18 ppm (tdd, 1H, J = 6.3, 2.4, 1.8 Hz) in the 1H NMR spectrum was assigned as the CHOH proton from its chemical shift, coupling constants, and COSY correlations. The assignment of the 2α configuration was made by conformational analysis with the aid of Chem3D MM2 calculations and NOE measurements. The preferred conformation of the A ring is a half-chair with the C4 methyl group axial. Irradiation of the CHOH peak (4.18 ppm) resulted in a significant enhancement (3.8%) of the C2 vinyl proton at 5.33 ppm. In the favored conformation the H1–H2β dihedral angle is about 5°, whereas the H1–H2α dihedral angle is about 150°. The relatively large NOE reveals close spatial proximity of H1 and H2, indicating the pseudo axial position of the OH group, i.e. 2α-OH (2S). (For NMR assignments, see supplemental materials analytical and NMR information.)
Epiaristolochen-2β-ol ((2R,4R,5R,7R)-Eremophila-9(10),11(12)-dien-2-ol)
The structure and stereochemistry were deduced from 1H NMR, APT, COSY, and MS data. The proton NMR peak at 3.77 ppm (tt, 1H, J = 11.4, 5.0 Hz) was assigned to the carbinyl proton CHOH at C2 based on its chemical shift, coupling constants, and COSY correlations. The two large couplings indicate axial orientations of H1, H2, and H3, and the two intermediate couplings arise from further axial-equatorial interactions between H1–H2 and H2–H3. The preferred conformation of the A ring of epiaristolochene is a half-chair with the vicinal methyl groups in axial positions (31). Thus the hydroxyl group at C-2 was assigned an equatorial disposition. (For NMR assignments, see supplemental materials analytical and NMR information.)
4-Epieremophilen-2β-ol ((2R,4R,5R,7R)-Eremophila-1(10),11(12)-dien-2-ol)
All protons on the A ring were assigned unambiguously from 1H NMR and COSY spectra. The carbinyl proton CHOH appears as a ddd with J values at 9.1, 5.5, and 3.3 Hz. These magnitudes are consistent with axial-axial and axial equatorial couplings to the adjacent CH2, and smaller coupling with the vinyl proton at C-1. It follows that the hydroxyl group is located in the equatorial allylic position at C-2. (For NMR assignments, see supplemental materials analytical and NMR information.)
Computational Studies
Homology modeling was performed with Modeler 6.2 (32) by threading the HPO amino acid sequence (GenBank accession number EF569601) onto the structural coordinates for the mammalian CYP2B4 protein (Protein Data Bank code 1SUO) (33). The ligands premnaspirodiene, valencene, EA, and EE were created using Chemdraw, energy minimized with MOPAC in Chem3D (CambridgeSoft Corporation, Cambridge, MA), and subsequently used in docking simulations. Docking calculations were conducted with the GOLD software package (Genetic Cambridge Crystallographic Data Centre, Cambridge, UK) (34). Visualizations and three-dimensional rendered figures were prepared with PyMOL (www.pymol.org).
Site-directed Mutagenesis
Mutagenesis of CYP71D20 (EAH) and HPO cDNAs used standard conditions given in QuikChange protocols (Stratagene) (35). The full-length HPO or EAH cDNAs inserted into the BamHI/EcoRI restriction sites of the pYeDP60 vector (15) were used in combination with Pfu Turbo DNA polymerase (Stratagene) and the primer pairs noted in supplemental Tables S1 and S2. Site-specific mutations were confirmed by automated DNA sequencing.
RESULTS
Isolation of a Member of the CYP71D Gene Family from H. muticus
With previous success in structurally and functionally mapping molecular determinants for catalytic specificity in two evolutionary related solanaceous sesquiterpene synthases, namely tobacco (Nicotiana tabacum) 5-epi-aristolochene synthase and henbane (H. muticus) premnaspirodiene synthase (HPS) (36), comparative studies naturally extend to the final oxidative step(s) of each pathway. The goal of the current studies was therefore the isolation of a terpene hydroxylase(s) responsible for oxidation of C-2 of premnaspirodiene, followed by biochemical comparison to the previously isolated EAH from tobacco (14, 15). Given the close phylogenetic relationship between tobacco and henbane supported by the high degree of amino acid and DNA sequence identity between tobacco 5-epi-aristolochene synthase and HPS, nearly 80%, it was reasonable to expect that a similar degree of sequence similarity exists between EAH and the expected premnaspirodiene-specific CYP gene. Genes encoding CYP71D family members functionally characterized as terpene hydroxylases were aligned and used to design DNA primers (supplemental Fig. S1) for reverse transcriptase-PCR with mRNA isolated from elicitor-induced henbane cell cultures.
Eight independent PCR products representing different combinations of primer pairs were obtained and shown to encompass overlapping regions and identical DNA sequences. The longest amplification product, ~1.6 kbp, contained obvious start and stop codons built into the PCR primers, which were separated by the expected length of a CYP71D homologue (Fig. 1). Subsequent 5′ and 3′ rapid amplification of cDNA ends and genomic cloning experiments verified the termini sequences of the full-length cDNA (data not shown). The isolated H. muticus cDNA encoded a predicted protein possessing typical carboxyl-terminal CYP domains (PERF, GGGGRRRICPG (14)) and was 80% identical at the amino acid level to tobacco EAH with 91-amino acid differences and a 2-amino acid insertion. Moreover, the putative henbane CYP was 53% identical at the protein sequence level to limonene C3 hydroxylase (37), 50% identical to limonene C6 hydroxylase (37), and 70% identical to 71D16 (38). Based upon Nelson et al. (39), the identified henbane CYP gene was classified as CYP71D55.
FIGURE 1. Amino acid sequence alignment between HPO and EAH.
The shaded boxes identify amino acid differences between these two enzymes, and the domains corresponding to the 6 putative SRS as defined by Gotoh (16) are outlined and annotated. Amino acids 368, 482, 484, and 486 are noted as well because their potential involvement in catalysis was subsequently investigated by mutagenesis.
Functional Characterization of HPO
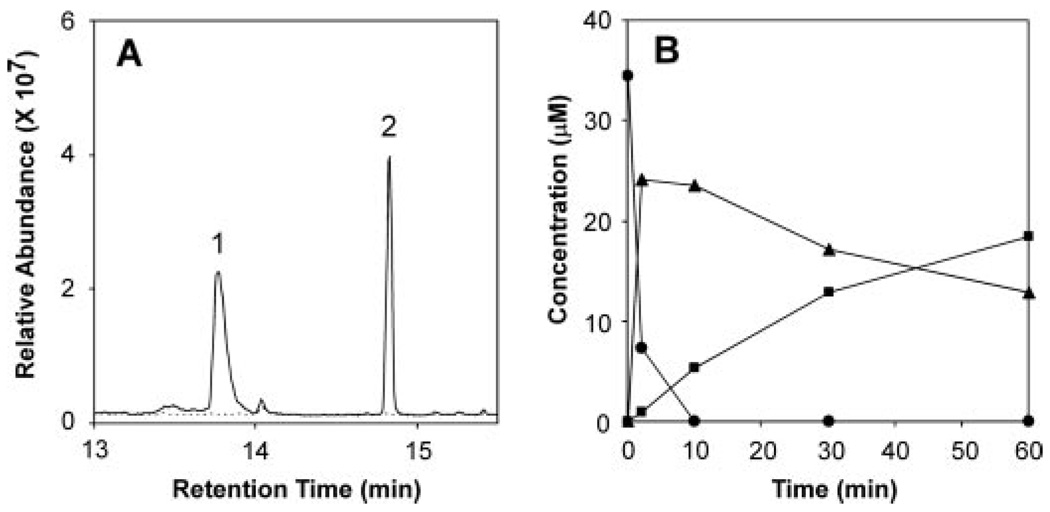
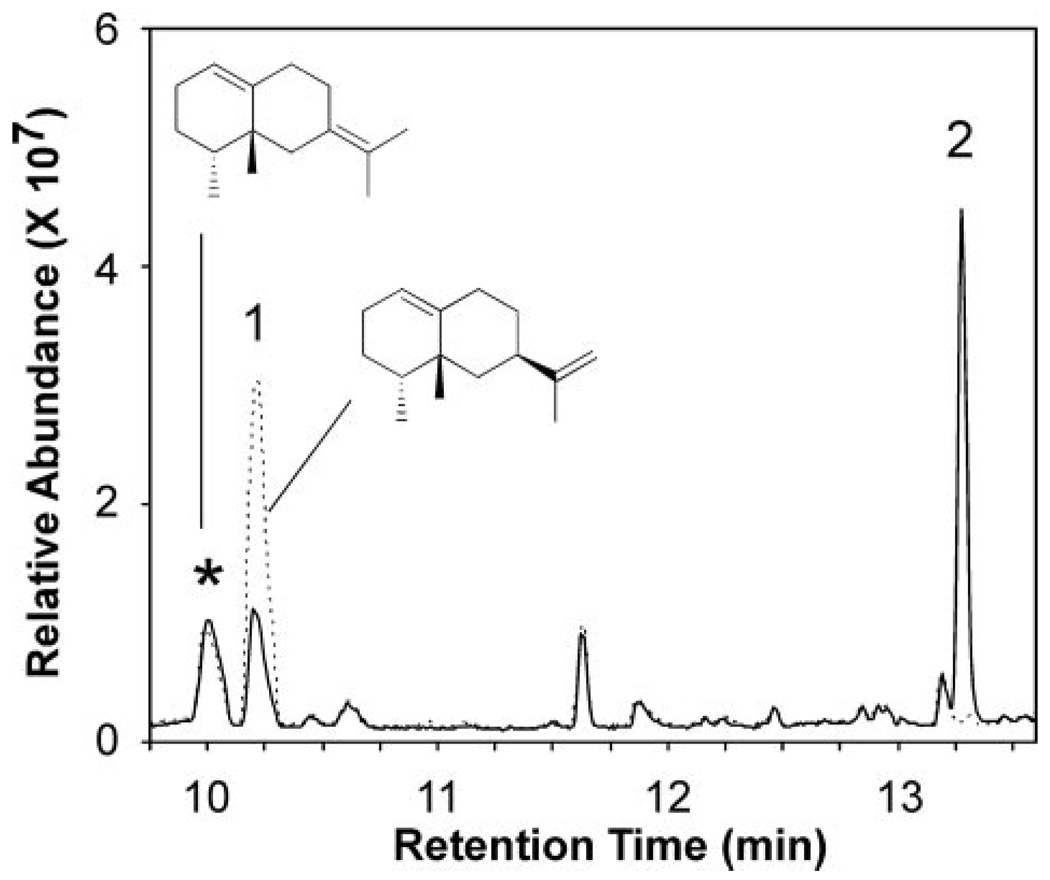
Protein expression of the corresponding cDNA in yeast and biochemical characterization of microsomes isolated from the yeast strains provided functional identification of CYP71D55. No sesquiterpene metabolizing activity was observed with microsomes isolated directly from yeast strain WAT11 or WAT11 harboring an “empty” expression vector. In contrast, microsomes isolated from yeast overexpressing the CYP71D55 cDNA exhibited a typical CO difference spectrum with a minor absorbance peak at ~418 nm and a dominant absorbance peak at 450 nm (supplemental Fig. 6A). When incubated with pure premnaspirodiene, the isolated microsomes produced oxidized isoprenoid product(s) in a time- and NADPH-dependent manner as shown by GC-MS analysis (Fig. 2). A dominant reaction product, identified as solavetivol (peak 1) appeared during the first few minutes of the incubations and was followed by a slower appearing linear accumulation of solavetivone (peak 2). Based on chiral GC analysis, only solavetivol accumulated with little if any episolavetivol under the conditions analyzed (data not shown). No other products were observed. A clear stoichiometric relationship between the loss of premnaspirodiene and the initial accumulation of mono-hydroxylated solavetivol was followed by a slow loss of the solavetivol peak with an equivalent gain in the solavetivone peak (Fig. 2B). Based on the biochemical oxidation of premnaspirodiene to solavetivone, the CYP71D55 enzyme was subsequently designated as Hyoscyamus muticus premnaspirodiene oxidase (HPO).
FIGURE 2. Reaction product analysis of HPO incubations with premnaspirodiene.
Shown are total ion chromatograms for reaction products generated by HPO in the presence (solid line) and absence of NADPH (dotted line). Assays were incubated with 40 mm premnaspirodiene for 30 min before profiling the reaction products by GC/MS (A). Time-dependent consumption of substrate and accumulation of reaction products was monitored upon incubating HPO with premnaspirodiene (●) for 2, 10, 30, and 60 min, and the total ethyl acetate extractable reaction products were analyzed by GC-MS. Solavetivol (▲) and solavetivone (■) were the only NADPH-dependent reaction products detected (B). (Relevant mass spectra are shown in supplemental Fig S2.)
As described previously for EAH, kinetic studies of recombinant HPO were consistent with a successive hydroxylation mechanism (15) (Table 1). However, in the case of HPO, the initial hydroxylation reaction was kinetically favored with an overall catalytic efficiency 3-fold greater than the second hydroxylation reaction. In contrast, the second hydroxylation step of EAH is 8-fold more efficient than the first hydroxylation, leading to the rapid accumulation of the final di-hydroxylated reaction product and very little buildup of the mono-hydroxylated intermediate. For HPO, the absence of any solavetivol conversion by microsomes isolated from control yeast lines lacking the HPO gene provided further confirmation that the second hydroxylation reaction was indeed mediated by HPO and not a contaminating yeast dehydrogenase or similar oxidative activity.
TABLE 1. Comparison of kinetic constants for EAH and HPO hydroxylases.
Constants were calculated by nonlinear regression fits to the Michaelis-Menten equation.
| Enzyme | Substrate | Product | Km | kcat | kcat/Km |
|---|---|---|---|---|---|
| µm | S−1 | ||||
| EAHa | EA | Capsidiol | 19.2 ± 2.9 | 0.5 ± 0.04 | 0.03 |
| 1β-(OH)EA | Capsidiol | 1.7 ± 0.1 | 0.6 ± 0.01 | 0.35 | |
| HPO | PSD | Solavetivol | 14.0 ± 1.9 | 2.1 ± 0.10 | 0.15 |
| PSD | Solavetivone | 1.7 ± 0.2 | 0.1 ± 0.01 | 0.06 | |
| Solavetivol | Solavetivone | 1.2 ± 0.1 | 0.1 ± 0.01 | 0.08 | |
| Valencene | α-Nootkatol | 11.5 ± 1.9 | 0.1 ± 0.01 | 0.01 | |
| Valencene | β-Nootkatol | 7.4 ± 1.2 | 1.9 ± 0.07 | 0.26 | |
| EA | 2β-(OH)EA | 3.3 ± 0.3 | 0.2 ± 0.01 | 0.06 | |
| EE | 2β-(OH)EE | 7.8 ± 1.0 | 0.8 ± 0.03 | 0.10 | |
| Cedr-8-ene | Cedr-8-en-15-ol | 26.5 ± 1.1 | 0.1 ± 0.01 | 0.00 | |
EAH kinetic data are from Takahashi et al. (15); EA, 5-epi-aristolochene; PSD, premnaspirodiene; EE, epi-eremophilene.
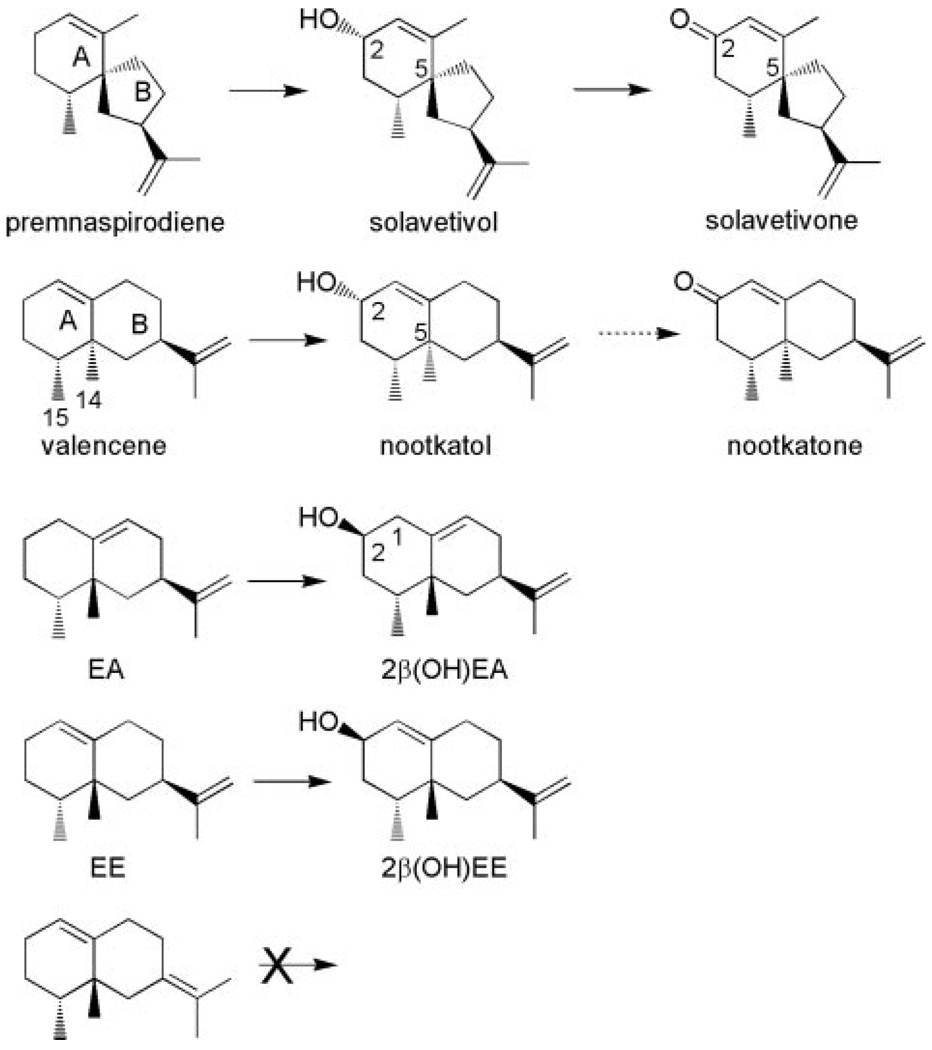
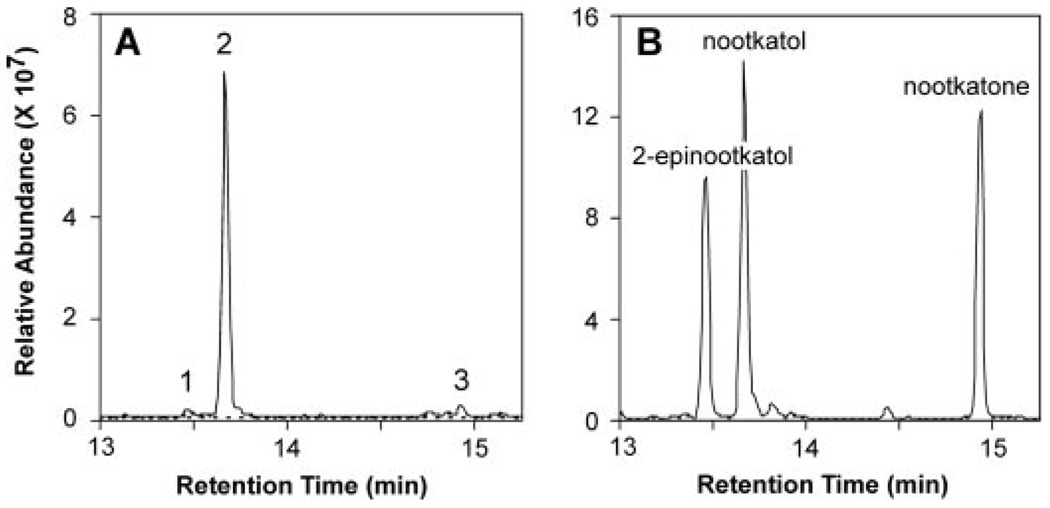
The insertion of a carbonyl function into the C-2 position of premnaspirodiene is analogous to one of the possible reaction mechanism proposed for the biosynthesis of the oxidized sesquiterpene nootkatone (Scheme 2), a citrus flavorant of industrial importance (25). Interestingly, HPO readily catalyzed the mono-hydroxylation of valencene to nootkatol (Fig. 3), but failed to convert the resultant mono-hydroxylated product to the ketone form or additional hydroxylation products with appreciable catalytic efficiency. Important to note, the catalytic efficiency (kcat/Km) of the valencene to nootkatol conversion was 70 times greater than the efficiency of valencene to 2-epinootkatol, and 70% more efficient than the native reaction of premnaspirodiene to solavetivol (Table 1). Nootkatone formation was observed at very high concentrations (>30 µm) of nootkatol, but the reaction rate was too low to obtain accurate kinetic values.
SCHEME 2. Summary of the regio- and stereo-specific hydroxylations of the indicated sesquiterpene hydrocarbons by HPO.
FIGURE 3. Reaction product analysis of HPO incubations with valencene.
Shown are total ion chromatograms for reaction products generated by HPO in the presence (solid line) and absence of NADPH (dotted line). Assays were incubated with 40 µm valencene for 10 min before extracting the reaction products into ethyl acetate. Total ion chromatograms of the reaction products (A) in comparison to those for standards of 2-epinootkatol, nootkatol, and nootkatone (B). (Relevant mass spectra are shown in supplemental Fig. S3.)
Given the robust catalytic activity of HPO with valencene (an eremophilane sesquiterpene) relative to premnaspirodiene (a vetispirane sesquiterpene), the ability of HPO to oxidize other members of the eremophilane class of sesquiterpenes were examined. In addition, whereas other vetispirane-type sesquiterpenes, especially various isomers of premnaspirodiene, would be desirable for testing, they are not readily available commercially, synthetically, or biosynthetically. 5-Epi-aristolochene differs from valencene in the orientation of the C-14 and C-15 methyl substituents and the placement of the double bond in the B ring rather than the A ring. HPO catalyzed the conversion of 5-epi-aristolochene (EA) to four mono-hydroxylated products with 2β-hydroxy-EA (2β-(OH)EA) comprising greater than 80% of the reaction products, 1β-(OH)EA about 5%, 3α-(OH)EA less than 2%, and an unknown mono-hydroxy-lated product of ~20% (Fig. 4). Substrate affinity for EA was 2 to 4 times lower than that for valencene or premnaspirodiene, respectively, and the turnover rate for EA was only about 1/10th that for either of these substrates (Table 1). Epi-eremophilene (EE), which has the same overall configuration as EA except that the double bond is in the A ring rather than the B ring, was also catalytically converted to a single dominant reaction product, 2β-hydroxy EE (Fig. 5). In contrast to EA, the HPO enzyme exhibited a substrate affinity for EE more similar to the values obtained for valencene and premnaspirodiene, but the turnover rate was significantly higher than for EA (Table 1). Interestingly, a contaminant in the EE preparation, a regioisomer in the position of the double bond in the isopropenyl substituent (* peak in Fig. 5, Scheme 2), was not consumed in the incubations with the HPO, and hence did not compete with EE as a substrate for HPO. Cedrene, another commercially available sesquiterpene hydrocarbon, was also tested for its suitability as an HPO substrate, but any catalytic conversions required very high substrate concentrations, and the observed turnover rate was very low relative to premnaspirodiene.
FIGURE 5. Reaction product analysis of HPO incubations with EE.
Shown are total ion chromatograms for reaction products generated by HPO in the presence (solid line) and absence of NADPH (dotted line). Assays were incubated with the mixture of 30 µm EE (peak 1) and 10 µm of a double bond isomer of EE (*) for 10 min before profiling the reaction products by GC-MS. (Relevant mass spectra are shown in supplemental Fig. S5.)
Molecular Dissection of the Oxygenase/Hydroxylase Activity of HPO
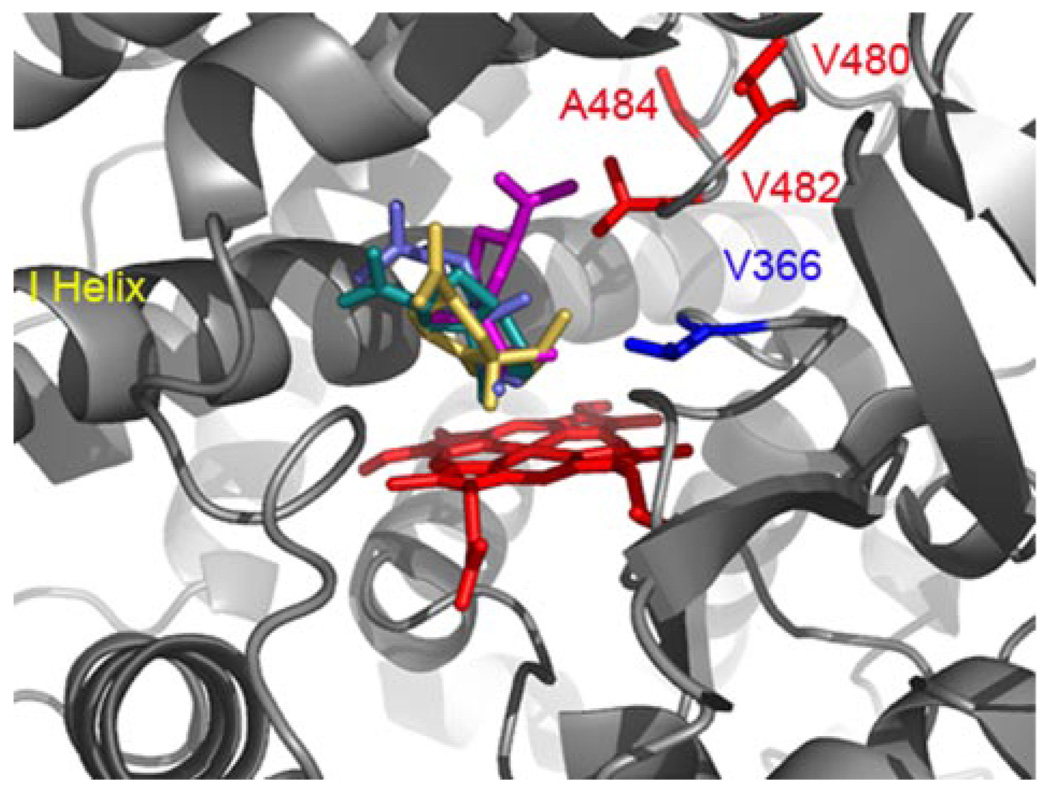
A combination of sequence alignments, molecular modeling, and site-directed mutagenesis were used previously to identify residues that contributed to catalytic specificity within putative substrate recognition sequences (SRS) 5 and 6 as defined by Gotoh (16) of EAH (15). In particular, Ser-368 and Ile-486 were previously shown to be critical for the second, successive hydroxylation reaction catalyzed by EAH. Interestingly, sequence comparison with the corresponding regions of HPO (Fig. 1) identified residue differences in HPO at positions 366, 480, 482, and 484 (equivalent to EAH 368, 482, 484, and 486) (Fig. 1). These four positions appear poised to influence substrate recognition within the active site pocket of HPO based on modeling and docking studies of premnaspirodiene, valencene, EA, or EE into an HPO model calculated using the coordinates for the substrate closed form of CYP2B4 (33) (Fig. 6).
FIGURE 6. Substrate docking into a modeled HPO active site.
A homology model of HPO was created using the mammalian P450 2B4 as template (PDB 1SUO), and used for docking experiments with premnaspirodiene (teal), valencene (blue), EA (gold), and EE (magenta) as described under “Experimental Procedures.” Distinct binding modes were predicted from docking simulations that positioned the β-face of each ligand within 3.5 Å of the heme center for oxidation at C-2 of the respective substrate. Residue differences between HPO and EAH within SRS 5 (blue) and SRS 6 (red) are labeled and highlighted accordingly.
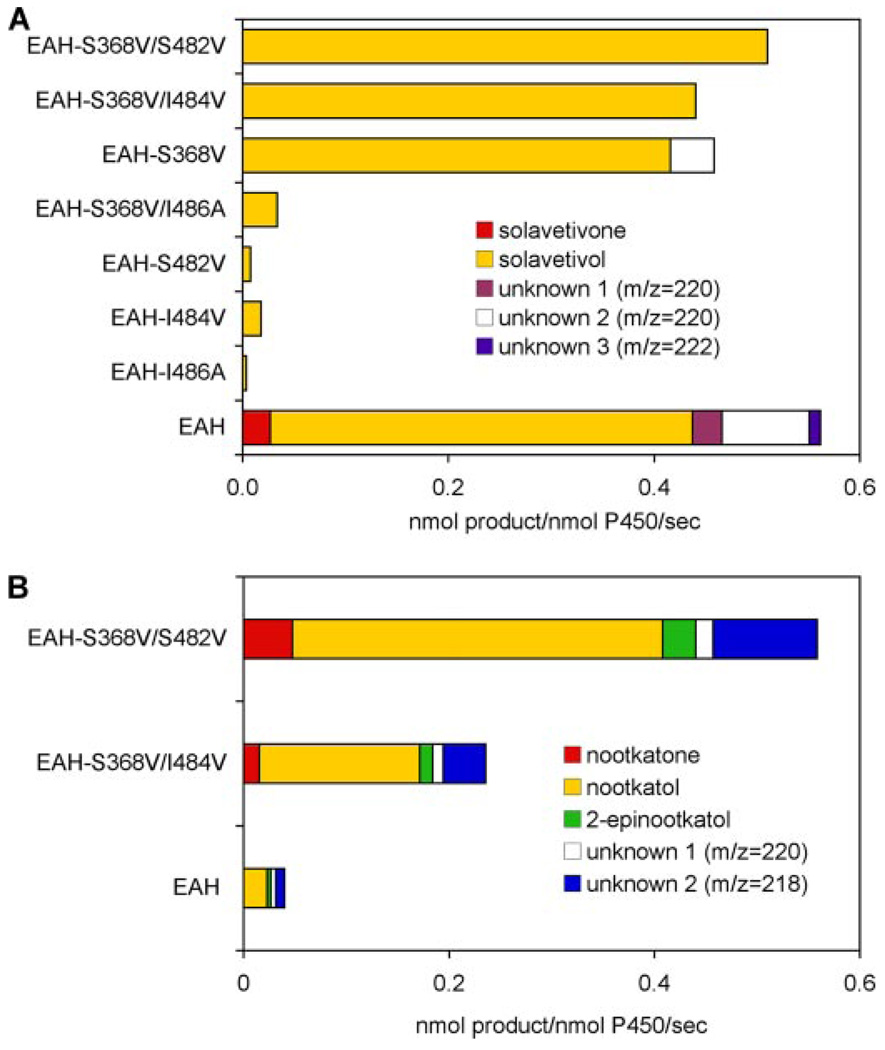
To assess the functional contribution of these residues to HPO, the corresponding residues of HPO and EAH were inter-converted and the capacity of each mutant enzyme of EAH and HPO to hydroxylate premnaspirodiene was determined. Greenhagen et al. (20) previously demonstrated that wild type EAH was capable of utilizing premnaspirodiene as a substrate in coupled in vitro assays, but the reaction product(s) were not identified. EAH in fact catalyzes the conversion of premnaspirodiene to solavetivol (74%), solavetivone (4%), and three other mono-hydroxylated products (based on a parent ion of 220 m/z in GC/MS analysis) comprising in total 21.5% of the product profile of EAH (Fig. 7A). The catalytic efficiency (kcat/Km) of EAH for solavetivol biosynthesis was, however, only one-third that of HPO measured here (Table 2). Mutating the amino acid residues at positions 482, 484, and 486 within SRS6 of EAH to the corresponding amino acids found in HPO essentially abolished all enzyme activity toward premnaspirodiene, whereas for the S368V mutant, the only difference between HPO and EAH in SRS5 narrowed the reaction specificity to ~92% solavetivol and 8% for one of the other unidentified mono-hydroxylated products. Catalytic efficiency of the S368V mutant for solavetivol was still only one-third that of the HPO enzyme (Table 2). Interestingly, whereas combinations of the S368V mutation with I484V or S482V slightly improved the catalytic efficiency of EAH for solavetivol biosynthesis (Table 2), the double mutations eliminated biosynthesis of the unknown mono-hydroxylated product, thus creating mono-specific enzymes yielding only solavetivol (Fig. 7A).
FIGURE 7. Assessing the importance of residue differences between EAH and HPO within SRS 5 and 6 for catalysis and product specificity.
The indicated residues in EAH were converted to those corresponding residues in HPO (S368V, S482V, I484V, I486A), the mutant genes were expressed in yeast, and CO difference spectroscopy was performed to normalize for the amount of properly folded CYP enzyme (supplemental Fig. S6) before the enzyme assays. Assays were incubated with 40 µm premnaspirodiene for 1 min before profiling the reaction products by GC-MS (A). Only solavetivol and solavetivone were observed as reaction products of the mutant enzymes. Select EAH mutants were also incubated with 40 µm valencene for 2 (EAH-S368V/ S482V) or 5 min (EAH-S368V/I484V and EAH) before profiling the reaction products by GC-MS (B).
TABLE 2. Comparison of kinetic constants for EAH mutants to those for HPO.
Constants were calculated by nonlinear regression fits to the Michaelis-Menten equation.
| Enzyme | Substrate | Product | Km | kcat | kcat/Km |
|---|---|---|---|---|---|
| µm | S−1 | ||||
| HPOa | PSD | Solavetivol | 14.0 ± 1.9 | 2.1 ± 0.10 | 0.15 |
| EAH | PSD | Solavetivol | 9.2 ± 1.1 | 0.4 ± 0.02 | 0.04 |
| EAH, S368V | PSD | Solavetivol | 11.4 ± 1.2 | 0.6 ± 0.02 | 0.05 |
| EAH, S368V/I484V | PSD | Solavetivol | 7.6 ± 1.2 | 0.4 ± 0.02 | 0.05 |
| EAH, S368V/S482V | PSD | Solavetivol | 6.5 ± 1.2 | 0.6 ± 0.03 | 0.09 |
Data are from Table I; PSD, premnaspirodiene; EA, 5-epi-aristolochene.
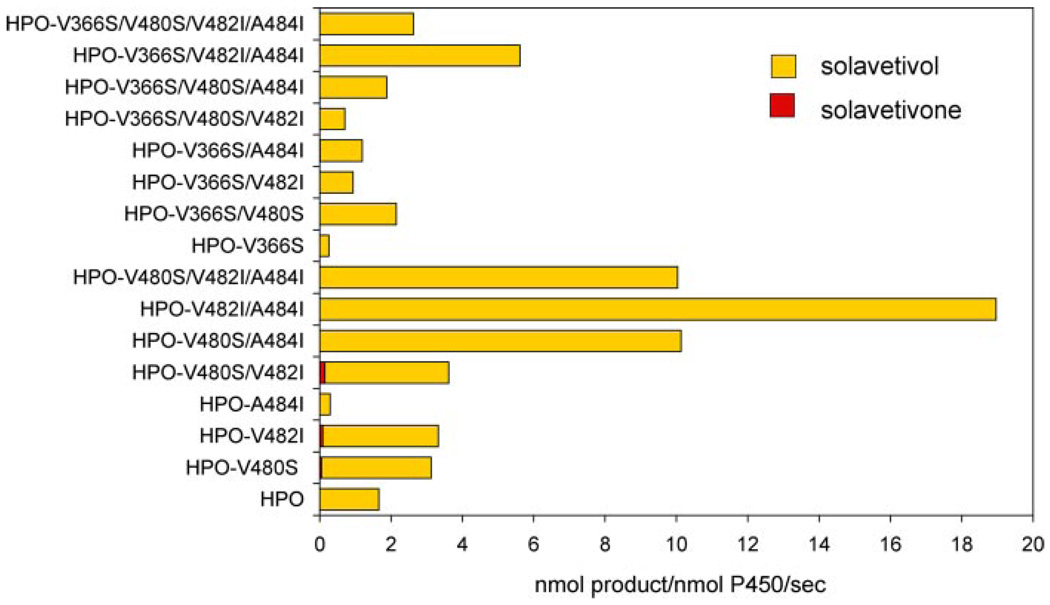
The results of mutating position 368 in combination with 482 and 484 in EAH suggested that these residues might possess greater influence on the turnover rates and catalytic efficiency than dictating substrate selectivity or product specificity per se. Therefore, one might expect that mutation of the reciprocal positions in SRS 5 and 6 of HPO would not alter the product specificity of HPO with premnaspirodiene as substrate, but rather alter the turnover rates for the reactions. In comparison to the observations with EAH, single mutations V480S and V482I in HPO (corresponding to S482V and I484V in EAH) improved the kcat for the conversion of premnaspirodiene to solavetivol about 2-fold, but the A484I mutation resulted in a catalytically compromised enzyme (Fig. 8). Surprisingly, double and triple mutations within the 480–484 region yielded enzymes with 5–10-fold improved turnover rates for premnaspirodiene without compromising the catalytic specificity for solavetivol (Fig. 8). Km values for premnaspirodiene were unchanged or reduced ~40% in these mutants (Table 3). In contrast to the narrowing of catalytic specificity by the S368V EAH mutant, the reciprocal V366S mutation in HPO dramatically reduced overall enzyme activity (Fig. 8). Combinations of the S366V mutation with additional point mutations in SRS6 restored enzyme activity up to 100% of that observed with the wild type HPO, and inclusion of double and triple SRS6 mutations further improved overall activity 200 to 300%. None of these mutant enzymes yielded products other than solavetivol under the reaction conditions used here (reaction times of 1 to 10 min), and solavetivone after extended incubation times.
FIGURE 8. Assessing the importance of residue differences between HPO and EAH within SRS 5 and 6 for catalysis and product specificity.
The indicated residues in HPO were converted to those corresponding residues in EAH (V366S, V480S, V482I, A484I) in various combinations, the mutant genes were expressed in yeast, and CO difference spectroscopy was used to normalize for the amount of properly folded CYP enzyme (supplemental Fig. S6) before the enzyme assays. Assays were incubated with 40 µm premnaspirodiene for 1 min before profiling the reaction products by GC-MS. Only solavetivol and solavetivone were observed as reaction products.
TABLE 3. Comparison of kinetic constants for HPO mutants.
Constants were calculated by nonlinear regression fits to the Michaelis-Menten equation.
| Enzyme | Substrate | Product | Km | kcat | kcat/Km |
|---|---|---|---|---|---|
| µm | S−1 | ||||
| HPOa | PSD | Solavetivol | 14.0 ± 1.9 | 2.1 ± 0.10 | 0.15 |
| Valencene | β-Nootkatol | 7.4 ± 1.2 | 1.9 ± 0.07 | 0.26 | |
| EA | 2β-(OH)EA | 3.3 ± 0.3 | 0.2 ± 0.01 | 0.06 | |
| HPO, V482I | PSD | Solavetivol | 6.2 ± 0.8 | 3.8 ± 0.17 | 0.61 |
| Valencene | β-Nootkatol | 6.9 ± 1.0 | 5.1 ± 0.23 | 0.74 | |
| EA | 2β-(OH)EA | 17.9 ± 2.5 | 1.4 ± 0.10 | 0.08 | |
| HPO, V480I/A484I | PSD | Solavetivol | 8.4 ± 0.8 | 12.7 ± 0.43 | 1.51 |
| Valencene | β-Nootkatol | 19.2 ± 1.3 | 5.7 ± 0.18 | 0.30 | |
| EA | 2β-(OH)EA | 2.5 ± 0.2 | 0.7 ± 0.01 | 0.28 | |
| HPO, V482I/A484I | PSD | Solavetivol | 13.0 ± 0.9 | 20.7 ± 0.64 | 1.59 |
| Valencene | β-Nootkatol | 12.1 ± 1.9 | 15.9 ± 0.95 | 1.31 | |
| EA | 2β-(OH)EA | 4.3 ± 0.9 | 2.8 ± 0.14 | 0.65 | |
Data are from Table I; PSD, premnaspirodiene; EA, 5-epi-aristolochene.
Parallel improvements in the catalytic constants for other substrates were also observed for the HPO and EAH mutants and without compromising the regio- or stereo-specificities of the enzyme reactions. For instance, mutations within the SRS6 region of HPO improved the turnover rate of premnaspirodiene to solavetivol as well as the turnover rates of valencene to nootkatol and 5-epi-aristolochene to 2β-(OH)EA, but the reactions were accompanied by somewhat greater changes in the Km values for the alternative substrates than those observed for premnaspirodiene (Table 3). Regardless, the V482I/A484I HPO mutant possessed a 5-fold improvement in its catalytic efficiency for nootkatol biosynthesis and a 10-fold improvement for 2β-(OH)EA biosynthesis. None of these mutations changed the overall reaction product profiles of the HPO enzymes. Similarly, the S368V/I484V and S368V/S484V mutations in EAH, which narrowed the reaction specificity of the mutant enzymes to solavetivol with premnaspirodiene as substrate (Fig. 7A), also improved the turnover rates for valencene 6- and 15-fold, respectively, and the resulting mixture was accompanied by a proportional increase in all six reaction products observed with the wild type EAH enzyme (Fig. 7B).
DISCUSSION
The current studies were undertaken, in part, to define further the biosynthetic pathways for sesquiterpene phytoalexin production in plants, and furthermore, to probe in depth the fundamental catalytic properties of the specialized enzymes forming essential steps in these plant biosynthetic pathways. We previously reported that the pathogen-inducible accumulation of capsidiol in tobacco was due to the induction of a two-step branch pathway involving the sequential activity of a sesquiterpene synthase and a sesquiterpene hydroxylase (Scheme 1). The second step catalyzed by EAH was further shown to be unusual in comparison with other well characterized small molecule CYPs by virtue of the sequential and stereospecific hydroxylations it introduces at distal positions on the bicyclic 5-epi-aristolochene hydrocarbon core. In the present work, we focused on the isolation and characterization of a second sesquiterpene hydroxylase, HPO, a structurally related but functionally distinct CYP from EAH, that could be used to dissect the catalytic features of these specialized enzymes.
HPO shares similarities and differences with EAH. Like EAH, HPO exhibits a very high degree of catalytic fidelity for the initial hydroxylation at a single site on the sesquiterpene ring system with greater than 95% stereochemical specificity (Fig. 2). The initial hydroxylation reaction catalyzed by HPO, however, occurs with at least a 10-fold greater turnover rate than that exhibited by EAH for its initial substrate. Moreover, both enzymes, whereas catalyzing sequential oxidation reactions, do so with opposing rates for the second oxidation step (15). Notably, EAH and HPO exhibit similar Km values for their respective hydroxylated intermediates (~1.7 µm), but the turnover number for EAH is ~5 times greater than the turnover number for HPO. These kinetic differences create distinctive differences in the reaction profiles of these two phylogenetically related enzymes. Little if any mono-hydroxylated product is observed when EAH is incubated with substrate concentrations near the Km value for EA, whereas mono-hydroxylated product dominates the HPO reaction profile under these same in vitro conditions.
HPO is able to hydroxylate vetispirane and eremophilane sesquiterpene substrates readily with comparable kinetic efficiencies and specificities, but aspects of selectivity suggest features of the substrate molecules and the enzyme that may be of importance for catalysis (Scheme 2). HPO catalyzes the introduction of the first hydroxyl at the equivalent regio position in both the vetispirane and eremophilane substrates. However, this hydroxyl group can be inserted in one of two stereochemical orientations relative to the plane of the A ring in the respective substrates. Regio-specificity was independent of the overall hydrocarbon scaffold (spirovetivane versus eremophilane) and of the position of the double bond in the A or B rings (opposite orientation in nootkatol and 2β-(OH)EE). However, in comparisons of the product specificities of HPO with valencene and epi-eremophilene substrates, the best measure for predicting the stereo-specificity of the first hydroxylation appears to be the stereo-chemical orientation of the vicinal methyls at C-14 and C-15, which must play a role in how these different substrates orient in the active site near the heme center. The importance of methyl groups for the positioning of terpene substrates relative to the reactive oxo-iron-heme has been noted previously for P450cam and used to identify the corresponding amino acid residues within the active site that may facilitate binding of the α-pinene substrate (40).
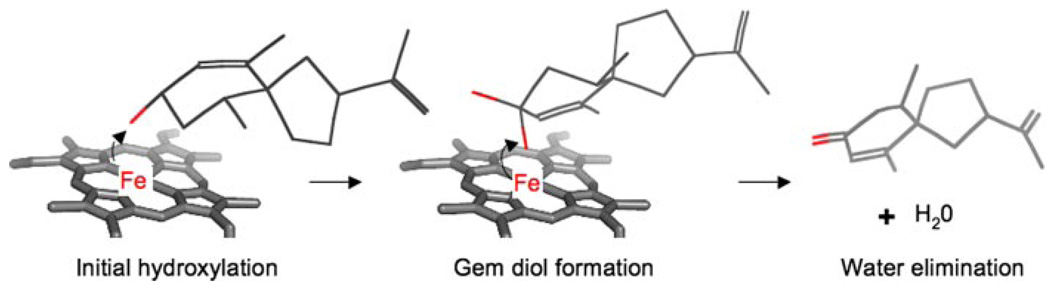
Similar consideration of substrate specificities also provides hints as to those features of the enzyme important for the second hydroxylation reaction. Whereas solavetivol is readily converted to solavetivone, nootkatol, the only eremophilene intermediate with a hydroxyl group oriented similarly to solavetivol, was marginally converted to nootkatone only upon long incubation times at high substrate concentrations. Structural features of the mono-hydroxylated spirane substrate are obviously crucial for efficient turnover to the fully oxidized ketone containing sesquiterpenoid end product, as are corresponding features intrinsic to the HPO enzyme necessary to accommodate this class of substrates but not the mono-hydroxylated eremophilenes. The solavetivol intermediate, for instance, must be bound in a very different orientation from that for the initial hydrocarbon as illustrated in Scheme 3, essentially flipping the intermediate and positioning the opposite face of the initially hydroxylated carbon in close proximity to the reactive oxoiron-heme center. This reorientation may be accommodated by rotation of the initial hydroxylated intermediate in the pocket, but is more apt to result from the dissociation and re-association of the intermediate in a completely different configuration more favorable for either a double proton abstraction or formation of a gem-diol intermediate (18,19). Thus, future modeling of the mono-hydroxylated intermediates in the active site of HPO should focus on distinguishing residues positioned to facilitate binding of the solavetivol structure as well as those that might hinder or prevent binding of nootkatol. Equally important will be determining if carbonyl formation occurs via a double proton abstraction or formation of an unstable gem-diol intermediate mechanism. Distinguishing between these two possibilities should, however, be relatively straightforward by preparing 18O-labeled solavetivol and monitoring the retention of label in the final solavetivone product (18). A double proton abstraction mechanism predicts complete retention of 18O in the final carbonyl, whereas a gem-diol mechanism would result in one-half of the 18O exchanging as a consequence of a second atmospheric 16O-hydroxyl being introduced, followed by equal elimination of [16O]H2O or [18O]H2O (Scheme 3).
SCHEME 3. Proposed mechanism for the successive hydroxylation of premnaspirodiene by HPO, followed by water elimination and carbonyl formation.
The oxo-iron-heme reaction center of HPO is shown schematically relative to the hydroxylated intermediate, solavetivol, and a transient gem diol intermediate leading to the final reaction product solavetivone. Red sticks denote oxygens. Adapted from Bellucci et al. (18).
Significant improvements in the catalytic rates of CYPs have been reported previously and can arise from changes in the rates of substrate association, the rate of heme reduction via coupling to the cytochrome reductase, the intrinsic physical-chemical features of the reaction itself or intermediate/product dissociation rates from the active site. However, many mutants affecting catalysis in CYPs reside outside the active site pocket and appear to influence catalysis in indirect ways. Pan et al. (41), for instance, demonstrated that position 115, which maps out-side of the active pocket to SRS1 of the swallowtail CYP6B1, correlated with improved metabolic turnover of furanocouma-rins, plant secondary products that are normally toxic except to insects able to catabolize them. Kumar et al. (42) reported the identification of four residues within the mammalian 2B1 CYP via a random mutagenesis approach that improved turnover rates from 2–20-fold but increased the Km values for several substrates 2–5-fold, yielding efficiency gains (kcat/Km) of up to 10-fold. All the residues identified (183, 202, 209, and 334) are positioned outside of the putative active site cavity, several actually appearing to lie on the surface of the protein, and thus likely to exert their influence over catalysis at a distance. Likewise, He et al. (43) demonstrated that mutants V117A and R372H, independently, increased the specific activity of CYP2A13 ~10-fold for coumarin 7-hydroxylation without altering the Km for the substrate. Using a successive random mutagenesis approach, Kim and Guengerich (44) observed a 10-fold improvement in the hydroxylation efficiency of a surrogate substrate by CYP1A2 containing 3 mutations distal to the active site. The authors suggested that these residues likely influence catalysis via energetically coupled interactions in a manner analogous to the cooperative networks of residues described by Hilser et al. (45). The notion of residues not in direct contact with substrate dramatically influencing catalytic characteristics in CYPs was also described by Glieder et al. (46), who observed upwards of a 100-fold increase in turnover rates, increased substrate acceptability, and changes in regiochemical hydroxylations after five rounds of random mutagenesis of the soluble bacterial BM-3 enzyme. Only 1 of the 11 mutated residues in the final mutant enzyme was associated with a putative substrate recognition and/or binding domain.
In contrast to these previous reported studies, the sites we identified within HPO and EAH affecting catalytic turnover rates map to SRS domains 5 and 6 (16) and in the molecular model shown in Fig. 6 appear to reside within the active site pocket. Furthermore, given the position, orientation, and hydrophobic nature of the residues found at these four positions, it would seem more likely that these residues directly influence catalysis through their contribution to the active site geometry, rather than by a direct interaction with the reaction intermediates. For instance, the HPO V482I/A484I mutant may result in a larger lip structure extending into the pocket and arching over the substrate ring systems, in essence forcing the β-face of C-2 of premnaspirodiene, or C-2 of the eremophilene substrates, into close proximity of the reactive heme center. Alternatively, these mutations might indirectly influence the spin state of the oxo-iron-heme center as suggested by the work of Glieder et al. (46), and thus enhance the turnover rate via modulation of the intrinsic reactivity of the heme center. Measurements of the spin states of the wild type and mutant enzymes in the presence of substrates should allow one to discern the contribution of these mutations in the latter hypothesis. However, determining the crystal structure of the wild type and mutant enzymes will likely be necessary before a complete appreciation for how these enzymes influence catalysis is realized.
In summary, we have identified a CYP enzyme capable of catalyzing the last biochemical step in the biosynthesis of solavetivone, and have laid the groundwork for identifying those residues or regions of HPO, and its evolutionarily related cousin EAH, that are responsible for the regio- and stereo-specificity of these enzymes. The residues within SRS 5 and 6 identified here are clearly important for catalytic robustness, but it is somewhat puzzling that these more “optimized” residues are not found in the respective wild type enzymes. Perhaps overexpressing the mutant enzymes in transgenic plants will reveal unexpected phenotypic consequences of the mutant enzymes including the possible turnover of other yet to be identified plant metabolites. The current results also point to the other residue differences between HPO and EAH that lie outside of SRS 5 and 6 as critical modulators of the regio- and stereo-specificity of these multifunctional plant oxygenases.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health Grants GM54029 (to J. P. N. and J. C.) and GM13956 (to R. M. C.) and National Science Foundation Grant 07-21203 (to J. C.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2, Figs. S1–S7, additional references and data.
The abbreviations used are: EAS, 5-epi-aristolochene synthase; FPP, farensyl diphosphate; HPO, Hyoscyamus premnaspirodiene oxygenase; HPS, Hyoscyamus premnaspirodiene synthase; EAH, 5-epi-aristolochene hydroxylase; EA, 5-epi-aristolochene; EE, 4-epi-eremophilene; 2β-(OH)EA, 2β-hydroxy-5-epi-aristolochene; SRS, substrate recognition sites; CYP, cytochrome; GC-MS, gas chromatography-mass spectrometry.
REFERENCES
- 1.VanEtten HD, Mansfield JW, Bailey JA, Farmer EE. Plant Cell. 1994;6:1191–1192. doi: 10.1105/tpc.6.9.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dakora FD, Phillips DA. Physiol. Mol. Plant Pathol. 1996;49:1–20. [Google Scholar]
- 3.Hansen B, Halkier B. Planta. 2005;221:603–606. doi: 10.1007/s00425-005-1553-1. [DOI] [PubMed] [Google Scholar]
- 4.Balunas MJ, Kinghorn AD. Life Sci. 2005;78:431–441. doi: 10.1016/j.lfs.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Oksman-Caldentey KM, Inze D. Trends Plant Sci. 2004;9:433–440. doi: 10.1016/j.tplants.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Engstrom K, Widmark AK, Brishammar S, Helmersson S. Potato Res. 1999;42:43–50. [Google Scholar]
- 7.Yao MK, Desilets H, Charles MT, Boulanger R, Tweddell RJ. Mycorrhiza. 2003;13:333–336. doi: 10.1007/s00572-003-0267-0. [DOI] [PubMed] [Google Scholar]
- 8.Yokose T, Katamoto K, Park S, Matsuura H, Yoshihara T. Biosci. Biotech. Biochem. 2004;68:2640–2642. doi: 10.1271/bbb.68.2640. [DOI] [PubMed] [Google Scholar]
- 9.Kuroyanagi M, Arakawa T, Mikami Y, Yoshida K, Kawahar N, Hayashi T, Ishimaru H. J. Nat. Prod. (Lloydia) 1998;61:1516–1519. doi: 10.1021/np980214i. [DOI] [PubMed] [Google Scholar]
- 10.Threlfall DR, Whitehead IM. Phytochemistry. 1988;27:2567–2580. [Google Scholar]
- 11.Vögeli U, Chappell J. Plant Physiol. 1988;88:1291–1296. doi: 10.1104/pp.88.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cane DE. Chem. Rev. 1990;90:1089–1103. [Google Scholar]
- 13.Facchini PJ, Chappell J. Proc. Natl. Acad. Sci. U. S.A. 1992;89:11088–11092. doi: 10.1073/pnas.89.22.11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ralston L, Kwon ST, Schoenbeck M, Ralston J, Schenk DJ, Coates RM, Chappell J. Arch. Biochem. Biophys. 2001;393:222–235. doi: 10.1006/abbi.2001.2483. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi S, Zhao YX, O’Maille PE, Greenhagen BT, Noel JP, Coates RM, Chappell J. J. Biol. Chem. 2005;280:3686–3696. doi: 10.1074/jbc.M411870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gotoh O. J. Biol. Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 17.Back K, Chappell J. J. Biol. Chem. 1995;270:7375–7381. doi: 10.1074/jbc.270.13.7375. [DOI] [PubMed] [Google Scholar]
- 18.Bellucci G, Chiappe C, Pucci L, Gervasi PG. Chem. Res. Toxicol. 1996;9:871–874. doi: 10.1021/tx9600053. [DOI] [PubMed] [Google Scholar]
- 19.de Kraker J-W, Franssen MCR, Dalm MCF, de Groot A, Bouwmeester HJ. Plant Physiol. 2001;125:1930–1940. doi: 10.1104/pp.125.4.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenhagen BT, Griggs P, Takahashi S, Ralston L, Chappell J. Arch. Biochem. Biophys. 2003;409:385–394. doi: 10.1016/s0003-9861(02)00613-6. [DOI] [PubMed] [Google Scholar]
- 21.Zhao YX, Schenk DJ, Takahashi S, Chappell J, Coates RM. J. Org. Chem. 2004;69:7428–7435. doi: 10.1021/jo049058c. [DOI] [PubMed] [Google Scholar]
- 22.Ayers AR, Ebel J, Valent B, Albersheim P. Plant Physiol. 1976;57:760–765. doi: 10.1104/pp.57.5.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. J. Biol. Chem. 1997;272:19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- 24.Omura T, Sato R. J. Biol. Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 25.de Kraker JW, Schurink M, Franssen MCR, Konig WA, de Groot A, Bouwmeester HJ. Tetrahedron. 2003;59:409–418. [Google Scholar]
- 26.Shaffer GW, Eschinasi EH, Purzycki KL, Doerr AB. J. Org. Chem. 1975;40:2181–2185. [Google Scholar]
- 27.Kaspera R, Krings U, Nanzad T, Berger RG. Appl. Microbiol. Biot. 2005;67:477–483. doi: 10.1007/s00253-004-1794-0. [DOI] [PubMed] [Google Scholar]
- 28.Sakamaki H, Itoh K, Taniai T, Kitanaka S, Takagi Y, Chai W, Horiuchi CA. J. Mol. Catal. B Enzym. 2005;32:103–106. [Google Scholar]
- 29.Weyerstahl P, Marschall H, Splittgerber U, Wolf D. Flavor Frag. J. 2000;15:153–173. [Google Scholar]
- 30.Buckingham JE. Dictionary of Natural Products. London: Chapman and Hall; 1994. [Google Scholar]
- 31.Birnbaum GI, Stoessl A, Grover SH, Stothers JB. Can. J. Chem. 1974;52:993–1005. [Google Scholar]
- 32.Fiser AS, Sali A. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 33.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. J. Biol. Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 34.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 35.Rising KA, Starks CM, Noel JP, Chappell J. J. Am. Chem. Soc. 2000;122:1861–1866. [Google Scholar]
- 36.Greenhagen BT, O’Maille PE, Noel JP, Chappell J. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9826–9831. doi: 10.1073/pnas.0601605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lupien S, Karp F, Wildung M, Croteau R. Arch. Biochem. Biophys. 1999;368:181–192. doi: 10.1006/abbi.1999.1298. [DOI] [PubMed] [Google Scholar]
- 38.Wang EM, Wang R, DeParasis J, Loughrin JH, Gan SS, Wagner GJ. Nat. Biotechnol. 2001;19:371–374. doi: 10.1038/86770. [DOI] [PubMed] [Google Scholar]
- 39.Nelson DR, Kamataki T, Waxman DJ, Guengerich FP, Estabrook RW, Feyereisen R, Gonzalez FJ, Coon MJ, Gunsalus IC, Gotoh O, Okuda K, Nebert DW. DNA Cell Biol. 1993;12:1–51. doi: 10.1089/dna.1993.12.1. [DOI] [PubMed] [Google Scholar]
- 40.Bell SG, Chen XH, Sowden RJ, Xu F, Williams JN, Wong LL, Rao ZH. J. Am. Chem. Soc. 2003;125:705–714. doi: 10.1021/ja028460a. [DOI] [PubMed] [Google Scholar]
- 41.Pan LP, Wen ZM, Baudry J, Berenbaum MR, Schuler MA. Arch. Biochem. Biophys. 2004;422:31–41. doi: 10.1016/j.abb.2003.09.047. [DOI] [PubMed] [Google Scholar]
- 42.Kumar S, Chen CS, Waxman DJ, Halpert JR. J. Biol. Chem. 2005;280:19569–19575. doi: 10.1074/jbc.M500158200. [DOI] [PubMed] [Google Scholar]
- 43.He XY, Shen J, Hu WY, Ding XX, Lu AYH, Hong JY. Arch. Biochem. Biophys. 2004;427:143–153. doi: 10.1016/j.abb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 44.Kim D, Guengerich FP. Biochemistry. 2004;43:981–988. doi: 10.1021/bi035593f. [DOI] [PubMed] [Google Scholar]
- 45.Hilser VJ, Dowdy D, Oas TG, Freire E. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9903–9908. doi: 10.1073/pnas.95.17.9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glieder A, Farinas ET, Arnold FH. Nat. Biotechnol. 2002;20:1135–1139. doi: 10.1038/nbt744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.