Abstract
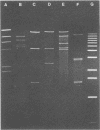
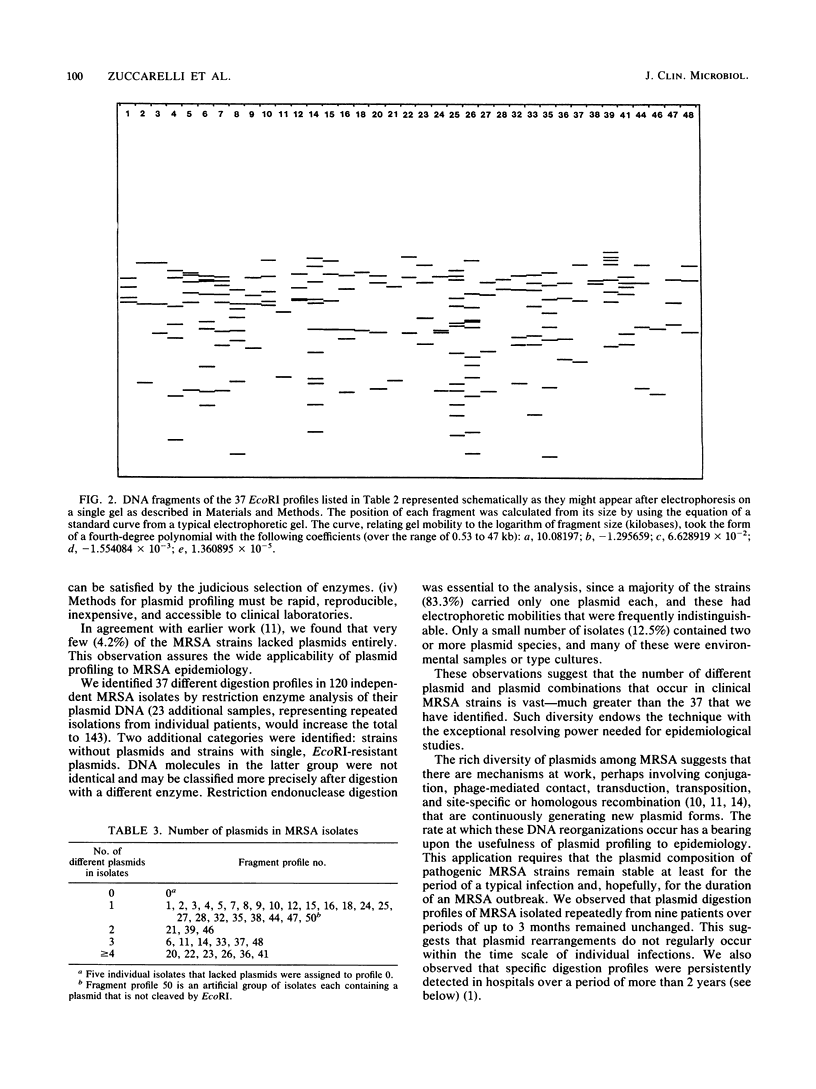
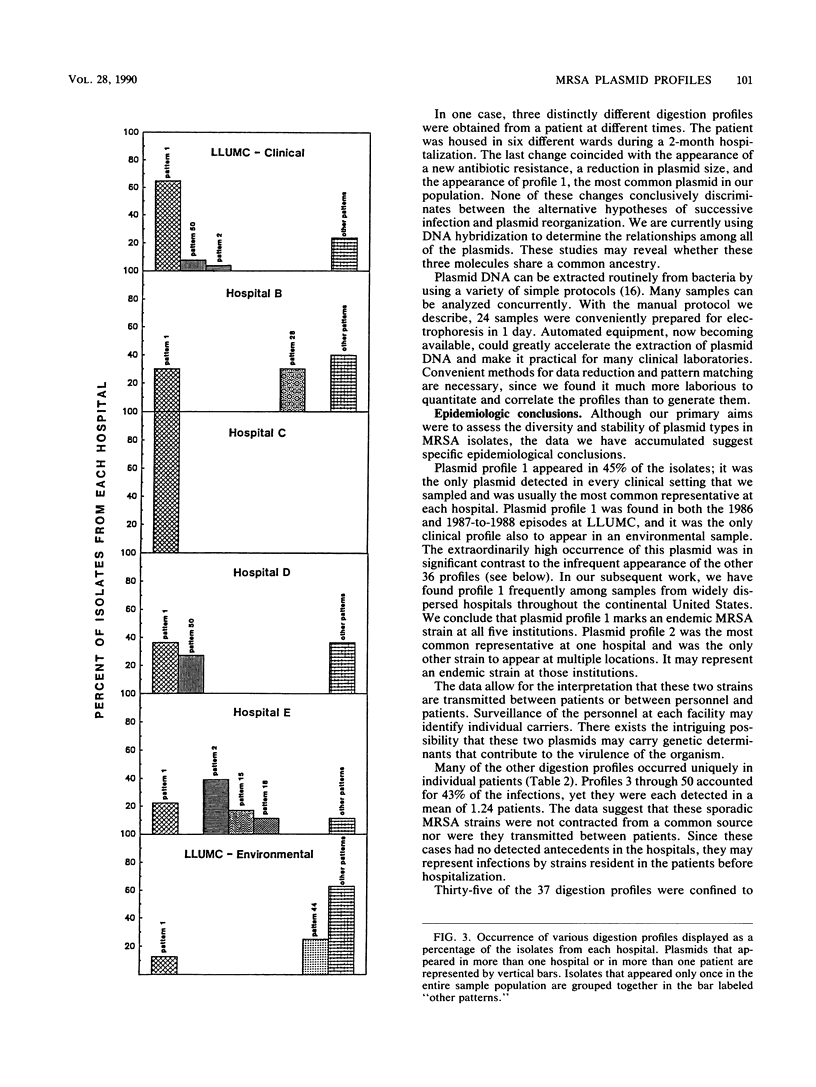
Nosocomial infections caused by methicillin-resistant Staphylococcus aureus (MRSA) are a significant epidemiological problem. Detecting the sources of epidemic strains and preventing their access to patients, however, depend upon the availability of techniques to reliably distinguish among MRSA strains. We evaluated restriction enzyme analysis of plasmid DNA for use as an epidemiological marker of MRSA strains. The diversity of plasmid types was assessed by examining 120 clinical and environmental MRSA isolates from five southern California hospitals and from the American Type Culture Collection. Thirty-seven distinctive EcoRI digestion patterns were observed. We characterized each strain by the number of plasmids it contained and the sizes of the fragments that were generated by EcoRI. Very few of the isolates (4.2%) lacked plasmids, and some (6.7%) contained DNA that was not digested by EcoRI. Several isolates (12.5%) contained two or more plasmids. We were able to assess the stability of MRSA plasmid types by tracking epidemic strains over a 2-year period. We also examined successive isolates from 10 individual patients during their hospitalization. In all but one case, the patient's plasmid profiles remained unchanged. We conclude that the diversity and stability of MRSA plasmid types make them excellent epidemiological markers. In support of this conclusion, we found that our data provided significant epidemiological insights. Two epidemic strains, accounting for more than half of the infections, were identified in the five hospitals. The remaining cases were sporadic, caused by MRSA strains that appeared very infrequently and that may have originated from sources outside the hospitals.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Archer G. L., Mayhall C. G. Comparison of epidemiological markers used in the investigation of an outbreak of methicillin-resistant Staphylococcus aureus infections. J Clin Microbiol. 1983 Aug;18(2):395–399. doi: 10.1128/jcm.18.2.395-399.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon A. E., Jorgensen K. A., Wilson K. H., Kauffman C. A. Emergence of nosocomial methicillin-resistant Staphylococcus aureus and therapy of colonized personnel during a hospital-wide outbreak. Infect Control. 1987 Apr;8(4):145–150. doi: 10.1017/s0195941700065802. [DOI] [PubMed] [Google Scholar]
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966 Apr;45(4):493–496. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley R. W., Hightower A. W., Khabbaz R. F., Thornsberry C., Martone W. J., Allen J. R., Hughes J. M. The emergence of methicillin-resistant Staphylococcus aureus infections in United States hospitals. Possible role of the house staff-patient transfer circuit. Ann Intern Med. 1982 Sep;97(3):297–308. doi: 10.7326/0003-4819-97-3-297. [DOI] [PubMed] [Google Scholar]
- Hartstein A. I., Morthland V. H., Eng S., Archer G. L., Schoenknecht F. D., Rashad A. L. Restriction enzyme analysis of plasmid DNA and bacteriophage typing of paired Staphylococcus aureus blood culture isolates. J Clin Microbiol. 1989 Aug;27(8):1874–1879. doi: 10.1128/jcm.27.8.1874-1879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloos W. E., Orban B. S., Walker D. D. Plasmid composition of Staphylococcus species. Can J Microbiol. 1981 Mar;27(3):271–278. doi: 10.1139/m81-043. [DOI] [PubMed] [Google Scholar]
- Kozarsky P. E., Rimland D., Terry P. M., Wachsmuth K. Plasmid analysis of simultaneous nosocomial outbreaks of methicillin-resistant Staphylococcus aureus. Infect Control. 1986 Dec;7(12):577–581. doi: 10.1017/s0195941700065413. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Locksley R. M., Cohen M. L., Quinn T. C., Tompkins L. S., Coyle M. B., Kirihara J. M., Counts G. W. Multiply antibiotic-resistant Staphylococcus aureus: introduction, transmission, and evolution of nosocomial infection. Ann Intern Med. 1982 Sep;97(3):317–324. doi: 10.7326/0003-4819-97-3-317. [DOI] [PubMed] [Google Scholar]
- Lyon B. R., May J. W., Skurray R. A. Analysis of plasmids in nosocomial strains of multiple-antibiotic-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1983 Jun;23(6):817–826. doi: 10.1128/aac.23.6.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T. F. Resistance of bacteria to antibacterial agents: report of Task Force 2. Rev Infect Dis. 1987 May-Jun;9 (Suppl 3):S244–S260. doi: 10.1093/clinids/9.supplement_3.s244. [DOI] [PubMed] [Google Scholar]
- Rhinehart E., Shlaes D. M., Keys T. F., Serkey J., Kirkley B., Kim C., Currie-McCumber C. A., Hall G. Nosocomial clonal dissemination of methicillin-resistant Staphylococcus aureus. Elucidation by plasmid analysis. Arch Intern Med. 1987 Mar;147(3):521–524. [PubMed] [Google Scholar]
- Schaberg D. R., Zervos M. Plasmid analysis in the study of the epidemiology of nosocomial gram-positive cocci. Rev Infect Dis. 1986 Sep-Oct;8(5):705–712. doi: 10.1093/clinids/8.5.705. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Nagano Y. Rapid procedure for isolation of plasmid DNA and application to epidemiological analysis. J Clin Microbiol. 1984 Oct;20(4):608–613. doi: 10.1128/jcm.20.4.608-613.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsmuth K. Molecular epidemiology of bacterial infections: examples of methodology and of investigations of outbreaks. Rev Infect Dis. 1986 Sep-Oct;8(5):682–692. doi: 10.1093/clinids/8.5.682. [DOI] [PubMed] [Google Scholar]
- Walsh T. J., Vlahov D., Hansen S. L., Sonnenberg E., Khabbaz R., Gadacz T., Standiford H. C. Prospective microbiologic surveillance in control of nosocomial methicillin-resistant Staphylococcus aureus. Infect Control. 1987 Jan;8(1):7–14. doi: 10.1017/s0195941700066923. [DOI] [PubMed] [Google Scholar]
- Wiegand R. C., Godson G. N., Radding C. M. Specificity of the S1 nuclease from Aspergillus oryzae. J Biol Chem. 1975 Nov 25;250(22):8848–8855. [PubMed] [Google Scholar]