Abstract
Alternative splicing (AS) is a mechanism by which multiple types of mature mRNAs are generated from a single pre-mature mRNA. In this study, we completely sequenced 1800 full-length cDNAs from Arabidopsis thaliana, which had 5′ and/or 3′ sequences that were previously found to have AS events or alternative transcription start sites. Unexpectedly, these sequences gave us further evidence of AS, as 601 out of 1800 transcripts showed novel AS events. We focused on the combination patterns of multiple AS events within individual genes. Interestingly, some specific AS event combination patterns tended to appear more frequently than expected. The two most common patterns were: (i) alternative donor–0∼12 times of exon skips–alternative acceptor and (ii) several times (∼8) of retained introns. We also found that multiple AS events in a transcript tend to have the same effects concerning the length of the mature mRNA. Our current results are consistent with our previous observations, which showed changes in AS profiles under different conditions, and suggest the involvement of hypothetical cis- and trans-acting factors in the regulation of AS events.
Keywords: Arabidopsis, alternative splicing, bioinformatics, full-length cDNA
1. Introduction
Alternative splicing (AS) is a mechanism by which multiple forms of mature mRNAs are generated from a single pre-mature mRNA. The most recent genome-wide analysis of AS events in Arabidopsis thaliana found that >4700 transcribed pre-mature mRNAs were alternatively spliced.1 The number of known AS events in Arabidopsis has recently been increasing. AS events are important because they diversify the proteome. The impacts of AS events on the human transcriptome and proteome are also well known. In humans, ∼70% of all genes are thought to be alternatively spliced.2,3 It is thought that many kinds of proteins are translated from the limited number of genes encoded by the human genome. Compared with humans, there are fewer known AS events in plants. AS events in Arabidopsis occur in around 20% of all known genes.1 Although the number of AS events is small, some important AS events have been described in Arabidopsis. For example, the floral promoter FCA, disease resistance protein RPS4 and splicing factor atRSZ33 are known to undergo AS.4–6
AS events are not only important sources for the diversification of the proteome, but they also have an important role in post-transcriptional regulation. In a previous study, we reported that the pattern of AS events on a genome-wide scale (called AS profiles) was affected by stress conditions and tissue types, especially by cold stress.7 In a full-length cDNA library from cold-treated plants, cassette exons tend to be selected in mature mRNAs, and introns tend to be retained. Several recent studies have also found that stresses and/or tissue types affect the selectivity of AS events.8–10
Recently, several novel technologies have been established, which have improved our ability to study the transcriptome. For studying AS events on a genome-wide scale, a microarray approach, with probes designed for each exon11 or covering the whole genome,2,3 may prove useful. However, it is difficult to elucidate the entire sequence of AS variants with these methods. The best way to analyze the exact AS events for every transcript is still to make libraries of full-length cDNAs and sequence them. In 2004, we found 2092 AS events after analyzing ∼278 000 sequences from full-length cDNAs.7,12,13 However, most of these sequences were partial reads from 5′ or 3′ regions of the full-length cDNAs. Because our previous studies tended to analyze AS events in terminal regions, we could not address impacts of these AS events on the full-length sequences and their coding potentials. In the current study, we chose 1800 cDNAs that had AS events or transcriptional start sites (TSSs) in their 5′/3′ sequences, and sequenced them entirely. On the basis of these novel full-length cDNA sequences, we determined how many AS events occurred in each gene, how multiple AS events within a single gene affected each other and how AS events affected coding potentials.
2. Materials and methods
2.1. Selecting the full-length cDNAs for determination of the full-length sequences
We used clones from the RIKEN Arabidopsis full-length (RAFL) cDNA libraries.12,13 We classified the fully and partially read RAFL cDNA sequences based on their locations in the genome, AS events and TSSs. As a first step, we constructed transcription units (TUs) based on locations of the sequences in the genome.7 Next, we checked for the existence of AS events, which were classified as exon skip (ES), alternative donor (AD), alternative acceptor (AA) or retained intron (RI) for each TU. A set of sequences, which did not have any AS events, was classified into an AS group. In some cases, a pair of transcripts in a particular TU lacked overlapping exons. In these cases, although no AS events were found, we classified these two transcripts into different AS groups.
Finally, we checked the TSSs for each AS group. If a pair of the 5′ end read sequences or fully read sequences had TSSs with differences in length of >500 bp, we separated them into two different TSS groups. As a result, each transcript was classified into a group based on TU, AS and TSS. If a group lacked fully read transcripts, we chose one clone from this group as a target for determination of the full-length sequences.
2.2. End and full-length sequencing
Plasmid DNA extraction was performed using an automated DNA isolation system (PI-1100, Kurabo), and end sequencing of the 1800 clones was performed with BigDye terminator ver.3 sequencing kits and ABI 3730 automated capillary DNA sequencers (Applied Biosystems). For full-length sequencing, template DNA was individually prepared from each plasmid DNA using the rolling-circle amplification (RCA) method (TempliPhi DNA amplification kit, GE Healthcare). A pooled sample, consisting of 768 RCA products, was subjected to shotgun sequencing (ABI 3730). Another pooled sample (1032 RCA products) was sequenced using a Genome Sequencer 20 System (454 Life Sciences/Roche). The generated contig sequences were then incorporated into the end sequence data using the Phrap/Consed system.14 Primer walking was used to close gaps and re-sequence low-quality regions of the assembled data. The reconstructed cDNA sequences were completely covered by the Sanger reads. All sequences, excluding 55 sequences that were not mapped to the genome, were submitted to the DNA Data Bank of Japan (DDBJ) under the accession numbers AK316663–AK317782 and AK318618–AK319175.
2.3. Classification of transcripts as known or novel
All novel sequences were mapped to the genome sequence with BLAST and GeneSeqer15,16 using a previously described method.17 Next, the transcripts were classified as known or novel transcripts based on comparisons with annotated gene models. In addition to the four types of AS events (ES, AD, AA and RI), to make a detailed classification of known and novel transcripts, we also checked for alternative structure events, in which transcriptional initiation or termination occurred within the introns of other transcripts (AI and AT). Unlike the method in Section 2.1, we ignored TSS differences in this step, even if those differences were more than 500 bases, because differences found in TSSs could have occurred during the sequencing of potential partial cDNAs.
For transcript classification, we used novel sequences and annotated gene model in The Arabidopsis Information Resource (TAIR), version 7,18 which corresponded with the novel sequences. For these sequences, TU clustering and AS-group clustering were performed again, using a method similar to that described in Section 2.1. Every result was checked manually after the computational classification.
2.4. Analysis of the number of AS events per gene
To analyze the number of AS events per gene, we chose annotated gene loci with AS variants from TAIR. For the data set in the current study, we used novel sequenced transcripts and the annotated gene models, which were derived from corresponding loci with novel sequences. In this analysis, the number of AS events from each of the four types (ES, AD, AA and RI) was counted. The number of AS events was defined based on the type of AS event and the region affected by the AS event. We noted special cases of splice acceptors with more than three acceptor sites. In a case of an intron of three possible splice acceptor sites, two AS events were counted. One event was defined by the region affected between the first and the second acceptor sites, and the second event by the region between the second and third acceptor sites. If we had defined the AS events based on the combination of acceptor sites of the AS variant, there would have been three events. Therefore, the number of AS events would have increased dramatically due to the increased number of acceptor sites. The new method avoided this great increase in event numbers. There was also a similar rule about multiple AD sites. We compared the number of AS events found in each gene using a Mann–Whitney test and used R package (http://cran.r-project.org/) to calculate the P-values.
2.5. Analysis of the AS event combination patterns
In cases of transcripts with more than two AS events, we analyzed the pattern of AS combinations. For these analyses, we sorted AS events that were found in a transcript by the order of their locations. Then, we paired adjacent AS events according to their orders (i.e. first and second, second and third and so forth). Expectation values of each of the AS event combination patterns were calculated based on the probability of a single AS event. Statistical analyses were performed using binomial tests. We also used R package to calculate the P-values.
AS events could be classified based on their directions. We called AS events that made longer mRNAs ‘in-type’ events, which means regions affected by AS were included in the mRNAs. In the opposite cases, we called AS events that made shorter mRNAs ‘out-type’ events. We classified all the AS events as in- or out-type. Then, we counted the number of combinations for either the in- or out-types from adjacent AS events. Statistical analysis was performed using a chi-squared test with R package.
2.6. Prediction of coding sequences and analysis of the effects of as events on functional motifs
When transcripts were classified into known TUs, we determined their coding sequences (CDSs) based on their corresponding genes. For novel transcripts, we performed BLASTX searches against the annotated protein sequences encoded in mRNAs from the corresponding loci of TAIR.15 From the result, we determined the core CDSs and phases, and then extended the CDS to its up- and downstream regions until we found the farthest initial codons and the nearest termination codons. The results were checked manually after the computational prediction. For transcripts classified as novel TUs, we searched CDSs based on a BLASTX search results using all protein sequences in TAIR's database (ftp://ftp.arabidopsis.org).
To assess the impact of AS events on the encoded protein sequences, we performed motif search analyses for the protein sequences encoded by the novel AS variants and corresponding annotated gene model. In this case, we selected the most similar protein sequences generated from the corresponding loci that were used as reference sequences. The motif search was performed with Pfam.19
3. Results and discussion
3.1. Full sequencing of 1800 full-length cDNAs
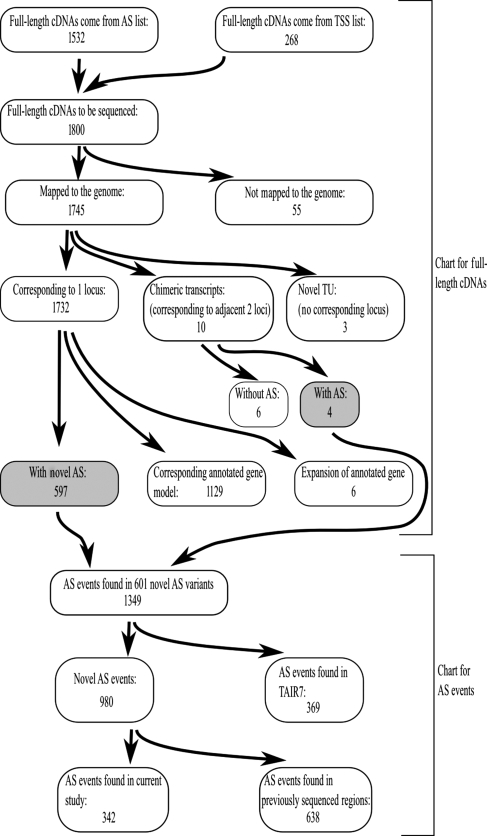
We determined the full-length sequences of 1800 RAFL cDNAs; 1657 reads (92%) had clear poly-A tails. The average length was 1495 nt, and the GC content was 42.9%. These features were quite similar to our previous sets of full-length cDNAs (average lengths were 1483 nt and GC contents were 42.8%).12,13 We mapped the transcripts with BLAST and GeneSequer.15,16 A total of 1745 transcripts were mapped to the Arabidopsis genome. We could not find the corresponding loci for the other 55 transcripts (Fig. 1).
Figure 1.
Flowchart and outline of the classifications used for current data set (upper part) and AS events found in the novel AS variants (lower part). The gray-highlighted boxes correspond to the transcripts with novel AS events. In total, 601 transcripts were classified as novel AS variants. And 342 AS events were novel ones found in the current study.
3.2. Classification of transcripts as known or novel
Out of the 1745 mapped transcripts, three lacked corresponding annotated genes in TAIR. They were classified as novel TUs. Another 14 transcripts were chimeric transcripts that were produced from two adjacent genes. In humans, a large number of such chimeric transcripts have been reported.3,20,21 The current data set also showed that Arabidopsis had some chimeric transcripts. A total of 1129 transcripts had corresponding gene models in the annotated gene set (Fig. 1, Supplementary Table S1). Six transcripts had additional exons compared with the annotated gene models in TAIR. The remaining 597 transcripts had novel AS events and/or alternative structure variants (as a whole we called them AS variants). Out of them, 572 transcripts contained novel AS events that had been unknown. The others were AS variants that consisted of novel combinations of known AS events. In addition to these 597 novel AS variants, four chimeric transcripts had novel exon–intron structures in comparison with annotated ones. In total, 601 transcripts were classified as novel alternatively spliced/structure variant transcripts (Fig. 1, Supplementary Table S2). AS events found in the 601 transcripts were summarized in Table 1. We checked relative frequencies of each AS event within all AS events found in the novel transcripts and the novel AS events only within the transcripts. In both cases, the frequencies of AS events were similar to the previous studies;1 RI events were the most abundant ones and AA events were the next.
Table 1.
The results of AS and alternative structure event prediction
| Event typea | Total events in novel transcripts |
Novel events in novel transcripts |
||
|---|---|---|---|---|
| Number of AS/structure events | Ratio | Number of AS/structure events | Ratio | |
| RI | 610 | 0.45 | 479 | 0.36 |
| AA | 279 | 0.21 | 178 | 0.13 |
| ES | 185 | 0.14 | 143 | 0.11 |
| AD | 161 | 0.12 | 111 | 0.08 |
| AI | 66 | 0.05 | 39 | 0.03 |
| AT | 48 | 0.04 | 30 | 0.02 |
| Total | 1349 | 980 | ||
aRI, retained intron; AA, alternative acceptor; ES, exon skip; AD, alternative donor; AI, alternative initiation of transcription; AT, alternative termination of transcription. AI and AT, the alternative structure events were checked for the classification of the transcripts.
3.3. Analysis of the number of AS events per gene
Six hundred and one genes were classified as novel AS variants, which was unexpected because we chose full-length cDNA clones that already had AS events in their sequenced 5′/3′ regions. These 5′/3′ sequences and AS events have already been registered to the GenBank database.7,12,13 Since the TAIR group makes efforts to continuously update the gene models, including the AS information,18 it was expected that the AS events in previously sequenced regions would have been included in TAIR's annotation and classified as known AS events. Thus, we thought that the 601 novel AS variants must contain additional AS events in their novel sequence regions. We compared the regions containing the AS events with the regions that were sequenced previously (as 5′/3′ sequences). Out of the 980 AS events without corresponding events in TAIR, 638 AS events were present in the previous 5′/3′ sequences (Fig. 1, Supplementary Table S2). We concluded that the main reason why we found so many novel AS events was that some of the AS events found in the previous 5′/3′ sequences were not reflected in TAIR's annotation. It might be due to TAIR's policy in the database update to exclude alternatively spliced mRNAs with premature termination codons.18
Despite that we found many AS events corresponded with the previously sequenced partial sequences, we still found 342 novel AS events in the newly sequenced regions. This result showed that transcripts with AS events tended to have additional AS events in other positions, suggesting that the number of AS events in a single gene should be much greater than what is currently known. Next, we compared the number of AS events per gene between the current data set and TAIR. The average number of AS events per gene was found to be 2.0 in the TUs that corresponded to the current transcripts. On the other hand, it was 1.4 for the genes that had more than two mature mRNA structures in TAIR's annotation. In the current data set, a higher fraction of genes had more than two AS events compared with the genes in TAIR's data set (Supplementary Fig. S1). These differences were statistically significant (P < 2.2E−16) with a Mann–Whitney test. Therefore, we concluded that Arabidopsis genes should have more AS events per gene. In the current analysis, a further sequencing effort showed many more AS events in Arabidopsis. This finding was consistent with a previous report, in which the frequency of AS events is correlated with the abundance of EST/full-length sequences.22 Further sequencing should show that more AS events occur in Arabidopsis.
3.4. Analysis of the pattern of AS event combinations in each gene
In the current study of AS events, specific patterns of AS event combinations preferentially occurred. To confirm this result, we focused on transcripts that had more than two AS events. Among the cDNAs characterized in this study, 354 transcripts had more than two AS events. The number of combinations between adjacent AS events was examined and compared with expectation values that were calculated from the frequencies of every AS event type found in the 354 transcripts (Table 2). The frequencies of the individual AS event types were similar between the 601 novel AS variants and the 354 AS variants with multiple AS events (Tables 1 and 2). The four combinations (AD–ES, AD–AA, ES–ES and ES–AA) were observed more frequently than expected, which was statistically significant (P < 0.01 in a binomial test; Table 2). The combination of RI–RI was also observed more frequently than expected, although this was not statistically significant (P = 0.106 in a binomial test). The prominence of these five combinations suggests the following two models for AS patterns: combinations of AD–ES (0∼12 times)–AA and multiple repeats of RI (2∼8 times) events. Out of our 601 novel AS variants, the former model was found in 80 transcripts and the latter was found in 125 transcripts (examples in Fig. 2). In a previous study, Wang and Brendel1 analyzed AS events in Arabidopsis and rice and concluded that most AS events were mutually independent. The reason why their conclusions and ours were different might be due to that we analyzed the sequences of full-length cDNAs in the current studies, whereas Wang and Brendel treated the data sets including a vast amount of partial read sequences. Analysis of full-length cDNAs showed possibilities that some AS events in a transcript were related each other.
Table 2.
Analysis of the combination of two adjacent AS events
| First AS events |
Second AS events |
||||||
|---|---|---|---|---|---|---|---|
| Number of the AS eventsa | Frequencya | RI | AA | ES | AD | ||
| RI | 509 | 0.45 | Expectation valueb | 156.5 | 82.1 | 58.1 | 54.4 |
| Observation value | 221 | 71 | 12 | 25 | |||
| P-valuec | 0.106 | — | — | — | |||
| AA | 267 | 0.23 | Expectation valueb | 82.1 | 43.1 | 30.5 | 28.6 |
| Observation value | 60 | 42 | 11 | 21 | |||
| P-valuec | — | — | — | — | |||
| ES | 189 | 0.17 | Expectation valueb | 58.1 | 30.5 | 21.6 | 20.2 |
| Observation value | 14 | 59 | 85 | 13 | |||
| P-valuec | — | 2.30E−05 | <2.2E −16 | — | |||
| AD | 177 | 0.15 | Expectation valueb | 54.4 | 28.6 | 20.2 | 18.9 |
| Observation value | 53 | 45 | 44 | 12 | |||
| P-valuec | — | 0.009 | 1.32E−05 | — | |||
aThese values were counted as novel AS variants with more than two AS events.
bExpectation values. Calculated based on the frequencies of independent events. Case number is the observation number: 788.
cResults of the binomial test. P-values are shown if the observation values are greater than the expectation values.
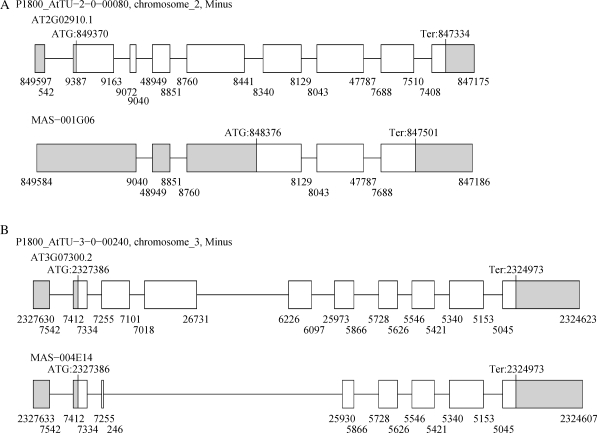
Figure 2.
Examples of multiple occurrences of AS events. (A) The AT2G02910 locus which encodes a predicted hydrolase that acts on carbon–nitrogen bonds. The transcript (MAS-001G06) retains introns 1, 2, 5 and 8 compared with AT2G02910.1. (B) The AT3G07300 locus which encodes a protein from the eukaryotic translation initiation factor 2B family. The transcript (MAS-004E14) shows an AD, two ES and an AA AS events. All four of these events were of the ‘out-type’.
It should be noted that 26 out of 125 transcripts with multiple RI events consisted of single exons. Most of these transcripts seemed to be mature mRNAs, because 21 out of 26 had clear poly-A tails. However, it was still possible that these transcripts without intron splicing were immature mRNAs, and therefore they were excluded. In this case, the probability of RI events was 0.40. As a result, the number of RI–RI combinations was 157, which was still greater than the expectation value (114.9). Fig. 2B shows an example of a combination pattern in which multiple, but not all, introns were retained in a single transcript. Thus, the model of multiple repeats of RI events was also enriched in the current data set.
We also classified the AS events by their direction, as in- and out-type events (see Section 2 for details). The direction of AS events showed that for two adjacent AS events, a single direction was preferable (Supplementary Table S3). This was clear when the adjacent events consisted of the same type of AS event (e.g. ES–ES or RI–RI, which was frequently observed in the analysis of the patterns of AS event combinations). This observation was supported by a chi-squared test with a P-value of <2.2E−16. Interestingly, this trend was still statistically significant in cases of combinations that were made up of two different AS events (P = 1.7E−07 in a chi-squared test; Supplementary Table S3).
For further evidence, we analyzed all public full-length cDNAs of Arabidopsis and rice, respectively (Supplementary Tables S4 and S5). Both results were similar to those of current study concerning the combination of AS events types and directions of the AS events. Thus, we concluded that transcriptome of land plants had preferred combinations of AS event types and directions.
3.5. Potential AS regulatory mechanisms
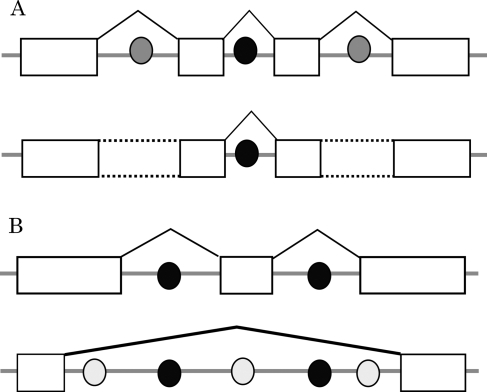
In the first step of intron splicing in plants, it is thought that the splicing machinery recognizes the intron sequences. In contrast, exon sequences are recognized in mammals.23 The present study demonstrates that specific patterns of combinations and directions of AS events are preferred in Arabidopsis. Our results are consistent with a model in which the splicing machinery recognizes intronic sequences.23 If trans-acting factors, which recognize weak splicing enhancers in introns (called intronic splicing enhancers, ISEs24), are abundant in the nucleus, then the recognized regions are spliced out (Fig. 3). This model was similar to the previous one which had explained abundant RI events in plants.1 However, we thought that this model could also explain the existence of another special type of combination of AS events; AD–ES–AA (Fig. 3B). When some trans factors recognize weak splicing signals which lay on close positions, one huge intron might be spliced out from a pre-mRNA.
Figure 3.
Hypothesis regarding the regulation of multiple AS events. (A) Model for cases of multiple RI events. The circles show the putative trans-acting factors. In this model, if the trans-acting factors, which recognize weak splicing signals (gray circles), are decreased, introns with the weak signals remain in the mRNA. (B) The AS model consisting of AD–ES–AA. The gray circles show putative trans-acting factors, which recognize weak splicing signals. If the concentration of the factors increases, several regions are recognized as introns and then the large region is spliced out as a single intron. It is not known if the trans- or cis-acting factors in the model are the same.
It is unclear whether or not the same trans-acting factors recognize both weak ISEs and constitutive splicing signals. However, some SR proteins are candidate trans-acting factors in the proposed model. SR proteins are splicing factors that are involved in constitutive and AS.25 It is known that the regulation of many SR proteins is complicated and can vary by tissue and stress conditions.6,10 It has also been noted that some SR proteins can recognize ISEs.24 On the basis of these evidences, it may be possible that SR proteins work as trans-acting factors. Future work must focus on elucidating the ISEs and trans-acting factors in this model.
It should also be noted out that our current results are consistent with our previous work on AS profiles. In the previous work, we found a tendency to select cassette exons, retain introns and select AD and acceptor sites to make longer mRNAs in plants during conditions of cold stress.7 All these tendencies could be explained by the fact that in-type events are more frequent during cold stress. In the current analysis, we had 32 transcripts with 111 AS events from a library of cold-treated plants. Out of the 111 AS events, 77 (69%) were in-type AS events. In 324 transcripts with multiple AS events, 569 of 1142 (50%) AS events were in-type events. These differences were statistically significant by a chi-squared test (P < 0.001). It may be possible that one or more trans-acting factors that recognize weak ISEs are down-regulated in response to cold stress. Interestingly, microarray analysis26 showed that expressions of many SR protein genes largely changed under cold stress conditions (Supplementary Table S6). Such regulation on SR proteins might change AS profiles in a large scale.
3.6. Analysis of the motifs found in the CDSs of AS variants
We also analyzed the CDSs encoded in the 601 novel AS variants. Of these, 52 transcripts had no clear CDS and 476 had shorter CDSs compared with the corresponding annotated gene models in TAIR. Out of the 476 transcripts with short CDSs, 96 transcripts had introns in their 3′ UTRs. Including them, we found 112 transcripts with introns in their 3′ UTRs from the data set of the 601 novel AS variants. Similar to mammals, it was reported that nonsense-mediated mRNA decay (NMD) mechanism exists in Arabidopsis. The NMD mechanism takes transcripts that have exon junctions downstream of the termination codon and causes them to decay.27 Although we may not know the actual CDS encoded by the transcripts, the number of transcripts with 3′ UTRs that contain introns appears to be large. In addition, comparison of functional motifs between the CDSs of AS variants and their reference proteins showed that most AS variants encoded proteins that lacked some functional domains. For 460 out of 549 genes that provided the novel AS variants, we could assign at least one functional domain for the CDS encoded in the AS variant and/or its reference transcript (Supplementary Table S7). In the protein sequences encoded by the novel AS variants, 392 (85%) had a reduced number of functional domains in comparison with their reference protein sequences (Supplementary Table S7). Only four transcripts had additional motifs when compared with the reference protein sequences. These results seem to show that most AS events decrease the functionality of protein products. We hypothesize that many AS events regulate the amount of functional protein by making mRNAs that are targeted by the NMD or by making mRNAs that encode non-functional proteins. This view is consistent with our previous results from a study on SR proteins in Arabidopsis, rice and moss.28 In this case, three independent AS events of mRNAs encoding SR proteins with incomplete RNA-binding domains were highly conserved in land plants. Similar models have also been proposed for AS events in animals.29,30 Our current results support the existence of a similar regulatory system in Arabidopsis.
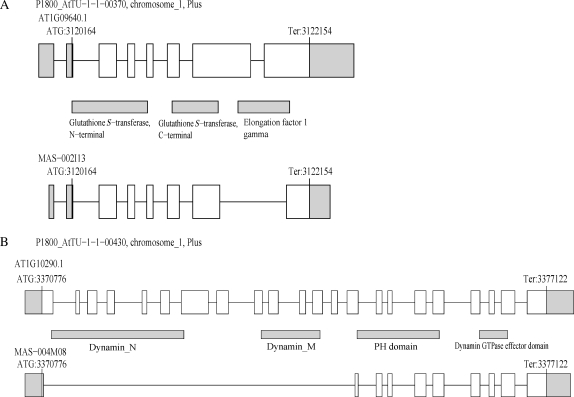
In the same time, some AS events possibly modulated functions of proteins with multiple domains. A locus AT1G09640 encodes a protein with glutathione S-transferase (GST) domains and elongation factor (EF) 1 gamma domain (Fig. 4A). It was thought that GST activities regulated assembly of a complex containing EF1 subunits and aminoacyl-tRNA synthetases.31 An AS variant encoded a protein with GST domains but not EF1 gamma domain (Fig. 4A), which might not be a member of the complexes or have some regulatory roles on assembly of the complex. Another example was found in the locus AT1G10290 encoding dynamin-like protein, ADL6. It was reported that ADL6 had a function in vesicle trafficking from the trans-Golgi network to the central vacuole.32 This protein has dynamin domains followed by pleckstrin homology domain which was required for interaction between the protein and lipid32 (Fig. 4B). An AS variant encodes a protein without dynamin domains, which should lack dynamin activities but may have activities of interaction with lipids. This AS isoform protein possibly affects activity of the intact proteins. These examples showed that some AS events can modulate functions of proteins especially for ones with multiple domains.
Figure 4.
Examples of AS events affecting functional motifs. (A) The AT1G09640 locus which encodes a protein with GST domains and EF1 gamma domain. An AS variant MAS-002I13 encodes a protein lacking EF1 gamma domain. (B) The AT1G10290 locus which encodes dynamin-like protein ADL6. An AS variant MAS-004M08 encodes a protein lacking dynamin domains. Such AS events might modulate protein functions.
We also checked the CDSs of the 14 chimeric transcripts. Of these, only one transcript (MAS-001H13) had an ORF that spanned between the first and second genes (Supplementary Fig. S2). The transcript was a fused mRNA that began with the AT3G63340 locus (encoding a protein phosphatase 2C-related protein) and ended with the AT3G63330 locus (encoding a protein kinase). Still it is hard to image a function for the protein product of this chimeric transcript, but it is likely that this chimeric protein would function in both adding and removing phosphate groups on target proteins.
3.7. Summary
In the current study, we fully sequenced 1800 full-length cDNAs. Even though this data set was not large, it suggests the possibility that many more AS events occur in Arabidopsis than expected. Furthermore, the analysis of multiple AS events in individual genes suggested a regulatory mechanism that controls AS profiles. We hypothesize that some splicing regulatory elements and trans-acting factors are involved in the regulation of AS profiles. Further computational and experimental approaches should help clarify these cis- and trans-acting factors. We also examined coding potentials and protein sequences in the current study. It was interesting that many transcripts seemed to be targeted by NMD and/or to encode potentially non-functional proteins. More integrated analyses of the regulation of AS profiles and coding potential should give us a greater understanding of the importance of AS in Arabidopsis and other plants.
Supplementary Data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
Support to characterize the full-length sequences of RAFL cDNA clones discussed in this manuscript was provided by the National Bio-Resource Project of the MEXT, Japan.
Acknowledgements
We thank the entire technical staff of the Sequence Technology Team at RIKEN GSC for their assistance. We also thank Mr Issei Sasaki (RIKEN BRC) for technical assistance.
Footnotes
Edited by Mikio Nishimura
References
- 1.Wang B. B., Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson J. M., Castle J., Garrett-Engele P., Kan Z., Loerch P. M., Armour C. D., Santos R., Schadt E. E., Stoughton R., Shoemaker D. D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 3.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 4.Macknight R., Duroux M., Laurie R., Dijkwel P., Simpson G., Dean C. Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell. 2002;14:877–888. doi: 10.1105/tpc.010456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X. C., Gassmann W. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol. 2007;145:1577–1587. doi: 10.1104/pp.107.108720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalyna M., Lopato S., Voronin V., Barta A. Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res. 2006;34:4395–4405. doi: 10.1093/nar/gkl570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iida K., Seki M., Sakurai T., Satou M., Akiyama K., Toyoda T., Konagaya A., Shinozaki K. Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res. 2004;32:5096–5103. doi: 10.1093/nar/gkh845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bove J., Kim C. Y., Gibson C. A., Assmann S. M. Characterization of wound-responsive RNA-binding proteins and their splice variants in Arabidopsis. Plant Mol. Biol. 2008;67:71–88. doi: 10.1007/s11103-008-9302-z. [DOI] [PubMed] [Google Scholar]
- 9.Tanabe N., Yoshimura K., Kimura A., Yabuta Y., Shigeoka S. Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs, atSR30 and atSR45a, in response to environmental stress. Plant Cell Physiol. 2007;48:1036–1049. doi: 10.1093/pcp/pcm069. [DOI] [PubMed] [Google Scholar]
- 10.Palusa S. G., Ali G. S., Reddy A. S. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 2007;49:1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x. [DOI] [PubMed] [Google Scholar]
- 11.Gardina P. J., Clark T. A., Shimada B., et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seki M., Narusaka M., Kamiya A., et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- 13.Yamada K., Lim J., Dale J. M., Chen H., Shinn P., Palm C. J., Southwick A. M., Wu H. C., Kim C., Nguyen M., et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 14.Gordon D., Abajian C., Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 15.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brendel V., Xing L., Zhu W. Gene structure prediction from consensus spliced alignment of multiple ESTs matching the same genomic locus. Bioinformatics. 2004;20:1157–1169. doi: 10.1093/bioinformatics/bth058. [DOI] [PubMed] [Google Scholar]
- 17.Iida K., Shionyu M., Suso Y. Alternative splicing at NAGNAG acceptor sites shares common properties in land plants and mammals. Mol. Biol. Evol. 2008;25:709–718. doi: 10.1093/molbev/msn015. [DOI] [PubMed] [Google Scholar]
- 18.Swarbreck D., Wilks C., Lamesch P., et al. The Arabidopsis information resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn R. D., Mistry J., Schuster-Böckler B., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akiva P., Toporik A., Edelheit S., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R. Transcription-mediated gene fusion in the human genome. Genome Res. 2006;16:30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babushok D. V., Ohshima K., Ostertag E. M., Chen X., Wang Y., Mandal P. K., Okada N., Abrams C. S., Kazazian H. H., Jr A novel testis ubiquitin-binding protein gene arose by exon shuffling in hominoids. Genome Res. 2007;17:1129–1138. doi: 10.1101/gr.6252107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brett D., Pospisil H., Valcárcel J., Reich J., Bork P. Alternative splicing and genome complexity. Nat. Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- 23.Lorković Z. J., Wieczorek Kirk D. A., Lambermon M. H., Filipowicz W. Pre-mRNA splicing in higher plants. Trends Plant Sci. 2000;5:160–167. doi: 10.1016/s1360-1385(00)01595-8. [DOI] [PubMed] [Google Scholar]
- 24.Reddy A. S. Plant serine/arginine-rich proteins and their role in pre-mRNA splicing. Trends Plant Sci. 2004;9:541–547. doi: 10.1016/j.tplants.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Kalyna M., Barta A. A plethora of plant serine/arginine-rich proteins: redundancy or evolution of novel gene functions? Biochem. Soc. Trans. 2004;32:561–564. doi: 10.1042/BST0320561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D'Angelo C., Bornberg-Bauer E., Kudla J., Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 27.Hori K., Watanabe Y. Context analysis of termination codons in mRNA that are recognized by plant NMD. Plant Cell Physiol. 2007;48:1072–1078. doi: 10.1093/pcp/pcm075. [DOI] [PubMed] [Google Scholar]
- 28.Iida K., Go M. Survey of conserved alternative splicing events of mRNAs encoding SR proteins in land plants. Mol. Biol. Evol. 2006;23:1085–1094. doi: 10.1093/molbev/msj118. [DOI] [PubMed] [Google Scholar]
- 29.Lewis B. P., Green R. E., Brenner S. E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birzele F., Csaba G., Zimmer R. Alternative splicing and protein structure evolution. Nucleic Acids Res. 2008;36:550–558. doi: 10.1093/nar/gkm1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koonin E. V., Mushegian A. R., Tatusov R. L., Altschul S. F., Bryant S. H., Bork P., Valencia A. Eukaryotic translation elongation factor 1 gamma contains a glutathione transferase domain–study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci. 1994;3:2045–2054. doi: 10.1002/pro.5560031117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S. H., Jin J. B., Song J., Min M. K., Park D. S., Kim Y. W., Hwang I. The intermolecular interaction between the PH domain and the C-terminal domain of Arabidopsis dynamin-like 6 determines lipid binding specificity. J. Biol. Chem. 2002;277:31842–31849. doi: 10.1074/jbc.M204770200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.