Abstract
Granins regulate secretory vesicle formation in neuroendocrine cells, and granin-derived peptides are co-released with neurotransmitters as modulatory signals at sympathetic sites. We report evidence for association between a regulatory polymorphism in Secretogranin II (SCG2) and hypertension in African-American subjects. The minor allele is ancestral in the human lineage and is associated with disease risk in two case-control studies and with elevated blood pressure in a separate familial study. Mechanistically, the ancestral allele acts as a transcriptional enhancer in cells that express endogenous Scg2, while the derived allele does not. ARIX (PHOX2A) and PHOX2B are identified as potential transactivating factors by oligonucleotide affinity chromatography and mass spectrometry and confirmed by chromatin immunoprecipitation. Each of these transcription factors preferentially binds the risk allele, both in vitro and in vivo. Population genetic considerations suggest positive selection of the protective allele within the human lineage. These results identify a common regulatory variation in SCG2 and implicate granin gene expression in the control of human blood pressure and susceptibility to hypertension.
Introduction
Chromogranin/secretogranin family proteins play a major role in the biology of catecholaminergic systems, including autonomic neurons and their physiologic targets (1). Chromogranin A (CHGA), chromogranin B (CHGB) and secretogranin II (SCG2) evolved from a common ancestral gene by duplication (2), suggesting closely related cellular and physiological functions. Heterologous expression of any one of the three “granins” is sufficient to induce secretory granules and regulated secretion in cells that otherwise lack both (3, 4) and depletion of endogenous CHGA impairs formation of granules in PC12 cells. Within secretory granules, granins are co-stored with catecholamine neurotransmitters and are co-released upon stimulation of regulated secretion. Proteolytic cleavage of granins at dibasic sites generates an array of bioactive peptides within secretory vesicles. In aggregate, these peptides have physiological effects on cardiovascular, endocrine, metabolic and immune systems through both direct and indirect modulation of the sympathetic nervous system (1).
Several lines of evidence suggest that granins play a role in the pathogenesis of hypertension through autonomic control of blood pressure. The secretoneurin peptide derived from SCG2 (human SCG2165-187) stimulates migration and proliferation of vascular smooth muscle cells (5) and acts as an endothelial cytokine to promote angiogenesis and vasculogenesis (6, 7), providing a direct link to vascular development and remodeling. The best studied granin, CHGA, is overexpressed in rodent models of both genetic and acquired hypertension (8-10) and plasma catestatin levels are reduced in human subjects with established hypertension and in at-risk siblings compared to controls (11-13). Functional polymorphisms in human CHGA affect gene expression and peptide potency and are under investigation for association with hypertension (14, 15).
Paired-like homeobox transcription factors PHOX2A (ARIX) and PHOX2B play essential roles in the autonomic nervous system. These paralogous genes are expressed in a largely overlapping pattern during development (16) but genetic studies point to unique requirements for each factor in vivo. Gene targeting of Phox2a in mice results in profound defects in sensory and autonomic ganglia, with complete loss of locus coeruleus (17). Subsequently, both exon skipping and missense mutations of PHOX2A have been found in human patients with congenital fibrosis of extraocular muscles (18). Ablation of Phox2b in mice blocks proper development of all autonomic and cranial sensory ganglia (19). Mutations in human PHOX2B have been found in patients with congenital central hypoventilation syndrome, Hirschsprung disease, and neuroblastoma (20-22). Phox2-responsive target genes may therefore be attractive candidate genes for less acute autonomic disorders, including some forms of hypertension.
Here we use comprehensive resequencing to identify a single nucleotide variant in the intron of SCG2 that shows association with hypertension in two African-American case-control studies and with blood pressure in a family study. Functional analysis of this polymorphism shows that the risk-conferring allele acts as a transcriptional enhancer while the protective allele, which is derived in the human lineage, does not. Gel shift, mass spectrometry and chromatin immunoprecipitation experiments show that the risk allele is preferentially bound by Phox2 transcription factors in nuclear extracts and in a cell culture model, establishing a molecular basis for functional differentiation of this allele.
Results
A common SCG2 SNP is associated with hypertension
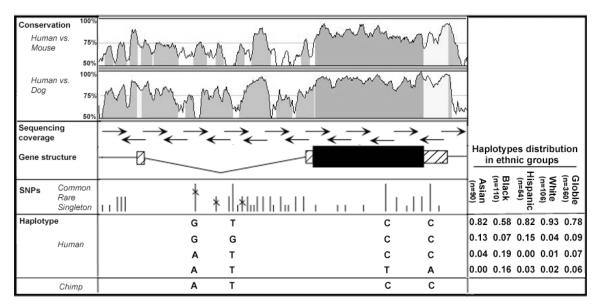
To identify SCG2 sequence variants that might alter gene function or activity, we resequenced the entire gene (∼6.5 kb) from 180 ethnically diverse human subjects (Figure 1). Among 35 SNPs and one mononucleotide-repeat polymorphism, only four variants have minor allele frequencies >5% among all subjects (Figure 1 and Supplemental Table S1). The most common SNP overall, intronic SNP_736, is only common among African American subjects. Nucleotide diversity across SCG2 in the general population (π=0.000201) (Supplemental Table S2) is several fold lower than genome-wide averages (π=0.0005-0.0007) reported by others (23, 24) and several loci we have examined in the same subjects (π=0.0008, 0.00075, 0.001 and 0.00095 for CHGA, DBH, PYY and NPY2R, respectively). SCG2 haplotypes inferred from the four most common SNPs show higher diversity among African American subjects, where all four haplotypes have frequencies ≥ 5%, while only one or two haplotypes appeared common in other populations (Figure 1). To determine the likely ancestral alleles at polymorphic sites we also resequenced SCG2 from two chimpanzees and specific segments from other nonhuman primates. Surprisingly, given the limited sequence diversity, three SNPs have minor alleles that appear to be ancestral based on comparison to nonhuman primates. All three of these derived SNPs are located in a region of intron 1 that is well conserved among available mammalian sequences (Figure 1 and 2B,C).
Figure 1. SCG2 sequence variation.
Top, sequence conservation is plotted for 100 bp sliding windows, visualized by VISTA. Arrows indicates initial resequencing reads for variant detection in clinical samples. Boxes indicate exons, coding region in black. Identified sequence variations are indicated by vertical bars. Variant sites whose minor alleles are common (>0.05), rare (<0.05, but seen more than once) or singleton in our data are indicated by bars of different height. X indicates SNPs for which the minor allele appears to be ancestral by comparison with other primates. Computationally constructed haplotypes from the four SNPs with minor allele frequency ≥0.05 (positions 736, 1253, 4366 and 5024 relative to the reported cap site) are indicated along with their distribution frequencies in four populations.
Figure 2. SCG2 intron haplotype network and sequences around SCG2 variant sites. show conservation of the minor allele in non-human primates.
(A) Each haplotype is represented by a circle whose area represents relative haplotype frequency. Branches in the minimum spanning tree represent one nucleotide substitution (except for the branch to the inferred last common ancestor with chimpanzees, LCA). Dashed line indicates alternative topologies of equal length. Each number represents a haplotype that is corresponding to haplotypes in Supplemental Table S5. (B, C) Alignments of orthologous sequences around SNP_736 (B) and SNP_964 (C). Asterisks mark the human polymorphism sites. Primate sequences were obtained by PCR-directed sequencing; rat and mouse sequences were obtained from UCSC public database. Sequences were aligned in ClustalW.
To detect associations between SCG2 variants and physiological outcomes we focused on African American subjects, where higher minor allele frequencies provide greater statistical power. We resequenced SCG2 from 329 additional African American subjects, for a total of 383, including 166 hypertensive and 217 normotensive individuals. Age-adjusted logistic regression analysis showed association of the most common polymorphism, SNP_736, with hypertension in these subjects (P=0.0049; Table 1), with the ancestral allele (A) predicting increased risk for hypertension. The P value remains significant after correction for multiple tests (P=0.013). Other SNPs in linkage disequilibrium with SNP_736 show weaker associations. Haplotype sliding window analysis (Supplemental Figure S1) indicated that the most significant association with hypertension is SNP_736 alone (1-1 SNP haplotype) or a polymorphism in complete LD but outside the sequenced interval. Longer haplotypes containing SNP_736 showed less significant associations. SNP_964 showed significant association with plasma secretoneurin levels in heterozygous subjects (no minor allele homozygotes were found), but not with hypertension.
Table 1.
Significant association of SCG2 polymorphisms with hypertension and plasma secretoneurin in African Americans
| SCG2 SNP (by location) |
Genotype | Cases | Controls | Multivariable OR (95% CI) |
P value | Secretoneurin Mean±SD (nM) |
P value |
|---|---|---|---|---|---|---|---|
| 736 | AA | 0.13 | 0.05 | 5.27 (1.88, 14.78) | 0.14±0.04 | ||
| GA | 0.40 | 0.35 | 1.51 (0.88, 2.59) | 0.14±0.06 | |||
| GG | 0.47 | 0.61 | 1.00 | 0.0049 | 0.14±0.06 | 0.9699 | |
| 964 | TT | 0 | 0 | nc | nc | ||
| CT | 0.09 | 0.07 | 1.55 (0.57, 4.21) | 0.17±0.07 | |||
| CC | 0.91 | 0.93 | 1.00 | 0.3855 | 0.14±0.06 | 0.0070 | |
nc, not calculable; OR, odds ratio. Results are age-adjusted.
To test whether admixture in the African American population could account for this association, we applied the method of Pritchard et al. (25) to genotypes at 33 SNP loci across the genome in the same sample populations (Supplemental Table S3). The results show no evidence for population stratification differentiating hypertensive and normotensive subjects in this data set (χ2 = 27.83, P = 0.7233).
Replication studies and meta-analysis support association
To ask whether, and under what conditions, this association could be replicated, we genotyped SCG2_736 in four additional subject populations, including different study designs and subject populations. Significant association was observed in a second African American cohort of unrelated subjects (n=465) from metropolitan San Diego for both recessive (P=0.022) and dominant (P=0.0098) models (Table 2). Taking the two case-control studies as independent observations supports P∼10-4 for either a dominant or recessive model.
Table 2.
Association studies in case-control subjects and population extremes
| Ascertainment (subjects typed) |
UCSD (n=386) |
P value | VA+Scripps (n=465) |
P value | Loyola (n=370) |
P value | Kaiser* (n=1315) |
P value |
|---|---|---|---|---|---|---|---|---|
| Study design | Case-control | Case-control | Population extremes |
Population extremes |
||||
| Population | African- American |
African- American |
African | European- American |
||||
| Geographic origin |
San Diego | San Diego | Nigeria | San Diego | ||||
| OR, Recessive: AA (95% CI) |
5.27 (1.88,14.78) |
0.0049 | 1.23 (0.60,2.53) |
0.0221 | 0.75 (0.36,1.56) |
0.503 | NA | NA |
| OR, Dominant: AA+GA (95% CI) |
1.87 (1.12,3.11) |
0.0164 | 1.67 (1.13,2.45) |
0.0098 | 1.06 (0.70,1.62) |
0.782 | 1.37 (0.80,2.34) |
0.249 |
only dominant models were analyzed due to low allele frequency.
OR, odds ratio.
Blood pressure phenotypes show multifactorial inheritance and can be strongly influenced by environment. To test SCG2_736 association with blood pressure phenotypes in subjects that have substantially different environment, allele frequencies, or both, we examined Nigerian (n=370) and European American (n=1315) subjects representing extremes (distribution tails) for blood pressure measurements within their respective populations. Neither population shows significant association (Table 2), although the allele frequency among European Americans was too low to be highly informative and we observed no minor allele homozygotes among these 1315 subjects.
As an additional test of the influence of SCG2_736 on heritable blood pressure phenotypes in African Americans, we examined familial subjects from the GenNet network of the Family Blood Pressure Program (n=781) for transmission disequilibrium (Table 3). This analysis shows association of SNP_736 to systolic blood pressure (FBAT, P = 0.032 under an additive model) and pulse pressure (FBAT, P = 0.0074 additive model, P = 0.0076 dominant or recessive models), though not with hypertension diagnosis as a dichotomous trait. However, age was a significant covariate (FBAT, P = 0.013 additive, 0.017 dominant or recessive) in this level of analysis (but see below).
Table 3.
Association with blood pressure in GenNet familial subjects (FBAT P values)
| Phenotype | “A” recessive | “A” dominant | Additive |
|---|---|---|---|
| SBP | 0.18 | 0.066 | 0.032 |
| DBP | 0.154 | 0.246 | 0.10 |
| PP | 0.285 | 0.0076 | 0.0074 |
SBP, systolic blood pressure; DBP, diastolic blood pressure; PP, pulse pressure. Similar results were obtained for raw or medica tion-adjusted values of each parameter, or considering only unmedicated subjects. P values shown are for raw values, all subjects.
All three African American cohorts show significant associations of the SCG2_736 A allele to elevated blood pressure or medically treated hypertension. While failure to replicate in the population extremes could be due to environmental or epistatic factors, we considered whether the association was significant for all groups taken together. To guard against bias due to sampling or study design we performed a meta-analysis of all unrelated subjects from all five cohorts. This analysis showed significant associations in both additive and dominant models (P=0.017 and 0.028, respectively) that survived correction for covariates (Table 4).
Table 4.
Meta-analysis of SCG2 A736G association with hypertension in all 5 cohorts
| Subjects | Genotype | OR (95% CI) | P value |
|---|---|---|---|
| All unrelated individuals both races N=2787 |
AA | 0.92 (0.63, 1.34) | 0.0172 |
| GA | 1.31 (1.07, 1.60) | ||
| GG | 1 | ||
| AA+GA | 1.24 (1.02, 1.51) | 0.0277 | |
| GG | 1 | ||
| All unrelated individuals blacks only N=1473 |
AA | 0.90 (0.61, 1.33) | 0.0273 |
| GA | 1.30 (1.05, 1.62) | ||
| GG | 1 | ||
| AA±GA | 1.23 (1.00, 1.51) | 0.0529 | |
| GG | 1 | ||
Adjusted for age and study. OR, odds ratio.
Two potential gender effects were also noted in our analysis. First, allele frequencies differ by gender in the combined sample (Supplemental Table S4), with male subjects having higher frequency of the minor allele than females. However, this is only significant within the Nigerian cohort, which does not show association to blood pressure phenotypes. More intriguingly, analysis of GenNet subjects by SOLAR shows significant, but divergent, associations with blood pressure variables in female and male subjects (Supplemental Table S5). Females show association to systolic pressure that is attenuated by adjustment for age and males showing association to pulse pressure that is enhanced by adjustment for age. Analysis of the combined data from each of the other cohorts — using a logistic regression model to relate SCG2 genotype to hypertension while accounting for study type and age effects — revealed gender specific effects of SCG2 genotype (Supplemental Table S6), with females showing a more pronounced effect (OR 1.60, 95% CI=1.15 to 2.22, P=0.005 under a dominant model).
Arrangement of SCG2 polymorphisms suggests positive selection pressure
Although the SCG2 intron is highly conserved across species, three intronic SNPs in human SCG2 have the relatively unusual property that the minor allele is ancestral in comparisons with non-human primates, including the two variants associated with blood pressure and plasma secretoneurin level (Figure 2). To ask whether the distribution of variants in SCG2 provides significant evidence for recent selection, we applied specific tests of neutrality (Supplemental Table S7). Both Fay and Wu’s H test and Fu’s Fs test are particularly sensitive to derived variants at high frequency and show significant departure from neutrality in the direction of positive selection for derived SCG2 alleles. Positive selection can also be identified by negative values of Tajima’s D in a sliding window of dense genotype data (26, 27). Published data available in the UCSC genome browser (26) show negative Tajima’s D values around SCG2 in each of three independent populations (Supplemental Figure S2), falling below the bottom 5% (African and Asian ancestry) and 1% (European ancestry) of empirical Tajima’s D distribution across the genome. Importantly, SCG2 is the only annotated gene in this interval.
To visualize relationships among extant haplotypes, we constructed haplotype networks using chimpanzee as the outgroup. Haplotypes were inferred by PHASE from five intronic SNPs common among African American subjects (Supplemental Table S8). The minimum spanning network shows all human haplotypes are closely related and weighted in frequency toward the most recently derived haplotypes, further suggesting an effect of directional selection (Figure 2A). African-derived chromosomes account for the majority of haplotypes 3-6, which are closer to the root of extant human haplotypes compared with the major haplotypes 1-2 in the general population. Haplotypes 3-6 include the SNP_736 A allele associated with hypertension in our subjects.
To better understand evolutionary constraints on the interval containing SNP_736 and SNP_964, we performed phylogenetic analysis (Figure 2B, C). We partially resequenced SCG2 orthologs from seven additional primates and obtained rat and mouse Scg2 sequences from public databases. Multiple sequence alignments (ClustalW) place the two SNPs in a highly conserved region. Human major alleles (G in SNP_736 and T in SNP_964) disrupt sites that are absolutely conserved among the 8 primates and two rodents. Together, the association and selection studies suggest functional consequences of SCG2 intron polymorphisms.
SNP_736 variation alters enhancer activity and binding by nuclear extracts
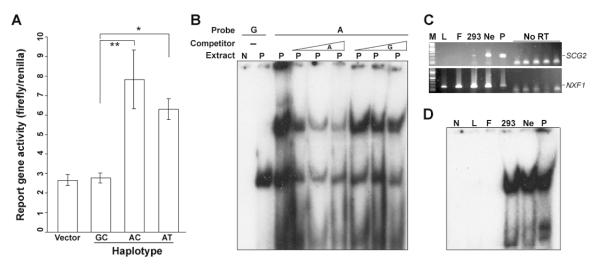
To test whether either SNP_736 or SNP_964 might affect transcriptional regulation of SCG2, we examined expression of reporter gene fusion constructs in PC12 chromaffin cells, which express endogenous rat Scg2. Only three haplotypes are observed spanning these two polymorphisms in human samples: GC, AC and AT. For each of these three haplotypes a 611 bp fragment (527 to 1137) was inserted into the upstream region of a basal SV40 promoter in the firefly luciferase reporter plasmid pGL3. Each construct was co-transfected with a control reporter (Renilla luciferase). Test plasmids with SNP_736 allele A (haplotypes AC and AT), showed significantly higher reporter activation than those with SNP_736 allele G or empty control vector (Figure 3A). SNP_736 allele G did not increase reporter gene activity relative to controls. SNP_964 alleles did not significantly influence expression.
Figure 3. SCG2 SNP_736 affects enhancer activity and nuclear extract factor binding.
(A) Luciferase reporter assays show allele-specific enhancer activity. PC12 cells were co-transfected with a reporter driven by basal SV40 promoter plus SCG2 intronic fragment from the indicated haplotype and a second reporter as a transfection control in three independent experiments. One-factor ANOVA with Bonferroni correction indicates significant differences for haplotype GC vs. AT (*, P= 0.038) and for haplotype GC vs. AC (**, P= 0.003). (B) EMSA shows allele-selective binding competition assays from PC12 cells. 5, 10, 20-fold concentrations of the indicated unlabeled competitor were present in the binding reaction. (C) RT-PCR detection of SCG2 (top panel) in human and rat cell lines. NXF1 (bottom) is shown as a positive control. (D) Electrophoretic mobility shift assay (EMSA) with a 19 bp probe centered on SNP_736 identifies a complex specific to SCG2-expressing cell types. M, size marker; N, no extract; L, EBV-transformed lymphoblasts; F, EBV-transformed fibroblasts; 293, Human embryonic kidney 293 cells; Ne, Human neuroblastoma cell line IMR-32; P, Rat pheochromacytoma cell line PC12.
To ask whether allelic variation at SNP_736 or SNP_964 affects protein-DNA interactions, we performed electrophoretic mobility shift assays (EMSAs) with allele-specific probes. Nuclear extracts from PC12 cells form stable complexes with either allele of SNP_736, but a slower-migrating complex is notably more abundant with the SNP_736 minor allele (A) probe than with the major allele (G). Unlabeled A allele also competes more effectively than G for probe binding (Figure 3B). Binding to SNP_964 probes did not show any allelic difference in either noncompetitive binding or heteroallelic competition assays (data not shown). To ask whether specific binding to SNP_736 A correlates with SCG2 gene expression, we assayed mobility shift activity in nuclear extracts from two additional cell lines that express SCG2 and two that do not (Figure 3C, D). Specific binding in the allele-dependent complex is only observed in the cell lines that express endogenous SCG2 mRNA.
Phox2A and Phox2B bind to SNP_736 A in nuclear extracts
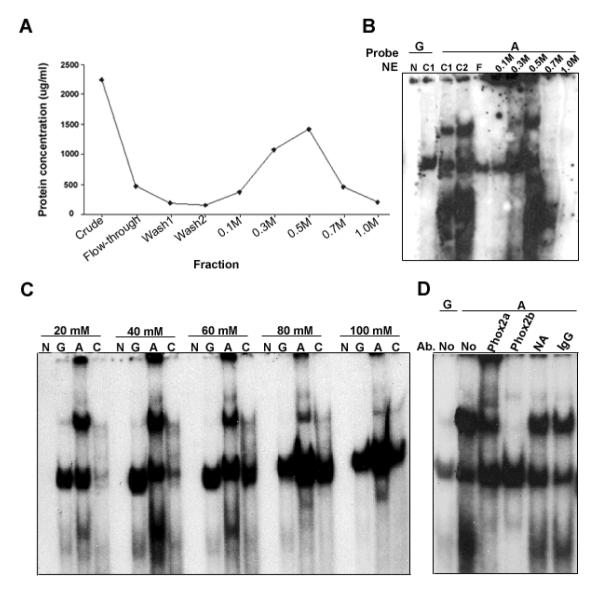
To identify nuclear factors that might mediate SNP_736 allele-dependent expression, we partially purified DNA-binding proteins by oligonucleotide affinity chromatography and identified enriched proteins by mass spectrometry. Nuclear extract from PC12 cells was first fractionated by heparin chromatography. A 0.5 M NaCl fraction contained most of the binding activity monitored by EMSA (Figure 4A, B). This fraction was incubated with magnetic beads containing allele-specific oligonucleotide pairs equivalent to the EMSA probes sequences, first for the major allele (G), then with the minor allele (A). Eluates from each set of beads were subjected to mass spectrometry for protein identification (Table 5). Abundant cellular proteins without known DNA binding activity (presumably non-specific contaminants) were not considered further. Several known transcription factors were identified from G allele eluates, but only the paired-like homeobox factors Phox2a (ARIX) and Phox2b were identified from A allele eluates. These closely related transcription factors are required for neuronal differentiation in developing sympathetic, parasympathetic and enteric ganglia (19, 28), and regulate expression of key enzymes in catecholamine synthesis, including tyrosine hydroxylase and dopamine-beta hydroxylase (28).
Figure 4. Properties of allele-specific binding activity and identification of Phox2a and Phox2b.
(A) Elution profile of nuclear extracts after heparin column chromatography. (B) Allele-selective SCG2 SNP_736 binding activity is enriched in 0.5M NaCl fraction. Equal protein (10μg) was loaded for each fraction. N, no extract; C1, Crude nuclear extract before dialysis; C2, Crude nuclear extract after dialysis; F, Flow-through fraction. (C) Allele-selective binding by PC12 nuclear extract is salt-sensitive. N, no extract; G, G allele probe; A, A allele probe; C, Control probe (TGTACCTAATCACATTTG). (D) EMSA activity is sensitive to factor-specific antibodies. Phox2a antibody produces a modest supershift fraction while pre-incubation with Phox2b antibody neutralizes allele-selective binding for SCG2 SNP_736. Each antibody result was replicated in at least two independent experiments. No, No antibody control; NA, Non-specific antibody (TFIIB).
Table 5.
Mass spectrometry of tryptic peptides from SNP_736-bound fractions
| Accession number |
Z-score | Peptide identifieda | Description |
|---|---|---|---|
| A allele eluate | |||
| IPI00195372 | 4.74 | K.MDSTEPPYSQK.R | Elongation factor 1-alpha 1 (Eef1a1) |
| 4.202 | K.IGGIGTVPVGR.V | ||
| IPI00200918 | 4.206 | K.EAAGEGPVLYEDPPDQK.T | Aapurinic/apyrimidinic endonuclease 1 (Apex1) |
| 2.704 | K.ICSWNVDGLR.A | ||
| 3.252 | K.GLDWVKEEAPDILCLQETK.C | ||
| IPI00205912 | 2.667 | R.AVVIVDDR.G | Non-POU domain-containing octamer-binding protein (Nono) |
| 6.009 | R.LFVGNLPPDITEEEMR.K | ||
| IPI00208193 | 5.656 | K.EVYQQQQYGSGGR.G | Nucleic acid binding factor pRM10 (Hnrpab) |
| 2.512 | K.FGEVVDCTIK.M | ||
| 5.143 | K.IFVGGLNPEATEEK.I | ||
| IPI00208271 | 6.082 | R.DHQPAPYSAVPYK.F | Paired mesoderm homeobox protein 2A (Phox2a) |
| 2.337 | R.TTFTSAQLK.E | ||
| 8.049 | R.VFAETHYPDIYTR.Eb | ||
| IPI00210090 | 1.419 | R.GYFEYIEENKYSR.A | SP120 (Hnrpu) |
| 7.625 | R.LQAALDNEAGGRPAMEPGNGSLDLGGDAAGR.S | ||
| 3.997 | K.SSGPTSLFAVTVAPPGAR.Q | ||
| IPI00627068 | 4.852 | R.FGQGGAGPVGGQGPR.G | NonO/p54nrb homolog (Sfpq) |
| 7.204 | K.YGEPGEVFINK.G | ||
| IPI00763263 | 4.839 | K.QQLSAEELDAQLDAYNAR.M | REF1-I (Thoc4) |
| 7.910 | R.SLGTADVHFER.K | ||
| 4.159 | K.QYNGVPLDGRPMNIQLVTSQIDTQR.R | ||
| G allele eluate | |||
| IPI00208193 | 5.500 | K.EVYQQQQYGSGGR.G | Nucleic acid binding factor pRM10 (Hnrpab) |
| 2.955 | K.FGEVVDCTIK.M | ||
| 4.535 | K.IFVGGLNPEATEEK.I | ||
| IPI00208271 | 1.275 | K.IDLTEAR.V | Paired mesoderm homeobox protein 2A (Phox2a) |
| 4.554 | R.TTFTSAQLK.E | ||
| 7.357 | R.VFAETHYPDIYTREELALK.Ib | ||
The N-terminal amino acid before “.” indicates the amino acid before the trypsin cleavage site, while the C-terminal amino acid after “.” indicates the amino acid after the trypsin cleavage site. Z-score for each peptide is listed (60). Keratin and trypsin were also found and not listed here.
Peptide shared by Phox2a and Phox2b.
Phox2 binding affinity can be sensitive to nucleotide substitutions adjacent to the ATTA/TAAT consensus binding site (29). SNP_736 lies directly adjacent to this site in the SCG2 intron and oligonucleotide probes (TCTTGGGTT[G/A]TAATTTGGC). To test whether the allelic difference in complex formation might be based on relative affinity rather than an absolute binding restriction, we examined the sensitivity of complex formation to salt concentration for each SCG2 SNP_736 allele and a previously reported Phox2 control probe (30). Consistent with an affinity-based difference, the SNP_736 G allele supports a complex of similar mobility to that formed on the A allele at 20 mM NaCl, but not at 60 mM, while all probes show diminished complex formation or stability at 100 mM NaCl in vitro (Figure 4C).
To confirm that these complexes include Phox2 proteins, we incubated nuclear extracts with antibodies against Phox2a or Phox2b prior to EMSA (Figure 4D). Phox2a antibody induces a modest supershift and reduces the ratio between the allele-dependent, slower migrating complex and the allele-independent, faster migrating complex (lane 3), while Phox2b antibody substantially inhibits the allele-dependent complex (lane 4). The specificity of each antibody was confirmed by western blot.
Phox2a and Phox2b show allele-dependent binding to SNP_736 in vivo
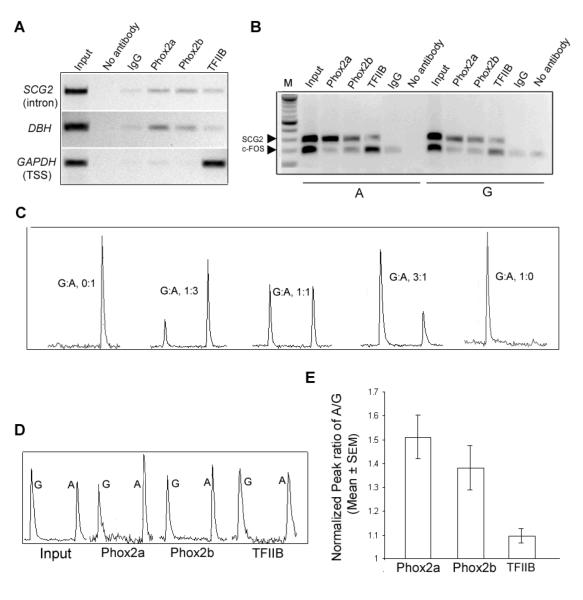
To test whether Phox2 proteins bind to the endogenous SCG2 SNP_736 intronic site, we performed chromatin immunoprecipitation (ChIP) in a human neuroblastoma cell line, IMR-32, which expresses SCG2. Both Phox2a and Phox2b antibodies enrich SCG2 intron DNA, compared with the DBH proximal promoter region as a positive control for Phox2 binding (29, 31) and the GAPD start site as a transcriptionally-active negative control (Figure 5A).
Figure 5. Allele-selective Phox2 binding in cells.
(A) Chromatin immunoprecipitation (ChIP) from IMR-32 human neuroblastoma cells. SCG2 intron site is selectively enriched by Phox2 antibodies compared with positive (DBH) and negative (GAPDH) controls. The general transcription factor TFIIB enriches each target site. (B) ChIP from PC12 cells transfected with human SCG2 reporter constructs carrying either SNP_736 allele (A or G). ChIP products were assayed by competitive PCR for transfected human SCG2 intron enhancer (top band) and endogenous rat c-fos promoter (bottom band) as an internal control. SCG2 was selectively enriched by both Phox2a and Phox2b antibodies and to a lesser extent by TFIIB. c-fos was consistently enriched only by TFIIB. (C) Pyrosequencing quantifies SNP_736 allele ratios. Peak height ratios on each pyrogram report G (left) and A (right) alleles in mixing experiments for the indicated proportions. (D) Representative pyrosequencing analysis of ChIP products and input chromatin from cells co-transfected with equal amounts of each SNP_736 allele. Both Phox2a and Phox2b antibodies selectively enrich the ancestral A allele. (E) Quantification of pyrosequencing peak height ratios of A/G in SCG2 intron SNP_736 enriched by ChIP with Phox2a or Phox2b antibodies, along with control antibody TFIIB after normalization to peak height ratio of A versus G in the ChIP input fraction, indicates allele-selective binding by endogenous Phox2 proteins in synthetically heterozygous cells.
To ask whether Phox2a and Phox2b bind the SCG2 intron sequence in an allele-dependent manner in vivo, we performed ChIP to enrich either allele from transfected PC12 cells. PC12 cells transfected with either allele alone were subjected to ChIP with Phox2a, Phox2b, TFIIB antibodies or a pre-immune IgG control. The ChIP-enriched DNA fraction was assayed by PCR for both the transfected human SCG2 intronic site and the endogenous rat c-fos promoter (32), as an internal control in the same reaction (Figure 5B). Both Phox2a and Phox2b antibodies significantly enrich human SCG2 intron containing either SNP_736 A or G allele, but not rat c-fos. As a positive control, antibody against the general transcription factor TFIIB enriches both the transfected SCG2 intron and the endogenous rat c-fos promoter.
To quantify the relative occupancy of SCG2 alleles by Phox2 factors, we compared allele ratios in ChIP-enriched fractions to total chromatin fractions from synthetically heterozygous cells by pyrosequencing (Figure 5C). Because we have been unable to identify human cell lines that both express endogenous SCG2 and are heterozygous with respect to SNP_736, we transfected PC12 cells with expression plasmids for both human SCG2 alleles as a model system. Comparison of Phox2a and Phox2b ChIP products to input in each experiment indicates a significant bias toward occupancy of the A allele (Figure 5D). After normalization to the non-enriched fraction in each experiment, ChIP-pyrosequencing indicates ∼50% higher occupancy of the A allele for either endogenous Phox2 protein in SCG2-expressing cells (Figure 5E).
Discussion
Sympathetic outflow is a key component of blood pressure regulation. We have examined association of sympathetic regulatory genes with human hypertension and intermediate phenotypes in the physiology of blood pressure control (10). Chromogranin/secretogranin family members play key roles in vesicular signaling at catecholaminergic synapses, including those at neuroeffector junctions. Physiological effects of granins are complex and quantitatively regulated at several steps, including feedback regulation through trans-synaptic peptide signaling. Thus, relatively subtle changes in genetic regulation of granin genes could play an important role in heritable sympathetic phenotypes.
Population genetic considerations, including differentiation away from ancestral alleles and a paucity of common variation in an ethnically diverse population, suggest that SCG2 may have been under selection favoring newly derived alleles in the human lineage. SCG2 is chromosomally isolated, with the nearest known genes 500 kb proximal and 100 kb distal, and is so far the only gene within the interval of negative Tajima’s D values. The most common minor allele (SNP_736A, 0.32 in African Americans, but <0.05 in other populations) is associated with blood pressure phenotypes in the direction of increased susceptibility to hypertension. This allele is ancestral in the human lineage and conserved in all other primates examined. Ancestral alleles that confer susceptibility to human common diseases are thought to reflect ancient adaptations, whereas protective derived alleles arise from recent adaptations to more contemporary environmental conditions (33, 34). Interestingly, half of the ancestral risk variant genes reviewed by Di Rienzo and Hudson (33) are associated with hypertension, suggesting blood pressure regulation mechanisms experienced significant alteration in human evolution. Similarly, Young et al. (34) reported evidence for selection of hypertension-resistant alleles during expansion of human populations out of Africa. Such adaptations in genes that interact with SCG2 in modulating sympathetic function, as well as environmental and cultural factors that impact blood pressure, may explain why we observe significant association with hypertension in African American, but not African (Nigerian) subjects.
Significant associations to a sequence polymorphism may indicate that a variant is functionally distinct or acts as a surrogate for one or more “true” functional variants in linkage disequilibrium. We have shown that the SCG2 variant best associated with hypertension alters a transcriptional enhancer through increased recruitment of Phox2 homeobox transcription factors in both biochemical and cellular assays. The variant site lies adjacent to the ATTA homeobox consensus motif. Previous work has shown that consensus-adjacent sites can affect homeobox protein binding (29) and that engineered optimization of binding affinity of natural Phox2 sites can influence transcriptional strength in vitro and in vivo (35). Our results show that a natural variation adjacent to a Phox2 binding site alters SCG2 gene expression, and imply by association that the magnitude of this difference is physiologically important in susceptibility to human hypertension.
Variation of SCG2 expression could influence susceptibility to hypertension through both direct and indirect actions on the cardiovascular system. The SCG2 fragment secretoneurin is a potent cytokine, with strong angiogenic and vasculogenic activities that make it an attractive candidate gene for genetic variation in sympathetic control of blood pressure (6, 7). Like insulin (36) and angiotensin II (37), secretoneurin also activates intracellular phosphatidylinositol 3-kinase/Akt pathways (7), which can regulate peripheral blood pressure by enhancing the production of nitric oxide (NO) (38, 39).
Different sites of SCG2 expression may have different effects on physiology. SCG2 is highly expressed in intestine, central nervous system, anterior pituitary, pancreas and adrenal gland, but can be differentially regulated at each site, including different levels of post-translational processing (40). Plasma secretoneurin, which shows association with SNP_964, is thought to mainly derive from SCG2 expression in intestine (40). By contrast, SCG2 association with blood pressure, which shows association with SNP_736, is likely to be more directly impacted by local/paracrine signaling by sympathetic neurons at neuroeffector junctions. Expression of PHOX2 genes predominantly overlaps that of SCG2 in the sympathetic system; thus variation in Phox2 binding at SNP_736 may have little effect on measured plasma levels, but a large impact on physiology through local control of secretoneurin or other SCG2-derived peptides.
Our findings provide the first evidence for an association between SCG2 alleles and hypertension, and link the risk-associated ancestral allele to quantitative regulation of SCG2 expression through a previously undescribed Phox2-responsive intronic enhancer. Phox2 also regulates other key catecholaminergic effectors such as tyrosine hydroxylase and dopamine beta-hydroxylase (41) and correspondingly, mutations in Phox2 factors produce severe phenotypes. By contrast, genetic variations that quantitatively alter expression of downstream physiologic regulators such as SCG2 may be an important mechanism for population and evolutionary adaptation to new physiological stressors and residual risk to late onset chronic disease.
Materials and Methods
Subjects and clinical characterization
Ethnically diverse volunteer subjects were drawn from urban southern California (San Diego). The protocol was approved by the University of California San Diego institutional review board. Subject characteristics are defined as in previous reports (15). Unrelated African Americans were from the San Diego area. One cohort (“UCSD”) was obtained from clinics within the UCSD and VA medical systems (n=383, average age ±SD = 44.8+/-11.9 years). The other cohort (“Kaiser/VA”) was obtained from combined African Americans samples from outpatient primary care clinics in the San Diego Kaiser Permanente system and the VA San Diego Healthcare System (n=527, 53.6±12.7). The diagnosis of hypertension was based on a previous diagnosis of hypertension by a physician and use of prescription antihypertensive medication, or having a systolic blood pressure (SBP)≥140 mm Hg or diastolic blood pressure (DBP)≥90 mm Hg measured on 2 occasions after they have been seated resting for ≥5 minutes. Ethnicity/race is self-assigned. The Kaiser white blood pressure extremes cohort consists of 660 male and 734 female white subjects (average age 58.1±13.4). These participants were ascertained for diastolic blood pressure in the upper and lower extremes among 25,599 males and 27,479 females in a database developed by Kaiser-Permanente of Southern California. The Nigerian blood pressure extremes cohort (“Loyola”) were ascertained from samples collected by R.S.C. in the International Collaborative Study on Hypertension in Blacks, as previously described (42). These subjects are the extreme quartiles for systolic blood pressure (SBP) adjusted by age, sex and body mass index. The SBP is 177.2±23.5 for the high blood pressure group (N=193) and 101.2±9.3 for low blood pressure group (N=190). Average age ±SD, 51.8±12.8. GenNet familial subjects comprise 781 African Americans from 274 pedigrees ascertained by R.S.C. on the basis of probands with hypertension, as described (43). Average age ±SD for unrelated subjects, 44.5±12.4
Molecular genetics
Genomic DNA was prepared from leukocytes in EDTA-anticoagulated blood, using PureGene extraction columns (Gentra Biosystems, Minnesota) as described (44). Promoter positions were numbered with respect to the mRNA cap (transcriptional initiation) site. PCR primers were selected with Primer3 (45). The nucleotide sequence was determined using standard reagents and a capillary sequencing instrument as described (15, 46). Genotypes for SCG2_736 SNP were scored by mass-tagged minisequencing of amplified DNA and MALDI-TOF mass spectrometry (Sequenom) and pyrosequencing (Pyrosequencing, Uppsala, Sweden). To ensure accurate assignment, genotypes were verified by visual inspection and suspect calls were excluded from further statistical analysis.
Statistical analysis
Haplotypes were estimated from un-phased genotypes by using the PHASE program (47). Haplotype homozygosities were confirmed by visual inspection. Neutrality tests, Tajima’s D, Fu and Li’s D and H test were performed as described (48-50). SCG2 intron haplotype networks were constructed in Arlequin (51), which computed a Minimum Spanning Tree (MST) from the matrix of pairwise distances calculated between all pairs of haplotypes. Association and admixture analysis was performed in the SAS software package (SAS software, version 9.0, SAS Institutes, Cary, NC). Differences in mean levels or proportions of continuous or categorical covariates among cases and controls were determined by analysis of variance or chi-square analysis, respectively. Logistic regression was used to evaluate associations between SCG2 SNPs and hypertension status. Standard linear regression analysis was used to test the significance of in vivo associations between SNPs and plasma SCG2 levels. Meta-analyses were performed by combining raw data from all studies and adjusting for study and age. Study and race were also highly correlated, therefore only study was used as a covariate in the meta-analyses. Unadjusted and age-adjusted associations were tested for all models. To correct for multiple testing, we used the Nyholt SnpSpD method to determine the “effective” number of independent SNPs (which was found to be ∼8 out of the 10 total SNPs that we studied) and applied a Bonferroni correction using this number (52).
Radioimmunoassays
Human plasma treated with EDTA as an anticoagulant was frozen and stored at -70°C prior to assay. 125I-radiolabeling of peptide was enabled by either an endogenous or adventitious (terminal) Tyr residue. The RIA was conducted as previously described (53), with an antibody directed against synthetic human secretoneurin (SCG2154-165).
Promoter haplotype/reporter activity assays
Human SCG2 intron/reporter plasmids were constructed essentially as described (54, 55). Haplotype-specific intronic fragments (611 bp) corresponding to SCG2 +527 to +1137 were amplified from genomic DNA of known homozygotes, cloned into the upstream site of an SV40 promoter / firefly luciferase reporter plasmid (pGL3-promoter, Promega, Madison, WI). Synthetic replacement was made by site-directed mutagenesis (QuickChange Stratagene) to produce the least common haplotype. The firefly and Renilla luciferase activities in the cell lysates were measured 48 hours after transfection, and the results were expressed as the ratio of firefly/Renilla luciferase activity (“Stop & Glo®”, Promega, Madison, WI). Each experiment was repeated a minimum of three times.
Electrophoretic mobility shift assays (EMSAs)
Nuclear extracts were prepared from EBV-transformed human lymphoblasts or fibroblasts (Coriell Cell Repositories), human embryonic kidney cell line 293 (ATCC), human neuroblastoma cell line IMR-32 (ATCC) and rat PC12 cells (ATCC) as described (56). Oligonucleotide pairs containing allelic variants for SNP_736 (TCTTGGGTT[G/A]TAATTTGGC and its complement) and SNP_964 (ATGCTCTAG[C/T]CTGAATTCA and its complement) were annealed and end-labeled by T4 polynucleotide kinase and gama-32P-ATP. Antibodies were added to 10 μg nuclear extract 10 min before adding 100 fmol labeled oligonucleotide. Binding reactions were for 20 minutes at room temperature. Binding buffer was 10 mM Tris-HCl pH 7.5, 40 mM NaCl, 10 mM KCl, 1 mM MgCl, 1 mM DTT, 1 mM EDTA and 2 μg poly(dI-dC) (Sigma), except as noted for salt-sensitivity assays. Reactions were separated on 6% PAGE and visualized by STORM phophorimager (Molecular Dynamics).
Oligonucleotide affinity chromatography
Nuclear extracts were prepared from PC12 cells grown in DME medium supplemented with fetal bovine and horse serum. Affinity purification was performed as described (57, 58) with modifications. 5′-Biotinylated versions of the oligonucleotide probes used for EMSA were captured with M280 streptavidin magnetic beads (Dynal Biotech, Oslo, Norway). All purification procedures were carried out at 4°C. Crude nuclear extracts (∼38 mg) were dialyzed and fractionated on heparin columns (Hitrap Heparin HP, Amersham Biosciences) equilibrated with Buffer D (20 mM HEPES pH 7.5, 10% Glycerol, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF) containing 75 mM NaCl. Bound proteins were eluted stepwise with 0.1, 0.3, 0.5, 0.7 and 1.0 M NaCl. The fraction with highest DNA-binding activity by EMSA (0.5 M NaCl, 7 mg protein) was dialyzed against 50 volumes of Buffer D containing 0.1 M NaCl and precleared three times with empty beads. Pre-cleared extract was then incubated with beads coated with G allele oligonucleotide twice before being incubated with beads coated with A allele oligonucleotide. Beads were washed six times with Buffer D Solution containing 100 mM NaCl. The first four washes also contained 0.1 mg/ml poly (dI:dC). The last two washes contained double-stranded and single-stranded salmon sperm DNA. The beads then were eluted in 50 μl of Buffer D containing 0.5 M NaCl. Eluates were subjected to mass spectrometry.
Mass spectrometry
0.5 μg of trypsin was added to the purified proteins for overnight digestion at 37°C. Tryptic peptides were analyzed by microcapillary reverse-phase HPLC micro electrospray tandem mass spectrometry ( LC-ESI-MS/MS) on a Finnigan LCQ quadrupole ion trap mass spectrometer. A 100
LC-ESI-MS/MS) on a Finnigan LCQ quadrupole ion trap mass spectrometer. A 100  m × 10 cm fused silica capillary column packed with the C18 resin (Michrom BioResources, CA) was used, and the flow rate was 250 nanoliter/min. MS/MS data were searched against the rat protein database with no restriction on the enzyme used for digestion using SEQUEST (59). Only tryptic peptides identified from the analysis were further considered to make sure that only significant fragment ions were accounted for.
m × 10 cm fused silica capillary column packed with the C18 resin (Michrom BioResources, CA) was used, and the flow rate was 250 nanoliter/min. MS/MS data were searched against the rat protein database with no restriction on the enzyme used for digestion using SEQUEST (59). Only tryptic peptides identified from the analysis were further considered to make sure that only significant fragment ions were accounted for.
Database search was performed using COMET and the rat protein database. No restriction on the enzyme used for proteolysis was applied. After completing the database search, only doubly tryptic peptides were further considered. We required at least two different tryptic peptides for each protein for that protein to be considered identified. Peptides used to identify proteins were manually inspected to verify correct identification. The sequence and Z score (60) of each of these peptides is given in the Table 5.
Chromatin immunoprecipitation assay (ChIP)
ChIP assays were performed as described (61) with slight modification. Cells (5-10 × 106) were crosslinked in 1% formaldehyde for 10 min and resuspended in RIPA lysis buffer. After sonication and centrifugation, samples were pre-cleared with salmon sperm DNA and protein A agarose slurry, then incubated with Phox2a, Phox2b or TF-IIB antibodies (Santa Cruz Biotechnology) at 4°C overnight. Immune complexes were captured with 40 μl of protein A and salmon sperm DNA slurry, washed extensively and eluted with 100 μl of elution buffer at room temperature. Eluates were pooled with 20 μl of 5 M NaCl and heated at 65°C overnight to reverse cross-links. DNA fragments were purified with QIAquick PCR purification kits (Qiagen). Human-specific SCG2 and rat-specific c-fos promoter primers were used for PCR amplification with 35 cycles. Amplicons did not plateau in yield.
Supplementary Material
Acknowledgements
This work was supported by program project grant P01 HL58120 from the National Heart, Lung and Blood Institute. Additional support was provided by The NHLBI Family Blood Pressure Program (FBPP); The NIA Longevity Consortium; the NIMH Consortium on the Genetics of Schizophrenia (COGS); and NIH grants HL074730, HL070137, and DK60702. W.Z. and H.Z. are supported by HG002604 from NHGRI and Ludwig Institute for Cancer Research.
Footnotes
Conflict of Interest Statement The authors declare no competing interests.
References
- 1.Taupenot L, Harper KL, O’Connor DT. The chromogranin-secretogranin family. N. Engl. J. Med. 2003;348:1134–1149. doi: 10.1056/NEJMra021405. [DOI] [PubMed] [Google Scholar]
- 2.Mahata SK, Kozak CA, Szpirer J, Szpirer C, Modi WS, Gerdes HH, Huttner WB, O’Connor DT. Dispersion of chromogranin/secretogranin secretory protein family loci in mammalian genomes. Genomics. 1996;33:135–139. doi: 10.1006/geno.1996.0171. [DOI] [PubMed] [Google Scholar]
- 3.Kim T, Tao-Cheng JH, Eiden LE, Loh YP. Chromogranin A, an “on/off” switch controlling dense-core secretory granule biogenesis. Cell. 2001;106:499–509. doi: 10.1016/s0092-8674(01)00459-7. [DOI] [PubMed] [Google Scholar]
- 4.Beuret N, Stettler H, Renold A, Rutishauser J, Spiess M. Expression of regulated secretory proteins is sufficient to generate granule-like structures in constitutively secreting cells. J. Biol. Chem. 2004;279:20242–20249. doi: 10.1074/jbc.M310613200. [DOI] [PubMed] [Google Scholar]
- 5.Wiedermann CJ. Secretoneurin: a functional neuropeptide in health and disease. Peptides. 2000;21:1289–1298. doi: 10.1016/s0196-9781(00)00271-0. [DOI] [PubMed] [Google Scholar]
- 6.Kirchmair R, Egger M, Walter DH, Eisterer W, Niederwanger A, Woell E, Nagl M, Pedrini M, Murayama T, Frauscher S, et al. Secretoneurin, an angiogenic neuropeptide, induces postnatal vasculogenesis. Circulation. 2004;110:1121–1127. doi: 10.1161/01.CIR.0000139884.81390.56. [DOI] [PubMed] [Google Scholar]
- 7.Kirchmair R, Gander R, Egger M, Hanley A, Silver M, Ritsch A, Murayama T, Kaneider N, Sturm W, Kearny M, et al. The neuropeptide secretoneurin acts as a direct angiogenic cytokine in vitro and in vivo. Circulation. 2004;109:777–783. doi: 10.1161/01.CIR.0000112574.07422.C1. [DOI] [PubMed] [Google Scholar]
- 8.Schober M, Howe PR, Sperk G, Fischer-Colbrie R, Winkler H. An increased pool of secretory hormones and peptides in adrenal medulla of stroke-prone spontaneously hypertensive rats. Hypertension. 1989;13:469–474. doi: 10.1161/01.hyp.13.5.469. [DOI] [PubMed] [Google Scholar]
- 9.Takiyyuddin MA, De Nicola L, Gabbai FB, Dinh TQ, Kennedy B, Ziegler MG, Sabban EL, Parmer RJ, O’Connor DT. Catecholamine secretory vesicles. Augmented chromogranins and amines in secondary hypertension. Hypertension. 1993;21:674–679. doi: 10.1161/01.hyp.21.5.674. [DOI] [PubMed] [Google Scholar]
- 10.O’Connor DT, Takiyyuddin MA, Printz MP, Dinh TQ, Barbosa JA, Rozansky DJ, Mahata SK, Wu H, Kennedy BP, Ziegler MG, et al. Catecholamine storage vesicle protein expression in genetic hypertension. Blood Press. 1999;8:285–295. doi: 10.1080/080370599439508. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy BP, Mahata SK, O’Connor DT, Ziegler MG. Mechanism of cardiovascular actions of the chromogranin A fragment catestatin in vivo. Peptides. 1998;19:1241–1248. doi: 10.1016/s0196-9781(98)00086-2. [DOI] [PubMed] [Google Scholar]
- 12.Mahata SK, Mahata M, Taylor C.V. Livsey, Taupenot L, Parmer RJ, O’Connor DT. The novel catecholamine release-inhibitory peptide catestatin (chromogranin A344-364). Properties and function. Adv. Exp. Med. Biol. 2000;482:263–277. doi: 10.1007/0-306-46837-9_21. [DOI] [PubMed] [Google Scholar]
- 13.O’Connor DT, Kailasam MT, Kennedy BP, Ziegler MG, Yanaihara N, Parmer RJ. Early decline in the catecholamine release-inhibitory peptide catestatin in humans at genetic risk of hypertension. J. Hypertens. 2002;20:1335–1345. doi: 10.1097/00004872-200207000-00020. [DOI] [PubMed] [Google Scholar]
- 14.Mahapatra NR, O’Connor DT, Vaingankar SM, Hikim AP, Mahata M, Ray S, Staite E, Wu H, Gu Y, Dalton N, et al. Hypertension from targeted ablation of chromogranin A can be rescued by the human ortholog. J. Clin. Invest. 2005;115:1942–1952. doi: 10.1172/JCI24354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen G, Mahata SK, Cadman P, Mahata M, Ghosh S, Mahapatra NR, Rao F, Stridsberg M, Smith DW, Mahboubi P, et al. Both rare and common polymorphisms contribute functional variation at CHGA, a regulator of catecholamine physiology. Am. J. Hum. Genet. 2004;74:197–207. doi: 10.1086/381399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development. 1997;124:4065–4075. doi: 10.1242/dev.124.20.4065. [DOI] [PubMed] [Google Scholar]
- 17.Morin X, Cremer H, Hirsch MR, Kapur RP, Goridis C, Brunet JF. Defects in sensory and autonomic ganglia and absence of locus coeruleus in mice deficient for the homeobox gene Phox2a. Neuron. 1997;18:411–423. doi: 10.1016/s0896-6273(00)81242-8. [DOI] [PubMed] [Google Scholar]
- 18.Nakano M, Yamada K, Fain J, Sener EC, Selleck CJ, Awad AH, Zwaan J, Mullaney PB, Bosley TM, Engle EC. Homozygous mutations in ARIX(PHOX2A) result in congenital fibrosis of the extraocular muscles type 2. Nat. Genet. 2001;29:315–320. doi: 10.1038/ng744. [DOI] [PubMed] [Google Scholar]
- 19.Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399:366–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- 20.Amiel J, Laudier B, Attie-Bitach T, Trang H, de Pontual L, Gener B, Trochet D, Etchevers H, Ray P, Simonneau M, et al. Polyalanine expansion and frameshift mutations of the paired-like homeobox gene PHOX2B in congenital central hypoventilation syndrome. Nat. Genet. 2003;33:459–461. doi: 10.1038/ng1130. [DOI] [PubMed] [Google Scholar]
- 21.Trochet D, Bourdeaut F, Janoueix-Lerosey I, Deville A, de Pontual L, Schleiermacher G, Coze C, Philip N, Frebourg T, Munnich A, et al. Germline mutations of the paired-like homeobox 2B (PHOX2B) gene in neuroblastoma. Am. J. Hum. Genet. 2004;74:761–764. doi: 10.1086/383253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trochet D, Hong SJ, Lim JK, Brunet JF, Munnich A, Kim KS, Lyonnet S, Goridis C, Amiel J. Molecular consequences of PHOX2B missense, frameshift and alanine expansion mutations leading to autonomic dysfunction. Hum. Mol. Genet. 2005;14:3697–3708. doi: 10.1093/hmg/ddi401. [DOI] [PubMed] [Google Scholar]
- 23.Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 24.Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, Messer CJ, Chew A, Han JH, et al. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001;293:489–493. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- 25.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am. J. Hum. Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlson CS, Thomas DJ, Eberle MA, Swanson JE, Livingston RJ, Rieder MJ, Nickerson DA. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 2005;15:1553–1565. doi: 10.1101/gr.4326505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 28.Lo L, Morin X, Brunet JF, Anderson DJ. Specification of neurotransmitter identity by Phox2 proteins in neural crest stem cells. Neuron. 1999;22:693–705. doi: 10.1016/s0896-6273(00)80729-1. [DOI] [PubMed] [Google Scholar]
- 29.Kim HS, Seo H, Yang C, Brunet JF, Kim KS. Noradrenergic-specific transcription of the dopamine beta-hydroxylase gene requires synergy of multiple cis-acting elements including at least two Phox2a-binding sites. J. Neurosci. 1998;18:8247–8260. doi: 10.1523/JNEUROSCI.18-20-08247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rychlik JL, Gerbasi V, Lewis EJ. The interaction between dHAND and Arix at the dopamine beta-hydroxylase promoter region is independent of direct dHAND binding to DNA. J. Biol. Chem. 2003;278:49652–49660. doi: 10.1074/jbc.M308577200. [DOI] [PubMed] [Google Scholar]
- 31.Yang C, Kim HS, Seo H, Kim CH, Brunet JF, Kim KS. Paired-like homeodomain proteins, Phox2a and Phox2b, are responsible for noradrenergic cell-specific transcription of the dopamine beta-hydroxylase gene. J Neurochem. 1998;71:1813–1826. doi: 10.1046/j.1471-4159.1998.71051813.x. [DOI] [PubMed] [Google Scholar]
- 32.Fass DM, Butler JE, Goodman RH. Deacetylase activity is required for cAMP activation of a subset of CREB target genes. J Biol Chem. 2003;278:43014–43019. doi: 10.1074/jbc.M305905200. [DOI] [PubMed] [Google Scholar]
- 33.Di Rienzo A, Hudson RR. An evolutionary framework for common diseases: the ancestral-susceptibility model. Trends Genet. 2005;21:596–601. doi: 10.1016/j.tig.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Young JH, Chang YP, Kim JD, Chretien JP, Klag MJ, Levine MA, Ruff CB, Wang NY, Chakravarti A. Differential susceptibility to hypertension is due to selection during the out-of-Africa expansion. PLoS Genet. 2005;1:e82. doi: 10.1371/journal.pgen.0010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang DY, Hwang MM, Kim HS, Kim KS. Genetically engineered dopamine beta-hydroxylase gene promoters with better PHOX2-binding sites drive significantly enhanced transgene expression in a noradrenergic cell-specific manner. Mol. Ther. 2005;11:132–141. doi: 10.1016/j.ymthe.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 36.Huang HN, Lu PJ, Lo WC, Lin CH, Hsiao M, Tseng CJ. In situ Akt phosphorylation in the nucleus tractus solitarii is involved in central control of blood pressure and heart rate. Circulation. 2004;110:2476–2483. doi: 10.1161/01.CIR.0000145116.75657.2D. [DOI] [PubMed] [Google Scholar]
- 37.Nagata D, Takeda R, Sata M, Satonaka H, Suzuki E, Nagano T, Hirata Y. AMP-activated protein kinase inhibits angiotensin II-stimulated vascular smooth muscle cell proliferation. Circulation. 2004;110:444–451. doi: 10.1161/01.CIR.0000136025.96811.76. [DOI] [PubMed] [Google Scholar]
- 38.Dimmeler S, Fleming I, Fisslthaler B, Hermann C, Busse R, Zeiher AM. Activation of nitric oxide synthase in endothelial cells by Akt-dependent phosphorylation. Nature. 1999;399:601–605. doi: 10.1038/21224. [DOI] [PubMed] [Google Scholar]
- 39.Fulton D, Gratton JP, McCabe TJ, Fontana J, Fujio Y, Walsh K, Franke TF, Papapetropoulos A, Sessa WC. Regulation of endothelium-derived nitric oxide production by the protein kinase Akt. Nature. 1999;399:597–601. doi: 10.1038/21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leitner B, Fischer-Colbrie R, Scherzer G, Winkler H. Secretogranin II: relative amounts and processing to secretoneurin in various rat tissues. J. Neurochem. 1996;66:1312–1317. doi: 10.1046/j.1471-4159.1996.66031312.x. [DOI] [PubMed] [Google Scholar]
- 41.Zellmer E, Zhang Z, Greco D, Rhodes J, Cassel S, Lewis EJ. A homeodomain protein selectively expressed in noradrenergic tissue regulates transcription of neurotransmitter biosynthetic genes. J. Neurosci. 1995;15:8109–8120. doi: 10.1523/JNEUROSCI.15-12-08109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooper R, Rotimi C, Ataman S, McGee D, Osotimehin B, Kadiri S, Muna W, Kingue S, Fraser H, Forrester T, et al. The prevalence of hypertension in seven populations of west African origin. Am. J. Public Health. 1997;87:160–168. doi: 10.2105/ajph.87.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.FBPP Investigators Multi-center genetic study of hypertension: The Family Blood Pressure Program (FBPP) Hypertension. 2002;39:3–9. doi: 10.1161/hy1201.100415. [DOI] [PubMed] [Google Scholar]
- 44.Herrmann V, Buscher R, Go MM, Ring KM, Hofer JK, Kailasam MT, O’Connor DT, Parmer RJ, Insel PA. Beta2-adrenergic receptor polymorphisms at codon 16, cardiovascular phenotypes and essential hypertension in whites and African Americans. Am. J. Hypertens. 2000;13:1021–1026. doi: 10.1016/s0895-7061(00)01188-2. [DOI] [PubMed] [Google Scholar]
- 45.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 46.Concepcion D, Seburn KL, Wen G, Frankel WN, Hamilton BA. Mutation rate and predicted phenotypic target sizes in ethylnitrosourea-treated mice. Genetics. 2004;168:953–959. doi: 10.1534/genetics.104.029843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider S, Roessli D, Excoffier L. Arlequin: A software for population genetics data analysis. 2.0 ed Genetics and Biometry Lab, Dept. of Anthropology, University of Geneva; Geneva: 2000. [Google Scholar]
- 52.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicol L, McNeilly JR, Stridsberg M, McNeilly AS. Differential secretion of gonadotrophins: investigation of the role of secretogranin II and chromogranin A in the release of LH and FSH in LbetaT2 cells. J. Mol. Endocrinol. 2004;32:467–480. doi: 10.1677/jme.0.0320467. [DOI] [PubMed] [Google Scholar]
- 54.Rozansky DJ, Wu H, Tang K, Parmer RJ, O’Connor DT. Glucocorticoid activation of chromogranin A gene expression. Identification and characterization of a novel glucocorticoid response element. J. Clin. Invest. 1994;94:2357–2368. doi: 10.1172/JCI117601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu H, Rozansky DJ, Webster NJ, O’Connor DT. Cell type-specific gene expression in the neuroendocrine system. A neuroendocrine-specific regulatory element in the promoter of chromogranin A, a ubiquitous secretory granule core protein. J. Clin. Invest. 1994;94:118–129. doi: 10.1172/JCI117297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 57.Masternak K, Barras E, Zufferey M, Conrad B, Corthals G, Aebersold R, Sanchez JC, Hochstrasser DF, Mach B, Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat. Genet. 1998;20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 58.Yaneva M, Tempst P. Affinity capture of specific DNA-binding proteins for mass spectrometric identification. Anal. Chem. 2003;75:6437–6448. doi: 10.1021/ac034698l. [DOI] [PubMed] [Google Scholar]
- 59.Yates JR, 3rd, Eng JK, McCormack AL. Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal. Chem. 1995;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]
- 60.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100024. 2005 0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spencer VA, Sun JM, Li L, Davie JR. Chromatin immunoprecipitation: a tool for studying histone acetylation and transcription factor binding. Methods. 2003;31:67–75. doi: 10.1016/s1046-2023(03)00089-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.