Abstract
Somatic mutation accumulation has been implicated as a major cause of cancer and aging. By using a transgenic mouse model with a chromosomally integrated lacZ reporter gene, mutational spectra were characterized at young and old age in two organs greatly differing in proliferative activity, i.e., the heart and small intestine. At young age the spectra were nearly identical, mainly consisting of G·C to A·T transitions and 1-bp deletions. At old age, however, distinct patterns of mutations had developed. In small intestine, only point mutations were found to accumulate, including G·C to T·A, G·C to C·G, and A·T to C·G transversions and G·C to A·T transitions. In contrast, in heart about half of the accumulated mutations appeared to be large genome rearrangements, involving up to 34 centimorgans of chromosomal DNA. Virtually all other mutations accumulating in the heart appeared to be G·C to A·T transitions at CpG sites. These results suggest that distinct mechanisms lead to organ-specific genome deterioration and dysfunction at old age.
Evidence that somatic mutations are causally related to the degenerative aspects of the aging process has been derived from human syndromes of accelerated aging, such as Werner syndrome. This disease is caused by a heritable mutation in the WRN gene (1), encoding both a helicase and an exonuclease (2) and thought to play a role in suppressing genomic instability (3). Indeed, cultured somatic cells from patients with Werner syndrome display an increased rate of somatic mutations and a variety of cytogenetic abnormalities, such as deletions and translocations (4). Also, other so-called progeroid syndromes, such as ataxia telangiectasia and Bloom syndrome, show increased genomic instability (5).
Recent evidence indicates that mouse models with inactivated genes involved in double-strand break repair show enhanced chromosomal instability (6) and prematurely exhibit symptoms of age-related degeneration in various tissues (7, 8). Telomere erosion in telomerase-deficient mice has been shown to result in an early initiation of genetic instability, accelerating the age-related loss of cell viability and increased tumor formation (9, 10). Hence, it appears that genetic defects promoting gross genomic instability are associated with symptoms of accelerated aging.
Aging affects both proliferative and postmitotic organs. Mutation accumulation in postmitotic cells is difficult to study because most methods require cycling cells. With the development of transgenic mouse models harboring bacterial reporter genes that can be retrieved from chromosomal DNA, the possibility has emerged to quantitate and characterize mutation accumulation in postmitotic tissues (11–13). Indeed, because reporter loci are not expressed in the mouse and are therefore neutral, they can be expected to reflect levels of genomic instability more faithfully than do endogenous expressed genes, which may suffer from a selection bias (14). By using a lacZ-plasmid transgenic reporter mouse model, we have previously reported an age-related increase in mutation frequency in the liver but not in the brain (15). Whereas in the liver a substantial fraction of the spontaneous mutations were large genome rearrangements, in the brain mainly point mutations were detected. These results suggested that postmitotic tissue is resistant to mutation accumulation in the nuclear genome and that cytogenetic alterations are limited to cells that are still mitotically capable.
In this paper, we present the first completely characterized mutational spectra, ranging from point mutations to large genomic rearrangements, in an actively proliferating and a postmitotic organ from young and old mice. The results indicate distinct differences between the heart and small intestine in both the type of mutations and their rate of accumulation. Surprisingly, genomic rearrangements were found to occur predominantly in the heart and to accumulate with age.
Materials and Methods
Plasmid Rescue and Mutant Frequency Determination.
Aging cohorts of male C57BL/6 pUR288-lacZ mice of line 60 were maintained in the animal facilities of the Beth Israel Deaconess Medical Center (Boston, MA) as described previously (15). The animals were killed by decapitation after asphyxiation. Heart and small intestine were removed, rinsed in PBS, placed in 1.5-ml microcentrifuge tubes, and frozen on dry ice. They were maintained at −80°C until used. DNA was extracted by routine phenol/chloroform extractions. Complete protocols for plasmid rescue and mutant frequency determinations with this model are given elsewhere (16). Briefly, between 10 and 20 μg of genomic DNA was digested with HindIII for 1 h in the presence of magnetic beads (Dynal) precoated with lacI-lacZ fusion protein. The beads were washed three times to remove the unbound mouse genomic DNA. Plasmids subsequently were eluted from the beads by isopropyl β-d-thiogalactoside. After circularization of the plasmids with T4 DNA ligase, they were ethanol-precipitated and used to electrotransform Escherichia coli C (ΔlacZ, galE −) cells. One-thousandth of the transformed cells was plated on the titer plate (with 5-bromo-4-chloro-3-indolyl β-d-galactoside) and the remainder on the selective plate (with phenyl β-d-galactoside). The plates were incubated for 15 h at 37°C. Mutant frequencies were determined as the number of colonies on the selective plates versus the number of colonies on the titer plate (times the dilution factor of 1,000). Each determination point is based on at least 300,000 recovered plasmids. The background mutant frequency of this system is about 1 × 10−5, as determined in E. coli, and consists mostly of false positive size-change mutants at HindIII star activity sites (17).
Mutant Characterization.
Mutant colonies were taken from the selective plates and grown overnight in 3 ml of LB medium. Then, 1 μl was directly plated on 5-bromo-4-chloro-3-indolyl β-d-galactoside to screen for galactose-insensitive host cells (18). The remainder of the cell culture was used for plasmid mini preparation (Wizard 9600; Promega). The purified plasmids were digested with PstI and AvaI and size-separated on 1% agarose gels. The mutant spectra were adjusted for HindIII star-activity mutants as described (17). For each animal, about 50 mutants were characterized. To check for possible cloning artifacts, each mouse sequence mutant routinely was redigested with HindIII. In all cases the plasmid linearized as one fragment only, indicating perfect ligation during the rescue procedure. Sequence reactions were performed with the CEQ dye terminator cycle sequencing kit (Beckman), according to the manufacturer's standard protocol, and analyzed with a CEQ 2000 DNA analysis system (Beckman). The primers used were the same as described earlier (18). To screen for size-change mutants with a breakpoint in the mouse genome, a PCR amplification was performed by using a forward primer specific for the last 18 bp of lacZ before the HindIII site (pUR3285-F, GGA TCC GTC GAC TCT AGA A) in combination with a reverse primer after the HindIII site (pUR3421-R, GCC ACC TGA CGT CTA AGA A). This procedure results in a 137-bp fragment only when both breakpoints are in the lacZ gene. As a positive control, a primer set specific for the ampicillin resistance gene (pUR4071-F, GTG ACA CCA CGA TGC CTG CA; and pUR4181-R, TAT CCG CCT CCA TCC AGT CT) was used to coamplify a 111-bp fragment in the same reaction.
Physical Mapping.
Primer sets specific for the cloned integration sites (InSiteA-F, GGT GAT GAC TAC ACC TGC ATC C; InSiteA-R, AAA GGA CCT CGT CAA GAG GG; InSiteB-F, GAG CCA GAG GCG TGG AAA; and InSiteB-R, CAG CTG CCT ATG AGG GTT CC) were used to screen a P1 library (Mouse ES-P1 library; Genome Systems, St. Louis). Four positive clones were returned, three for integration site A (clone addresses, plates 198, 217, and 266) and one for integration site B (clone address, plate 269). Primer pairs specific for the recovered mouse sequences were used to amplify the desired fragments by using the P1 clones as a template and total genomic mouse DNA as a positive control.
Genetic Mapping.
Primer sets specific for the integration sites and the recovered mouse sequences (primer sequences available on request) were used to find polymorphisms between mouse strains C57BL/6 and Mus spretus. The PCR products were sequenced with the forward and reverse primers that created the fragment. One or both of two backcross panels of 94 mice were screened for these polymorphisms. BSB panel 1 consisted of animals from the cross (C57BL/6 × M. spretus) × C57BL/6, and BSS panel 2 consisted of animals from the reciprocal backcross (C57BL/6 × M. spretus) × M. spretus (19). Mapping data are available online at www.jax.org/resources/documents/cmdata.
Results
Mutation Accumulation in Heart and Small Intestine.
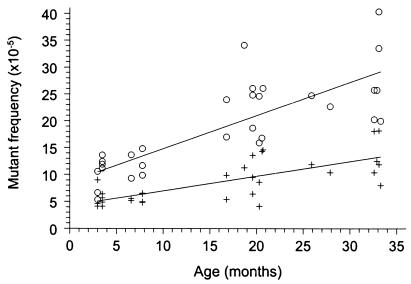
Spontaneous mutant frequencies determined in heart and small intestine of 31 mice, ranging in age from 3 to 33 months and homozygous for ±10 copies of the lacZ-plasmid at each integration site on chromosomes 3 and 4, were found to increase in both organs over time (Fig. 1). Linear regression analysis of the mutant frequencies per organ revealed an age-related increase of mutant frequencies of (0.27 ± 0.05) × 10−5 per month for heart (P = 0.0001) and (0.62 ± 0.09) × 10−5 per month for small intestine (P = 0.0001). The age-related increase in mutant frequency in small intestine was two times faster than that in heart, as examined by one-way ANOVA [F (1, 61) = 12.49; P = 0.0008]. A two-way ANOVA [F (2, 61) = 7.14; P = 0.0017] was used to compare the mean mutant frequencies per age group: 3–8 months (young), 16–21 months (middle), and 25–34 months (old). Significant age-related increases were found for heart (P = 0.0049) from young [(5.6 ± 1.3) × 10−5] to old [(12.7 ± 3.7) × 10−5] and for small intestine (P = 0.0001) from young [(11.0 ± 2.7) × 10−5] to middle [(22.8 ± 5.7) × 10−5] and from young to old [(25.6 ± 5.3) × 10−5). Furthermore, mean mutant frequencies in small intestine were significantly higher than those found in heart at all age groups (P ≤ 0.022).
Figure 1.
Spontaneous lacZ mutant frequencies in small intestine (○) and heart (+) of 3- to 33-month-old mice.
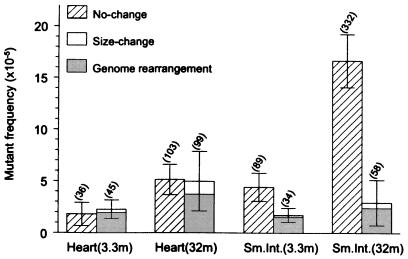
To investigate the nature of the mutational events in the aging heart, which may offer some insight into the molecular mechanisms involved in their formation, they were further analyzed at the molecular level. To obtain a first impression, a representative number of mutants from both old heart and old intestine were classified by restriction analysis. A total of 50 mutants of each organ per animal of 7 young (3 to 3.5 months) and 7 old (28 to 33 months) mice were taken from the selective plates and grown for plasmid preparation. After digestion with the restriction enzymes PstI and AvaI, fragments were separated on agarose gels. Mutant plasmids with similar migration patterns as wild-type control plasmids were assumed to be point mutations and termed “no-change mutants,” whereas those deviating from the wild-type restriction pattern represent deletions or other rearrangement events; these mice were termed “size-change mutants.” The results obtained revealed a striking difference between the two organs. As shown in Fig. 2, in the heart both point mutations and deletion mutations increased in about equal fractions. By contrast, virtually all mutations accumulated with age in the small intestine were point mutations.
Figure 2.
Mean frequencies of no-change mutants and size-change mutants in mouse heart and small intestine of 3.3- and 32-month-old animals (n = 7). Error bars indicate standard deviations of the means. Significant age-related changes for no-change mutants were found in both heart (P = 0.0049) and small intestine (P = 0.0022). Age-related changes for size-change mutants were significant for heart (P = 0.0073), but not for small intestine (P = 0.61; Wilcoxon Rank-Sum test). The shaded area indicates the fraction of size-change mutants with one break point in the mouse genome (genome rearrangements). The mutation load per cell, determined on the basis of a 3,000-bp lacZ target locus and a 6 × 109-bp diploid genome, is given in parentheses above each bar.
In an attempt to quantify the number of mutations on a per-cell basis, the mutant frequencies observed were extrapolated from the lacZ target locus to the overall diploid genome. With 332 mutational events per cell, the highest mutational load was observed for point mutations in the old intestine; the old heart appeared to contain about 200 mutations, almost equally distributed over the two classes of mutations (Fig. 2).
Molecular Characterization of Point Mutations.
If different processes are involved in mutation accumulation in heart and small intestine during aging, the molecular nature of the mutations could provide some insight into the mechanisms involved in their formation. We first focused on the no-change mutants, which presumably represent base pair substitutions and small deletions or insertions. For each organ from 4 young and 4 old mice, ±20 no-change mutants were randomly selected and sequenced. The complete set of data with sequence information on all mutations is available as Table 2, which is published as Supplementary Material on the PNAS web site at www.pnas.org. Most mutations proved to be unique, but not all. For example, the G·C to A·T transition at position 1,827 (a CpG site) was found to occur six times, three in heart and three in small intestine, indicating a mutational hot spot. A second mutational hot spot, found at position 1,387, is also a G·C to A·T mutation at CpG; this mutation was found five times. In addition to base changes and 1-bp deletions and insertions, two small deletions of 13 and 17 bp were found in the no-change spectrum of heart. Both deletions showed small regions of homology between the two breakpoints (see Table 2 published as Supplementary Material on the PNAS web site at www.pnas.org).
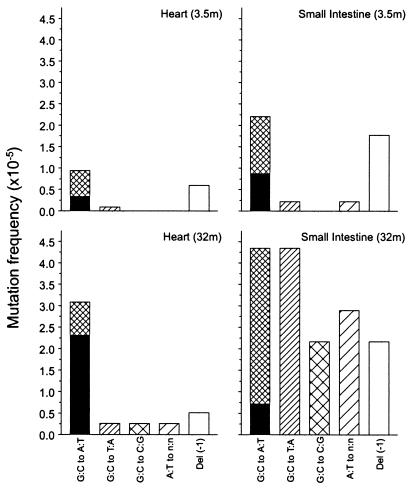
The point mutational spectra of the two organs at the two age levels are summarized in Fig. 3. At young age, the two spectra are not very different. Most mutations are either G·C to A·T transitions or 1-bp deletions. Mutations at A·T base pairs were rarely found except in small intestine at old age. At old age, the mutation spectra in heart and small intestine deviate significantly. Whereas in old heart only the frequency of G·C to A·T base pair substitutions at CpG sites increased substantially, in small intestine a more varied pattern of point mutations developed, with substantial increases in G·C to A·T transitions, G·C to T·A and G·C to C·G transversions, and some mutations at A·T base pairs. No increase was found for G·C to A·T transitions at CpG sites or 1-bp deletions.
Figure 3.
Point mutational spectra of heart and small intestine at young and old age. Bars indicate the frequency of each type of point mutation as indicated. The black areas in the G:C to A:T bars indicate the fraction of such mutations that had occurred at CpG sites.
Molecular Characterization of Size-Change Mutants.
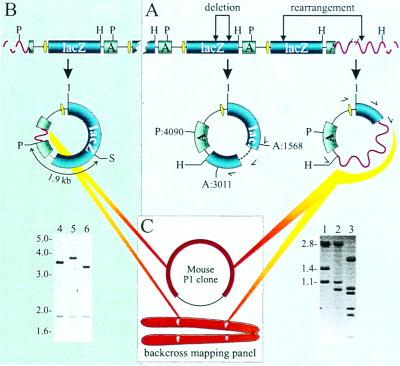
To investigate the nature of the size-change mutants in the old heart, their breakpoints were identified by sequence analysis. Breakpoints of internal deletions usually were identified by using one of the forward primers in the 5′ region of the lacZ gene (Fig. 4A). From a total of eight internal deletions in mutant lacZ plasmids obtained from old heart, none were associated with direct repeats at their breakpoints (Table 1). The small regions of homology that were present indicate that these mutations may have found their origin as illegitimate recombinations, possibly as a result of misrepair or misannealing during nonhomologous end-joining of double-strand breaks.
Figure 4.
(A) Breakpoint identification of size-change mutants. Internal lacZ deletion events (lane 2, no. 305) or rearrangements across the mouse flanking sequence (lane 3, no. 205) were recognized by their PstI (P) and AvaI (A) restriction patterns deviating from the wild-type pattern (lane 1, the 5,346-bp plasmid results in 2.8-, 1.4-, and 1.1-kb restriction fragment lengths). The multiple fragments seen in lane 3 are caused by PstI and/or AvaI restriction sites in the recovered mouse sequence. H indicates the unique HindIII restriction site. The single-sided curved arrows indicate the direction and the location of the primers used to sequence the breakpoints. (B) Cloning of the integration sites of pUR288 concatemers in C57BL/6 transgenic mice. PstI digestion yields outer plasmid copies with a disrupted ampicillin resistance gene, containing the flanking genomic mouse sequence until the first PstI site (see Molecular Characterization of Size-Change Mutants for details). Lanes 4 and 5 show the plasmid digests with the upstream flanking sequence of integration site A (on chromosome 3) and B (on chromosome 4), respectively. Lane 6 shows the wild-type pattern (3.4- and 1.9-kb fragments). S indicates the unique SacI restriction site. (C) Strategy for determining the size of mouse sequence mutations. Mouse sequences recovered in mutant plasmids first were tested for their presence on P1 clones containing the integration sites. When these results were negative, a backcross panel was used to map the location of the recovered mouse fragment genetically (see Molecular Characterization of Size-Change Mutants for details).
Table 1.
Sequenced size-change mutants from heart of old mice
| Mutant ID | Breakpoints (5′ → 3′) | Mutation size* | Location† |
|---|---|---|---|
| 207 | TGATGG/aaacca … ctgagc/GCCGGT | 125 bp | 3,124–3,248 |
| 294 | CGCTGG/aaggcg … ttgaag/TGGCGA | 184 bp | 2,649–2,832 |
| 305 | CGGCGG/agccga … aagcgg/TGAAGT | 528 bp | 1,688–2,215 |
| 180 | GCATCA/tcctct … gcgata/ACGAGC | 866 bp | 1,298–2,163 |
| 239 | GCCGTG/cgctgt … cgccgg/GCAACT | 914 bp | 1,380–2,293 |
| 283 | CTGGAG/gctgaa … gtcgaa/CGCTGG | 1,701 bp | 942–2,642 |
| 192 | GTATGT/tgtgtg … ctttca/CAGATG | 2,346 bp | 171–2,516 |
| 209 | CAGCAA/cgcggc … gccgaa/GCAGCG | 2,846 bp | 5,176–2,675 |
| 99 | GGCGTT/tcatct … nnnnnn/GATCAA | ND | 668-mouse |
| 158 | GGCGAT/gagcga … nnnnnn/ATGAGA | ND | 1,526-mouse |
| 167 | GGATTC/actggc … nnnnnn/TACTTG | ND | 236-mouse |
| 180 | AACGTC/gtgact … nnnnnn/CCATAC | ND | 259-mouse |
| 259 | AGTGGT/caaatg … nnnnnn/TACAGA | ND | 2,805-mouse |
| 273 | TACACG/ctgtgc … nnnnnn/AGGTCG | ND | 1,419-mouse |
| 216 | GGTCAT/ggatga … nnnnnn/ATTGGA | >P1 | 1,319-mouse |
| 233 | GTGGTA/cacgct … nnnnnn/ACTACT | >P1 | 1,415-mouse |
| 288 | TTTCCG/gcacca … nnnnnn/TGGGAT | >P1 | 411-mouse |
| 199‡ | TTCCGC/gaggtg … nnnnnn/CAAAAA | 31 cM | 1,218-chr3 |
| 205‡ | CTGGCC/gtcgtt … nnnnnn/ACCACA | 34 cM | 243-chr4 |
| 256‡ | AGCTGG/cacgac … nnnnnn/TCGCAT | Translocation | 59-chr10 |
| 271‡ | ATTATC/cgaacc … nnnnnn/TATCTG | Translocation | 1,396-chr11 |
Breakpoints 5′ and 3′ of the mutation are shown. The deleted nucleotides are shown in lowercase letters and the remaining nucleotides in capital letters. Underlined sequences indicate direct homology between breakpoints.
Mutation size given in: bp for mutations within the lacZ gene, “>P1” for mutations with breakpoints not present in P1 clones containing the integration sites, and cM or “translocation” for mutations with mapped breakpoints using the backcross panel. ND, not determined.
The nucleotide sequence is numbered according to SYNPUR288V (GenBank accession no. L09147). The numbers correspond to the first and the last deleted base respectively; “chr” stands for the chromosome number the breakpoint was mapped to; “mouse” stands for unlocated breakpoint in the mouse genome.
Approved Mouse Genome Database nomenclature locus names for mapped mouse sequences of mutants 199, 205, 256, and 271 are D3Vij1, D4Vij3, D10Vij1, and D11Vij1, respectively. Other assigned names are Tgn(LacZpl)60aVij, Tgn(LacZpl)60bVij, D4Vij1, and D4Vij2 for cloned upstream region of integration sites A and B and recovered mouse sequences of brain and liver mutants, respectively. Backcross mapping data for these loci are available at www.jax.org/resources/documents/cmdata.
Internal deletions in lacZ seemed to comprise only a small fraction of the total. Indeed, most size-change mutants appeared to involve the mouse flanking sequence. In our previous study on liver and brain, such mutational events were detected by hybridization analysis of the restriction enzyme patterns with radiolabeled mouse genomic DNA (15). As we recognized at the time, this result was an underestimate because single-copy mouse sequences cannot be detected in this way. Sequence analysis of a number of size-change mutants using the reversed primer outside the lacZ gene revealed mouse sequences at its 3′ end, immediately after the HindIII site (which was used to excise the plasmids from genomic DNA; Fig. 4A). To obtain quantitative information on the fraction of size-change mutants with one breakpoint outside the plasmid cluster, a simple PCR test was designed covering the last 18 bp of the lacZ gene before the HindIII excision site. The results indicate that in heart and small intestine at both young and old age as much as 75–89% of all size-change mutants had one breakpoint in the mouse genome with only a small fraction of internal lacZ deletions (Fig. 2). To further characterize size-change mutants having one breakpoint in the mouse genome, a combination of physical and genetic mapping was applied, using the cloned integration site as one anchor point and the mouse sequence in the mutants as the other.
The two integration sites on chromosomes 3 and 4 originally were determined by fluorescense in situ hybridization (results not shown). To clone the integration site, a strategy was applied (Fig. 4B) that involved the excision of plasmid copies from genomic DNA by PstI and their transfer, after magnetic bead purification, into E. coli without the use of ampicillin selection. Among the lacZ-positive (blue) colonies, 2 of 10 should harbor lacZ-plasmids from the ends of each cluster. The transgenic mouse originally had been made by microinjection of lacZ-plasmid linearized at the PstI site, which is in the middle of the ampicillin resistance gene. Hence, it was expected that the cluster-end copies would be truncated at the breakpoints. Their excision with PstI would then result in lacZ-plasmids of larger size, because of the inclusion of a mouse sequence up until the first PstI site in the flanking sequence (Fig. 4B). This prediction turned out to be correct. Among 69 blue colonies derived from the agar plate without ampicillin, five plasmids of increased length were found. On digestion with PstI and SacI, the five plasmids with cloned fragments could be subdivided into two groups that both had their 3.4-kb wild-type size band increased in size (Fig. 4B). This finding indicated that for both integration sites, only the upstream flanking sequence had been cloned. Sequence analysis confirmed that as predicted the ampicillin resistance genes at these two sites indeed were truncated (results not shown) during integration of the PstI linearized plasmids when this mouse model was originally constructed (12).
To map the rearrangement mutations in the mouse genome, a combined physical and genetic mapping strategy was applied (Fig. 4C). First, PCR primers were developed on the basis of the mouse sequence information of the two integration sites and used to screen a mouse P1 library. A total of four independent clones were obtained; three for integration site A and one for integration site B (results not shown). Next, PCR primers were designed for a unique part of the mouse sequences identified by sequencing size-change mutants with the forward primer outside the lacZ gene (Fig. 4A). We reasoned that if these mutants represented rearrangements with breakpoints within these P1 clones, they should positively identify them by yielding the same PCR fragment. PCR assays for a total of seven size-change mutants from the heart were designed and tested, but none yielded a product from the P1 clones. We then tested three size-change mutants from the brain and eight from the liver, which did yield the expected PCR fragment in two mutants, one from the brain and one from the liver. This finding indicated that these mutants had breakpoints within the P1 clone overlapping the integration site. Somewhat surprisingly, the far majority of the size-change mutants tested appeared to involve more than the about 80 kb of the P1 clones overlapping the integration sites, which would be in keeping with our observation that of a total of 50 of these size-change mutants from brain, liver, and heart, all sequences adjacent to the HindIII sites near the breakpoint in the mouse-flanking sequence were unique (results not shown). If a considerable number of mutants would involve relatively small events, i.e., thousands of base pairs, one would expect occasionally to find the same mouse sequence next to the HindIII site.
To further characterize the size-change mutants that could not be mapped within the P1 clones, we switched from physical to genetic mapping. By using a C57BL/6 × M. spretus backcross panel (19), the locations of both integration sites and breakpoints of the mutants could be determined relative to the positions of a large number of polymorphic marker loci typed in the panel. To confirm the validity of this approach, we first mapped the PCR fragment specific for the two integration sites. To do this, suitable polymorphisms were identified in these fragments. As expected, the mapping results indicated that integration site A is on chromosome 3 and integration site B is on chromosome 4 (results not shown), which confirmed the previously obtained fluorescense in situ hybridization mapping results. Then, polymorphic sites were identified for a set of four size-change mutants from the heart, one from the brain, and one from the liver. The liver mutant mapped to the same 1.1-centimorgan (cM) interval as integration site B, which may roughly correspond to an interval of 2 million base pairs. The brain mutant appeared to be far larger and could be localized 11 cM proximal to integration site B on chromosome 4. Of the heart mutants, one mapped to chromosome 3 (31 cM proximal to integration site A), one mapped to chromosome 4 (34 cM distal to integration site B), one mapped to chromosome 10, and one mapped to chromosome 11 (Table 1). The latter two indicate translocations rather than large deletions. Thus, this series of experiments has established that most size-change mutations in the old heart involving the mouse flanking sequence are very large, i.e., possibly up to 60–70 million base pairs.
Discussion
Increased mutation frequencies in organs and tissues of transgenic reporter mouse models have been demonstrated previously by us (15) and by others (20, 21). The results from Ono et al. (20) and Stuart et al. (21) in terms of the presence or absence of an age-related increase in mutation frequency are not different from ours, i.e., an increase in liver and heart and virtually no increase in brain. Where they differ, however, is in our finding of organ-specificity in the mutational spectra as they develop with age. Indeed, in the bacteriophage λ-based models used by both Ono et al. (20) and Stuart et al. (21), the majority of spontaneous mutations are at CpG sites, possibly reflecting deamination events. In addition, because of the nature of the bacteriophage λ vector, virtually no deletions, except very small ones, were detected. This difference might be explained by the large size of the reporter gene construct in the bacteriophage λ models, i.e., a total of about 2 million base pairs of prokaryotic DNA (40 copies of the ≈50-kb λ vector in both models). In the plasmid-based system, used in our present study, only about 50,000 bp of vector DNA is present (about 10 copies of 5 kb) per integration site. It is possible that large amounts of prokaryotic DNA influence the process of mutagenesis in mammalian cells in a way that obscures natural differences between organs in, for example, cellular activities or DNA repair capacity. Another possible explanation would be the difference in integration sites. However, although data from our own laboratory have indicated the presence of mutational hot spots in transgenic reporter mice (22), most integration sites seem to be identical in terms of both mutation frequencies and spectra. Indeed, the aforementioned bacteriophage λ models do not differ dramatically in this respect and with our own plasmid-based model we have observed identical mutation frequencies and spectra in a number of organs at three different loci (ref. 15 and unpublished results).
Previously, we reported age-related mutation accumulation in liver but not in brain of adult mice (15). At the time these findings were interpreted in terms of a possible relationship between organ-specific proliferative activity and mutation formation (23). Although our present findings are in keeping with a generally higher rate of mutation accumulation in highly proliferative tissue, the observation of an age-related increase in mutant frequency in the aging heart does not support the concept that postmitotic organs are resistant to mutation accumulation with age. Hence, mutation accumulation could not have only adverse effects in highly proliferative organs during aging, but also may underlie some of the functional decline associated with postmitotic organs. In this context, three questions remain to be addressed. First, what is the source of the process that drives the induction of mutations during the aging process in postmitotic tissue? Second, what are the mechanisms that give rise to their formation? And, third, what could be the impact of such mutations as a possible causal factor in the functional decline and increased disease incidence at old age?
Source of Mutation Accumulation with Age.
Oxidative stress has been implicated as a major causal factor in aging and disease (24, 25). Although an age-related increase in oxidative DNA lesions, such as 8-hydroxyguanine, has been reported, potential artifacts as the real source of the damage proved difficult to rule out (26, 27). One major problem is the large variety of DNA lesions that can be induced by oxygen free radicals. This essentially constrains the use of signature mutations as an indication of oxidative stress, as has been done successfully for some environmental agents and mutation spectra in tumors. Our own data on the old heart show mainly size-change mutations, mostly at the chromosomal level, and G·C to A·T transitions. Although this finding is far from conclusive, oxidative stress has been found associated with chromosomal aberrations and DNA deletion mutations (28, 29), whereas the C to T mutation is considered as a potential oxygen radical signature (30). However, other spontaneous sources of mutations, such as deamination of 5-methylcytosine, almost certainly contribute. Indeed, mismatch repair has been proposed as a possible mechanism to convert the mismatch resulting from the deamination into a stable mutation in a replication-independent manner (31).
Mechanism of Mutation Formation in the Aging Heart.
To explain how mutations can be formed and accumulate during aging in a postmitotic organ in the absence of DNA replication, it is necessary to invoke a process such as error-prone DNA repair. Because a major part of the mutation spectrum in the old heart involves large genome rearrangements, it is tempting to speculate that misrepair or misannealing of double-strand breaks or crosslinks by nonhomologous end-joining is responsible for many of the mutations observed. Indeed, mice deficient in double-strand break repair pathways display symptoms of accelerated aging (7, 8) as well as high levels of chromosome instability (6). And patients with Werner syndrome, a segmental progeroid syndrome defective in suppressing illegitimate recombinations (3, 32), show a high level of genome rearrangement type of mutations in their blood lymphocytes (4). Thus far, recombinational repair has been associated with meiosis or mitosis (33). However, there is evidence for mutation-generating mechanisms in nondividing cells (31) and there is no real reason why reannealing of double-strand breaks should be restricted to meiosis or mitosis. Hence, despite the fact that some of the mutations observed in the old heart may in fact find their origin in early cell division or may have occurred in the noncardiomyocytes in the heart, which are not postmitotic, it is conceivable that most of them are associated with a replication-independent error-prone repair mechanism.
Role of Mutation Accumulation in the Aging Process.
Aging is associated with cell and tissue degeneration at various levels, ultimately resulting in functional decline, disease, and death. Cardiac function is impaired in old humans and rodents, most notably peak aortic velocity and aortic acceleration (34). It is conceivable that mutation accumulation can contribute to this age-related functional decline by causing cells to die or to function less effectively. In this sense, an old heart represents a mosaic of cells at various stages of dysfunction on a trajectory to death. How could mutations occurring at low frequency contribute to cell dysfunctioning? Based on our estimate from the lacZ-plasmid reporter locus, an old heart may harbor an average of about 100 size-change mutations and about 100 point mutations per genome (Fig. 2). It is unlikely that 100 randomly induced point mutations will have a major effect on cell functioning. Indeed, cellular systems are robust and insensitive to many mutations. However, as many as 100 genome rearrangements, some of which involving millions of base pairs of DNA could seriously affect normal regulation, i.e., through gene dosage or position effects (35). In actively proliferating cell compartments, one of the highly visible effects of genome instability would be loss of proliferative homeostasis, i.e., hyperplasia, neoplasia, and tissue atrophy. Less visibly, in postmitotic cells a variety of functional pathways could be affected, leading to a mosaic of cells at different stages on a trajectory that would finally lead to cell death.
In 1963, Howard Curtis (36) noted the difficulty in proving or disproving the somatic mutation theory of aging and the need to develop a quantitative measure of mutations in somatic cells. In this sense he was especially referring to the need to monitor organs with cells that almost never divide, the argument being that such cells are incapable to select against mutants, permitting a buildup of dysfunctional cells. The in vivo model system, applied in this present study, allows testing this hypothesis directly by screening for a broad range of mutagenic events, ranging from point mutations to large genomic rearrangements. The picture that is now beginning to emerge is more complex than expected and reveals a pattern of mutation accumulation that is highly organ-specific. It predicts an equally high level of variation in aging rate, both among and within the different organ systems. The latter, especially, is still very difficult to analyze by current mutation analysis systems. Indeed, within each organ system both the susceptibility to mutations and their consequences could differ from cell to cell, a possibility—not addressed in our present study—that could further increase the observed organ-specific heterogeneity.
Supplementary Material
Acknowledgments
We thank Heidi Giese, Craig. L. Hopkins, and Albert Pastink for technical assistance and helpful discussion, and Shuko Lee, statistician at the Aging Research and Education Center, for her help in statistical evaluations. This research was supported by grants from the National Institutes of Health.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Yu C E, Oshima J, Fu Y H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 2.Shen J C, Gray M D, Oshima J, Kamath-Loeb A S, Fry M, Loeb L A. J Biol Chem. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 3.Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Proc Natl Acad Sci USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuchi K, Martin G M, Monnat R J., Jr Proc Natl Acad Sci USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turker M S, Martin G M. In: Principles of Geriatric Medicine and Gerontology. 4th Ed. Hazzard W R, Blass J P, Ettinger W H Jr, Halter J B, Ouslander J G, editors. New York: McGraw–Hill; 1999. pp. 21–44. [Google Scholar]
- 6.Difilippantonio M J, Zhu J, Chen H T, Meffre E, Nussenzweig M C, Max E E, Ried T, Nussenzweig A. Nature (London) 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weeda G, Donker I, de Wit J, Morreau H, Janssens R, Vissers C J, Nigg A, van Steeg H, Bootsma D, Hoeijmakers J H J. Curr Biol. 1997;7:427–439. doi: 10.1016/s0960-9822(06)00190-4. [DOI] [PubMed] [Google Scholar]
- 8.Vogel H, Lim D S, Karsenty G, Finegold M, Hasty P. Proc Natl Acad Sci USA. 1999;96:10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H W, Blasco M A, Gottlieb G J, Horner J W, II, Greider C W, DePinho R A. Nature (London) 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 10.Rudolph K L, Chang S, Lee H W, Blasco M, Gottlieb G J, Greider C W, DePinho R A. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- 11.Gossen J A, de Leeuw W J F, Tan C H T, Lohman P H M, Berends F, Knook D L, Zwarthoff E C, Vijg J. Proc Natl Acad Sci USA. 1989;86:7971–7975. doi: 10.1073/pnas.86.20.7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gossen J A, Martus H-J, Wei J Y, Vijg J. Mutat Res. 1995;331:89–97. doi: 10.1016/0027-5107(95)00055-n. [DOI] [PubMed] [Google Scholar]
- 13.Boerrigter M E T I, Dollé M E T, Martus H-J, Gossen J A, Vijg J. Nature (London) 1995;377:657–659. doi: 10.1038/377657a0. [DOI] [PubMed] [Google Scholar]
- 14.da Cruz A D, Curry J, Curado M P, Glickman B W. Environ Mol Mutagen. 1996;27:165–175. doi: 10.1002/(SICI)1098-2280(1996)27:3<165::AID-EM1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 15.Dollé M E T, Giese H, Hopkins C L, Martus H-J, Hausdorff J M, Vijg J. Nat Genet. 1997;17:431–434. doi: 10.1038/ng1297-431. [DOI] [PubMed] [Google Scholar]
- 16.Vijg J, Boerrigter M E T I, Dollé M E T. In: Methods in Aging Research: Section D. Yu B P, editor. Boca Raton, FL: CRC; 1999. pp. 621–635. [Google Scholar]
- 17.Dollé M E T, Snyder W K, van Orsouw N J, Vijg J. Environ Mol Mutagen. 1999;34:112–120. [PubMed] [Google Scholar]
- 18.Dollé M E T, Martus H-J, Novak M, van Orsouw N J, Vijg J. Mutagenesis. 1999;14:287–293. doi: 10.1093/mutage/14.3.287. [DOI] [PubMed] [Google Scholar]
- 19.Rowe L B, Nadeau J H, Turner R, Frankel W N, Letts V A, Eppig J T, Ko M S, Thurston S J, Birkenmeier E H. Mamm Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- 20.Ono T, Ikehata H, Nakamura S, Saito Y, Hosoi Y, Takai Y, Yamada S, Onodera J, Yamamoto K. Mutat Res. 2000;447:165–177. doi: 10.1016/s0027-5107(99)00200-6. [DOI] [PubMed] [Google Scholar]
- 21.Stuart G R, Oda Y, de Boer J G, Glickman B W. Genetics. 2000;154:1291–1300. doi: 10.1093/genetics/154.3.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gossen J A, de Leeuw W J, Verwest A, Lohman P H, Vijg J. Mutat Res. 1991;250:423–429. doi: 10.1016/0027-5107(91)90198-w. [DOI] [PubMed] [Google Scholar]
- 23.Warner H R, Johnson T E. Nat Genet. 1997;17:368–370. doi: 10.1038/ng1297-368. [DOI] [PubMed] [Google Scholar]
- 24.Martin G M, Austad S N, Johnson T E. Nat Genet. 1996;13:25–34. doi: 10.1038/ng0596-25. [DOI] [PubMed] [Google Scholar]
- 25.Beckman K B, Ames B N. J Biol Chem. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 26.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 27.Collins A R. BioEssays. 1999;21:238–246. doi: 10.1002/(SICI)1521-1878(199903)21:3<238::AID-BIES8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 28.Hsie A W, Recio L, Katz D S, Lee C Q, Wagner M, Schenley R L. Proc Natl Acad Sci USA. 1986;83:9616–9620. doi: 10.1073/pnas.83.24.9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gille J J P, van Berkel C G M, Joenje H. Carcinogenesis. 1994;15:2695–2699. doi: 10.1093/carcin/15.12.2695. [DOI] [PubMed] [Google Scholar]
- 30.Reid T M, Feig D I, Loeb L L. Environ Health Perspect. 1994;102, Suppl. 3:57–61. doi: 10.1289/ehp.94102s357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacPhee D G. Cancer Res. 1995;55:5489–5492. [PubMed] [Google Scholar]
- 32.Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lustig A J. Nat Genet. 1999;23:130–131. doi: 10.1038/13755. [DOI] [PubMed] [Google Scholar]
- 34.Taffet G E, Pham T T, Hartley C J. J Gerontol. 1997;52:B285–B290. doi: 10.1093/gerona/52a.6.b285. [DOI] [PubMed] [Google Scholar]
- 35.Rasnick D, Duesberg P H. Biochem J. 1999;340:621–630. [PMC free article] [PubMed] [Google Scholar]
- 36.Curtis H J. Science. 1963;141:686–694. doi: 10.1126/science.141.3582.686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.