Abstract
The spindle checkpoint blocks cell cycle progression until chromosomes are properly attached to the mitotic spindle. Popular models propose that checkpoint proteins associate with kinetochores to produce a “wait anaphase” signal that inhibits anaphase. Recent data suggests that a two-state switch results from using the same kinetochore proteins to bind microtubules and checkpoint proteins. At least eight protein kinases are implicated in spindle checkpoint signaling arguing that a traditional signal transduction cascade is integral to spindle checkpoint signaling.
The Spindle Checkpoint and the Kinetochore
The spindle checkpoint is an evolutionarily conserved mechanism that regulates genome stability from yeast to humans (Reviewed in: (Lew and Burke, 2003; Cleveland et al., 2003; Musacchio and Salmon, 2007)). A single chromosome, detached from the mitotic spindle can activate the spindle checkpoint and inhibit the onset of anaphase. The chromosomal domain responsible for mitotic inhibition via the checkpoint is the kinetochore and popular models suggest that the kinetochore is a platform that produces a diffusible “wait anaphase” signal that inhibits mitosis. Ultimately, the checkpoint inhibits Cdc20, a specificity factor for the Anaphase Promoting Complex (APC), an E3 ubiquitin ligase that regulates the metaphase to anaphase transition (Lew and Burke, 2003; Cleveland et al., 2003). Conserved checkpoint proteins, originally identified in yeast, consist of Bub3, Mad1-3 and two kinases Bub1 and Mps1. The Ipl1 protein kinase (Aurora B in higher cells) was later identified as a component of the checkpoint but may function in a more limited way (Biggins and Murray, 2001). The spindle checkpoint is more complex in higher cells. Mad3 is associated with a Bub1-related kinase domain on the C-terminus and was named BubR1 (Taylor et al., 1998). BubR1 kinase activity is stimulated by the CENP-E plus end directed kinesin when it is not attached to microtubules (Mao et al., 2003). In addition, a complex of Rough Deal, Zeste-White 10 and Zwilch, abbreviated as RZZ , identified in Drosophila and absent in yeast, functions in the checkpoint as described below (Basto et al., 2000; Chan et al., 2000). There are homologs of all three proteins in higher cells and their checkpoint functions are conserved. There are four protein kinases (p38 MAP kinase, Nek2A, Tao1 and Prp4) required for the checkpoint in higher cells that are not present in yeast (Minshull et al., 1994; Lou et al., 2004; Draviam et al., 2007, Montembault et al,,2007). Despite the obvious potential for protein kinases as mediators of an intracellular signal transduction pathway, the roles of the protein kinases in the spindle checkpoint have been deemphasized.
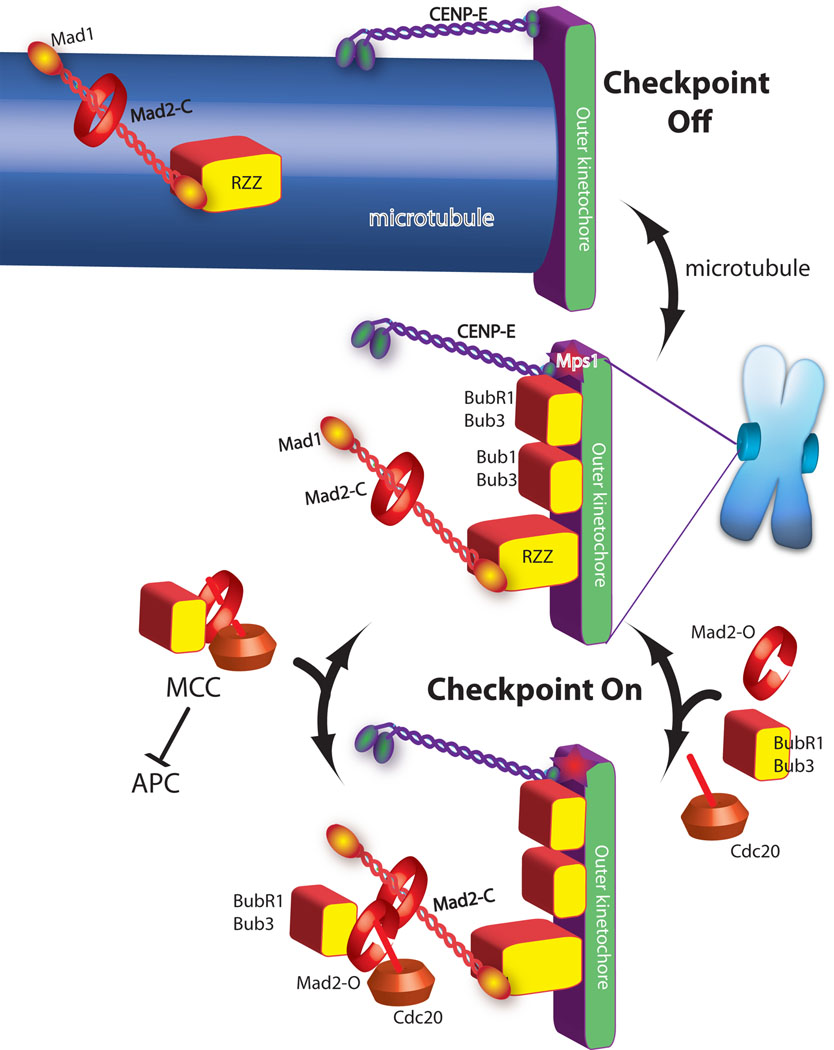
Models for the role of the kinetochore in the spindle checkpoint are derived from diverse experimental systems (yeast to human) and incorporate two important observations. The first is that checkpoint proteins (in systems where it can be measured) dynamically associate with unattached kinetochores (Shah et. al. 2004; Howell et al. 2004). The second derives from in vitro assays for APC regulation showing that a complex of checkpoint proteins called the mitotic checkpoint complex (MCC), consisting of Mad3/BubR1, Mad2, Bub3 and Cdc20, is a potent inhibitor of the ubiquitin ligase activity of the APC . A popular model (Figure 1) that is meant to be “universal” is that the dynamic association of checkpoint proteins with kinetochores of unattached chromosomes reflects the catalytic assembly and then release of MCC which then diffuses from the unoccupied kinetochore to inhibit the APC (Cleveland et al., 2003; Musacchio and Salmon, 2007). The key step is believed to be the formation of Mad2-Cdc20 complexes and an elegant model for how this is catalyzed by Mad1 has been proposed (Musacchio and Salmon, 2007). The current model for the role of the kinetochore in the spindle checkpoint is incomplete for several reasons. First, MCC formation does not require a kinetochore in yeast and perhaps human cells (Fraschini et al., 2001; Rancati et al., 2005). Second, the model does not explain why metazoans have increased the complexity of the signal by employing proteins such as the kinesin motor CENP-E and RZZ (Cleveland et al., 2003; Musacchio and Salmon, 2007). Third, the model does not describe how checkpoint proteins associate with the kinetochore. Finally, the model downplays the role that the protein kinases play in checkpoint signaling.
Figure 1. A current model of spindle checkpoint signaling highlights the importance of MCC formation as the step catalyzed by the kinetochore.
Green and purple are proteins required for microtubule binding. Checkpoint proteins (red and yellow shapes) are recruited to kinetochores that are not binding microtubules (central image). In the absence of microtubules the checkpoint proteins act to catalyze the assembly of the MCC checkpoint complex (bottom image), which diffuses from the kinetochore to inhibit the anaphase promoting complex (APC). Mad2 has both open and closed states and although it is a globular protein it is drawn as a ring to emphasize transitions between these two states. Mad2closed on kinetochores binds Mad2open, which in turn binds Cdc20, Bub1 and Bub3 to form MCC.
Microtubule attachment inhibits the signal by two mechanisms. The checkpoint proteins Mad1/Mad2 and RZZ are shown being “stripped” by dynein, which carries them away from the kinetochore by walking towards the minus end of the microtubule (top image). CENP-E activates BubR1 kinase activity unless it binds microtubules, which is also important for silencing the checkpoint.
Recent data has lead to a refined model where checkpoint proteins associate with the same proteins that bind microtubules producing a competition between signaling and microtubule binding. There is a “super complex” of proteins called “KMN” (a complex of KNL-1/AF15Q14/Spc105/Blinkin, Mis12 complex and Ndc80 complex) which is a critical microtubule-binding interface in the kinetochore required for microtubule attachments (Emanuele et al., 2007). KMN has at least two direct microtubule interacting proteins; the Ndc80/Hec1 subunit of the Ndc80 complex and KNL-1/Blinkin (Cheeseman et al., 2006). Each of these proteins has been implicated in checkpoint signaling and may directly associate with checkpoint proteins. These associations suggest that there is a relationship in higher cells, as there is in yeast, between microtubule attachment and checkpoint signaling. Moreover these microtubule attachment complexes are not strongly associated in solution but only come together at kinetochores (Emanuele et al., 2005). This assembly mechanism could assure that checkpoint signals are produced only at kinetochores.
Dual role of KMN as microtubule attachment site and spindle checkpoint signal generator
An important suggestion of a molecular link between spindle attachment and checkpoint activity in the kinetochore came from yeast mutants that were defective for the four proteins of the evolutionarily conserved Ndc80 kinetochore complex (Ndc80, Nuf2 Spc24, and Spc25). These cells are unable to attach chromosomes to the spindle and are also checkpoint defective (He et al., 2001; Janke et al., 2001; Wigge and Kilmartin, 2001; McCleland et al., 2003). Mutations that eliminate proteins from other kinetochore complexes Okp1 (COMA complex), Mtw1 (MIND complex), Duo1 (Dam1 complex) and Stu1 are checkpoint proficient (Gardner et al., 2001; Cheeseman et al., 2001; Kosco et al., 2001). The relationship between kinetochore-microtubule binding and checkpoint signaling is strengthened by the observation that Mps1, a protein kinase and checkpoint protein, is kinetochore-associated and implicated in regulating microtubule attachment (Jones et al., 2005). In addition, the C. elegans homolog of the yeast kinetochore protein Spc105 (KNL-1) binds microtubules in vitro and the phenotype of temperature sensitive spc105 mutants in yeast could be interpreted as lacking the spindle checkpoint which further supports a molecular link between kinetochore-microtubule attachments and checkpoint signaling (Nekrasov et al., 2003; Cheeseman et al., 2006).
In metazoans, all four members of the Ndc80 complex are required for congression of chromosomes to the metaphase plate and anaphase segregation of sister chromatids (Martin-Lluesma et al., 2002; DeLuca et al., 2002; McCleland et al., 2003; McCleland et al., 2004). The N-terminus of Ndc80 (Hec1 in human cells) has an unstructured tail followed by a globular head that binds microtubules and has limited homology to the microtubule plus end binding protein EB1 (Wei et al., 2007). Together these data suggest that the Ndc80 complex may be the key microtubule interface in the kinetochore. The Ndc80 complex is also required to generate a spindle checkpoint signal in higher cells as in yeast (Martin-Lluesma et al., 2002; McCleland et al., 2003; McCleland et al., 2004; Meraldi et al., 2004).
KMN may function in the checkpoint as a scaffold that brings checkpoint proteins together. A two-hybrid interaction between Ndc80 and Mad1 has been reported, and the Ndc80 complex is required for kinetochore assembly of Mad1, Mad2, Mps1, and RZZ (Martin-Lluesma et al., 2002; McCleland et al., 2003; McCleland et al., 2004; Stucke et al., 2004). This suggests that Ndc80 is a platform for Mad1 and perhaps Mps1 binding, although direct physical interactions between the proteins has not been shown. The role of Ndc80 in both kinetochore-microtubule attachment and checkpoint signaling suggests that recognition of microtubule binding by the checkpoint may be as simple as a mutually exclusive binding of the Ndc80 complex for microtubules and Mad1. However Mad1 association with kinetochores is not dynamic (Shah et. al. 2004; Howell et al. 2004), suggesting that this is unlikely to be a shared binding site. Rather models suggesting either a conformational change promoted by microtubule binding that displaces Mad1 or activation of attachment sensitive kinases are more consistent with these data.
A recent publication has documented interactions of other checkpoint proteins with a different member of KMN. The human homolog of KNL-1 (referred to as blinkin for Bub-linking) is the platform that directs Bub1 and BubR1 to kinetochores (Kiyomitsu et al., 2007). Bub1 and BubR1 have TPR domains that interact with the blinkin N-terminus. Most importantly, siRNA of blinkin abolishes the spindle checkpoint while maintaining kinetochore assembly. Point mutations that eliminate the interaction between the TPR domain of the Bubs and blinkin cannot localize Bub proteins to the kinetochore or generate checkpoint signals. Similarly loss of the N-and middle domains of blinkin prevents checkpoint signaling and mis-localize the Bub proteins. Given that the blinkin homolog in C. elegans, KNL-1, binds to microtubules, these data strongly implicate blinkin in both microtubule binding and checkpoint signaling through the Bub proteins.
The third member of KMN, Mis12, is required for checkpoint signaling (McAinsh, Meraldi et al., 2005). Moreover, Mis12 recruits a third checkpoint complex RZZ to kinetochores through an interaction with Zwint, a protein originally identified because of its interaction with Zw10 (Emanuele et al., 2005). RZZ regulates binding of the microtubule-based motor Dynein to kinetochores and is also required to generate the checkpoint signal. Bub1 and BubR1 properly localize in human cells after RZZ knockdown, but Mad1 and Mad2 do not (Chan et al., 2000; Wang et al., 2004; Kops et al., 2005). Chromosomes align and undergo anaphase movements suggesting that the KMN complexes are intact. These data suggest that KMN is not sufficient for Mad1 binding in higher cells and there is an additional requirement for RZZ. Overall, KMN is implicated as a central coordinator of microtubule attachment and checkpoint signaling in the kinetochore.
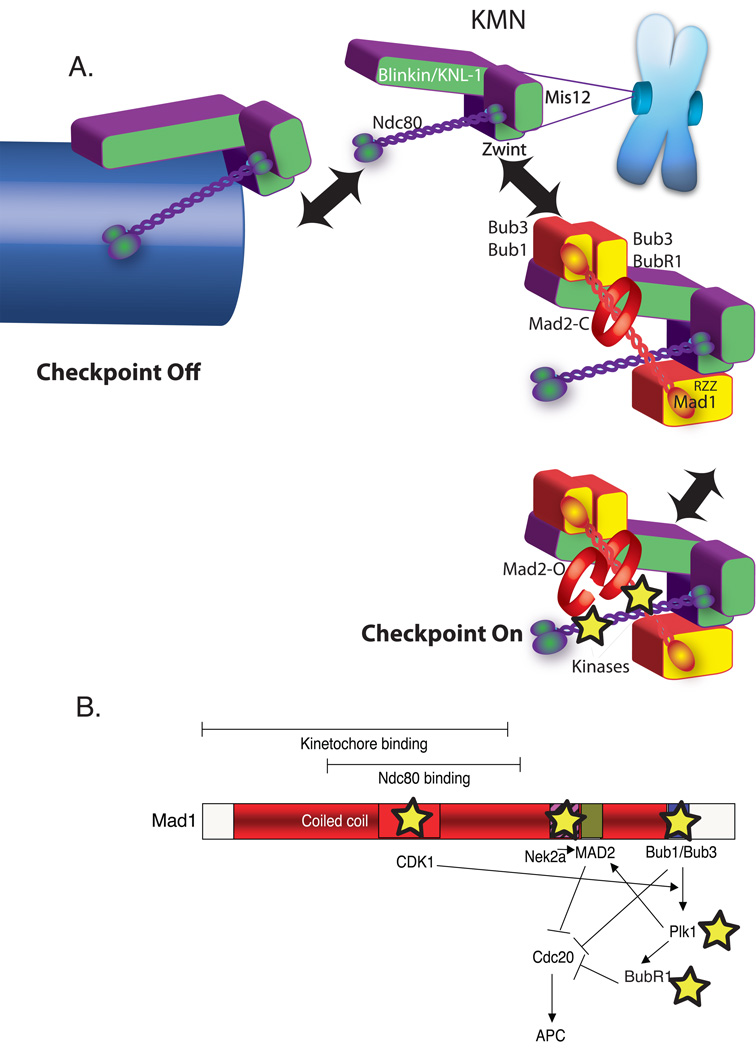
The event that is both evolutionarily conserved and most tightly correlated with spindle checkpoint signaling is Mad1 recruitment to the kinetochore. Mad1 levels are dramatically higher on unaligned kinetochores than those at the metaphase plate (Murray et al., 1999). How Mad1 is specifically recruited to unattached kinetochores is an important question for the field but a number of indirect experiments have implicated KMN as the binding site. Mad1 has at least three independent interactions with kinetochore proteins: KNL-1 (via Bub1), Ndc80 and RZZ (Figure 2A). Kinetochores bind either Mad1 or microtubules suggesting that Mad1 association is a microtubule-regulated step. There is a complex of Bub1, Bub3 and Mad1 in budding yeast suggesting that there is an indirect association between Mad1 and KNL-1 through Bub1 (Brady and Hardwick, 2000). Thus a simple model for the spindle checkpoint is that KMN binds either microtubules or Mad1 but cannot bind both simultaneously (Figure 2A). Microtubule attachment would block the generation of the signal at various points. First we propose that when Ndc80 and KNL-1 bind microtubules they lose the capacity to bind Mad1. Microtubule binding would also block the CENP-E/BubR1 interaction, shutting off BubR1 kinase activity (not shown in the model), and allow dynein to carry RZZ, Mad1 and Mad2 away from KMN.
Figure 2. An updated model for spindle checkpoint signaling.
A) Dual role of KMN in the kinetochore as both microtubule anchor and a scaffold to generate spindle checkpoint signals. Overall color scheme is the same as Figure 1. KMN (purple and green) is located in the outer plate of the kinetochore where both binds microtubules (arrow left) and spindle checkpoint protein (arrow right). KMN contains at least two microtubule-binding interfaces one in the KNL-1 subunit and another in the Hec1 subunit of the Ndc80 complex and approximately eight KMNs generate a binding pocket (not shown). In the absence of microtubules KMN has both direct and indirect interactions with checkpoint proteins. KNL-1 binds Bub3/Bub1 and Bub3/BubR1. The Ndc80 subunit can bind a coiled-coil region of Mad1 in a two-hybrid assay. Finally through the Zwint protein, the Mis12 complex binds RZZ, which can strip Mad1 from kinetochores. The kinetochore activates the checkpoint by acting as a scaffold to recruit and activate Bub1 and BubR1 kinases as well as other kinases recently implicated in checkpoint signaling (stars). B) A schematic Map of the Mad1 protein highlighting kinase interactions (stars) and a potential signal transduction network initiated after Mad1 recruitment to the kinetochore.
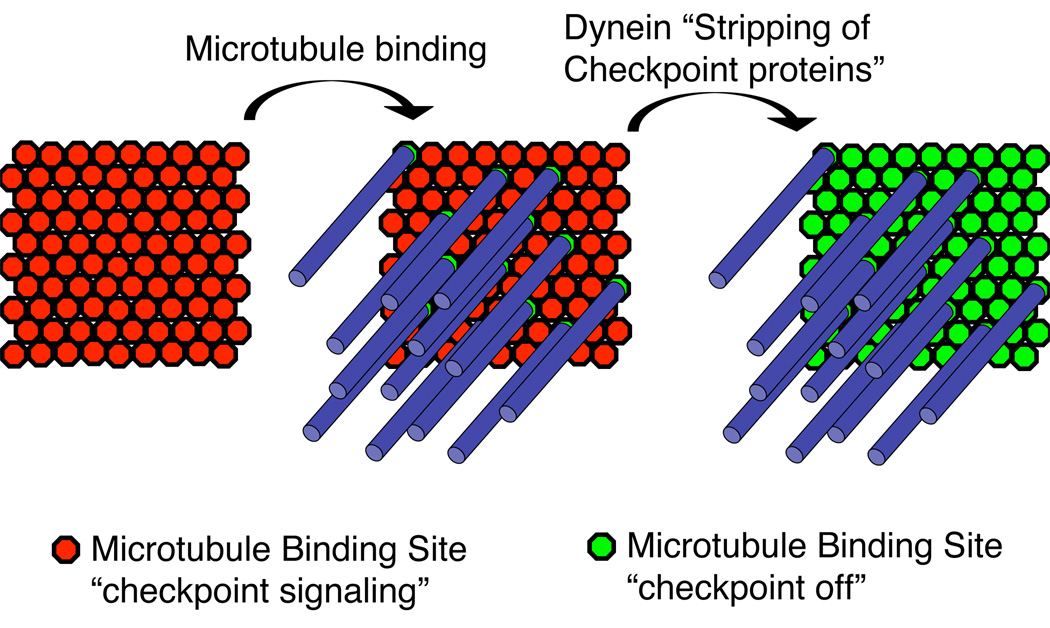
A simple model of mutually exclusive binding of Mad1 or a microtubule makes sense for yeast kinetochores that bind a single microtubule (Winey et al., 1995). This simple model, when applied to mammalian kinetochores, seems at odds with the observation that there is Mad2 localized to kinetochores with bound microtubules when dynein activity is inhibited (Howell et al., 2001). Yeast have approximately 8 copies of the Ndc80 complex and 6–7 copies of the Mis12 complex per kinetochore suggesting that a microtubule binding site has approximately 8 copies of KMN (Joglekar et al., 2006). Vertebrate kinetochores are more complicated than yeast and bind approximately 30 microtubules (McEwen et al., 1998; Howell et al., 2001; Dong et al., 2007). Interestingly, Xenopus kinetochores contain approximately 800 copies of KMN which should be sufficient to bind 100 microtubules (Emanuele et al., 2005). Therefore vertebrates have an additional requirement to remove checkpoint proteins, not only from binding sites with microtubules attached but potentially from unbound sites as well (Figure 3). Mutually exclusive binding of microtubules or checkpoint proteins to KMN may happen in vertebrate kinetochores but would not effectively silence the checkpoint. This may explain why higher cells evolved an additional dynein-dependent mechanism to strip checkpoint proteins in order to silence the checkpoint. This is supported by the observation that Mad2 is reduced at kinetochores where microtubules are bound and dynein is inhibited as compared to kinetochores without bound microtubules (Howell et al., 2001).
Figure 3. A possible role for Dynein in silencing the checkpoint signals by removing checkpoint proteins associated with KMN complexes that are not binding microtubules.
Since there is more KMN than is necessary to bind microtubules there may be two independent steps in checkpoint silencing. First, microtubule binding sterically prevents checkpoint protein binding to potential microtubule binding sites. Second, dynein “strips” checkpoint proteins from surrounding KMN complexes that are not associated with microtubules.
A central role for protein kinases in spindle checkpoint signaling
KMN as a dual site for microtubule binding and checkpoint protein binding accommodates the popular idea that the role of the kinetochore is to assemble MCC (Cleveland et al., 2003; Musacchio and Salmon, 2007). However this model is not necessarily correct. It is clear in yeast that Mad2-Cdc20 and MCC are essential for the checkpoint (Hardwick et al., 2000; Fraschini et al., 2001; Poddar et al., 2005). However, Mad2-Cdc20 complexes form in mitosis independently of both the checkpoint and the kinetochore in yeast. MCC also exists in the absence of kinetochores in mitotic Xenopus egg extracts although the amount of the complex increases in checkpoint signaling conditions (Chen, 2002). MCC has also been purified from HeLa cells arrested in S-phase (thymidine starved) but the interpretation is potentially complicated by a possible a lack of synchrony (Sudakin et al., 2001).
If MCC assembly is not integral to spindle checkpoint signaling, then what role does KMN play in signaling? Perhaps KMN acts as a platform to localize and activate kinases to generate a phosphorylation cascade. In checkpoint signaling Xenopus extracts, Bub1 and BubR1 are highly phosphorylated on chromatin while soluble protein is not phosphorylated (Chen, 2004), strongly suggesting that there are local phosphorylation events. The spindle checkpoint requires a large number of protein kinases many of which have recently been identified. These include Bub1, BubR1, Mps1, Aurora B, Tao1, Nek2a, p38 Map Kinase,Prp4, CDK1 and possibly Plk1 (Minshull et al., 1994; Kallio et al., 2002; Lew and Burke, 2003; D'Angiolella et al., 2003.; Cleveland et al., 2003; Hauf et al., 2003; Lou et al., 2004; Qi et al., 2006; Baumann et al., 2007 Montembault et al., 2007). The role of kinases in the checkpoint has been disputed since kinase inactive forms of Bub1 and BubR1 still generate signals (Sharp-Baker and Chen, 2001; Fernius and Hardwick, 2007) although the requirement for BubR1 kinase is disputed (Mao et al., 2005). However, when one considers that several kinases are involved it is reasonable that a signal may still be produced after the loss of some kinases because of redundancy.
An exciting model that requires additional experimentation is that Mad1 binding may bring Nek2a and CDK1 to kinetochores (Figure 2B) perhaps completing a signal transduction circuit to inhibit mitosis in response to a lack of microtubule occupancy in the kinetochore. Mad1 is a long coiled-coil protein that interacts tightly with Mad2 and recruits it to kinetochores. Mad1 also binds the Nek2a kinase that is required for signaling (Lou et al., 2004) and has a CDK1 binding site in its N-terminal region that is required for signaling (Jonathon Pines personal communication). We suggest that Mad1 recruitment initiates three independent pathways that inhibit Cdc20 (Figure 2B). Bub1 kinase phosphorylates and inhibits Cdc20 directly (Tang et al., 2004). Moreover, CDK1 priming phosphorylation on Bub1 targets Polo kinase to kinetochores; an event that allows Mad2 and BubR1 localization thus concentrating all of the components of the MCC (Qi et al., 2006). Moreover after phosphorylation by CDK1 in vitro, Cdc20 interacts with Mad2 rather than APC (D’Angiolella et al., 2003). This model for the spindle checkpoint is appealing from the standpoint that it uses kinase cascades as the mechanism to amplify the signal from a single unattached kinetochore and uses redundant pathways that converge to inhibit Cdc20.
Our goal is to point out potential shortcomings of current thinking and stimulate new directions. Our model extends many of the key aspects of the old model. Both envision that the spindle checkpoint is a simple two-state switch. When microtubules are absent the Mad1 scaffold assembles on KMN-RZZ and the checkpoint is on. When microtubules bind, RZZ is removed, the scaffold is displaced and the checkpoint is off. However, by incorporating KMN as the Mad1 docking site, our model needs only a slight modification for yeast where single microtubules bind kinetochores and RZZ is not present. It has been largely assumed that the rapid half-lives of checkpoint proteins argues that the proteins are being modified at kinetochores and soluble active complexes are released. Our model provides a simpler explanation for why some checkpoint proteins have short half-lives in kinetochores. If they associated tightly to kinetochores then microtubules would be precluded from binding.
Although most parts of our model are based on experimental evidence there are some untested elements. This includes biochemical evidence that Mad1 acts as a scaffold to activate kinases and brings Nek2a and CDK to kinetochores. It is critical to move beyond simply localizing proteins to the kinetochore, but rather to continue move toward mechanistic dissection of kinase activation and substrate identification. A beautiful example of such lines of experimentation are the elegant experiments in Xenopus that established that Map Kinase phosphorylation of MPS1 is required for its localization to the kinetochore (Zhao and Chen, 2006). Similarly, CENP-E binds to BubR1 only under checkpoint signaling conditions and this activates kinase activity (Mao et al., 2005). The last several years have witnessed an explosion in identifying and characterizing kinetochore and checkpoint proteins. We are only beginning to understand how the checkpoint is organized in the kinetochore. The future is very promising for the spindle checkpoint and kinetochore fields and the prospect for understanding, in molecular terms, the role of the kinetochore in the spindle checkpoint is exceedingly bright.
References
- Basto R, Gomes R, Karess RE. Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila. Nature Cell Biology. 2000;2:939–943. doi: 10.1038/35046592. [DOI] [PubMed] [Google Scholar]
- Baumann C, Korner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–114. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady DM, Hardwick KG. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 2000;10:675–678. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- Chan GK, Jablonski SA, Starr DA, Goldberg ML, Yen TJ. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat Cell Biol. 2000;2:944–947. doi: 10.1038/35046598. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 2001;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH. Phosphorylation and activation of Bub1 on unattached chromosomes facilitate the spindle checkpoint. EMBO J. 2004;23:3113–3121. doi: 10.1038/sj.emboj.7600308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- D'Angiolella V, Mari C, Nocera D, Rametti L, Grieco D. The spindle checkpoint requires cyclin-dependent kinase activity. Genes Dev. 2003;17:2520. doi: 10.1101/gad.267603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam VM, Stegmeier F, Nalepa G, Sowa ME, Chen J, Liang A, Hannon GJ, Sorger PK, Harper JW, Elledge SJ. A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat Cell Biol. 2007;9:556–564. doi: 10.1038/ncb1569. [DOI] [PubMed] [Google Scholar]
- Emanuele M, Burke DJ, Stukenberg PT. A Hec of a microtubule attachment. Nat Struct Mol Biol. 2007;14:11–13. doi: 10.1038/nsmb0107-11. [DOI] [PubMed] [Google Scholar]
- Emanuele MJ, McCleland ML, Satinover DL, Stukenberg PT. Measuring the Stoichiometry and Physical Interactions between Components Elucidates the Architecture of the Vertebrate Kinetochore. Mol Biol Cell. 2005;16:4882–4892. doi: 10.1091/mbc.E05-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernius J, Hardwick KG. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS. Genet. 2007;3:e213. doi: 10.1371/journal.pgen.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R, Beretta A, Sironi L, Musacchio A, Lucchini G, Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 2001;20:6648–6659. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RD, Poddar A, Yellman C, Tavormina PA, Monteagudo MC, Burke DJ. The spindle checkpoint of the yeast Saccharomyces cerevisiae requires kinetochore function and maps to the CBF3 domain. Genetics. 2001;157:1493–1502. doi: 10.1093/genetics/157.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Lechner J, Shevchenko A, Shevchenko A, Magiera MM, Schramm C, Schiebel E. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 2001;20:777–791. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, Huneycutt BJ, Pearson CG, Zhang C, Morgan G, Shokat K, Bloom K, Winey M. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr. Biol. 2005;15:160–165. doi: 10.1016/j.cub.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Kallio MJ, McCleland ML, Stukenberg PT, Gorbsky GJ. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr Biol. 2002;12:900–905. doi: 10.1016/s0960-9822(02)00887-4. [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Kops GJ, Kim Y, Weaver BA, Mao Y, McLeod I, Yates JR, 3, Tagaya M, Cleveland DW. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol. 2005;169:49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosco KA, Pearson CG, Maddox PS, Wang PJ, Adams IR, Salmon ED, Bloom K, Huffaker TC. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell. 2001;12:2870–2880. doi: 10.1091/mbc.12.9.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Burke DJ. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 2003;37:251–282. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- Lou Y, Yao J, Zereshki A, Dou Z, Ahmed K, Wang H, Hu J, Wang Y, Yao X. NEK2A interacts with MAD1 and possibly functions as a novel integrator of the spindle checkpoint signaling. J Biol Chem. 2004;279:20049–20057. doi: 10.1074/jbc.M314205200. [DOI] [PubMed] [Google Scholar]
- Mao Y, Abrieu A, Cleveland DW. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 2003;114:87–98. doi: 10.1016/s0092-8674(03)00475-6. [DOI] [PubMed] [Google Scholar]
- Mao Y, Desai A, Cleveland DW. Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. J Cell Biol. 2005;170:873–880. doi: 10.1083/jcb.200505040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–114. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland ML, Kallio MJ, Barrett-Wilt GA, Kestner CA, Shabanowitz J, Hunt DF, Gorbsky GJ, Stukenberg PT. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr. Biol. 2004;14:131–137. doi: 10.1016/j.cub.2003.12.058. [DOI] [PubMed] [Google Scholar]
- McEwen BF, Hsieh CE, Mattheyses AL, Rieder CL. A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma. 1998;107:366–375. doi: 10.1007/s004120050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Minshull J, Sun H, Tonks NK, Murray AW. A MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. Cell. 1994;79:475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- Montembault E, Duterte S, Prigent C, Giet R. Prp4 is a spindle assembly checkpoint protein required for Mps1, Mad1 and Mad2 localization to kinetochores. J. Cell Biol. 2007;179:601–609. doi: 10.1083/jcb.200703133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray D, Mirzayans R, Chen RH. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. British Journal of Cancer. 1999;81:959–965. [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:4931–4946. doi: 10.1091/mbc.E03-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poddar A, Stukenberg PT, Burke DJ. Two complexes of spindle checkpoint proteins containing Cdc20 and Mad2 assemble during mitosis independently of the kinetochore in Saccharomyces cerevisiae. Eukaryot. Cell. 2005;4:867–878. doi: 10.1128/EC.4.5.867-878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Tang Z, Yu H. Phosphorylation- and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol Biol Cell. 2006;17:3705–3716. doi: 10.1091/mbc.E06-03-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancati G, Crispo V, Lucchini G, Piatti S. Mad3/BubR1 Phosphorylation During Spindle Checkpoint ActivationDepends on both Polo and Aurora Kinases in Budding Yeast. Cell Cycle. 2005;4 doi: 10.4161/cc.4.7.1829. [DOI] [PubMed] [Google Scholar]
- Sharp-Baker H, Chen RH. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J Cell Biol. 2001;153:1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- Stucke VM, Baumann C, Nigg EA. Kinetochore localization and microtubule interaction of the human spindle checkpoint kinase Mps1. Chromosoma. 2004;113:1–15. doi: 10.1007/s00412-004-0288-2. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Shu H, Oncel D, Chen S, Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell. 2004;16:387–397. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu X, Ding X, Dou Z, Yang Z, Shaw AW, Teng M, Cleveland DW, Goldberg ML, Niu L, Yao X. Human Zwint-1 specifies localization of Zeste White 10 to kinetochores and is essential for mitotic checkpoint signaling. J Biol Chem. 2004;279:54590–54598. doi: 10.1074/jbc.M407588200. [DOI] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kilmartin JV. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chen RH. Mps1 phosphorylation by MAP kinase is required for kinetochore localization of spindle-checkpoint proteins. Curr Biol. 2006;16:1764–1769. doi: 10.1016/j.cub.2006.07.058. [DOI] [PubMed] [Google Scholar]