Synopsis
Exosomes constitute a discrete population of nanometer-sized (30-150 nm) vesicles formed in endocytic compartments and released to the extracellular environment by different cell types. In this work we demonstrated by electron microscopic, western blotting and proteomic analyses that primary hepatocytes secrete exosome-like vesicles containing proteins involved in metabolizing lipoproteins, endogenous compounds as well as xenobiotics. These new findings contribute to improve our knowledge about biology's hepatocyte and may have important diagnostic, prognosis and therapeutic implications in liver diseases
Exosomes represent a discrete population of vesicles that are secreted from various cell types to the extracellular media. Their protein and lipid composition are a consequence of sorting events at the level of the multivesicular body, a central organelle which integrates endocytic and secretory pathways. Characterization of exosomes from different biological samples has shown the presence of common as well as cell-type specific proteins. Remarkably, the protein content of the exosomes is modified upon pathological or stress conditions. Hepatocytes play a central role in the body response to stress metabolizing potentially harmful endogenous substances as well as xenobiotics. In the present study we described and characterized for first time exosome secretion in non-tumoral hepatocytes, and using a systematic proteomic approach, we establish the first extensive proteome of a hepatocyte-derived exosome population which should be useful in furthering our understanding of the hepatic function and in the identification of components that may serve as biomarkers for hepatic alterations. Our analysis identifies a significant number of proteins previously described among exosomes derived from others cell types as well as proteins involved in metabolizing lipoproteins, endogenous compounds and xenobiotics, not previously described in exosomes. Furthermore, we demonstrated that exosomal membrane proteins can constitute an interesting tool to express non-exosomal proteins into exosomes with therapeutic purposes.
Keywords: Celullar Sub-proteome, Liver Physiology, Hepatocyte Biology, Exosomes, Biomarker Discovery, CD63, Exosome Display
Introduction
Over the last several years a unique type of secreted subendocytic compartments called “exosomes” have gained renewed interest from the research community as they unveil promising potential for human diagnostic and therapeutic applications. Exosomes constitute a discrete population of nanometer-sized (30-150 nm) vesicles formed in the endocytic compartments called multivesicular bodies (MVBs) during endosome maturation by inward budding of their limiting membrane 1-4. Exosome protein and lipid composition is a consequence of sorting events at the level of these organelles that are known to be important intermediates of the endocytic and biosynthetic pathways 5. MVBs are involved in transporting proteins for degradation in lysosomes, although in many cell types MVBs fuse with the plasma membrane, thus releasing their internal vesicles into the extracellular environment 6, 7. Exosome secretion has been described for reticulocytes 8, dendritic cells 9, lymphocytes 10, 11, mast cells 12, 13, platelets 14-16, intestinal epithelial cells 17, 18, adipocytes 19, neurological cells 20-24 and cancer cells 25-27, including hepatoma cell lines HepG2 28 and Huh7 29. Exosomes have also been successfully purified from many body fluids such as epididymal fluid and seminal plasma 30, broncoalveolar fluid 31, pleural effusions 32, ascites 33, 34, amniotic fluid 35, blood 36 and urine 37.
Immunoelectron microscopy, western blot and mass spectrometry analysis of different exosome preparations have demonstrated the presence of common as well as cell-type specific proteins. Thus, exosomes from lymphocytes, dendritic cells, masts and intestinal epithelial cells are enriched in MHC class I and II proteins whereas exosomes from platelets contain von Willebrand factor 14, and cytotoxic T cells-derived exosomes contain perforin and granzymes as specific molecules 38. The common proteins include chaperones, tetraspanins, adhesion molecules, rab proteins, cytoskeletal proteins and metabolic enzymes 1. Remarkably, their protein content is also modified upon infection by mycobacterias 39, several viruses, exposition to heat stress 40 as well as in tumor cells 41, 42 and in blood sepsis conditions 15. A major goal in the field of clinical proteomics is to identify disease biomarkers in biological fluids that can be measured relatively inexpensively for the early diagnosis of disease. It has been recognized that exosomes exist in serum 36, providing a potential approach to capitalize on exosomes as disease biomarkers in readily attained blood samples. The liver is one of the most vascular organs in the body and its main cellular component, the hepatocyte, plays a central physiological role metabolizing nutrients, potentially harmful endogenous substances, and xenobiotics. Hepatocytes are also important in modern drug discovery programs as they constitute the chosen cellular model for the study of metabolism and pharmacotoxicological effects of drugs 43. In addition, in end-stage liver disease hepatocytes are being utilized within liver devices to “bridge” patients until they either recover or receive a liver transplant 44. Therefore, a better understanding of the relationship between hepatocytes and their extracellular enviroment, would contribute critically to search for the identification of candidate biomarkers, unravel drug clearance processes, and improve bioartificial liver technology.
Herein, we examined the delivery of exosomes into two cellular hepatic models; a mouse hepatic non-tumoral and non-transformed cell line, and in primary cultures of rat hepatocytes. Electron microscopic analysis revealed that hepatocytes secrete rounded vesicles structures with a diameter size similar to that of previously described exosomes 11. The identity of these vesicles was further confirmed as exosomes by western blot and mass spectrometry analysis. We establish the first extensive proteome of a hepatocyte-derived exosome population. Our analysis identifies several common exosomal proteins as well as proteins not previously described in exosomes, such as proteins involved in metabolizing lipoproteins, endogenous compounds as well as xenobiotics. Furthermore, we report the use of exosomal marker proteins as a tool to express non-exosomal proteins into exosomes.
Experimental Procedures
Reagents
All media and reagents for tissue culture were purchased from Invitrogen (Carlsbad, CA). All other reagents were of analytical grade and primarily acquired from Sigma-Aldrich (St. Louis, MO). Monoclonal antibodies (mAbs) were purchased from the indicated vendors: anti-early endosome antigen 1 (Eaa1), anti-β1-integrin (clone 18), anti-AIP1/Alix (clone 49), anti-rat Cd63 (clone AD1), and anti-BiP/Grp78 from BD Biosciences (Mountain View, CA), anti-ICAM1 (clone 1A29) from Abcam (Cambridge, UK), and anti-GFP (clones 7.1 and 13.1) from Roche Applied Science (Indianapolis, IN). Rabbit polyclonal antibodies were acquired from the indicated vendors: anti-Pmp70 from Invitrogen (Carlsbad, CA), anti-MFG-E8 (M-135), anti-ASGPR ½ (FL-291) and anti-Prohibitin (H-80) from Santa Cruz Biotech., Inc (Santa Cruz, CA), anti-Caveolin from BD Biosciences (Mountain View, CA), and anti-Tsg101 from Abcam (Cambridge, UK). Hamster Armenian anti-mouse Cd81 (clone Eat2) was purchased from Serotec (Oxford, UK). Goat anti-Apo E was purchased from Santa Cruz Biotech., Inc. (Santa Cruz, CA). Horseradish peroxidase (HRP)-conjugated secondary antibody was from GE Healthcare (Buckinghamshire, UK). The vector expressing EGFP protein was pEGFP-N3 (BD Biosciences, Montain View, CA) and the pEGFP-N3-derived plasmid encoding the full-length of human CD63 protein fused to EGFP (CD63-EGFP) was previously described 45.
Cell culture and transfection
Rat hepatocytes (DPK-HCWP-RS, purity >95%) obtained from livers of nine-week-old healthy Sprague-Dawley male rats were purchased from Dominion Pharmakine (Derio, Bizkaia, Spain). MLP-29 cells were grown in complete DMEM medium [Dulbecco’s modified Eagle medium supplemented with 10% (v/v) fetal bovine serum, 0.1 mg/ml streptomycin and 100 units/ml penicillin]. For stable transfection, MLP-29 cells were grown to 60-70% confluency and transfected with EGFP- or CD63-EGFP-encoding plasmids using FuGENE 6 (Roche Diagnostics, Manheim, Germany) and OptiMEN medium (Invitrogen, Carlsbad, CA) as described by the manufacte’s datasheet. The medium was changed to complete DMEM 5-hours post-transfection, and to complete DMEM containing 2 g/liter G418, 24-hours later. Cells displaying resistance to G418 and expressing EGFP or CD63-EGFP were enriched by preparative cell sorting on a BD FACSAria flow cytometer (Becton Dickinson).
Production and Isolation of Exosomes
For exosome production, MLP-29 cells or fresh rat hepatocytes suspension were plated to non-collagenized (MLP-29) or collagen-coated (rat hepatocytes) 150-mm dishes, at 15-30 million cells per dish. Cells were cultured in complete DMEM medium for 24-h (MLP-29 cells) or for 4-h (rat hepatocytes) at 37°C and 5% of CO2, washed two times with Dulbecco`s modified phosphate-buffered saline (PBS) and incubated 48-h (MLP29 cells) or 36-h (rat hepatocytes) in 25 mM HEPES-containing complete DMEN medium previously depleted of contaminiting vesicles (ie, exosomes) by overnight centrifugation at 110,000 x g 9. Exosomes were isolated as previously described 11; briefly, culture supernant was collected and centrifugated at 500 x g for 10 min to remove lifted cells. The resultant supernant was subjected to filtration on 0.22 μm pore filters, followed by ultracentrifugation at 10,000 x g and 100,000 x g for 30 min and 60 min, repectively. The resulting pellets were resuspended in PBS, pooled, and again ultracentrifuged at 100,000 x g for 60 min. The final pellet of exosomes (referred as “Ex”) was resuspended in PBS to between 1/700th and 1/2000th the original volume of the culture supernant, and the aliquoted solution was stored at -80°C. Mean protein content of exosome preparations were 0.36±0.18 (n=4) and 4.5±2.1 (n=4) pg/cell for MLP-29 cells and rat hepatocytes, respectively.
For rat hepatocyte derived-exosomes a further purification step was performed as previously described 46. Briefly, the PBS-suspended “Ex” preparation was diluted in 30 ml of PBS and underlayered on top of a density cushion composed of 20 mM Tris/30% sucrose/deuterium oxide (D2O) pH 7.4 (4 ml) forming a visible interphase. The samples were ultracentrifuged at 100,000 x g and 4°C for 75 min in a SW-32 Ti swinging bucket rotor (Beckman Coulter, Fullerton, CA). The ultracentrifuged-tubes were pierced on the side with an 18-gauge needle and 3.5 ml were withdrawn from the bottom. Exosomes contained in the 30% sucrose/D2O cushion were colllected, diluted a minimum of 10 times with PBS and centrifuged at 100,000 x g and 4°C for 60 min. The final exosome pellet of higher purity (referred as “Eh”) was resuspended in PBS to half of the initial “Ex”-volume, aliquoted and stored at -80°C.
Western blot analysis
To prepare cell lysates (referred as “C”), 106 trypsinated-cells were lysed during 15 min on ice in the presence of 100 μl of lysis buffer [300 mM NaCl, 50 mM Tris pH 7.4, 0.5% Triton X-100 and proteases inhibitors]. After clarification of the samples by centrifugation at 20.000 x g, the supernant was transfered to a fresh eppendorf tube. The protein concentration of the cell lysates “C”, purified “Ex” (MLP29 cells) and “Eh” (Rat hepatocytes) exosomes were determined by means of Bradford protein assay (Bio-Rad, Hercules, CA) using BSA as the standard. SDS-sample buffer was added and samples were incubated for 5 min at 37°C, 65°C and 95°C and separated on 4-12% precasted acrylamide gels (Invitrogen, Carlsbad, CA). After transfer to PVDF membranes (Millipore, Bedford, MA) and blocked overnight (5% milk and 0.05% Tween-20 in PBS), primary antibody was added for 1 hour, followed by PBS washing and the application of secondary HRP-conjugated antibody. With the exception of Cd63, Cd81, Icam-1 and Mfg-E8, all proteins were detected under reducing conditions. Chemioluminiscence detection of bands was performed with ECL Plus reagent (GE Healthcare, Buckinghamshire, UK).
Electron Microscopy
For cryo-electron microscopy, exosome preparations were directly adsorbed onto glow-discharged holey carbon grids (QUANTIFOIL, Germany). Grids were blotted at 95% humidity and rapidly plunged into liquid ethane with the aid of VITROBOT (Maastricht Instruments BV, The Netherlands). For negative staining, 2.5 μl drops of purified exosomes were adsorbed onto glow-discharged carbon-coated copper grids, washed with distilled water, and stained with freshly prepared 2.0% uranyl acetate in aqueous suspension. Samples were imaged using a JEM-1230 transmission electron microscope (JEOL, Japan) equipped with a thermionic tungsten filament and operated at an acceleration voltage of 120 kV. Images were taken with a pixel size of 0.34 nm using the ORIUS SC1000 (4008×2672 pixels) cooled slow-scan CCD camera (GATAN, UK).
Proteomic nanoLC-MS/MS analysis and Database search
Proteins (between 2 and 40 μg) rat hepatocytes-derived exosomes were separated on 4-12% SDS-PAGE gels, fixed and stained with Coomassie blue (Bio-Rad, Hercules, CA). Each gel lane was trimmed in pieces and each was subjected to trypsin digestion. The recovered peptides were subsequently vacuum-dried. Nanoflow LC-MS/MS analysis was performed using a nanoflow UPLC system coupled to a QTOF Premier mass spectrometer (Waters, Manchester, UK). Samples were loaded in 1% aqueous formic acid (FA) and analyzed by reverse phase LC-MS/MS in a UPLC reverse phase chromatography system (Waters). Tryptic peptides were desalted on a Symmetry C18 trapping cartridge (Waters) and further separated on an analytical column (Atlantis C18, 75 μm id × 3 μm, Waters) with an integrated electrospray ionization emitter tip (SilicaTips for Micromass ZSpray NanoFlow, 10 μm diameter, New Objective). Peptides were eluted at a flow rate of 250 nL/min from the analytical column directly to electrospray ionization emitter tip by using a 30 min gradient from 0 to 30% solvent B (solvent A: 1% aqueous FA and solvent B: 100% acetonitrile, 1%FA). Data was acquired in the data dependent acquisition mode (DDA), in which a full scan mass spectrum (m/z: 300-1500) was followed by MS/MS (m/z: 50-1995) in the three most abundant multi-charged ions (+2 and +3) every 4 s. Argon was used as the collision gas. Collision energies were interpolated linearly as a function of a charge state and m/z of each peptide. Dynamic exclusion was incorporated for 30 seconds. A scan of the reference compound (Glufibrinopeptide B) was acquired every ten scans of the analyte through the entire run. Raw data was processed using ProteinLynx Global Server v2.2.5 (Waters). The resulting pkl file was searched against v52 of SwissProt sequence database (Rattus: 5830 sequences) with rat as taxonomy using an in-house Mascot server (Version 2.2.03, Matrix Sciences, London, UK). One miss cleavage was allowed; carbamidomethyl was chosen as fixed modification and oxidation as variable modification. A peptide mass tolerance of 20 ppm and 0.1 Da of fragment mass tolerance were allowed. The data converted to XML file by using PrideConverter tool (http://code.google.com/p/pride-converter/) are accessible through the PRIDE database (http://www.ebi.ac.uk/pride/). The search against the decoy database gave a false-discovery rate (FDR) on protein level of 4.9% when using a protein threshold score of 30, so for this study just proteins with scores greater than 30 were considered (Table S1, supplemental data). Furthermore, only proteins with at least one specific peptide (with a Mascot colour code of red and bold) were included in the study. The sequences of all peptides considered and the percentage coverage for the identified proteins are provided in Table S2 (supplemental data). An additional file is provided for those proteins identified based on a single peptide containing infomation about precursor mass, charge and tandem mass spectrum associated to them (Figure S1, supplemental data).
GO slim categorization
Gene ontology (GO) annotations for proteins in UniProt knowledgebase (http://www.ebi.ac.uk/GOA/index.html) was used for categorization of the proteins identified by mass spectrometry. In order to calculate the statistical significance of enrichment or depletion of specific GO categories all rat proteins from the UniProt knowledgebase were extracted from the same database used for the mass spectrometry data search. The probabilities were calculated using the binomial distribution. The probability to observe x proteins in a specific category of K random proteins from the list consisting of all proteins in UniProt knowledgebase is given by:
| (1) |
Where N is the total number of rat proteins the UniProt knowledgebase. M is the number of genes belonging to a specific category. The P-value (significance) for over-represented categories can be calculated as:
| (2) |
This can be interpreted as the probability to obtain x proteins when redrawing a list of K proteins randomly from the whole database.
KEGG pathways Analysis
The KEGG webservice API was used to extract KEGG pathways for rat proteins. All rat proteins from UniProt knowledgebase were used as reference and equation (2) was used to test for pathways that have significantly enriched or depleted number of proteins.
Results
Exosome-like vesicles secreted by the cell line MLP-29
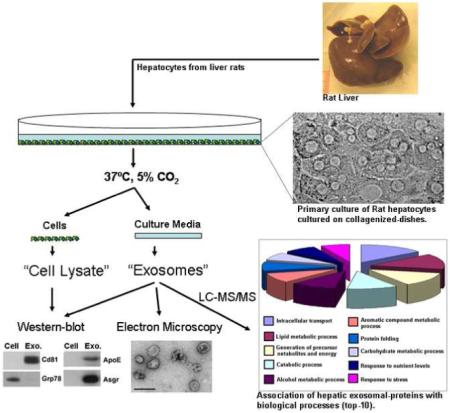
First, we examined the capacity to release exosome to the extracellular medium of the murine hepatic cell line MLP-29, which is an epithelial homogeneous cellular clone obtained by limiting dilution from a mouse embryonic liver cell line. We chose this cell line because besides displaying hepatocyte-like features such as albumin production, they can be propagated indefinitely in vitro, but are not transformed as measured by the soft agar assay and do not form tumors in immunocompromised mice47, 48. MLP-29 cells were cultured in conditioned medium and the vesicles that were secreted during a 48-hour incubation period were isolated as described in the Experimental Procedures. Cryo-electron microscopic analysis revealed that the isolated extracellular particles consisted primarily of rounded membrane vesicles (Figure 1a), with a mean diameter of 40.6±11.2 nm (n=116), which was compatible with the size previously reported for exosomes 1. Subsequently, equal amounts of protein extracts from cells or secreted-vesicles were analyzed by western blotting using antibodies specific for Tsg101 (a well-defined exosomal marker) and Grp78 (endoplasmic reticulum marker) proteins. This latter protein was clearly detected in cell lysates, but was barely detected in the vesicles (Figure 1b). On the contrary, under conditions in which Tsg101 was undetectable in cell lysates, a specific, clear band was obtained in the secreted-vesicles (Figure 1b), indicating that this exosomal marker is highly enriched within these vesicles arguing positively for the exosomal definition of these vesicles.
Figure 1.
Characterization of exosome-like vesicles from murine hepatocytes. (a) Cryo-electron micrograph of extracellular vesicles secreted by MLP-29 cells. Bar, 100 nm. (b) Western blot analysis of protein extracts prepared from MLP-29 cells (“C”) or from MLP-29-secreted vesicles (“Ex”). Proteins (10 μg) were analyzed by western blotting using antibodies against Tsg101 (exosome marker) or Grp78 (endoplasmic reticulum marker) proteins.
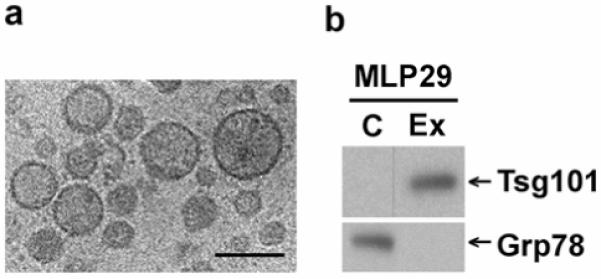
To further examine the presence of exosomes in this mouse hepatic cell line we generated stably MLP-29 cells ectopically-expressing either the exosomal marker CD63-tagged enhanced green fluorescent protein (EGFP), or the EGFP protein alone. As previously reported for others cell lines, EGFP protein presented a cytosolic distribution (Figure 2a) whereas CD63-EGFP protein presented a perinuclear-localized vesicular staining pattern (Figure 2b) compatible with late endosomal and lysosomal intracellular distribution, as previously reported for this fusion protein 49. As shown in Figure 2c, GFP-positive cells represented 72% of the cellular line expressing EGFP protein whereas in the case of the CD63-EGFP-transfected cell line, positive cells represented only 16%. Median fluorescence intensities of EGFP and CD63-EGFP expressing cells were 605 and 477, respectively. Thus, the control cell line which was expressing cytosolic EGFP alone has more positive cells, and the positive-cells express a greater quantity of the ectopic protein than does the CD63-EGFP-expressing cell line. Subsequently, exosomes secreted during a 48-h incubation period for each of the cell lines were isolated and the presence of EGFP and CD63-EGFP proteins were tested by Western blot analysis using an antibody against the GFP tag (Figure 2d). A unique band of the expected size was detected in cellular extracts prepared from cells expressing EGFP alone (Figure 2d). In the case of CD63-EGFP-expressing cells the majority of the GFP signal corresponded to a band of the expected size, however, we also detected a band at the level of EGFP alone (Figure 2d, asterisk) that we believe was due to the still present translational start point from the EGFP portion of the fusion protein. In the exosome preparations, CD63-EGFP protein was clearly detected in preparations from cells expressing the fusion protein supporting the existence of this secreted subcellular compartment (ie. exosomes) in this mouse hepatic cell line. On the contrary, we failed to detect any band of the EGFP protein size, either in cells expressing EGFP alone, or fused to CD63, indicating that this protein is unable to reach that compartment by itself, and excluding cell lysis as a major contributing factor. The purity of the exosome preparation was examined by using antibodies against proteins Tsg101, Grp78, and Pallidin (Pldn), a cytosolic and early endosomal membrane-associated protein 50, 51. While the presence of these two latter proteins was undetectable (Figure 2d) indicating at most a limited level of cytosolic, endoplasmic reticulum and early endosomal contamination, the presence of Tsg101 was highly enriched in the exosome preparations. Furthermore this result supports that CD63 could be a useful tool to target a desired protein to secretion into this subendocytic compartment (ie. exosomes).
Figure 2.
Targeting of Enhanced Green Fluorescent Protein (EGFP) to exosomes mediated by fusion to CD63 protein. (a) EGFP- or (b) CD63-EGFP-expressing MLP-29 cells were grown on glass coverlips, fixed using 2% (w/v) formaldehyde, washed two times with PBS and mounted on glass slides using Fluoromount-G (Southern Biotech, Birmingham, AL) containing 0.7 mg/liter of DAPI. Fluorescent samples were examined on a Leica TCS SP multiphoton confocal microscope. Images from GFP (green) and DAPI (blue) fluorescence were acquired using a 40X objetive and the Leica Confocal Software. Note the cytosolic distribution of EGFP versus the vesicular localization around the nucleus of CD63-EGFP protein. Bars, 40 μm. (c) MLP-29 cellular clones expressing EGFP or CD63-EGFP proteins were cultured in 150-mm dishes, trypsinized and washed two times with ice-cold PBS. GFP fluorescence was acquired using BD FACSDiva flow cytometry software version 5.0 in a FACScanto II flow cytometer. A threshold for GFP fluorescence was established based on the fluorescence observed for the parental cell line MLP-29. Between parentheses are indicated both the percentage of cells with GFP fluorescence above the threshold and the median GFP fluorescence intensity determined in this percentage of cells. (d) Protein extracts prepared from cells (“C”, 12μg protein) and exosomes (“Ex”, 3 μg protein) of MLP29 cellular clones expressing EGFP- or CD63-EGFP proteins were electrophoretically separated on 4-12% SDS-PAGE gel and analyzed by western blotting using antibodies against the following proteins: EGFP tag, Tsg101 (exosomal marker), Grp78 (endoplasmic reticulum marker) and Pallidin (“Pldn”, cytosolic and early-endosomal protein).
Characterization of exosomes secreted by rat hepatocytes
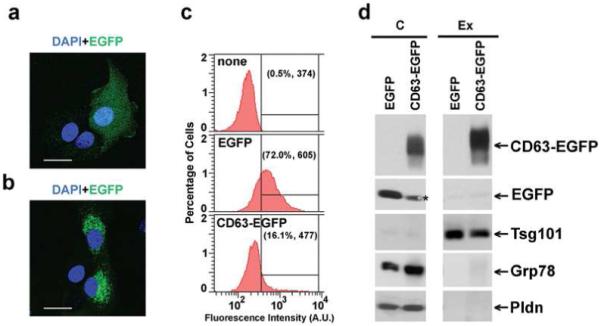
To further characterize exosomes of hepatic origin in a more physiological model; we examined their production by primary cultures of rat hepatocytes. Exosomes secreted to the media over a 36-h period by these cells were isolated as described in Experimental Procedures. The isolated-material was analyzed by electron microscopy, western blotting and LC-MS/MS. Negative staining- and cryo-electron microscopy of this material revealed the presence of multiple rounded vesicles-like particles (Figure 3). Quantitative analysis of cryo-electron micrographs (Figure 3c), illustrates that the mean diameter of these vesicles in two independent preparations were 57.6±23 nm (n=101) and 49.5±17 nm (n=73), consistent with the size of exosomes isolated from MLP-29 cells and from other sources1.
Figure 3.
Ultrastructural analysis of vesicles secreted by rat hepatocytes. Representative (a) negative stainning- and (b) cryo- electron micrographs of vesicles secreted by primary culture of rat hepatocytes. Bars, 200 nm. (c) Quantitative analysis of vesicle diameter was performed in cryo-electron micrographs acquired and processed by using Digital Micrograph Software. Distributions of percentage of cells, number of vesicles analyzed (n), and mean and standard deviation of two independent preparations are indicated.
Western blot analysis (Figure 4) shows the enrichment of numerous exosomal protein markers in these vesicles. Equal protein amounts of extracts prepared from cells or exosomes were subjected to western blot analysis using antibodies specific for known exosomal proteins including: Tsg101, Aip1/Alix, β1-Integrin, Cd81, Cd63, Icam-1 and Mfg-E8, or specific to other subcellular compartments like caveolae (Caveolin-1), early-endosome (Eaa1), endoplasmic reticulum (Grp78), peroxisome (Pmp70) or mitochondria (Prohibitin1 and mtPmp70). As despicted in Figure 4, while the presence of the non-exosomal markers in cellular extracts was clearly detected at the expected sizes, their presence in the exosome preparations was undetectable under the same conditions, meaning that the contamination of material derived from these compartments was minimal. However, the clear enrichment of exosomal proteins in the latter preparation supports the exosome definition for these secreted vesicles. Similar results were obtained in the western-blot analysis of exosomes isolated from three additional preparations of primary rat hepatocytes (data not shown). These results along with those obtained from MLP-29 cells strongly suggest that hepatocytes are in the group of cell-types that deliver exosomes to the extracellular environment.
Figure 4.
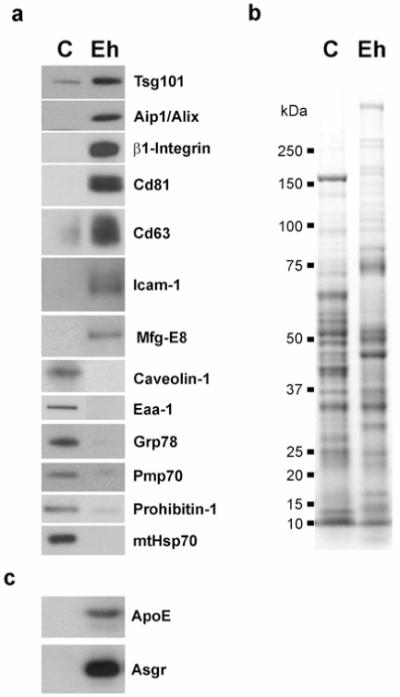
Western blot analysis and Coomassie Blue staining of exosomes secreted by rat hepatocytes. (a) Protein extracts (10 μg) prepared from rat hepatocytes (“C”) or from exosomes-derived from those cells (“Eh”) were analyzed by western blotting using antibodies against either exosomal markers (Tsg101, Aip1/Alix, β1-Integrin, Cd81, Cd63, Icam-1, Mfg-E8) or protein markers from other subcellular compartments; caveolae (Caveolin1), early endosome (Eaa1), endoplasmic reticulum (Grp78), peroxisome (Pmp70), mitochondria (Prohibitin-1, mtHsp70). Note the enrichment of exosomal markers in the exosome preparation compared to the cellular lysate. (b) Coomassie staining pattern of protein extracts (10 μg) prepared from rat hepatocytes (“C”) or from exosomes-derived from those cells (“Eh”). Notice that the pattern of bands presents in both samples is different. (c) Protein extracts (10 μg) prepared from rat hepatocytes or from exosomes-derived from those cells were loaded on a 4-12% SDS gel followed by western blotting using antibodies against ApoE and Asgr proteins.
For proteomic analysis, protein extracts of exosomes isolated from four independent preparations of rat hepatocytes were separated on SDS-PAGE gels and stained using Coomassie Blue. As expected the pattern of bands obtained for exosome preparation was different from that of cell lysates as is shown in Figure 4b for one of the preparations. To further refine the analysis of the proteins, gel lanes were sliced into pieces which were subsequently subjected to in-gel trypsinization, and the extracted peptides were analyzed by nano-spray LC-MS/MS. Overall, 251 unique proteins were identified (Table S1, supplemental material). A list of those proteins identified in at least two independent preparations or described in exosomes from other sources is provided in Table 1. These included transmembrane proteins, such as tetraspanins Cd63, Cd81 and Cd82, transporters and receptors such as the asialoglycoprotein receptor (Asgr). The exosomal enrichment of this hepatic-specific receptor was further confirmed by western-blot analysis (Figure 4c). In addition, the LC-MS/MS analysis identified multiple proteins known to be involved in the endosomal pathway, such as annexins, small GTP-binding proteins and several cytoskeletal (actin, tubulin), cytoskeletal binding-proteins (moesin, ezrin, cofilin-1) and motor proteins (myosins). We identified numerous cytosolic proteins that were presumably trapped in the lumina of forming exosomes during their biogenesis. Furthermore we identified many proteins such as cytochromes and UDP-glucuronosyltransferases involved in detoxification processes of both exogenous and endogenous compounds, such as hormones, ammonia and pharmaceutical agents. A significant group of identified proteins were secreted proteins, such as coagulation-related proteins, serum albumin and apolipoproteins which are released to the blood stream mainly by the liver. The presence and enrichment of apolipoprotein E (ApoE) in hepatic exosomes was further confirmed by western blot analysis (Figure 4c).
Table 1. Proteins identified by LC-MS/MS in rat hepatocytes-derived exosomes*.
| Secreted proteins |
| Serum albumin |
| ApoE† |
| Serum paraoxonases -1 and -3 |
| Fibronectin precursor |
| Fibrinogen beta chain precursor |
| Complement C4 |
| Coagulation factor II / Prothrombin |
| Hemoglobin subunits α-1/2 |
| Clusterin, Kininogen-1 and Serotransferrin |
| Polymeric-immunoglobulin receptor |
| Dipeptidyl peptidase 4 / CD26 |
| Transmembrane Proteins |
| CD63†, CD81† and CD82 |
| Lamp-1 |
| Membrane-associated progesterone receptor |
| Receptor expression-enhancing protein 6 |
| Sodium/bile acid cotransporter |
| Sodium/potassium-transporting ATPase 1α |
| Asialoglycoprotein receptor / ASGR† |
| Cytoskeleton-related proteins |
| Tubulins: α-2, α-6, β-2A, β-2C, β-3, β-5 |
| Actin, cytoplasmic 2 and Actin-like protein 2 |
| Myosin-9, -11 |
| Moesin and Ezrin |
| Cofilin-1 |
| Dynein heavy chain, cytosolic |
| Small GTPases |
| Ras-related protein: Rab-1A, -1B, -2A, , -7, -11A, -12, -13, -14, -35 |
| Ras-related protein Rap-1b precursor |
| ARF-1, -3, GTPase HRas precursor |
| Signal transduction |
| Guanine nucleotide-binding protein sub beta 2-like 1 |
| Guanine nucleotide-binding G(I)/G(S)/G(T) beta 2 |
| Guanine nucleotide-binding G(s) α ,and G(i) -α2 |
| 14-3-3 proteins: β/α, ε, γ, ζ,/δ |
| Major vault protein |
| Adhesion, Membrane transport and fusion |
| Lactadherin / Milk fat globule-EGF factor 8† |
| Annexins: A2, A4, A5, A6 |
| Clathrin heavy chain |
| Coatomer subunits beta and beta’ |
| Transmembrane emp24 domain-containing protein 2 |
| PDC6CI / AIP1/ Alix† |
| Cation-dependent mannose-6-phosphate receptor |
| Transitional endoplasmic reticulum ATPase |
| Protein synthesis and post-traductional |
| 40S ribosomal protein SA, S3, S4(X), S6 |
| 60S ribosomal protein L6 and L7 |
| EF1α1, EF1α2 and EF2 |
| Protein Folding |
| Heat shock proteins: HS70L, HSP71, TRAP1, HSP90-α and -β |
| Grp78 |
| Calnexin |
| T-complex protein 1 subunits α, β, δ, ε, γ |
| Protein disulfide-isomerase A3 |
| Calregulin / Calreticulin |
| Protein Processing and degradation |
| Aminopeptidase N / CD13 |
| Ubiquitin |
| Lipid Metabolism |
| Fatty acid synthase and ATP-citrate synthase |
| Long-chain specific acyl-CoA |
| Long-chain-fatty-acid-CoA ligase -1 and -6 |
| Peroxisomal bifunctional enzyme |
| Acyl-coenzyme A oxidase 2, peroxisomal |
| Hydroxymethylglutaryl-CoA synthase, mito. |
| Fatty aldehyde dehydrogenase |
| Carbohydrate and Amino-acid Metabolism |
| Alpha-enolase |
| Fructose-1,6-bisphosphatase 1, aldolase A |
| Glyceraldehyde-3-phosphate dehydrog. |
| Pyruvate kinase isozymes R/L |
| Pyruvate carboxylase, mitochondrial |
| Phosphoglycerate kinase 1 |
| 4-trimethylaminobutyraldehyde |
| L-lactate dehydrogenase A chain |
| Malate dehydrogenase, cytoplasmic |
| Cystathionine gamma-lyase |
| C-1-tetrahydrofolate synthase, cytoplasmic |
| 10-formyltetrahydrofolate dehydrogenase |
| Formimidoyltransferase-cyclodeaminase |
| Adenosylhomocysteinase |
| Betaine--homocysteine S-methyltransferase |
| Cysteine sulfinic acid decarboxylase |
| Arginase-1 |
| Argininosuccinate synthase and lyase |
| Carbamoyl-phosphate synthase |
| Hormone Metabolism |
| 3-oxo-5-beta-steroid 4-dehydrogenase |
| Corticosteroid 11-beta-dehydrogenase isozyme 1 |
| Retinol dehydrogenase -2, -3 |
| All-trans-retinol 13,14-reductase precursor |
| Xenobiotics / Endogenous compounds Metabolism |
| Cytochromes P450: 2A1, 2B3, 2C11, 2D1 2D3, 2D10, 2D18, 2D26 |
| UDP-glucuronosyltransferases: 2B2, 2B3 and 2B5 |
| NADPH-cytochrome P450 reductase |
| Alcohol dehydrogenase 1 and Epoxide hydrolase 1 |
| Microsomal glutathione S-transferase 1 |
| Sulfotransferase 1A1 |
| Thiosulfate sulfurtransferase |
| Dimethylaniline monooxygenase [N-oxide-forming] |
| Nuclear Proteins |
| Histones: H2A-1, H2B-1, H2B-1A, H4 |
| Importin beta-1 subunit |
| Miscellaneous |
| ATP synthase subunits α and β |
| Cytochrome b5 |
| Ferritin heavy chain |
| Ferritin light chain 1 |
Of the 251 proteins identified from hepatic exosomes 244 could be assigned to Genome Ontology (GO) categories which are defined by the GO consortium 52. We found that some functional activities such as oxidoreductase, GTPase, hydrolase, iron ion and lipid binding were significantly over-represented suggesting that these activites may be essential for the hepatic exosomes in order to perform their functions (Figure 5). Furthermore, there was a significant enrichment in proteins associated with intracellular transport, protein folding, stress response, cellular homeostasis and lipid metabolism (Figure 5), supporting a possible role of exosomes in those processes. An additional KEGG pathway analysis where 214 proteins from our exosome proteome were mapped to the Kyoto Encyclopedia for Genes and Genomes (KEGG) pathway database supported the implication of hepatic exosomes in the lipid metabolism. When we used this approach a significantly higher number of proteins than randomly expected fall into the fatty acid metabolism pathway, whose function are to modulate lipid homeostasis in the body. There was a lower than expected number of hepatic-derived exosomal proteins falling into GO categories related to translation, transcription, cell cycle or apoptosis (Figure 5), suggesting that the role of hepatic exosomes in those processes is not significant.
Figure 5.
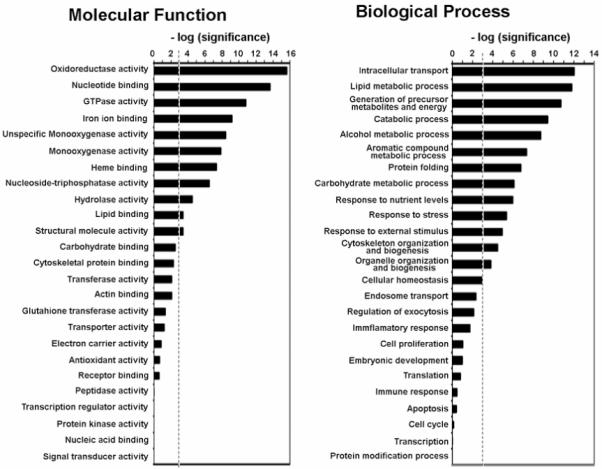
Genome Ontology (GO) Analysis. 244 exosomal proteins identified in this study were classified in molecular function and biological process categories defined by GO consortium. GO categories with a higher or lower number of proteins assigned than expected by random were identified by stadistical analysis. The enrichment and depletion statistics was calculated according to the equation (2), described in Experimental Procedures, using all entries in Swiss-Prot rat protein database as reference set. We represented the [-log(P)], where P is the significance calculated according to the mentioned equation, for the categories indicated. A threshold (discontinued line) was established as P=0.001. Categories with significance above the threshold have significantly greater number of identified protein than expected by random.
Proteins identified in this study include at least 109 proteins (Table S1) that have previously reported to be present in exosomes derived from other sources, further supporting the conclusion that the vesicles secreted to the extracellular medium by hepatocytes consist largely of exosomes.
Discussion
In this study, we were able to isolate vesicles from the extracellular media of two hepatic non-tumoral and non-transformed cellular models, a mouse liver cell line MLP29 and primary rat hepatocytes. The data is strongly supportive of the notion that these vesicles correspond to bona fide exosomes. Their round morphology and size distribution (30-150 nm) match those seen in exosomes from other sources, and conforms not only to the size criterion for exosomes proposed by Thery and colleagues 53, but is also consistent with biophysical properties (e.g. flotation at a cushion of 30% sucrose) that are a specific attribute of exosomes. Finally, we determined that their protein composition is also in agreement with an exosomal origin. The size variability found among the exosome population could indeed reflect the existence of exosomes with different composition or grade of maturation, alternatively some of the variation could be artificially generated during the purification process.
Besides the 109 proteins already known to be present in these vesicles, we have detected many proteins not previously described in exosome from other origins that could be considered as specific of hepatocyte-derived populations. We have detected the ASGR receptor which mediates uptake and intracellular degradation of desialylated glycoproteins, and remarkably, this protein is expressed exclusively in hepatocytes 54. Its enrichment in our exosome preparations (Figure 4c) provides a specific marker for exosomes of hepatic origin what may be useful to discriminate and purify hepatocyte-derived exosomes in complex biological samples like the blood in which a mixed population of exosomes secreted by different cell-types are present. We have also identified proteins involved in the metabolism of atherogenic lipoproteins such as apolipoproteins (ApoE and ApoAV) and paraoxonases (PON1 and PON3). The present model for lipoproteins removal from blood and lymph involves the receptor-mediated endocytosis pathway 55. In this process ApoE associates with the lipoprotein molecule in the extracellular space and binds the ApoE-containing lipoprotein to specific membrane receptors mediating its endocytosis and posterior intracellular metabolism 56. Several studies demonstrate that apoE is internalized within hepatocytes, and then it is either targeted for degradation, or routed out of the cell 56-58. It has also been shown that some apolipoproteins such as apo[A]s and apoC can be assembled extracellularly by a mechanism still incompletely understood 59, 60. Further studies are required to determine the role of hepatic exosomes in lipoprotein metabolism, although our proteomic data suggests that they may act as a vehicle for secretion to the extracellular space of the recycled lipoprotein-associated proteins.
Our proteomic analysis also identified members of several protein families such as cytochromes P450, UDP-glucuronosyltransferases and glutathione S-transferases (Tables 1 and S1), that serve important roles in the cellular detoxification process 61. The presence of these enzymes in hepatic exosomes suggests that it might be possible to transfer these activities in an encapsulated-membrane structure (ie. exosomes) to other cell-types of the body. In agreement with this hyphothesis, the extra-hepatic detection of many cytochrome P450 enzymes has been reported in the respiratory and gastrointestinal tracts including the lung, stomach, small intestine and colon 62, as well as in brain tissues 63.
During exosome formation cytoplasm is included into the vesicle and many cytosol-localized metabolic enzymes are engulfed. As a result, exosome secretion delivers discrete packets of cytosol, providing a potential means for non-invasive detection and analysis of alterations in protein-expression during pathological conditions. Hepatocytes are exceptionally active in fat and carbohydrate metabolism, thus it was not surprising to detect in our proteomic analysis multiple enzymes involved in these processes. The phenomenon of increased expression of glucose transporters and glycolitic enzymes in tumor cells was described as the Warburg effect 64 and is one of the most universal characteristic of solid tumours 65. Thus, increased amounts of glycolytic enzymes in exosomes could reflect an ongoing tumoral process.
Although no attempt was made in this study to investigate abnormalities, our analysis identified several homologues of human proteins involved in specific diseases. By searching in Genetic Association Database (GAD), 32 proteins out of the 251 proteins detected in hepatic exosomes have been associated with diseases (Table 2). Proteins identified in our analysis such as catechol O-methyltransferase, prothrombin, annexins A2 and A5, insulin receptor protein 1, aldolase B, fatty acid synthase, alcohol dehydrogenase 1, dimethylaniline monooxygenase 3, ERGIC-53, cystathionine beta-synthase, glutathione S-transferase Mu 1, paraoxonase 1 and 3 and apopoliproteins A and A5 have been associated among others with metabolic and cardiovascular diseases. In addition, two of the proteins found in our study, CD81 and Regucalcin, have been reported to be differentially expressed between healthy individuals and patients with non-alcoholic steatohepatitis 66. Further characterization of these exosome-detected proteins under normal and pathological conditions could help to define new biomarkers for specific disorders.
Table 2. Association of exosomes-detected proteins with diseases*.
| Diseases Classes | ||||||||
|---|---|---|---|---|---|---|---|---|
| Metabolic | Cardiovascular | Renal | Immune | Hematological | Developmental | Cancer | Neurological | Phamacogenetics |
| APOA5 | APOA5 | APOE | APOA5 | MYH9 | TSC2 | APOA5 | APOA5 | APOA5 |
| APOE | APOE | PON1 | APOE | FAS | RAB3A | APOE | APOE | APOE |
| PON1 | PON3 | PIGR | PON1 | ALDOA | IRS1 | PON1 | PON3 | PON1 |
| THRB | PON1 | THRB | GSTM1 | CBS | PIGR | PON1 | IRS1 | |
| LAMP1 | GNAI2 | DPP4 | BHMT2 | MYH11 | LMAN1 | CBS | ||
| IRS1 | IRS1 | IRS1 | GSTM1 | IRS1 | MEP1A | GSTM1 | ||
| ANXA2 | ANXA5 | ANXA5 | FAS | FAS | COMT | |||
| FAS | LMAN1 | LMAN1 | PGK1 | ALDH2 | ||||
| ALDOB | FAS | FAS | ALDH2 | CBS | ||||
| GCKR | ALDH2 | ALDH2 | CBS | GSTM1 | ||||
| ALDH2 | CBS | ADK | MGST1 | COMT | ||||
| CBS | GSTM1 | MGST1 | GSTM1 | |||||
| GSTM1 | FMO3 | GSTM1 | FMO3 | |||||
| FMO3 | COMT | COMT | ||||||
| COMT | RENBP | |||||||
The batch of 251 proteins identified in this study was searched into the Genetic Association Database (GAD; http://geneticassociationdb.nih.gov/).
The expanding role in biological functions and relevance of exosomes remain unclear at this time. Data from different laboratories indicates that exosome release is a mechanism capable of transferring material and signals to other cells, and it could also be a mechanism to remove metabolic by-products of enzymatic and signalling events, which must ultimately be disposed of by other cells. Further studies are required to define the physiological role of hepatocyte-derived exosomes, although data from GO and KEGG pathways databases points to an important role of these exosomes in lipid metabolism.
Supplementary Material
Acknowledgment
We thank Dr. R. Finnell and Dr. E. González for critical reading of the manuscript, Dr. E. Medico for providing the MLP-29 cell line and Dr. E.C. Dell’Angelica for providing the mAb anti-Pldn. This work was supported by grants from the Fondo de Investigaciones Sanitarias (Instituto de Salud Carlos III, 06/0621 to J.M.F.P.), PN I+D SAF 2005-00855 (to J.M.M.), NIH grants (AA13847 and AT-1576 to S.C.L. and J.M.M.), HEPADIP consortium (HEPADIP-EULSHM-CT-205), and BBVA foundation. CIBERehd is funded by the Instituto de Salud Carlos III. Mass spectrometry analysis was performed at CIC bioGUNE Proteomics Core Facility, member of Proteored.
Abbreviations
- MVBs
multivesicular bodies
- PBS
phosphate-buffered saline
- EGFP
enhanced green fluorescent protein
- GO
Genome Ontology
References
- 1.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Current Opinion in Cell Biology. 2004;16(4):415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Johnstone RM. Revisiting the road to the discovery of exosomes. Blood Cells Mol Dis. 2005;34(3):214–9. doi: 10.1016/j.bcmd.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 3.van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem (Tokyo) 2006;140(1):13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 4.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319(5867):1244–7. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 5.Novikoff AB, Essner E, Quintana N. Golgi Apparatus and Lysosomes. Fed Proc. 1964;23:1010–22. [PubMed] [Google Scholar]
- 6.Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. The biogenesis and functions of exosomes. Traffic. 2002;3(5):321–30. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 7.Schorey JS, Bhatnagar S. Exosome Function: From Tumor Immunology to Pathogen Biology. Traffic. 2008 doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33(3):967–78. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 9.Thery C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147(3):599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanchard N, Lankar D, Faure F, Regnault A, Dumont C, Raposo G, Hivroz C. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J Immunol. 2002;168(7):3235–41. doi: 10.4049/jimmunol.168.7.3235. [DOI] [PubMed] [Google Scholar]
- 11.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183(3):1161–72. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raposo G, Tenza D, Mecheri S, Peronet R, Bonnerot C, Desaymard C. Accumulation of major histocompatibility complex class II molecules in mast cell secretory granules and their release upon degranulation. Mol Biol Cell. 1997;8(12):2631–45. doi: 10.1091/mbc.8.12.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 14.Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94(11):3791–9. [PubMed] [Google Scholar]
- 15.Janiszewski M, Do Carmo AO, Pedro MA, Silva E, Knobel E, Laurindo FR. Platelet-derived exosomes of septic individuals possess proapoptotic NAD(P)H oxidase activity: A novel vascular redox pathway. Crit Care Med. 2004;32(3):818–25. doi: 10.1097/01.ccm.0000114829.17746.19. [DOI] [PubMed] [Google Scholar]
- 16.Robertson C, Booth SA, Beniac DR, Coulthart MB, Booth TF, McNicol A. Cellular prion protein is released on exosomes from activated platelets. Blood. 2006;107(10):3907–11. doi: 10.1182/blood-2005-02-0802. [DOI] [PubMed] [Google Scholar]
- 17.Van Niel G, Mallegol J, Bevilacqua C, Candalh C, Brugiere S, Tomaskovic-Crook E, Heath JK, Cerf-Bensussan N, Heyman M. Intestinal epithelial exosomes carry MHC class II/peptides able to inform the immune system in mice. Gut. 2003;52(12):1690–7. doi: 10.1136/gut.52.12.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Niel G, Raposo G, Candalh C, Boussac M, Hershberg R, Cerf-Bensussan N, Heyman M. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology. 2001;121(2):337–49. doi: 10.1053/gast.2001.26263. [DOI] [PubMed] [Google Scholar]
- 19.Aoki N, Jin-no S, Nakagawa Y, Asai N, Arakawa E, Tamura N, Tamura T, Matsuda T. Identification and characterization of microvesicles secreted by 3T3-L1 adipocytes: redox- and hormone-dependent induction of milk fat globule-epidermal growth factor 8-associated microvesicles. Endocrinology. 2007;148(8):3850–62. doi: 10.1210/en.2006-1479. [DOI] [PubMed] [Google Scholar]
- 20.Faure J, Lachenal G, Court M, Hirrlinger J, Chatellard-Causse C, Blot B, Grange J, Schoehn G, Goldberg Y, Boyer V, Kirchhoff F, Raposo G, Garin J, Sadoul R. Exosomes are released by cultured cortical neurones. Mol Cell Neurosci. 2006;31(4):642–8. doi: 10.1016/j.mcn.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G. Cells release prions in association with exosomes. Proc Natl Acad Sci U S A. 2004;101(26):9683–8. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potolicchio I, Carven GJ, Xu X, Stipp C, Riese RJ, Stern LJ, Santambrogio L. Proteomic analysis of microglia-derived exosomes: metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J Immunol. 2005;175(4):2237–43. doi: 10.4049/jimmunol.175.4.2237. [DOI] [PubMed] [Google Scholar]
- 23.Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci U S A. 2006;103(30):11172–7. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vingtdeux V, Hamdane M, Loyens A, Gele P, Drobeck H, Begard S, Galas MC, Delacourte A, Beauvillain JC, Buee L, Sergeant N. Alkalizing drugs induce accumulation of amyloid precursor protein by-products in luminal vesicles of multivesicular bodies. J Biol Chem. 2007;282(25):18197–205. doi: 10.1074/jbc.M609475200. [DOI] [PubMed] [Google Scholar]
- 25.Hegmans JP, Bard MP, Hemmes A, Luider TM, Kleijmeer MJ, Prins JB, Zitvogel L, Burgers SA, Hoogsteden HC, Lambrecht BN. Proteomic analysis of exosomes secreted by human mesothelioma cells. Am J Pathol. 2004;164(5):1807–15. doi: 10.1016/S0002-9440(10)63739-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riteau B, Faure F, Menier C, Viel S, Carosella ED, Amigorena S, Rouas-Freiss N. Exosomes bearing HLA-G are released by melanoma cells. Hum Immunol. 2003;64(11):1064–72. doi: 10.1016/j.humimm.2003.08.344. [DOI] [PubMed] [Google Scholar]
- 27.Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, Angevin E, Amigorena S, Zitvogel L. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7(3):297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 28.Calzolari A, Raggi C, Deaglio S, Sposi NM, Stafsnes M, Fecchi K, Parolini I, Malavasi F, Peschle C, Sargiacomo M, Testa U. TfR2 localizes in lipid raft domains and is released in exosomes to activate signal transduction along the MAPK pathway. J Cell Sci. 2006;119(Pt 21):4486–98. doi: 10.1242/jcs.03228. [DOI] [PubMed] [Google Scholar]
- 29.Masciopinto F, Giovani C, Campagnoli S, Galli-Stampino L, Colombatto P, Brunetto M, Yen TS, Houghton M, Pileri P, Abrignani S. Association of hepatitis C virus envelope proteins with exosomes. Eur J Immunol. 2004;34(10):2834–42. doi: 10.1002/eji.200424887. [DOI] [PubMed] [Google Scholar]
- 30.Gatti JL, Metayer S, Belghazi M, Dacheux F, Dacheux JL. Identification, proteomic profiling, and origin of ram epididymal fluid exosome-like vesicles. Biol Reprod. 2005;72(6):1452–65. doi: 10.1095/biolreprod.104.036426. [DOI] [PubMed] [Google Scholar]
- 31.Admyre C, Grunewald J, Thyberg J, Gripenback S, Tornling G, Eklund A, Scheynius A, Gabrielsson S. Exosomes with major histocompatibility complex class II and co-stimulatory molecules are present in human BAL fluid. Eur Respir J. 2003;22(4):578–83. doi: 10.1183/09031936.03.00041703. [DOI] [PubMed] [Google Scholar]
- 32.Bard MP, Hegmans JP, Hemmes A, Luider TM, Willemsen R, Severijnen LA, van Meerbeeck JP, Burgers SA, Hoogsteden HC, Lambrecht BN. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am J Respir Cell Mol Biol. 2004;31(1):114–21. doi: 10.1165/rcmb.2003-0238OC. [DOI] [PubMed] [Google Scholar]
- 33.Andre F, Schartz NEC, Movassagh M, Flament C, Pautier P, Morice P, Pomel C, Lhomme C, Escudier B, Le Chevalier T, Tursz T, Amigorena S, Raposo G, Angevin E, Zitvogel L. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360(9329):295. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 34.Navabi H, Croston D, Hobot J, Clayton A, Zitvogel L, Jasani B, Bailey-Wood R, Wilson K, Tabi Z, Mason MD, Adams M. Preparation of human ovarian cancer ascites-derived exosomes for a clinical trial. Blood Cells, Molecules, and Diseases. 2005;35(2):149–152. doi: 10.1016/j.bcmd.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S, Hager HD, Abdel-Bakky MS, Gutwein P, Altevogt P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007;72(9):1095–1102. doi: 10.1038/sj.ki.5002486. [DOI] [PubMed] [Google Scholar]
- 36.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17(7):879–87. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 37.Pisitkun T, Shen R-F, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proceedings of the National Academy of Sciences. 2004;101(36):13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters PJ, Borst J, Oorschot V, Fukuda M, Krahenbuhl O, Tschopp J, Slot JW, Geuze HJ. Cytotoxic T lymphocyte granules are secretory lysosomes, containing both perforin and granzymes. J. Exp. Med. 1991;173(5):1099–1109. doi: 10.1084/jem.173.5.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatnagar S, Schorey JS. Exosomes Released from Infected Macrophages Contain Mycobacterium avium Glycopeptidolipids and Are Proinflammatory. J. Biol. Chem. 2007;282(35):25779–25789. doi: 10.1074/jbc.M702277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clayton A, Turkes A, Navabi H, Mason MD, Tabi Z. Induction of heat shock proteins in B-cell exosomes. J Cell Sci. 2005;118(Pt 16):3631–8. doi: 10.1242/jcs.02494. [DOI] [PubMed] [Google Scholar]
- 41.Andreola G, Rivoltini L, Castelli C, Huber V, Perego P, Deho P, Squarcina P, Accornero P, Lozupone F, Lugini L, Stringaro A, Molinari A, Arancia G, Gentile M, Parmiani G, Fais S. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med. 2002;195(10):1303–16. doi: 10.1084/jem.20011624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gutwein P, Stoeck A, Riedle S, Gast D, Runz S, Condon TP, Marme A, Phong MC, Linderkamp O, Skorokhod A, Altevogt P. Cleavage of L1 in exosomes and apoptotic membrane vesicles released from ovarian carcinoma cells. Clin Cancer Res. 2005;11(7):2492–501. doi: 10.1158/1078-0432.CCR-04-1688. [DOI] [PubMed] [Google Scholar]
- 43.Gomez-Lechon MJ, Donato MT, Castell JV, Jover R. Human hepatocytes in primary culture: the choice to investigate drug metabolism in man. Curr Drug Metab. 2004;5(5):443–62. doi: 10.2174/1389200043335414. [DOI] [PubMed] [Google Scholar]
- 44.Strain AJ, Neuberger JM. A bioartificial liver--state of the art. Science. 2002;295(5557):1005–9. doi: 10.1126/science.1068660. [DOI] [PubMed] [Google Scholar]
- 45.Falcon-Perez JM, Dell’Angelica EC. Zinc transporter 2 (SLC30A2) can suppress the vesicular zinc defect of adaptor protein 3-depleted fibroblasts by promoting zinc accumulation in lysosomes. Exp Cell Res. 2007;313(7):1473–83. doi: 10.1016/j.yexcr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamparski HG, Metha-Damani A, Yao JY, Patel S, Hsu DH, Ruegg C, Le Pecq JB. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods. 2002;270(2):211–26. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 47.Medico E, Mongiovi AM, Huff J, Jelinek MA, Follenzi A, Gaudino G, Parsons JT, Comoglio PM. The tyrosine kinase receptors Ron and Sea control “scattering” and morphogenesis of liver progenitor cells in vitro. Mol Biol Cell. 1996;7(4):495–504. doi: 10.1091/mbc.7.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muller M, Morotti A, Ponzetto C. Activation of NF-kappaB is essential for hepatocyte growth factor-mediated proliferation and tubulogenesis. Mol Cell Biol. 2002;22(4):1060–72. doi: 10.1128/MCB.22.4.1060-1072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lebrand C, Corti M, Goodson H, Cosson P, Cavalli V, Mayran N, Faure J, Gruenberg J. Late endosome motility depends on lipids via the small GTPase Rab7. Embo J. 2002;21(6):1289–300. doi: 10.1093/emboj/21.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Pietro SM, Falcon Perez JM, Tenza D, Setty SR, Marks MS, Raposo G, Dell’Angelica EC. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol Biol Cell. 2006;17(9):4027–38. doi: 10.1091/mbc.E06-05-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falcon-Perez JM, Starcevic M, Gautam R, Dell’Angelica EC. BLOC-1, a Novel Complex Containing the Pallidin and Muted Proteins Involved in the Biogenesis of Melanosomes and Platelet-dense Granules. J. Biol. Chem. 2002;277(31):28191–28199. doi: 10.1074/jbc.M204011200. [DOI] [PubMed] [Google Scholar]
- 52.Creating the gene ontology resource: design and implementation. Genome Res. 2001;11(8):1425–33. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 54.Spiess M. The asialoglycoprotein receptor: a model for endocytic transport receptors. Biochemistry. 1990;29(43):10009–18. doi: 10.1021/bi00495a001. [DOI] [PubMed] [Google Scholar]
- 55.Cooper AD. Hepatic uptake of chylomicron remnants. J Lipid Res. 1997;38(11):2173–92. [PubMed] [Google Scholar]
- 56.Farkas MH, Swift LL, Hasty AH, Linton MF, Fazio S. The recycling of apolipoprotein E in primary cultures of mouse hepatocytes. Evidence for a physiologic connection to high density lipoprotein metabolism. J Biol Chem. 2003;278(11):9412–7. doi: 10.1074/jbc.M208026200. [DOI] [PubMed] [Google Scholar]
- 57.Heeren J, Weber W, Beisiegel U. Intracellular processing of endocytosed triglyceride-rich lipoproteins comprises both recycling and degradation. J Cell Sci. 1999;112(Pt 3):349–59. doi: 10.1242/jcs.112.3.349. [DOI] [PubMed] [Google Scholar]
- 58.Swift LL, Farkas MH, Major AS, Valyi-Nagy K, Linton MF, Fazio S. A recycling pathway for resecretion of internalized apolipoprotein E in liver cells. J Biol Chem. 2001;276(25):22965–70. doi: 10.1074/jbc.M100172200. [DOI] [PubMed] [Google Scholar]
- 59.Koschinsky ML, Marcovina SM. Structure-function relationships in apolipoprotein(a): insights into lipoprotein(a) assembly and pathogenicity. Curr Opin Lipidol. 2004;15(2):167–74. doi: 10.1097/00041433-200404000-00009. [DOI] [PubMed] [Google Scholar]
- 60.Nestruck AC, Rubinstein D. The synthesis of apoproteins of very low density lipoproteins isolated from the Golgi apparatus of rat liver. Can J Biochem. 1976;54(7):617–28. doi: 10.1139/o76-091. [DOI] [PubMed] [Google Scholar]
- 61.Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet. 2002;360(9340):1155–62. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- 62.Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 2003;43:149–73. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 63.Meyer RP, Gehlhaus M, Knoth R, Volk B. Expression and function of cytochrome p450 in brain drug metabolism. Curr Drug Metab. 2007;8(4):297–306. doi: 10.2174/138920007780655478. [DOI] [PubMed] [Google Scholar]
- 64.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 65.Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trends Biochem Sci. 1999;24(2):68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- 66.Rubio A, Guruceaga E, Vazquez-Chantada M, Sandoval J, Martinez-Cruz LA, Segura V, Sevilla JL, Podhorski A, Corrales FJ, Torres L, Rodriguez M, Aillet F, Ariz U, Arrieta FM, Caballeria J, Martin-Duce A, Lu SC, Martinez-Chantar ML, Mato JM. Identification of a gene-pathway associated with non-alcoholic steatohepatitis. J Hepatol. 2007;46(4):708–18. doi: 10.1016/j.jhep.2006.10.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


